Abstract
The universal role of calcium (Ca2+) as a second messenger in cells depends on a large number of Ca2+-binding proteins (CBP), which are able to bind Ca2+ through specific domains. Many CBPs share a type of Ca2+-binding domain known as the EF-hand. The EF-hand motif has been well studied and consists of a helix-loop-helix structural domain with specific amino acids in the loop region that interact with Ca2+. In Toxoplasma gondii a large number of genes (approximately 68) are predicted to have at least one EF-hand motif. The majority of these genes have not been characterized. We report the characterization of two EF-hand motif-containing proteins, TgGT1_216620 and TgGT1_280480, which localize to the plasma membrane and to the rhoptiy bulb, respectively. Genetic disruption of these genes by CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) resulted in mutant parasite clones (Δtg216620 and Δtg280480) that grew at a slower rate than control cells. Ca2+ measurements showed that Δtg216620 cells did not respond to extracellular Ca2+ as the parental controls while Δtg280480 cells appeared to respond as the parental cells. Our hypothesis is that TgGT1_216620 is important for Ca2+ influx while TgGT1_280480 may be playing a different role in the rhoptries.
Keywords: EF-hand domain, plasma membrane, rhoptry, calcium entry
INFECTION with the Apicomplexan parasite Toxoplasma gondii causes toxoplasmosis, a widespread disease that could result in devastating pathologies for fetuses from recently infected mothers and for immunocompromised patients, i.e. those with AIDS (Tenter et al. 2000). Approximately 30% of the world human population as well as a large number and variety of warm-blooded animals are infected with T. gondii (Hill et al. 2005). During its lytic cycle, Toxoplasma actively invades host cells, creating a parasitophorous vacuole (PV), where it divides to finally exit in search of a new host cell. Parasite invasion is an active process involving several discrete steps (Frenal et al. 2017). This lytic cycle is directly linked to the pathology caused by Toxoplasma infection (Blader et al. 2015).
Calcium ions (Ca2+) act as second messengers for multiple cell signaling pathways (Clapham 2007). Ca2+ reversibly binds to Ca2+-binding proteins over a wide range of affinities. This binding is important for buffering or for transmission of information (Clapham 2007). The localization and specificity of Ca2+ signals are directly linked to this wide range of binding affinities. The most studied and most common domain that binds Ca2+ is the EF-hand-motif. The EF-hand structure consists of about 30 amino acids in a helix-loop-helix sequential arrangement (Yanez et al. 2012, Grabarek 2006). This structural motif has been found in a large number of protein families (Grabarek 2006) with diverse functions that include Ca2+ buffering in the cytosol, signal transduction between cellular compartments (Yanez et al. 2012) and muscle contraction (Lewit-Bentley and Rety 2000). The most extensively studied family of Ca2+-binding proteins is the calmodulin family with a large number of members (Chin and Means 2000).
In T. gondii, Ca2+ signaling is important for invasion, motility, and egress (Arrizabalaga and Boothroyd 2004) and EF-hand domain-containing proteins are directly involved in the regulation of these cellular processes (Lourido and Moreno 2015).
In the present study, a bioinformatic analysis of Toxoplasma predicted EF-hand containing proteins (ToxoDB) (Gajria et al. 2008) resulted in the identification of 68 proteins with 8 of them containing transmembrane (TM) domains. We selected two genes (TgGT1_216620 and TgGT1_280480) encoding proteins with TM domains for further characterization based on their potential participation in Ca2+ signaling. TgGT1_216620 contains four EF-hand motifs predicted to face the extracellular surface of the cell and seven transmembrane domains. TgGT1_280480 contains two EF-hand-motifs predicted to face the intraluminal surface of the membrane and four transmembrane domains. TgGT1_280480 also includes a coat protein I (COPI) domain within its transmembrane domains.
MATERIALS AND METHODS
Cell culture and strains
T. gondii tachyzoites of RH and tetracyclin-regulated transactivator expressing strains (TatIΔku80) (Sheiner et al. 2011) were grown in Human telomerase reverse transcriptase immortalized foreskin fibroblasts (hTERT) (BD Biosciences, Franklin Lakes, NJ) with DMEM media containing 1% FBS. For subsequent experiments, extracellular tachyzoites were harvested after natural egress and then passaged through a 3 μm membrane (Whatman) followed by washing and dilution with phosphate-buffered saline (PBS, pH 7.4) or buffer A with glucose (BAG, 116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 5.5 mM glucose and 50 mM HEPES, pH 7.4).
