Abstract
Induction of B-cell immunity against infection depends on the initiation of the germinal center (GC) reaction in secondary lymphoid organs. Ex vivo recapitulation of the GC reaction in 2D cultures results in transient cell growth, with poor yield and short-term survival. Furthermore, no reported 2D ex vivo system can modulate the kinetics of a GC-like phenotype or the rate of antibody class switching. This protocol describes a methodology for developing immune organoids that partially mimic the B-cell zone of a lymphoid tissue, for efficient and rapid generation of B cells with a GC-like phenotype from naive murine B cells. The organoid is composed of a bioadhesive protein, gelatin, that is transformed into an ionically cross-linked hydrated network using biocompatible silicate nanoparticles (siNps). We explain how to establish the immune organoid culture to sustain immune cell proliferation and transformation into a GC-like phenotype. Starting with cell encapsulation in digested lymphoid tissues, clusters of proliferating B cells with a GC-like phenotype can be generated in the organoids at controlled rates, within ~1 week. The culture methodology described here is currently the only one that allows the accelerated induction of a GC-like phenotype in B cells and supports a controllable immunoglobulin class-switching reaction. This method can be easily implemented in a typical tissue culture room by personnel with standard mammalian cell culture expertise.
INTRODUCTION
Antibody-based immunotherapy approaches have been growing rapidly to treat various pathological conditions, including infections1,2, cancers3,4, and autoimmune diseases2,5. Antibodies are produced following the activation of B cells and differentiation into plasma and memory cells, which occurs in secondary lymphoid organs (spleen and lymph nodes)6. Humoral B-cell immunity against infections depends on the induction of the GC reaction in secondary lymphoid organs to ensure that the antibodies can transform into high-affinity binders, allowing them to interact with antigens from the infectious agent with high affinity6. B-cell follicles are composed of a dense stromal network of B-cell-activating follicular dendritic cells (FDCs)7,8, naive B cells, and CD40 ligand (CD40L)-presenting follicular T helper (TFH) cells9–12. Within the lymphoid microenvironment, an integrin αvβ3-binding Arg-Gly-Asp (RGD) motif is also presented by vitronectin within GCs13,14 and by laminin a5 within the marginal zone of B-cell follicles15. Other adhesive ligands implicated in a GC response include the α4β1 integrin (often called very late antigen 4)16. B-cell activation requires interactions between antigen-primed B cells and TFH cells via CD40L and secretion of interleukin (IL)-4 (refs. 6,8), which is critical for subsequent events leading to B-cell differentiation and antibody production. Expression of CD40L and cytokines such as IL-4 represents T-cell-derived signals associated with GC responses, affinity maturation, and long-lived plasma cells, while still generating some short-term antibody responses6. GC-like B cells are naturally prone to apoptosis, unless rescued by antiapoptotic signals17–19; therefore, researchers have developed methods of activating and differentiating B cells in vitro by culturing B cells in the presence of prosurvival ligands or a feeder layer presenting one or more biological signals, such as CD40L or B-cell-activating factor (BAFF; Fig. 1a). However, using this method, cell growth is transient and most cells die within a short time period20. Moreover, these approaches do not reflect the complexity of the lymphoid microenvironment, thus preventing them from becoming a physiologically relevant model of the immune system. Despite the success of animal models in explaining GC biology6,21–25, to date, no platform technology exists to generate GC-like B cells ex vivo with a control over the kinetics of the GC-like reaction. Mouse models with multiple genetic alterations are useful; however, it can be challenging to decouple the external factors that influence B-cell activation and the differentiation processes.
Figure 1 |.
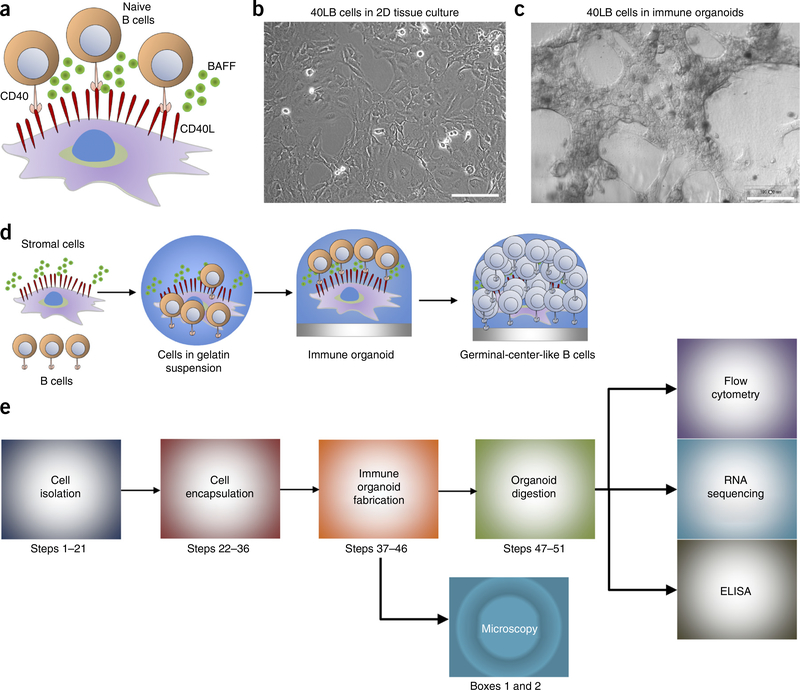
Overview of immune organoid. (a) Schematic representation of the interaction between primary B cells and 40LB stromal cells that presents membrane-bound CD40L and secretes soluble BAFF. B cells interact with CD40L by engaging the CD40 surface receptor. (b) Microscopy image of 40LB stromal cells grown in 2D (Step 22). Scale bar, 100 μm. (c) Microscopy image of 3D immune organoids (Steps 32–46). Organoids were imaged using a Nikon TE2000E fluorescence microscope. Scale bar, 100 μm. (d) Schematic of immune organoid development with GC-like B-cell differentiation processes occurring within the 3D immune organoids over time and (e) the workflow for the organoid culture system and the relevant biological assays (bottom panel).
We have developed an immune organoid that generates an ex vivo GC-like reaction and provides exciting opportunities to systematically study the immune cell process of differentiation into a GC-like phenotype. In this protocol, we detail how to establish the organoid culture system and analyze the cell population. In our studies, the BALB/c 3T3 fibroblasts are stably transduced with CD40L and BAFF (hereafter we refer to this transgenic cell line as 40LB), and function as a substitute for TFH cells and FDCs. The 40LB cell system provides the CD40L signal, which normally comes from TFH cells; hence, the system mimics a T-cell-dependent response. Importantly, the 40LB cells used in our immune organoids have been previously shown to induce phenotypically GC-like B cells from naive B cells in 2D cultures in vitro20. However, in this model, control over the kinetics of a GC-like reaction could not be achieved, and 3D spatial localization of cells undergoing in vitro differentiation cannot be studied. Immune organoids can be maintained for >10 d; however, with longer durations, organoids begin shrinking in size because of the protease-mediated degradation of the gelatin and mitomycin-C-treated 40LB stromal cells, which are therefore no longer viable.
Comparison with other methods
Comparison with in vivo and classic 2D cocultures.
To date, researchers have relied on live-animal models to understand immune cell development and functioning, and to screen immunotherapies against diseases. However, in vivo approaches are costly; they take a long time to conduct; and they do not allow for control over the rate of the immune reaction. Precise control of the kinetics of the immune reaction is not possible because extracellular and intracellular signals presented in vivo cannot be easily tuned for magnitude and persistence. Although activation of B cells with antibodies (anti-Ig or anti-CD40) can be achieved in vitro in 2D cultures through stimulation of CD40L and IL-4, the resulting cell growth is transient, with poor cell yield and short-term survival20. To the best of our knowledge, no 2D in vitro system has demonstrated the ability to modulate the kinetics of GC-like reactions or antibody class switching9–12. As B cells (and lymphocytes in general) are prone to apoptosis in the absence of stimulatory signals, prior studies have infected B cells with Epstein-Barr virus (EBV) for immortalization or used EBV-transformed cell lines26–28. These EBV-transformed cell lines are, however, difficult to clone29, and in some cases they lose their ability to secrete specific antibodies30, therefore limiting the scope of these B cells.
Comparison with existing in vitro lymph node tissue engineering approaches.
Current 3D scaffolds have shown GC-like formation only upon subsequent implantation in vivo, allowing exploitation of the host microenvironment31–34. Studies by Suematsu and Watanabe34 used collagen sponge scaffolds carrying a thymus-derived lymphotoxin (LT)-β receptor and a stromal cell line expressing vascular cell adhesion molecule-1, which is transduced to express murine LT-α and used as an implantable scaffold for synthetic lymph node formation. When implanted in mice, the scaffold-based organoid formed an organized secondary-lymphoid-like structure with compartmentalized zones of B-cell and T-cell clusters, GCs, and networks of follicular DCs. Using composite macroporous PEG hydrogel scaffolds infused with collagen, Stachowiak et al.35 engineered a hydrogel mimicking a lymphoid organ to study immune cell migration. In another study, using colloidal crystal templating, interconnected arrays of porous hydrogel were formed33. These PEG gels, infused with collagen, promoted intrascaffold migration of encapsulated T cells and DCs, with T-cell migration dependent on the connecting pore size, but no GC formation. These studies did not show evidence of controlling the rate of GC reaction ex vivo or in vivo, and no studies have been reported to demonstrate that differentiating B cells can survive ex vivo for successful conversion into GCs or a GC-like phenotype.
The organoid culture system described here is based on a 3D matrix adaptation of a coculture system that was previously published for in vitro generation of GC-like B cells20. We encapsulated the relevant cells using a gelatin matrix that was cross-linked with SiNPs36,37. We adapted the protocol to a 96-well plate culture system, enabling us to establish a high-throughput platform that facilitates the improved generation of the GC reaction. RGD-presenting gelatin is used here because RGD-containing glycoprotein vitronectin matrix is found in GCs, and RGD-binding integrin αvβ3 is known to be upregulated in GC B cells13. However, RGD is not the only integrin-binding ligand present in the lymphoid niche, as integrin α4β1-binding adhesion proteins are upregulated on the surface of FDCs during the GC response13,38,39. Gelatin, derived by denaturing type 1 collagen, has exposed many more RGD sites that bind to integrin αvβ3 as compared with collagen. Although gelatin may also interact with other integrin types, such as α2β1, as observed in type 1 collagen, previous studies have shown that gelatin binds α2β1 with a much lower affinity than that of the native collagen40. In our published data, we have shown the ability of this platform to facilitate accelerated generation of the GC-like phenotype (CD19+GL7+ B cells); rapid formation of class-switched CD19+IgG1+ B cells; high numbers of CD19+ B cells; integrin-mediated B-cell survival; and uniform stromal network formation41. Inhibition of gelatin-αvβ3 integrin interactions using a commercially available Cilengitide inhibitor resulted in partial reduction of B-cell proliferation41, which suggests the possibility of other integrin-ligand interactions, such as integrin α4β1, being important for the process of GC-like reactions.
Applications and limitations of the method
The engineering of an ex vivo immune organoid has the potential to overcome the limitations of using live-animal models, 2D systems, and existing 3D models. These immune organoids will enable researchers to reproduce immunological events with tunable parameters to better understand mechanisms of GC-like B-cell phenotype development and antibody class switching, as well as factors that may accelerate malignant lymphoid transformations and differentiation of B-cells into a GC-like B-cell phenotype20,42–47. At the same time, the 3D organoids may enable studies that will aid in understanding the spatial localization of B cells undergoing controlled differentiation ex vivo in the presence of CD40L and other factors24. We anticipate that in the long term immune organoids will grant researchers the ability to identify factors that have the potential to enhance antibody responses against diseases47,48, mechanistic understanding of B-cell hematological malignancies49–60, and immunity in the elderly61, and that will enable translation of immunotherapeutics31,46–48,55,62–71.
Immune organoids may aid in further mechanistic studies of B-cell differentiation and organization, as discussed elsewhere24,72. For example, pre-B cells express the CD83 surface marker once they express a functional B-cell receptor (BCR). Although naive B cells express only low levels of CD83, activated B cells (both in vitro and in vivo) rapidly upregulate CD83 expression levels72. Yet there are several outstanding questions in the field of immunology and hematological malignancies because the function of CD83 during GC responses is not entirely clear24,72. We provide previously unpublished data that indicate that 3D immune organoids can induce a significant increase in CD83 expression in CD19+ B cells as compared with 2D cocultures on day 4 of differentiation, which corresponds to an ~35-fold increase over naive B cells on day 0 (P < 0.05; Supplementary Fig. 1). The modularity of the organoids and the ability to upregulate CD83 may aid in mechanistic studies of spatial organization24, with possible complementary analyses such as flow cytometry, imaging, histology, and RNA sequencing. However, the immune organoids described here have not yet demonstrated the antigen-specific antibody secretion, affinity maturation, or the formation of dark and light zones of the GC process.
One of the limitations encountered in the immune organoid culture is the autofluorescence of SiNPs in the FL2 channel of the flow cytometer. As a result, the use of fluorophores (e.g., phycoerythrin) or chemical compounds (e.g., doxorubicin) requires compensation or, preferably, sorting of nanoparticles by FACS. The SiNPs can introduce autofluorescence in certain red channel dyes, such as phalloidin; however, this is not a limitation with antibodies conjugated with far-red-emitting dyes. Green autofluorescence may also be observed if the organoids contain ripples of gelatin, which can be avoided by uniform pipetting of polymers while forming the organoids.
Overview of the procedure
The following step-by-step protocol describes the process of establishing the immune organoids (Fig. 1b-e). The Reagent Setup section describes the development of 40LB stromal cells, biomaterials used to engineer organoids, and reagents used for culturing cells, as well as for processing the organoids. The main Procedure (Steps 1–46) describes the isolation of mouse splenocytes, the purification of B cells, the material preparation, and encapsulation of cells into the organotypic culture. Organoids are cultured in cytokine-supplemented cell culture medium that is replaced every 4 d. Cell extraction from the organoid (Steps 47–59) involves degradation of the organoid with collagenase, followed by staining the cell suspension with antibodies. The organoids are easy to prepare using various components and are adaptable for multiple analysis methods (Fig. 1e). We also describe protocols for flow cytometry, whole-organoid staining (Box 1), and histology (Box 2). The flow cytometry experiment described here is suitable for determining the expression of surface marker proteins in a cell population extracted from organoids. Such an analysis identifies the percentage or number of cells expressing a particular phenotype (such as the CD19+Gl7+Fas+ phenotype of GC-like B cells) within a mixed cell population. Quantitative analysis of surface markers (through fluorescently labeled antibodies) describes the differentiation of naive B cells into a GC-like phenotype. Whole-organoid staining, unlike flow cytometry, does not require cell extraction for staining. This analysis method is semiquantitative and is best performed using a confocal microscope because of the 3D structure of organoids. Whole-organoid staining is a powerful technique that provides information on cell morphology, spatial localization, cell surface markers, and cellular interaction with components of the surrounding microenvironment (e.g., extracellular matrix and other cells). Histology analysis of organoids can provide the microscopic anatomical morphology of cells while preserving their location within the organoids. Histology is a complementary analysis to confocal and flow cytometry, and is often used to study morphological changes in B cells undergoing differentiation41,73 or B-cell malignancies74.
Box 1 | Whole-organoid staining protocol ● TIMING variable (culture) plus 28 h
This procedure can be performed after Step 46. Whole-organoid staining is performed to nondestructively examine the 3D structure of organoids and obtain information on cell morphology, spatial localization, cell surface markers, and cellular interaction with components of the surrounding microenvironment (e.g., extracellular matrix and other cells).
Procedure
-
Prepare four organoids on one SigmaCote-coated glass coverslip (25 × 25 mm) for each experimental group, and culture them in organoid growth conditions (2 ml of culture medium) in a standard six-well plate (Fig. 5a). This analysis can be performed at any time point after organoids are formed, per the experimental design; however, we performed studies after 4 d in culture.
▲ CRITICAL STEP If ripples develop during organoid formation, then autofluorescence can be observed during confocal imaging. Ripples in gelatin organoids can be avoided by uniform pipetting of polymers while forming the organoids.
Gently remove the medium by aspirating with a 1-ml pipette tip, being careful not to aspirate the organoids. Wash twice with 1 ml of PBS++.
-
Add 1 ml of 4% (wt/vol) paraformaldehyde to each well to fix the samples, and incubate for 4 h in the dark at RT. Gently aspirate the paraformaldehyde and replace it with 1 ml of PBS. Incubate each gel for 15 min, and repeat the washing step two times.
■ PAUSE POINT At this step, it is possible to pause for 24–48 h before staining. To do so, immerse the hydrogel in fresh PBS, cover the plate with foil, and place it in the 4 °C fridge overnight before going to the next step.
Gently remove the medium by aspirating it with a 1-ml pipette tip, being careful not to aspirate the organoids, and add 2 ml of permeabilizing solution (1% (vol/vol) Triton X-100 in PBS (1:100 dilution)) to the well. Incubate the organoids in permeabilizing solution for 1 h at RT.
Gently wash the organoids in 1 ml of PBS and incubate in 2 ml of blocking buffer (20% (vol/vol) goat serum in PBS (1:5 dilution)) for 30 min at RT.
After aspirating the blocking buffer, incubate the organoids in 1 ml of primary antibody staining solution (0.2% antibody solution in blocking buffer (1:500 dilution)) for 12 h in the dark at 4 °C.
Gently remove the medium by aspirating it with a 1-ml pipette tip, being careful not to aspirate the organoids, and wash twice for 15 min with 1 ml of PBS.
-
After removing PBS, incubate the organoids in 1 ml of secondary antibody staining solution (0.2% antibody solution in blocking buffer (1:500 dilution)) for 4 h in the dark at 4 °C. During the final 15 min of staining with secondary antibodies, add the nuclear stain DAPI (stock 5 mg/ml in dH2O) to the staining solution at a 1:1,000 dilution.
▲ CRITICAL STEP If preconjugated antibodies are used, then incubate for 12-h overnight in the dark at 4 °C.
Gently remove the medium by aspirating it with a 1-ml pipette tip, being careful not to aspirate the organoids, and wash with PBS two times, with 15 min of incubation each time.
Perform imaging using confocal microscopy82.
Box 2 | Organoid histology protocol ● TIMING variable (culture) plus 36 h
Histology analysis of organoids can assess the microscopic morphological differences in cells while preserving their location within the organoids. In this procedure, organoids are dehydrated through a series of graded ethanol washes to displace the water, and then infiltrated with paraffin wax, followed by sectioning and staining.
Procedure
-
Prepare four organoids on one SigmaCote-coated glass coverslip (25 × 25 mm) for each experimental group and culture them in organoid growth conditions (2 ml of culture medium) in a standard six-well plate. This analysis can be performed at any time point after organoids are formed, per the experimental design; however, we performed studies after 4 d in culture.
▲ CRITICAL STEP If ripples develop during organoid formation, then autofluorescence can be observed during immunofluorescence imaging. Ripples in gelatin organoids can be avoided by uniform pipetting of polymers while forming the organoids.