C-terminal tagging
C-terminal endogenous tagging was done by ligation independent cloning as previously described (Huynh and Carruthers 2009). Briefly, a homology region of approximately 1 kb covering the 3’ region of TgGT1_216620 or TgGT1_280480 genes, excluding the STOP codon, was amplified by PCR using T. gondii RH genomic DNA as template. The PCR product was cloned into the p3HA.LIC.CAT plasmid (Huynh and Carruthers 2009) generously provided by Boris Striepen (University of Georgia). After sequencing for plasmid validation, the constructs were linearized with AflII and PstI, respectively, and transfected into the TatIΔku80 strain. This was followed by selection and by limiting dilution in the presence of chloramphenicol (20 μM). Positive clones were selected by PCR using primers located upstream of the homology region (forward) and downstream into the p3HA.LIC.CAT plasmid (reverse). These primers are shown in Table S2 as 216620_HA_Val_F, 280480_HA_Val_F and HA_Val_R.
Generation of knockouts using CRISPR/Cas9 system
Δtg216620 and Δtg280480 were generated by insertion of the entire DHFR cassette into the first ~300 nucleotides of the gene to disrupt the coding sequence, following the CRISPR/Cas9 knockout method reported previously (Shen et al. 2014). A fragment corresponding to the single guide RNA (sgRNA) was cloned into pSAG1:CAS9::U6:sgUPRT (Addgene #54467, Cambridge, MA) using the Q5 site-directed mutagenesis kit (NEB). The resulting pSAG1:CAS9::U6:sg216620 or pSAG1:CAS9::U6:sg280480 was co-transfected with the DHFR cassette (in proportion 3:1) to tachyzoites of the RH strain. After pyrimethamine selection (1 μM), parasites were sub-cloned by limiting dilution and screened for positive clones using PCR (216620_CRISPR_Val_F, 216620_CRISPR_Val_R, 280480_CRISPR_Val_F and 280480_CRISPR_Val_R, Table S2).
Quantitative Reverse Transcriptase PCR
Total RNAs of Δtg216620, Δtg280480, and RH as parental control were extracted from freshly lysed parasites using Trizol® reagent (Sigma, St Louis, MO) following manufacturer’s instructions. The RNA samples were further treated with DNAse I for 10 min at 37 °C (New England Biolabs, Ipswich, Massachusetts) to remove contaminating DNA. Four micrograms of purified mRNA were used to synthesize cDNA using the superscript III first-strand synthesis system according to the manufacturer’s protocol (Thermo Fisher Scientific-Life Technologies, Waltham, MA). Quantitative PCR (qPCR) was performed using iQ™SYBR Green master mix (BioRad, Hercules, CA), with 5 μM of each primer and 100 ng of reverse-transcribed cDNA to a final volume of 10 μL (Primers are described in Table S2). The qRT-PCR was carried out on a CFX96™ PCR Real-Time detection system (C1000Touch™ Thermal cycler, BioRad, Hercules, CA). Relative quantification software (CFX Maestro™ software) was used for the analysis. The relative expression levels of the TgGT1_216620 and TgGT1_280480 were calculated as the fold change using the formula 2−ΔΔCt (Livak et al. 1995). The expression of the tubulin gene was used as reference for normalization and the expression of the gene in the RH strain was used as control. For each experiment, reactions were performed in triplicate and at least 3 biological experiments were done.
Plaque assays
Plaque assays were performed as described previously (Liu et al. 2014). Two hundred freshly egressed tachyzoites were used to infect a confluent monolayer of hTERT fibroblasts followed by 8 days of growth. Monolayers were fixed and stained with 5x crystal violet and plaque sizes analyzed with FIJI (Schindelin et al. 2012) by measuring the area of fifteen plaques per biological replicate. At least 3 biological experiments were done for knockouts and controls.