Gently remove the medium by aspirating it with a 1-ml pipette tip, being careful not to aspirate the organoids. Wash with 1 ml of PBS++ for 5 min and aspirate it. Repeat the PBS++ washing procedure twice.
Fix the organoids in 4% (wt/vol) paraformaldehyde at RT for 4 h, but do not exceed 24 h.
Gently remove the paraformaldehyde by aspirating it with a 1-ml pipette tip, being careful not to aspirate the organoids. Wash with 1 ml of PBS++ for 15 min and aspirate it. Repeat the PBS++ washing procedure twice.
Using a scalpel blade, separate the organoids from the coverslips.
-
6. Using tweezers, gently place the organoids into the mold for paraffin embedding.
▲ CRITICAL STEP Ensure that the organoids are placed near the corners of the mold and far from each other.
-
Process and embed the organoids by washing them in 70% (vol/vol) alcohol for 45 mins; next, wash them in 80% (vol/vol) alcohol for 45 mins and then in 95% (vol/vol) alcohol for 45 mins, followed by one 45-min wash in 100% (vol/vol) alcohol and two 60-min washes in 100% (vol/vol) alcohol. Place the organoids in a clearing reagent (xylene or substitute) for 60 min twice, followed by a 60-min wash in paraffin 1 (Paraplast X-TRA), a 60-min wash in paraffin 2 (Paraplast X-TRA), and a 60-min wash in paraffin 3 (Paraplast X-TRA).
▲ CRITICAL STEP This procedure is similar to a standard paraffin-embedding protocol for tissues. These procedures are usually performed by histology or pathology laboratories; however, they can be performed by researchers themselves per the following protocol.
▲ CRITICAL STEP Variation in histology quality may occur; therefore, it is recommended that processing and embedding in paraffin be performed by experienced individuals.
Prepare a 40 °C clean dH2O bath next to the microtome.
9. Place the organoid block on ice (4 °C) for 30 min to make the paraffin harder and therefore easier to slice.
-
Section paraffin blocks at the desired thickness (usually 5–8 μm) on a microtome. Carefully pick up the ribbon by using tweezers, and allow it to float on a 40 °C water bath containing dH2O. For details on microtome adjustment, sectioning, and troubleshooting, users are referred to the steps described by Fischer et al.83.
▲ CRITICAL The lower limit for most paraffin-embedded blocks is ~3 μm in thickness. For thinner sections (2 μm), a glycol-methacrylate-based embedding protocol may be used84,85.
-
Slowly transfer the sections from the water bath onto a Superfrost Plus slide by dipping a clean microscope slide under the meniscus of the water bath and pulling the slide out at an ~45° angle. Allow the slides to dry overnight, and store the slides at RT until they are ready for use.
! CAUTION The slides can be stored indefinitely; however, the immunoreactivity of the cells may decrease if the section is not used within 2 weeks.
Before deparaffinization, place the slide on its edge and bake for 15 min at 60 °C to melt the pariffin wax.
Deparaffinize the sections by immersing them for ~5 min with agitation in xylene (or a xylene substitute). Repeat this step two times. Transfer the slides to 100% (vol/vol) alcohol for 5 min and wash once with 70% (vol/vol) alcohol for 5 min.
Rinse the slides in PBS++ two times for 5 min each time.
Perform H&E staining (Fig. 5b) using standard staining protocols. For immunohistochemistry, antigen retrieval may be necessary and a standard protocol may be applied86.
Experimental design
Biomaterial optimization and cell encapsulation.
The biomaterials used in this protocol are gelatin and SiNP, which are both commercially available. However, before starting the experiment proper, endotoxin levels must be tested on biomaterials to avoid any undesirable endotoxin contaminants. Bacterial endotoxin studies can be conducted using a limulus amebocyte lysate assay41,75. It is desirable to keep the endotoxin levels below 0.5 EU/ml at all times. Before commencing the protocol, it is also important to optimize the cell encapsulation step (Steps 39–42). If one is not careful, it can be challenging to achieve 100% cell encapsulation during the mixing process, because the gelatin and SiNP mixture rapidly turns into a viscous solution. In addition, for maximum B-cell proliferation and differentiation, it is critical to encapsulate 40,000 or more 40LB cells per 10 μl of organoids. However, users may vary the seeding density to adapt the protocol to their own experimental situation. We observed that 20,000 cells per organoid formed small clusters over 48 h, whereas 40,000 and 60,000 cells per organoid resulted in a dense, tightly connected cellular network41. Clustering of 40LB cells can reduce the proliferation and differentiation of B cells while at the same time providing a control over the kinetics of the B-cell reaction. Finally, organoids must be formed in an untreated tissue culture 96-well plate or SigmaCote siliconizing-reagent-coated tissue-culture-treated plates to introduce hydrophobic coating.
Culture medium.
The immune organoids are grown in cell culture medium supplemented with FBS and recombinant cytokines (IL-4). To achieve a successful GC reaction and for optimal organoid maintenance, it is essential to use medium containing freshly added cytokines.
Mouse age.
To establish the organoids, splenocytes are isolated from either male or female C57BL6 mice that are 6–8 weeks old; these are then negatively selected to obtain purified B cells. Older mice can be used, and we have tested mice aged 10–20 weeks with no observable change in the resulting activation and differentiation events. We have also successfully worked with both freshly isolated and cryopreserved splenocytes. Although we have not compared differences in the induction of a GC-like phenotype when frozen samples were used as compared with freshly isolated splenocytes, one caveat of using frozen primary splenocytes is the reduced cell viability upon thawing.
Control samples.
The separation of cells from hydrogel debris is based on separating cells that are positive for specific cell surface markers, in contrast to the autofluorescence of SiNPs observed in the FL2 channel. To specifically stain for B cells and to determine the purity of the B-cell population used, we use fluorophore-conjugated antibodies against CD19. For specific staining of differentiated cells, we use additional antibodies targeted against GL7 and Fas, to observe the formation of the GC-like B-cell phenotype, or those targeted against IgM and IgG1, to analyze the generation of class-switched B cells. For staining controls, we used unstained, isotype, or single-color-stained samples. The staining is performed in a fresh buffer containing FBS as a blocking agent to prevent nonspecific binding between cell surface markers and antibodies.
Cellular analysis.
The cells extracted from the enzymatically degraded organoids can be stained using antibodies for direct analysis via flow cytometer or can be sorted into specific cell populations for downstream analysis such as DNA or RNA isolation76. For the latter method, it may be necessary to pool cells from multiple organoids to achieve the sufficient cell number. For instance, a single 10-μl organoid is sufficient for flow cytometry analysis; however, RNA isolation may require pooling of ~96 organoids to obtain ~500,000 billion cells.
MATERIALS
REAGENTS
BALB/3T3 clone A31 (ATCC, cat. no. CCL-163) ! CAUTION The cell line should be regularly checked to ensure that it does not have cross-contaminants and is not infected with mycoplasma.
C57BL/6J mice, 10–20 weeks of age, male or female (Jackson Laboratory, cat. no. 000664) ! CAUTION Any experiment involving mice must conform to relevant institutional and national regulations. In our studies, all procedures were approved by Cornell University’s Institutional Animal Care and Use Committee.
pBABE-puro (Addgene, plasmid no. 1764)
pCA-T-intron(Neo)-G(LNL) (Addgene, plasmid no. 36908)
RPMI 1640 medium (Thermo Fisher Scientific, cat. no. 11875–093)
DMEM, high glucose (Thermo Fisher Scientific, cat. no. 11965–092)
FBS, qualified and of US origin (Thermo Fisher Scientific, cat. no. 26140–079)
Normal goat serum (Abcam, cat. no. ab7481)
Penicillin-streptomycin, 10,000 U/ml (P/S; Thermo Fisher Scientific, cat. no. 15140–122)
Mitomycin C (Amresco, cat. no. J594–2MG) ! CAUTION Mitomycin C is carcinogenic and can cause acute toxicity. Storage and disposal must be performed in accordance with the vendor’s guidelines and any institutional guidelines, such as those of your institution’s environment, health, and safety department.
Recombinant Murine IL-4 (PeproTech, cat. no. 214–14)
Gelatin from porcine skin (Sigma-Aldrich, cat. no. G2500–500G)
Laponite XLG (BYK Additives & Instruments)
PBS, pH 7.4 (Thermo Fisher Scientific, cat. no. 10010–023)
Collagenase Type 2 (Worthington Biochemical, cat. no. LS004174)
Red blood cell lysis buffer (G-Biosciences, cat. no. 786–650)
EDTA, disodium salt dihydrate solution, reagent grade, 0.25 M (BDH, cat. no. BDH7590–2)
EasySep Mouse B Cell Isolation Kit (Stemcell Technologies, cat. no. 19854)
Trypsin-EDTA, 0.25% (wt/vol; Thermo Fisher Scientific, cat. no. 25200056)
Dulbecco’s PBS with calcium and magnesium (PBS++; Corning, cat. no. 21–030)
DOTAP Liposomal Transfection Reagent (Roche, cat. no. 11202375001)
HEPES (Thermo Fisher Scientific, cat. no. 15630080)
Sodium chloride (NaCl; Sigma-Aldrich, cat. no. S7653–250G)
Murine CD40L cDNA sequence (RefSeq, NM_011616.2)
Murine BAFF cDNA nucleotide sequence (RefSeq, NM_033622.1)
DAPI (4′,6-diamidino-2-phenylindole; stock: 5 mg/ml in dH2O; Thermo Fisher, cat. no. D1306)
Paraformaldehyde aqueous solution, 16% (wt/vol) (Electron Microscopy Sciences, cat. no. 15710)
Xylenes, histological grade (Sigma-Aldrich, cat. no. 534056–500ML)
Paraplast X-TRA, for tissue embedding (Sigma-Aldrich, cat. no. P3808–1KG)
EQUIPMENT
Heracell 240i CO2 incubator (Thermo Scientific, cat. no. 51026280)
1300 Series Class II Type A2 Biological Safety Cabinet (Thermo Scientific, cat. no. 1335)
BD Accuri C6 Flow Cytometer (BD, cat. no. 653118)
Sorvall Legend X1 Centrifuge (Thermo Scientific, cat. no. 75004221)
Sorvall Legend Micro 17 Microcentrifuge (Thermo Scientific, cat. no. 75002431)
EasySep Magnet (Stemcell Technologies, cat. no. 18000)
Water bath (Thermo Scientific, cat. no. 11–100-49SH)
Analytical balance (Mettler Toledo, cat. no. 11144917)
Hot plate (Fisher Scientific, cat. no. 15–460-10Q)
Vortex mixers (Fisher Scientific, cat. no. 02215365)
WaterPro PS Polishing Systems (Labconco, cat. no. 9000520)
Falcon cell strainer, 70-μm pore size (Corning, cat. no. 352350)
Acrodisc syringe filter, 0.2-μm pore size membrane (Pall, cat. no. 28143–310)
Corning Costar clear polystyrene 96-well plates, untreated (Corning, cat. no. 3370)
Centrifuge tubes, 15-ml (VWR, cat. no. 89004–368)
Centrifuge tubes, 50-ml (VWR, cat. no. 89004–364)
Falcon Polystyrene Tubes with 12 × 75 mm size and 5-ml volume (Corning, cat. no. 60819–295)
Snaplock microtube, 0.6-ml volume (Axygen Scientific, cat. no. MCT060C)
Snaplock microtube, 2-ml volume (Axygen Scientific, cat. no. MCT200C)
Parafilm (Bemis, cat. no. PM992)
Eppendorf Research Plus single-channel 0.1– 2.5-μl pipette (Eppendorf, cat. no. 3120000011)
Eppendorf Research Plus single-channel 2–20-μl pipette (Eppendorf, cat. no. 3120000038)
Eppendorf Research Plus single-channel 20–200-μl pipette (Eppendorf, cat. no. 3120000054)
Eppendorf Research Plus single-channel 100–1,000-μl pipette (Eppendorf, cat. no. 3120000062)
Syringe with Luer-Lok tip, 5-ml (BD, cat. no. 309646)
Syringe with Luer-Lok tip, 10-ml (BD, cat. no. 309604)
Pipette tips for 0.1- to 2.5-μl pipettes (VWR, cat. no. 89079–464)
Pipette tips for 2– 20-μl and 20- to 200-μl pipettes (VWR, cat. no. 89079–458)
Pipette tips for 100– 1,000-μl pipettes (VWR, cat. no. 89003–422)
Tissue culture plate, six-well format, with treated surface (Corning, cat. no. 353934)
Tissue culture plate, 96-well format, with treated surface (Corning, cat. no. 353227)
Tissue culture plate, 60-mm-diameter, with treated surface (Corning, cat. no. 353002)
Glass coverslips (25 × 25 mm; Fisherbrand, cat. no. 12–540c)
Sigmacote (Sigma-Aldrich, cat. no. SL2–100ML)
Microtome
VWR Superfrost Plus Micro Slide (75 × 25 mm (3 × 1 inch); VWR, cat. no. 48311–703)
REAGENT SETUP
Splenocyte isolation from mice
Collect spleen from mice77 and store it in the splenocyte isolation buffer. Although it is preferred to use freshly isolated spleen for the organoid culture, we have successfully observed the differentiation process into GC-like B cells using cryopreserved splenocytes (frozen in 90% (vol/vol) FBS and 10% (vol/vol) DMSO) and/or spleens incubated overnight in lymphocyte culture medium at 4 °C. Frozen splenocytes may have reduced cell viability upon thawing, as compared with freshly isolated cells from spleen. ▲ CRITICAL If the cells are already undergoing apoptosis, it can affect the organoid culture. Apoptosis in splenocytes can be reduced by keeping the cell suspension on ice (4 °C) and reducing the time from cell isolation to encapsulation in organoids. ▲ CRITICAL Mouse experiments must be performed in compliance with local institutional guidelines and regulations. All procedures in this study were approved by Cornell University’s Institutional Animal Care and Use Committee.
40LB stromal cell line
Transfect BALB/c3T3 fibroblasts with the CD40L gene and then with the BAFF gene using any method of choice. The protocol here makes use of a 40LB cell line that was generated by Kitamura and colleagues20. First, clone mouse CD40L cDNA into a puromycin-selection vector (e.g., pBABE-puro) and transfect the BALB/c3T3 fibroblasts with the vector by lipofection. In studies by Kitamura and colleagues, a pApuro2 expression vector (a gift from T. Kurosaki, RIKEN) was used20. After the transfection process, select puromycin-resistant stable clones (40L cells) and subsequently pick a naive B cell clone that proliferates most extensively in the presence of IL-4 for BAFF transfection. Clone mouse BAFF cDNA into a neomycin selection vector (e.g., pCA-T-intron(Neo)-G(LNL)) and transfect the selected clone of the 40L cells with the vector. Select G418-resistant clones (40LB cells) and expand them for future use20. Although any transfection method can be used, the lipofection performed in this protocol used DOTAP Liposomal Transfection Reagent78. All transfection reagents should be freshly prepared. Briefly, prepare the DOTAP/nucleic acid mixture by combining 50 μl of 0.1 μg/μl DNA diluted in HEPES buffer saline (HBS) and 100 μl of 30% (vol/vol) DOTAP diluted in HBS buffer. Incubate the DOTAP/nucleic acid mixture for 15 min at room temperature (RT; 25 °C) and add it to 6 ml of cell culture medium to prepare the transfection medium. Incubate the cells to be transfected with the transfection medium for 6 h, and select the transfected clone with antibiotic-supplemented medium. The 40LB cells can be stored indefinitely in liquid nitrogen at −196 °C in 10% (vol/vol) DMSO and 90% (vol/vol) FBS; we have used these cells up until 30 passages. ▲ CRITICAL HBS buffer saline is filter-sterilized cell-culture-grade 20 mM HEPES solution with a pH of 7.4 and 150 mM NaCl. For best results, this buffer should be kept at 4 °C and used within 60 d of first use. ▲ CRITICAL It is critical to evaluate CD40L expression on a regular basis using flow cytometry, after every five passages and for every batch that is thawed from frozen vials.
Naive B lymphocyte culture medium
Prepare RPMI 1640 medium with 10% (vol/vol) FBS and 1% (vol/vol) P/S by combining 45 ml of medium with 50 ml of FBS and 0.5 ml P/S. Store the culture medium at 4 °C and prepare a small aliquot of medium to be warmed up before use in the experiments.
Stromal cell culture medium
Prepare DMEM medium with 10% (vol/vol) FBS and 1% (vol/vol) P/S by combining 45 ml of medium with 50 ml of FBS and 0.5 ml of P/S. Store the culture medium at 4 °C for up to 2 months and prepare a small aliquot of medium to be warmed up before use in the experiments.
Gelatin stock solution
Prepare a 6% (wt/vol) gelatin stock solution by dissolving 600 mg of gelatin in 10-ml of lymphocyte culture medium. Mix the reconstituted gelatin by vigorous shaking, incubate it in a 37 °C water bath for 15 min for complete dissolution, and centrifuge at 200g for 5 min at RT, to remove the bubbles that appear due to the mixing process. Heat the gelatin solution for another 15 min and pass it through a syringe filter (0.2-μm-pore-size membrane) to sterilize the material. Prepare 2-ml aliquots of the sterile gelatin in 2-ml microtubes and store them at 4 °C for up to 1 month.
SiNP stock solution
Prepare fresh 3% (wt/vol) SiNP stock solution by dissolving 30 mg of Laponite XLG in 1 ml of sterile H2O in a 2-ml microtube Vortex the resulting solution at maximum speed for 1–2 min until the solution turns clear with no visible particles on the tube wall. Use freshly made SiNP to create a set of 80–100 organoids.
Recombinant IL-4 stock solution
Spin down the tube containing lyophilized IL-4 at 10,000g for 1 min at RT and add sterile 0.1% (wt/vol) BSA solution in deionized H2O to prepare a 0.1 mg/ml IL-4 stock solution. Prepare aliquots of the cytokine solution and keep them at −20 °C for long-term storage (up to 12 months). IL-4 solution must be prepared fresh before its addition to the cell culture medium.
Mitomycin C stock solution
Add sterile PBS to the vial containing mitomycin C powder to prepare a 1 mg/ml mitomycin C stock solution. Prepare aliquots of the solution and store them at −20 °C for long-term storage (no longer than 2 months). Protect the solution from light throughout the entire process.
Splenocyte isolation buffer
Prepare splenocyte isolation buffer with PBS++ containing 2% (vol/vol) FBS, 1% (vol/vol) P/S, and 5 mM EDTA. The buffer can be made by mixing 38 ml of PBS++ with 800 μl of FBS, 400 μl of P/S, and 800 μl of EDTA. Keep the buffer at 4 °C for long-term storage (no longer than 4 weeks).
B-cell purification buffer
Prepare B-cell purification buffer, which is PBS++ with 2% (vol/vol) FBS, 1% (vol/vol) P/S, and 1 mM EDTA. The buffer can be made by mixing 40 ml of PBS++ with 800 μl of FBS, 400 μl of P/S, and 160 μl of EDTA. Keep the buffer at 4 °C for long-term storage (no longer than 4 weeks).