Western blot analysis
For western blots of the TgGT1_216620-3HA expressing parasites, a membrane protein extraction protocol previously described (Fang et al. 2006) was used. Briefly, 2 × 108 tachyzoites were suspended in lysis buffer (20 mM Hepes-Tris pH 7.4, 1 mM EDTA and 1:500 protease inhibitors (P8849 Sigma, St. Louis, MO)), the pellet was centrifuged at 1,000 g for 5 min followed by freeze-thaw. The final pellet was re-suspended in 100 μl of lysis buffer and centrifuged 10 min at 15,000 g. Finally, the protein concentration from supernatant and pellets were measured and boiled in sample buffer (BioRad, Hercules, CA). An SDS 7% gel was used to resolve samples. Extended running times (2 h and 30 min) at gradually increasing voltage (100-200 V) were used to allow the large proteins to slowly migrate into the gel. To improve transferring of high molecular weight proteins to the membrane, transfer was done at 70 V for 3 h. For westerns of the TgGT1_280480-3HA tagged line total lysates were prepared using CelLytic™ Cell M lysis reagent (Sigma, St. Louis, MO) followed by the addition of benzonase, and one volume of 1 mM EDTA with 2% SDS. Suspension was boiled with loading buffer containing 2-Mercaptoethanol. Samples were run in 12% gels and transferred to nitrocellulose membranes.
Immunoblot analysis was performed as reported previously (Chasen et al. 2017). Nitrocellulose membranes with transferred protein were developed using rat anti-HA monoclonal antibody (1:200) (Roche, Basel, Switzerland) or anti-αTubulin antibody (1:30,000). The Odyssey Clx LICOR system was used for detection, using goat anti Rat IRDye®680LT (1:10,000) or goat anti-mouse IRDye®800WC (1:10,000) for secondary antibodies.
Immunofluorescence microscopy
Extracellular parasites were harvested, filtered, and washed once with BAG and 50 μl aliquots containing 2 × 104 parasites were overlaid on coverslips previously treated with 1 mg/ml poly-L-Lysine. Intracellular tachyzoites were grown on hTERT cells previously prepared on coverslips and infected with freshly lysed parasites in regular growth media for 18 h. Both extracellular and intracellular parasites were then fixed with 3% paraformaldehyde for 15 min, followed by permeabilization with 0.25% Triton X-100 for 10 min and blocking with 3% bovine serum albumin (BSA) at room temperature for 1 h. Primary antibodies were mouse anti-HA 1:200 or rat anti-HA 1:100 and secondary antibody goat anti-mouse or goat anti-rat Alexa- Fluor488. Parallel reactions of the anti-HA antibody against parental lines were used for evaluating the unspecific background, which is subtracted from the IFAs of HA-tagged lines. For co-localization studies the antibodies used were: anti-SAG1 (provided by John Boothroyd, Stanford University, CA) at 1:10,000 dilution to label the plasma membrane, anti-GAP45 (a gift from Drew Etheridge, University of Georgia) to label the periphery of the parasites, anti-ROP7 (a gift from Peter Bradley, University of California, Los Angeles, CA) at 1:2,000 dilution and anti-TgCA_RP (carbonic anhydrase related protein) at 1:500 (Chasen et al. 2017) for labeling rhoptries. Secondary antibodies were goat anti-mouse or goat anti-rat Alexa-Fluor596 (Thermo Fisher Scientific-Life Technologies, Waltham, MA) used at 1:1,000 for 1h. Slides were mounted with FluoromountG® (SoutherBiotech, Birmingham, AL) containing 1 μg/mL 4’,6-diamino-2-phenylindole (DAPI). Images were taken with an Olympus IX-71 inverted fluorescence microscope with a Photometric CoolSnapHQ CCD camera driven by DeltaVision software (Applied Precision, Seattle, WA).