Collagenase stock solution
Prepare 10 U/ml of syringe-filtered collagenase II solution in serum-free RPMI medium. Incubate collagenase powder at RT for 15 min, weigh 2 mg of collagenase, and place it in a 2-ml microtube. Prepare 1,000 U/ml of concentrated collagenase solution by adding serum-free RPMI (volume depends on the concentration of collagenase from the vendor) to the tube, and mixing the solution gently via pipetting and inversion. Pass the solution through a syringe filter with a 0.2-μm-pore-size membrane), and dilute it with additional serum-free medium to obtain a final concentration of 10 U/ml. Store the solution at −20 °C for no more than 1 week, and avoid multiple free-thaw cycles.
FACS buffer
Prepare FACS buffer with PBS++ containing 2% (vol/vol) FBS and 5 μM EDTA. The buffer can be made by mixing 40 ml of PBS++ with 800 μl of FBS, 400 μl of P/S, and 0.8 μl of EDTA. Store the buffer at 4 °C for long-term use (no longer than 4 weeks).
PROCEDURE
Isolation of splenocytes from murine spleens ● TIMING 40–60 min
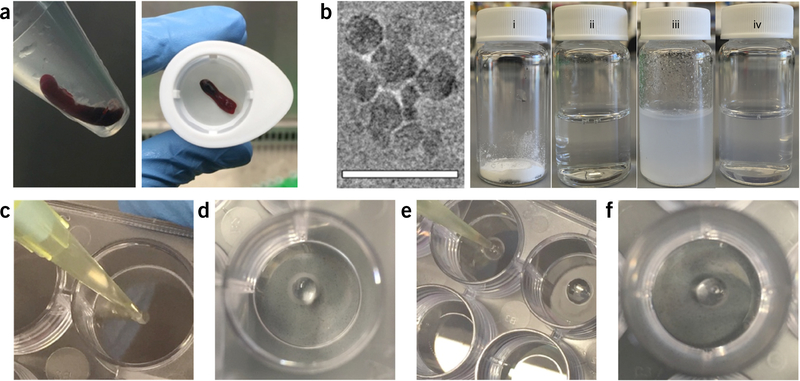
▲ CRITICAL The procedure for isolating splenocytes and preparation of organoids is summarized in Figure 2.
-
1|
Place a cell strainer on top of a 50-ml centrifuge tube (Fig. 2a).
-
2|
Put 5 ml of splenocyte isolation buffer into the tube.
-
3|
Put a freshly isolated spleen on top of the strainer.
Figure 2 |.
Organoid preparation. (a) mouse spleen stored in buffer after isolation (left) and placed inside a cell strainer before homogenization (right). (b) Transmission electron microscopy image of SiNP indicating the size and shape of nanoparticles (left, scale bar, 50 nm). The images were captured under 200-keV voltage using JEOL JEM-2010 TEM. SiNP preparation process (right) from (i) the SiNP powder form, (ii) water, (iii) appearance of cloudy suspension after addition of water to SiNP powder due to SiNP-mediated physical exfoliation of the aqueous phase, and (iv) formation of a clear homogeneous SiNP solution upon vortexing. (c-f) Generation of organoid by preparation of the SiNP droplet inside the well (c,d), injection of gelatin solution into the first droplet (e), and appearance of the formed organoid hydrogel, which stays intact in cell culture conditions (f). The organoid formation procedure is detailed in Steps 32–46.
▲ CRITICAL STEP Ensure that the spleen is placed in the middle of the strainer such that the cell suspension/slurry from the spleen will not get stuck on the side of the strainer.
-
4|
Wet the cell strainer by rinsing with 5 ml of splenocyte isolation buffer.
-
5|
Mash the spleen with the plunger of a 10-ml syringe.
-
6|
Flush the dissociated spleen with 10 ml of splenocyte isolation buffer.
▲ CRITICAL STEP Make sure that thorough washing is performed such that no spleen suspension is stuck to the strainer.
-
7|
Centrifuge the cell suspension at 200g for 5 min at RT, and resuspend the splenocytes in 10 ml of red blood cell lysis buffer.
-
8|
Incubate the cells in the dark at RT for 10 min.
-
9|
Centrifuge the cell suspension at 200g for 5 min, at RT.
▲ CRITICAL STEP Observe the cell pellet: if it has turned white, that indicates that the red blood cell lysis is complete. Repeat the process if a red streak is still visible at the top of the cell pellet.
-
10|
Resuspend the splenocytes with 5 ml of splenocyte isolation buffer. Ensure that there are no cell clumps. If cell aggregates can be seen in the solution, pass the splenocyte through a cell strainer again.
Purification of B cells from whole splenocytes ● TIMING 20 min
-
11|
Centrifuge the cell suspension at 200g for 5 min, at RT.
-
12|
Resuspend the splenocyte with 0.5 ml of B-cell purification buffer.
-
13|
Transfer the cell suspension to a 5-ml polystyrene tube.
-
14|
Add 25 μl of normal rat serum from the B Cell Isolation Kit and 25 μl of EasySep Mouse B Cell Isolation Cocktail.
-
15|
Mix gently and incubate the mixture for 10 min at RT
-
16|
Vortex the EasySep Streptavidin RapidSpheres for 30 s such that complete mixing occurs and the solution looks uniform, with no separation between the beads and the solution.
-
17|
Add 37.5 μl. of EasySep Streptavidin RapidSpheres to the cell suspension.
-
18|
Mix gently and incubate the mixture for 2.5 min at RT.
-
19|
Add 1.6 ml of B-cell purification buffer for a final volume of 2.5 ml.
-
20|
Mix the cell suspension gently and place the tube inside the magnet at RT and incubate for 2.5 min.
-
21|
Gently transfer the cell suspension to a new 50-ml tube, keep it on ice, and count the cells using a standard hemocytometer technique
preparation of the feeder layer using 40LB stromal cells ● TIMING 80 min
-
22|
Prepare a 0.01 mg/ml mitomycin C solution by mixing 120 μl of mitomycin C stock solution with 12 ml of stromal cell culture medium.
▲ CRITICAL STEP It is important to dissolve the stock thoroughly because mitomycin C powder can fall out of the solution during storage, and multiple pipetting can be needed to mix it with the cell culture medium.
-
23|
Prepare two 100-mm tissue culture plates of confluent stromal cell culture with 90–95% confluency, as described in the Reagent Setup.
-
24|
Replace the cell culture medium of the 40LB stromal cells with fresh mitomycin C solution.
-
25|
Incubate the cells at 37 °C in a standard cell culture incubator for 45–60 min.
-
26|
Carefully remove the culture medium containing mitomycin C and transfer the waste solution to a designated waste bottle where it will be stored until it can be disposed of safely.
! CAUTION Mitomycin C is carcinogenic, and it can cause acute toxicity. Storage and disposal must be in accordance with the vendor’s guidelines, and any institutional guidelines, such as those of your institution’s environment, health, and safety department.
-
27|
Wash cells with 10 ml of PBS and decant the first wash into the mitomycin C waste bottle.
-
28|
Wash cells with 10 ml of PBS for the second time and aspirate the buffer as usual.
-
29|
Detach the cells by adding 6 ml of trypsin-EDTA and incubating them for 6 min.
-
30|
Deactivate the trypsin-EDTA with 12 ml of serum-supplemented stromal culture medium, centrifuge the cell suspension at 200g for 5 min at RT, and resuspend the cells in 12 ml of serum-supplemented stromal culture medium.
-
31|
Transfer the cell suspension to a new, clean 50-ml tube and keep the suspended stromal cells at 4 °C until encapsulated within the organoids (encapsulation must be performed within 2 h of this process to reduce stromal cell apoptosis).
Establishment of immune organoids using primary murine B cells and gelatin matrix ● TIMING 60–80 min for 60 organoids
-
32|
Prepare 3% (wt/vol) SiNP stock solution and vortex the nanoparticle suspension in water (Fig. 2b). Keep the resulting nanoparticle solution at RT.
▲ CRITICAL STEP Vortexing of SiNP solution until the particles are fully dissolved is critical. Failure to achieve complete dissolution can result in clumps of nanoparticles.
-
33|
Place a glass beaker filled with water on a hot plate, and set the heating dial such that the water temperature is maintained ~37 °C at all times.
-
34|
Heat a 2-ml gelatin aliquot using the warmed water in the beaker until the gelatin is flowing.
-
35|
Mix B cells (from Step 21) and stromal cells (from Step 31) with gelatin such that the resulting solution has 5% (wt/vol) gelatin, 4,000 B cells per μl, and 4,000–12,000 40LB stromal cells per μl.
▲ CRITICAL STEP Gently mix the cell-gelatin suspension such that a uniform mixture is achieved without the formation of air bubbles, which can reduce the effective amount of material that can be used in the organoid-making process.
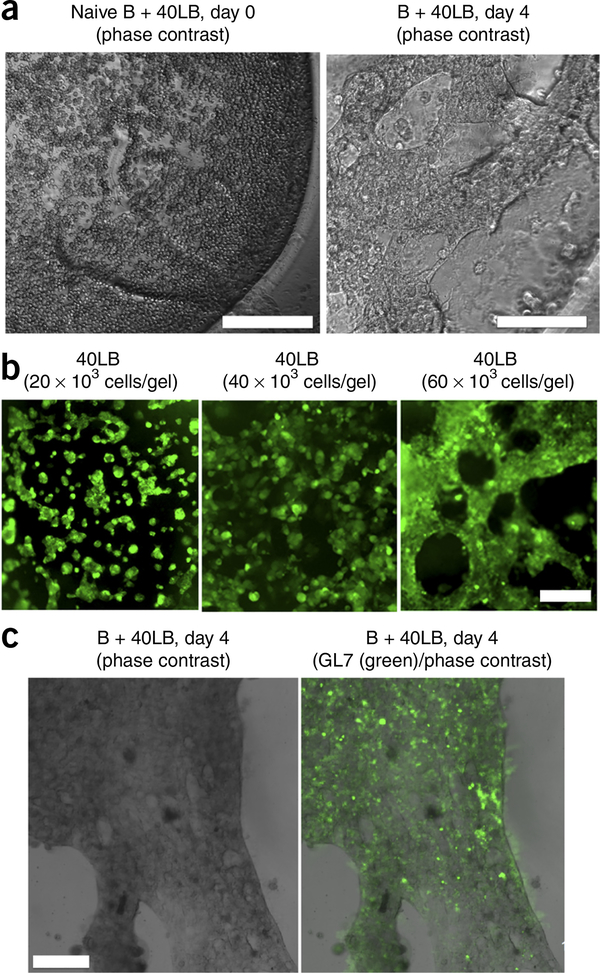
▲ CRITICAL STEP For maximum B-cell proliferation and differentiation, it is critical to encapsulate ≥40,000 40LB cells per 10 μl. of organoids by using 5 μl of a solution of ≥8,000 40LB cells per μl.. In our experience41, the use of a solution containing 40,000 cells per 10 μl of organoids resulted in dense, tightly connected cellular network (Fig. 3).
Figure 3 |.
Ex vivo induction of GC reaction within the immune organoid. (a) Naive B cells with 40LB cells coencapsulated with the immune organoids. On day 0, freshly encapsulated cells remain spherical. Organoid was imaged using a Nikon TE2000E microscope. (b) Immunofluorescence images of the 40LB stromal cell network formation in immune organoids with varying 40LB seeding density. (c) Immunofluorescence analysis of GC-like B cells (GL7+) in the presence of 40LB stromal cells inside the organoid, imaged on day 4. Organoids were stained with Alexa-Fluor-488-conjugated GL-7 antibody and imaged using a Zeiss 710 confocal microscope. Scale bars, 200 μm. b adapted with permission from ref. 41, Elsevier.
? TROUBLESHOOTING
-
36|
Keep the cell suspension on ice.
-
37|
Prepare a 10 ng/ml IL-4 solution by mixing 2 μl of the thawed IL-4 stock solution with 20 ml of lymphocyte culture medium.
-
38|
Heat the cell-gelatin suspension for 2 min using the warmed water until gelatin starts flowing. This step can be checked by inverting the tube and seeing whether the gelatin suspension can move quickly. Keep the cell-gelatin suspension warmed throughout the organoid fabrication process.
-
39|
Mix the cell-gelatin suspension to ensure that cells are homogeneously mixed and not settled.
▲ CRITICAL STEP Make sure to mix gently without forming bubbles, and repeat the process after encapsulating cells in four to five organoids.
? TROUBLESHOOTING
-
40|
Pipette 5 μl of SiNP solution into the middle of each well as a droplet (Fig. 2c-f).
? TROUBLESHOOTING
-
41|
Pipette 5 μl of the cell-gelatin suspension into each SiNP droplet.
? TROUBLESHOOTING
-
42|
Place the pipette tip in the middle of each droplet and pipette gently three to five times to mix the solution.
▲ CRITICAL STEP It is important to ensure that the tip is in the middle of the droplet so that efficient mixing can occur and most cells are contained within the droplet.
▲ CRITICAL STEP It is critical to maintain the organoid composition at 1.5% (wt/vol) SiNP with 2% (wt/vol) gelatin, because this composition results in robust spreading of 40LB cells, which is critical for expression of CD40L and cell-cell interactions41.
-
43|
Incubate the cells at RT for 10 min to complete the curing step.
? TROUBLESHOOTING
-
44|
Add 200 μl of 10 ng/ml IL-4-supplemented lymphocyte culture medium to each well.
-
45|
Repeat Steps 39–44 to continue making the next set of organoids.
-
46|
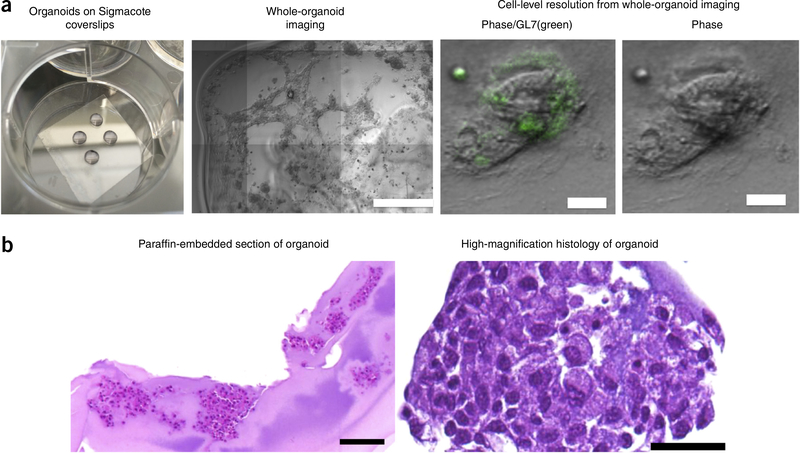
Replace the 10 ng/ml IL-4-supplemented lymphocyte culture medium every 4 d. Whole-organoid staining (Fig. 4a), as detailed in Box 1, or histology (Fig. 4b), as detailed in Box 2, can be performed on these samples. These analyses can be performed at any time point after the organoids are formed, per the experimental design; however, we performed studies after 4 d in culture.
Figure 4 |.
Whole immune organoid imaging. (a) naive B cells with 40LB cells were coencapsulated with the immune organoids, and after 4 d of differentiation organoids were fixed in paraformaldehyde. Organoids were stained with Alexa-Fluor-488-conjugated GL7 antibody and imaged using a Zeiss 710 confocal microscope. (Second panel from left) Low-magnification visualization of an organoid (scale bar, 500 μm) and (two right-hand panels) high-magnification visualization of 40LB cells colocalized with GL7-expressing B cells (scale bars, 200 μm). (b) Paraformaldehyde-fixed organoids were processed and paraffin-embedded, sectioned, and stained withH&E. Images represent H&E-stained sections of B cells in immune organoids (Scale bars, 50 μm).
Dissociation of organoids to extract the cells for cellular analysis ● TIMING 16–24 h for steps 47–49 and 90 min for steps 50 and 51
-
47|
Aspirate the medium from each well and wash each organoid with 200 μl of PBS. The organoid dissociation step can be performed at any time point after the organoids are fabricated, per the experiment design; however, we have performed studies after 2, 4, and 6 d in culture.
▲ CRITICAL STEP Make sure to perform both aspiration and rinsing gently to avoid perturbing the organoid; this can be done by placing the pipette tip at the well wall and pipetting slowly.
-
48|
Place 200 μl of collagenase solution into each well.
-
49|
Incubate the collagenase-covered organoid overnight in the CO2 incubator (5% CO2 and 37 °C) to ensure a thorough enzymatic degradation process.
-
50|
Pipette the supernatant five to ten times to dissociate the organoid.
▲ CRITICAL STEP Make sure that the pipetting is specifically directed toward the center of the organoid to ensure that the mechanical perturbation degrades most of the construct.
? TROUBLESHOOTING
-
51|
Pass the cell suspension through a cell strainer placed on top of a 0.6-ml centrifuge tube to remove large debris. The cell suspension should be passed through the side filters of a 70-μm strainer.
Cell staining for flow cytometry ● TIMING for 60 organoids: 90 min for Steps 52–57; 180 min for Steps 58 and 59
-
52|
Add 200 μl of FACS buffer to the strained cell suspension and spin it down at 200g for 5 min, at RT.
-
53|
Aspirate the supernatant gently and resuspend the cells in 100 μl of FACS buffer (note: each sample can be diluted with 200 or 300 μl of FACS buffer if the user is interested in using two or three antibody staining combinations. The diluted sample must be distributed over two or three 100-μl suspensions).
-
54|
Prepare the antibody solution by dissolving one or more fluorophore-conjugated antibody stock solutions in FACS buffer with a 1:250 or 1:500 dilution (or 1–2 μg of antibody per million cells).
-
55|
Combine 100 μl of antibody solution with 100 μl of cell suspension in a 0.6-ml Eppendorf tube, mix gently, and incubate for 45–60 min in the dark at 4 °C.
-
56|
Spin down the stained cell suspension at 200g for 5 min at RT and aspirate the supernatant gently.
-
57|
Add 100 μl of FACS buffer to each tube and store the cell suspension on ice protected from light.
-
58|
Analyze the cell suspension with a flow cytometer (or use a cell sorter to isolate a specific cell population for biological assays).
-
59|
Implement a gating strategy based on separating the cell population from debris in forward scatter - height (FSC-H) versus side scatter - height (SSC-H), segmenting the CD19+ cells from the rest of nanoparticles in FL2-H versus FL4-H (or the fluorescent channel of interest), and analyzing the purified cell population based on the additional stains added to the sample from the perspective of absolute cell number, percentage of cell population, and/or mean fluorescence intensity. The gating scheme is summarized in Figure 5. Antibodies are listed in Table 1. Sorted cells can be used for further analyses such as flow cytometry, RNA sequencing, ELISA, and protein phosphorylation assays.
Figure 5 |.