Cytosolic calcium measurements
Loading of Toxoplasma gondii tachyzoites with Fura-2 AM was done as previously described (Moreno and Zhong 1996). Briefly, freshly obtained parasites were washed twice with BAG by centrifugation (706 g for 10 min) and resuspended to a final density of 1 × 109 parasites/ml in loading buffer (BAG plus 1.5% sucrose, and 5 μM Fura-2 AM). The suspension was incubated for 26 min at 26 °C with mild agitation. Subsequently, parasites were washed twice with BAG to remove extracellular dye, re-suspended to a final density of 1 × 109/ml in BAG and kept on ice. For fluorescence measurements, 2 × 107 parasites/ml were placed in a cuvette with 2.5 ml of Ringer’s buffer. Fluorescence measurements were done in a Hitachi F-4500 fluorescence spectrophotometer using the Fura-2 AM conditions for excitation (340 and 380 nm) and emission (510 nm). The Fura-2 AM fluorescence response to Ca2+ was calibrated from the ratio of 340/380 nm fluorescence values after subtraction of the background fluorescence of the cells at 340 and 380 nm as described previously (Grynkiewicz et al. 1985). The Ca2+ release rate is the change in Ca2+concentration during the initial 20 s after compound addition (Pace et al. 2014).
RESULTS
EF-hand domain containing proteins with transmembrane domains
We searched the T. gondii database (ToxoDB) for EF-hand domain containing proteins. The strategy combined genes that are annotated in the InterPro and Pfam databases with genes that have the consensus EF-hand motif (DxDxDxxIxxxE). We selected genes that predicted to encode proteins with 1 or more TM domains. The list of genes retrieved is shown in Table S1. A total of 8 genes were found and we selected two for further characterization: TgGT1_216620 and TgGT1_280480, both genes have a negative phenotypic score in ToxoDB −1.9 and −0.75 respectively (Sidik et al. 2016).
Localization and characterization of TgGT1_216620
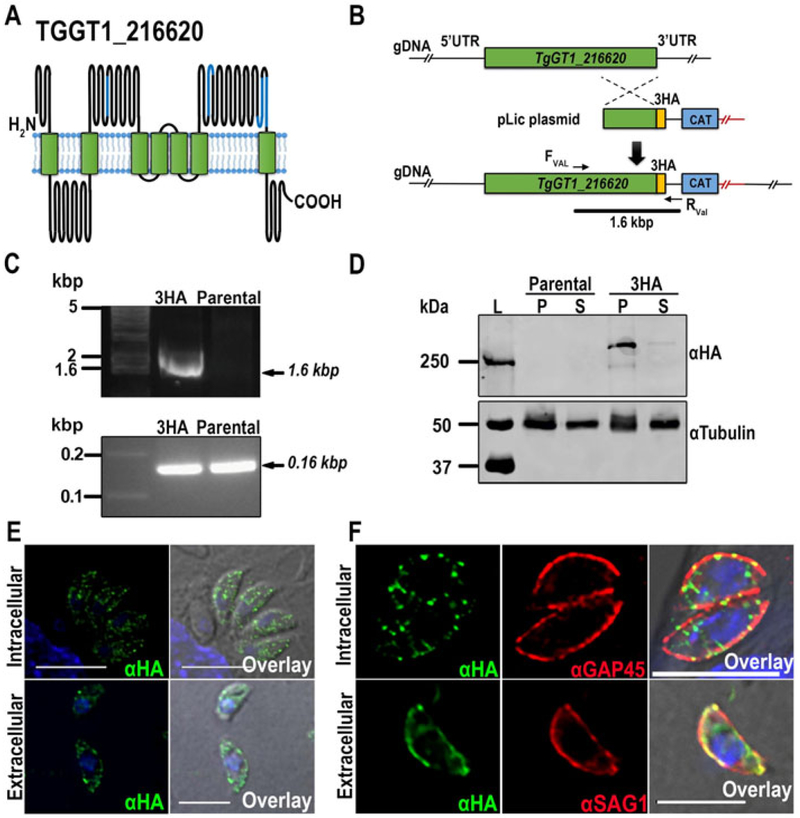
The TgGT1_216620 gene annotated in ToxoDB (Gajria et al. 2008) predicts a protein of 3,700 amino acids with a molecular weight of 412 kDa and seven TM domains (TMHMM server). TgGT1_216620 contains four EF-hand motifs at the positions: 1474-1491, 2386-2421, 3293-3330 and 3348-3383 that are predicted to face the extracellular side of the membrane. The predicted topology is shown in Fig 1A.
Figure 1.
Tagging and localization of TgGT1_216620. (A) Schematic model showing the predicted topology of TgGT1_216620. The EF-hand-motifs are marked in blue. The model was generated using the Protter web application (Omasits et al. 2014). (B) Scheme depicting the strategy used for the C-terminal tagging of TgGT1_216620 in the parental strain TatiΔku80. The genomic region and recombination fragment are shown in green, the C-terminal HA tag in yellow and the selection marker, chloramphenicol acetyl transferase (CAT), in blue. (C) Top panel, PCR analysis showing the correct insertion of 3HA tag (3HA) at the 3’ region of the TgGT1_216620gene. Bottom panel, PCR positive control of parental and 3HA tagged parasites. (D) Western blot analysis of total lysates from TgGT1_216620-3HA and TatIΔku80 parental cells, respectively. Top panel, two bands above ~250 kDa were detected in lysates of the tagged strain, but not in lysates of the parental strain. Bottom panel, loading controls developed with anti-tubulin. (E) Immunofluorescence analysis of intracellular and extracellular tachyzoites showing a punctuated localization of α-HA in both intracellular and extracellular parasites with a higher concentration at the plasma membrane. Scale bars = 5 μm (F) Top panel, IFAs showing co-localization of α-HA and α-GAP45 in intracellular parasites at the periphery. Bottom panel, co-localization of α-HA and α-SAG1, a plasma membrane marker, in extracellular tachyzoites (bottom). Scale bars = 5 μm
We investigated the localization of TgGT1_216620 by inserting a 3HA at the C-terminus of the TgGT1_216620 gene endogenous locus (Fig 1B), as described under materials and methods. The linearized construct was transfected into TatIΔku80 tachyzoites for chloramphenicol selection and further subcloning. The correct insertion of the tag was validated by PCR, which produced an expected fragment of 1.6 kbp (Fig. 1C). In addition, primers annealing inside the coding sequence of the gene were used as positive control and a band of the expected size of ~ 156 bp was obtained from both parental and TgGT1_216620-3HA DNA samples. (Figure. 1C, bottom). Expression of the tagged protein was verified by western blot analysis using anti-HA (Fig. 1D). Membrane fractions (P) of parasites expressing TgGT1_216620-3HA showed two bands above 250 kDa. We attempted to calculate the size of this protein by graphing migration distance versus log10 of the standard, which resulted in an approximate size of ~270 kDa for TgGT1_216620-3HA. The predicted size for this protein is 415 kDa, and we suspect that our calculation is inaccurate because this value is outside the molecular size range of the markers and outside the linearity of the standard curve. However, it is evident that TgGT1_216620 is significantly larger than that, so we could only predict that its size would be close to 415 kDa. The presence of two bands could be the result of internal processing. No band was observed in lysates of the parental TatiΔku80 cells or in supernatant (S) fractions (Fig 1D). Immunofluorescence analysis of cells expressing the tagged gene showed that TgGT1_216620 localized predominantly to the plasma membrane in extracellular tachyzoites (Fig. 1E) and have a plasma membrane and punctuated localization in intracellular tachyzoites (Fig 1E, 1F). Anti SAG1 antibodies were used for co-localization studies to confirm plasma membrane localization in extracellular tachyzoites (Fig 1F), and anti-GAP45 antibodies to label the periphery in intracellular parasites (Fig. 1F).