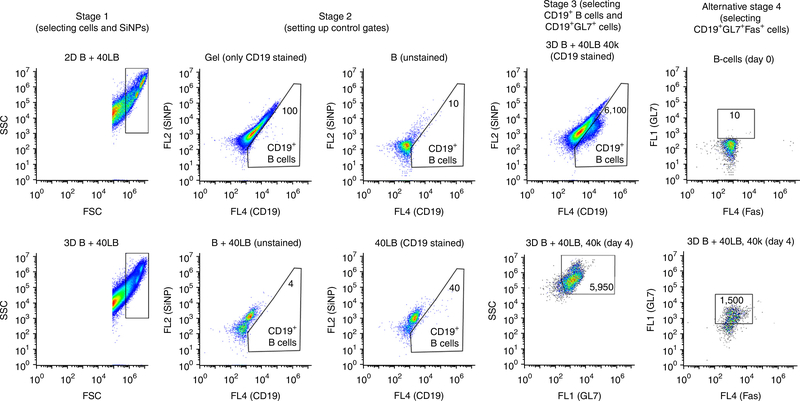
Immune organoid flow cytometry gating strategy. Stage 1 is performed to separate the cell population from debris based on size via gating implemented in the FSC versus SSC window. Stage 2 is performed to isolate CD19+ cells by preparing control gates in the FL2 versus CD19 window with the use of unstained sample, stained stromal cells, and stained blank gel without encapsulated cells. Gating was applied to separate SiNPs from APC-CD19+ B cells (FL4 channel). Numbers within the gate represent the number of CD19+ B cells for that particular group. Stage 3 is performed to quantify the FITC-GL7 subset (FL1 channel) of the overall CD19+ cells by setting up control gates in the SSC versus FL1 window based on primary naive B cells collected on day 0 and the gel-only control (CD19 and GL7 stained). Stage 4 is an alternative strategy to quantify the GL7+Fas+ subset of the overall CD19+ cells. Gating was applied to separate SiNPs from PE−Cy7−CD19+ B cells (FL3 channel). Gated CD19+ cells were analyzed for FITC-GL7 and APC–Fas staining. Numbers within the gate represent the number of CD19+GL7+Fas+ B cells for that particular group. Control gates in the FL4 versus FL1 window were based on primary naive B cells collected on day 0. Image adapted with permission from ref. 41, Elsevier.
TABLE 1 |.
Antibodies used in the protocol for flow cytometry or florescence microscopy.
| Target antigen | Fluorophore type | Clone | Supplier | Cat. no. |
|---|---|---|---|---|
| CD19 | PE-Cyanine7 | eBio1D3 (1D3) | eBioscience | 25-0193 |
| GL7 | Alexa Fluor 488 | GL-7 (GL7) | eBioscience | 53-5902 |
| Fas | APC | 15A7 | eBioscience | 17-0951 |
| IgM | FITC | eB121-15F9 | eBioscience | 11-5890 |
| IgG1 | APC | M1-14D12 | eBioscience | 17-4015 |
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2 |.
Troubleshooting table.
| Step(s) | Problem | Possible reason | Solution |
|---|---|---|---|
| 35 | Gelatin solution is too viscous | Gelatin is in the liquid state at the cell culture temperature of 37 °C, but it gradually becomes viscous and solidifies at RT | Warm the gelatin stock solution for at least 2 min before use and keep it warm for prolonged use during the experiments |
| 35, 39 | A small number of differentiated cells is found after organoid culture | Low encapsulation efficiency, with many cells located outside of the organoid hydrogel | Check after the hydrogel formation process to ensure that you have a cell-laden hydrogel core suspended in the left of a clear SiNP sphere, with minimal numbers of cells on the outside of the constructs |
| High data variation within groups | Lack of thorough mixing of the cell-gelation suspension before the encapsulation process | Mix the cell-gelatin suspension to prevent cells from settling at the bottom of the container. Complete mixing will also ensure that the cells are distributed evenly throughout the hydrogel | |
| 40 | SiNP solution is too viscous | SiNPs slowly become viscous over time; this process occurs more quickly with a more concentrated stock solution | Make sure to prepare fresh stock (for preparing 100 organoid samples at most) and vortex it before use |
| 41 | Organoid does not have a spherical structure | Incomplete cross-linking because of the lack of mixing between gelatin and SiNPs | Pipette the mixture up and down five times upon injecting the cell-gelatin suspension into the SiNP gel |
| 43 | No droplet formation, with the liquid spreading in the well | Failure to use an untreated 96-well plate | Use an untreated 96-well plate or apply SigmaCote siliconizing reagent within the wells of treated plates to introduce a hydrophobic coating |
| 50 | Low cell yield following organoid digestion process | Incomplete enzymatic degradation of the organoid | Increase digestion time and/or use fresh collagenase solution |
● TIMING
Steps 1–10, isolation of splenocytes from murine spleens: 40–60 min
Steps 11–21, purification of B cells from whole splenocytes: 20 min
Steps 22–31, preparation of the feeder layer using 40LB stromal cells: 80 min
Steps 32–46, establishment of immune organoids using primary murine B cells and gelatin matrix: 60–80 min for 60 organoids
Steps 47–51, dissociation of organoids to extract the cells for cellular analysis: 16–24 h for Steps 47–49 and 90 min for Steps 50 and 51
Steps 52–59, cell staining for flow cytometry: 90 min for Steps 52–57; 120 min for Steps 58 and 59 (for 60 organoids)
Box 1, whole-organoid staining protocol: variable (culture) plus 28 h
Materials preparation: 15 min
step 1, culture of organoids: variable; studies can be performed at any time point after the organoids are fabricated, per the experimental design; however, we have performed studies after 4 d in culture
steps 2 and 3, washing and fixation of organoids: 45 min for step 2; 4 h for step 3
steps 4 and 5, permeabilization and blocking: 70 min for step 4; 40 min for step 5
steps 6 and 7, primary antibody staining: 12 h for step 6; 40 min for step 7
steps 8 and 9, secondary antibody staining: 4 h for step 8; 40 min for step 9
Box 2, organoid histology protocol: variable (culture) plus 36 h
step 1, culture of organoids: variable; studies can be performed at any time point after the organoids are fabricated, per the experimental design; however, we have performed studies after 4 d in culture
steps 2–4, washing and fixation of organoids: 10 min for step 2; 4 h for step 3; at least 45 min for step 4
steps 5–8, processing and embedding: 10 h
steps 9–15, sectioning and deparaffinization: 5–6 h for four organoids for steps 9 and 10; 12 h for step 11; 40 min for steps 12–15
ANTICIPATED RESULTS
This protocol describes the generation of 3D immune organoid production from naive murine B cells and their subsequent analysis. The method is easy to implement in a typical tissue culture room using standard equipment. Furthermore, organoids can be examined at various time points to study a variety of developmental stages. The success rate for preparing individual organoid droplets can vary from 90 to 100% depending on the ability to generate droplets contained in the middle of each well, which could be attributed to variation in manual pipetting and other problems, as discussed in the Troubleshooting section. Organoids are typically not variable between wells within a preparation, although different batches of gelatin and SiNP made at different times can exhibit variability. For the organoid to demonstrate the expected result of improved GC reaction kinetics, it is important to use serum-supplemented cell culture medium (stored at 4 °C) and to add exogenous cytokines prepared freshly on the day of medium addition/replacement.
Within 24 h of seeding, 40LB cells will start spreading and form networks (Fig. 3a). We have observed that seeding densities of 20,000 cells/organoid formed small clusters over 48 h, whereas seeding densities of 40,000 and 60,000 cells per organoid resulted in dense, tightly connected cellular network (Fig. 3b)41. Clustering of 40LB cells can reduce the proliferation and differentiation of B cells. B cells will generally expand quickly once they are placed in immune organoids, and by day 4 of growth the entire organoid will have dark spots of proliferating cells35, which are difficult to examine using a standard tissue culture microscope. It is recommended to observe the organoid using a tissue culture microscope or dissecting microscope to visualize gross morphology; however, optimal resolution can be achieved only through confocal microscopy (Box 1). It is anticipated that the differentiating B cells will either colocalize with 40LB stromal cells or be embedded in the organoid matrix in close proximity to 40LB cells. To obtain fine-resolution images, the large size of immune organoids necessitates microscopic analysis by sectioning and immunohistochemical staining (Box 2). After 4–6 d, organoids should begin to exhibit GC-like phenotype differentiation, marked by GL7 and Fas expression41. Within the 3D organoid cultures, the CD19+GL7+ response will be dependent on 40LB seeding density (Fig. 3c) such that seeding organoids with 40,000 40LB cells will induce a >90- to 100-fold increase in CD19+GL7+ cell numbers (Fig. 4), whereas a 20,000 40LB cell seeding density is expected to result in an ~50- to 60-fold increase35. 2D cultures will result in a marginal 10- to 20-fold increase41.
Another hallmark of GC reaction is the antibody class switching that occurs in GC B cells to produce antibodies with distinct effector functions79–81. Activated B cells are known to undergo antibody class switching from IgM to IgG or other Igs. If allowed to develop further, organoids will progressively induce Ig class switching by days 6–8. Using flow cytometry, cells extracted from the organoids can be quantified to determine the change in surface IgM and IgG1 levels or those of another Ig subtype.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial support from a National Science Foundation (NSF) CAREER Award (DMR-1554275 (to A.S.)) and the National Institutes of Health (1R21CA185236–01, to A.S.). We thank D. Kitamura at Tokyo University of Science for providing 40LB cells. We thank L. Cerchietti and his laboratory at Weill Cornell Medical College of Cornell University for providing histology imaging expertise and A. Gaharwar and his laboratory at Texas A&M University for silicate nanoparticle imaging expertise. All procedures were approved by Cornell University’s Institutional Animal Care and Use Committee. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Shi W et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol 16, 663–673 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Saylor C, Dadachova E & Casadevall A Monoclonal antibody-based therapies for microbial diseases. Vaccine 27, G38–G46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabbatino F et al. Novel tumor antigen-specific monoclonal antibody-based immunotherapy to eradicate both differentiated cancer cells and cancer-initiating cells in solid tumors. Semin. Oncol 41, 685–699 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Weiner LM, Murray JC & Shuptrine C Antibody-based immunotherapy of cancer. Cell 148, 1081–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspi R Immunotherapy of autoimmunity and cancer: the penalty for success. Nat. Rev. Immunol 8, 970–976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nutt SL, Hodgkin PD, Tarlinton DM & Corcoran LM The generation of antibody-secreting plasma cells. Nat. Rev. Immunol 15, 160–171 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Schiemann B et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science 293, 2111–2114 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Heesters BA, Myers RC & Carroll MC Follicular dendritic cells: dynamic antigen libraries. Nat. Rev. Immunol 14, 495–504 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Cerutti A et al. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+ IgD+ B cell line. J. Immunol 160, 2145–2157 (1998). [PMC free article] [PubMed] [Google Scholar]
- 10.von Bergwelt-Baildon MS et al. Human primary and memory cytotoxic T lymphocyte responses are efficiently induced by means of CD40-activated B cells as antigen-presenting cells: potential for clinical application. Blood 99, 3319–3325 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Schultze JL et al. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J. Clin. Invest 100, 2757 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coughlin CM, Vance BA, Grupp SA & Vonderheide RH RNA-transfected CD40-activated B cells induce functional T-cell responses against viral and tumor antigen targets: implications for pediatric immunotherapy. Blood 103, 2046–2054 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Rodda LB, Bannard 0 & Cyster JG Integrin-mediated interactions between B cells and follicular dendritic cells influence germinal center B cell fitness 192, 4601–4609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen CD & Cyster JG Follicular dendritic cell networks of primary follicles and germinal centers: phenotype and function. Semin. Immunol 20, 14–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J et al. Extracellular matrix of secondary lymphoid organs impacts on B-cell fate and survival. Proc. Natl. Acad. Sci. USA 110, E2915–E2924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrasco YR & Batista FD B-cell activation by membrane-bound antigens is facilitated by the interaction of VLA-4 with VCAM-1. EMBO J. 25, 889–899 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YJ et al. Mechanism of antigen-driven selection in germinal centres. Nature 342, 929–931 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Valdez H et al. Human germinal center B cells express the apoptosis-inducing genes Fas, c-myc, P53, and Bax but not the survival gene bcl-2. J. Exp. Med. 183, 971–977 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein U & Dalla-Favera R Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol 8, 22–33 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Nojima T et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat. Commun 2, 465 (2011). [DOI] [PubMed] [Google Scholar]
- 21.De Silva NS & Klein U Dynamics of B cells in germinal centres. Nat. Rev. Immunol 15, 137–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulman Z et al. T follicular helper cell dynamics in germinal centers. Science 341, 673–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwickert TA et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature 446, 83–87 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Allen CD et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol 5, 943–952 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Berek C, Berger A & Apel M Maturation of the immune response in germinal centers. Cell 67, 1121–1129 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Pattengale P, Smith R & Gerber P Selective transformation of B lymphocytes by EB virus. Lancet 302, 93–94 (1973). [DOI] [PubMed] [Google Scholar]
- 27.Morello F et al. Differential gene expression of blood-derived cell lines in familial combined hyperlipidemia. Arterioscler. Thromb. Vasc. Biol 24, 2149–2154 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Amoli M, Carthy D, Platt H & Ollier W EBV Immortalization of human B lymphocytes separated from small volumes of cryo-preserved whole blood. Int. J. Epidemiol 37, i41–i45 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Traggiai E et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med 10, 871–875 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glasky MS & Reading CL Stability of specific immunoglobulin secretion by EBV-transformed lymphoblastoid cells and human-murine heterohybridomas. Hybridoma 8, 377–389 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Singh A & Peppas NA Hydrogels and scaffolds for immunomodulation. Adv. Mat 26, 6530–6541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purwada A, Roy K & Singh A Engineering vaccines and niches for immune modulation. Acta Biomater. 10, 1728–1740 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Irvine DJ, Stachowiak AN & Hori Y Lymphoid tissue engineering: invoking lymphoid tissue neogenesis in immunotherapy and models of immunity. Sem. Immunol 20, 137–146 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Suematsu S & Watanabe T Generation of a synthetic lymphoid tissue-like organoid in mice. Nat. Biotechnol 22, 1539–1545 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Stachowiak AN & Irvine DJ Inverse opal hydrogel-collagen composite scaffolds as a supportive microenvironment for immune cell migration. J. Biomed. Mater. Res. A 85, 815–828 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Gaharwar AK et al. Shear-thinning nanocomposite hydrogels for the treatment of hemorrhage. ACS Nano 8, 9833–9842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier JR et al. Bioactive nanoengineered hydrogels for bone tissue engineering: a growth-factor-free approach. ACS Nano 9, 3109–3118 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Victoratos P et al. FDC-specific functions of p55TNFR and IKK2 in the development of FDC networks and of antibody responses. Immunity 24, 65–77 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Goodnow CC, Vinuesa CG, Randall KL, Mackay F & Brink R Control systems and decision making for antibody production. Nat. Immunol 11, 681–688 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Dumin JA et al. Pro-collagenase-1 (matrix metalloproteinase-1) binds the alpha(2)beta(1) integrin upon release from keratinocytes migrating on type I collagen. J. Biol. Chem 276, 29368–29374 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Purwada A et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 63, 24–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerchietti LC et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J. Clin. Invest 120, 4569–4582 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerchietti LC et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17, 400–411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ame-Thomas P et al. Characterization of intratumoral follicular helper T cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia 26, 1053–1063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lenz G et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med 359, 2313–2323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cayrol F et al. Integrin αvβ3 acting as membrane receptor for thyroid hormones mediates angiogenesis in malignant T cells. Blood 125, 841–851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasturi SP et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470, 543–547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiarle R et al. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat. Med 14, 676–680 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Davis RE et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hailfinger S et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 106, 19946–19951 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenz G et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Lenz G et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J. Exp. Med 204, 633–643 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenz G et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA 105, 13520–13525 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagel D et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell 22, 825–837 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Tian YF et al. Integrin-specific hydrogels as adaptable tumor organoids for malignant B and T cells. Biomaterials 73, 110–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caganova M et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 123, 5009–5022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beguelin W et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su IH et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol 4, 124–131 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Fontan L et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell 22, 812–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clozel T et al. Mechanism-based epigenetic chemosensitization therapy of diffuse large B cell lymphoma. Cancer Discov. 3, 1002–1019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Linterman MA How T follicular helper cells and the germinal centre response change with age. Immunol. Cell Biol 92, 72–79 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Purwada A et al. Self-assembly protein nanogels for safer cancer immunotherapy. Adv. Healthc. Mater 5, 1413–1419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh A et al. An injectable synthetic immune-priming center mediates efficient T-cell class switching and T-helper 1 response against B cell lymphoma. J. Control. Release 155, 184–192 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Singh A, Suri S & Roy K In-situ crosslinking hydrogels for combinatorial delivery of chemokines and siRNA-DNA carrying microparticles to dendritic cells. Biomaterials 30, 5187–5200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh A et al. Efficient modulation of T-cell response by dual-mode, single-carrier delivery of cytokine-targeted siRNA and DNA vaccine to antigen-presenting cells. Mol. Ther 16, 2011–2021 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Moon JJ et al. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand Tfh cells and promote germinal center induction. Proc. Natl. Acad. Sci. USA 109, 1080–1085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Look M et al. Nanogel-based delivery of mycophenolic acid ameliorates systemic lupus erythematosus in mice. J. Clin. Invest 123, 1741–1749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou P et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin H et al. Vaccine site inflammation potentiates idiotype DNA vaccine-induced therapeutic T cell-, and not B cell-, dependent antilymphoma immunity. Blood 114, 4142–4149 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Chiarle R et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med 11, 623–629 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Chiarle R et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood 101, 1919–1927 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Krzyzak L et al. CD83 modulates B cell activation and germinal center responses. J. Immunol 196, 3581–3594 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Klein U et al. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA 100, 2639–2644 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ortega-Molina A et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med 21, 1199–1208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorbet MB & Sefton MV Endotoxin: the uninvited guest. Biomaterials 26, 6811–6817 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Round JL et al. Scaffold protein Dlgh1 coordinates alternative p38 kinase activation, directing T cell receptor signals toward NFAT but not NF-kappaB transcription factors. Nat. Immunol 8, 154–161 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Watarai H, Nakagawa R, Omori-Miyake M, Dashtsoodol N & Taniguchi M Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nat. Protoc 3, 70–78 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Moutai T, Yamana H, Nojima T & Kitamura D A novel and effective cancer immunotherapy mouse model using antigen-specific B Cells selected in vitro. PloS One 9, e92732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vuong BQ et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol 10, 420–426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shinkura R et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol 5, 707–712 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Liu N, Ohnishi N, Ni L, Akira S & Bacon KB CpG directly induces T-bet expression and inhibits IgG1 and IgE switching in B cells. Nat. Immunol 4, 687–693 (2003). [DOI] [PubMed] [Google Scholar]
- 82.Talay et al. IgE(+) memory B cells and plasma cells generated through a germinal-center pathway. Nat. Immunol 13, 396–404 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Fischer AH, Jacobson KA, Rose J & Zeller R Cutting sections of paraffin-embedded tissues. CSH Protoc 2008 10.1101/pdb.prot4987 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Lee TT et al. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater 14, 352–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitmire RE et al. Self-assembling nanoparticles for intra-articular delivery of anti-inflammatory proteins. Biomaterials 33, 7665–7675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Syrbu SI & Cohen MB An enhanced antigen-retrieval protocol for immunohistochemical staining of formalin-fixed, paraffin-embedded tissues. Methods Mol. Biol 717, 101–110 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.