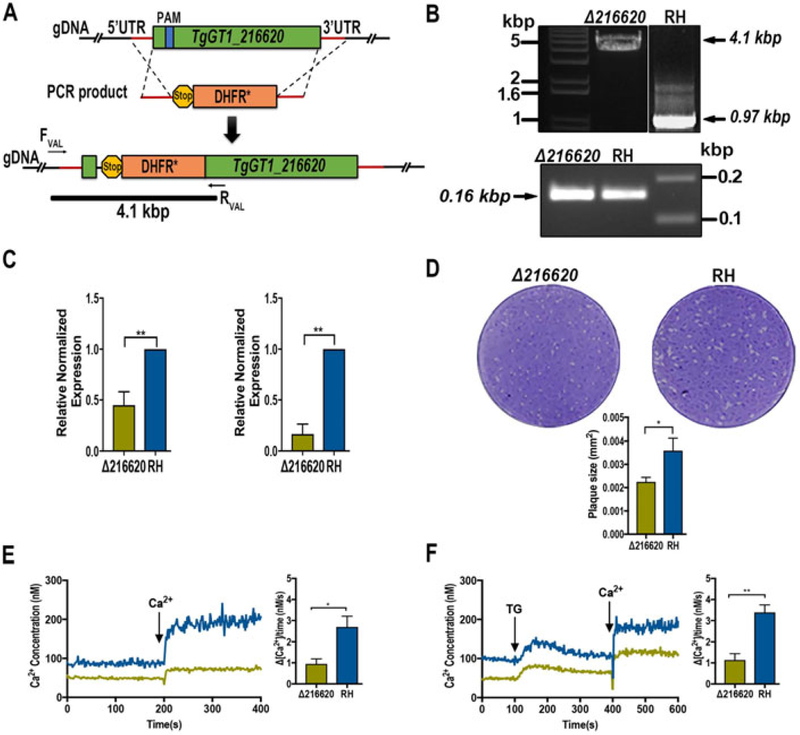
We used CRISPR/Cas9 to insert the selection marker gene DHFR to disrupt the transcription of the TgGT1_216620 gene (Sidik et al. 2014, Shen et al. 2014) (Fig. 2A, top). Insertion of this gene into the TgGT1_216620 locus interfered with the synthesis of the correct transcript and allowed the selection of clonal populations with pyrimethamine (Fig. 2A, bottom). A PCR product of 4.1 kbp obtained from the mutant parasites DNA indicates the insertion into the predicted locus. This fragment was only amplified from DNA of mutants but not from parental cells (Fig. 2B). Quantitative RT-PCR analysis showed a reduction of 55% in the levels of TgGT1_216620 RNA in Δtg216620 parasites when using primers targeting downstream the site of insertion of the drug cassette (Fig. 2C, left), and a decrease of 90% in mRNA levels using primers located upstream and downstream of the drug cassette (Fig. 2C, right). This result indicates that it is very unlikely that full transcripts of TgGT1_216620 are being made in the knockout strain line. Full growth of the mutant parasites was defective as shown by plaque assays (Fig. 2D, top). The size of the plaques formed by the Δtg216620 parasites was significantly smaller than the plaques formed by RH parasites (Fig 2D, bottom).
Figure 2.
Characterization of TgGT1_216620. (A) Scheme showing the strategy used for generation of TgGT1_216620 gene knockouts in the Toxoplasma gondii RH strain. The cartoon shows the genomic region and recombination fragment, protospacer (blue), and selection marker (orange; dihydrofolate reductase, DHFR). (B) Top panel, disruption of the TgGT1_216620 gene by insertion of a DHFR cassette. PCR reaction with primers upstream and downstream of insertion site produced a fragment of 4.1 kbp including the DHFR (Δtg216620]. PCR product obtained with template DNA from the parental strain produces a fragment of ~1 kbp corresponding to the original TgGT1_216620 gene. Bottom panel, PCR reaction of positive controls for Δtg216620 and parental parasites. (C) qPCR analysis of total RNA harvested from the Δtg216620 mutant strain and parental strain RH. Left panel, transcript levels obtained with primers downstream of the insertion site. Right panel, mRNA levels using primers upstream and downstream of the drug cassette insertion site. (D) Plaque assays showing growth of the Δtg216620 mutant strain compared with the RH strain. Plaque size measurements and statistic analysis n = 3, P<0.05. (E) Changes in cytosolic Ca2+ levels of Fura-2 AM loaded tachyzoites [Δtg216620 mutants and RH). Parasites were in suspension and Ca2+ (2 mM) was added at the time indicated. The bar graph to the right shows the quantification and statistical analysis from three biological experiments, each in duplicate. (F) Changes in cytosolic Ca2+ levels of Fura-2 AM loaded tachyzoites [Δtg216620 mutants and RH). Parasites were in suspension and 1 μM Thapsigargin (TG) and 2 mM Calcium (Ca2+) were added at the times indicated. The bar graph to the right shows the quantification and statistical analysis of three biological experiments, each in duplicate. P<0.05.
Because TgGT1_216620 localizes to the periphery of the parasite and its sequence predicts the presence of Ca2+ binding domains, we tested if mutant parasites had a defect in Ca2+ influx. Δtg216620 cells were loaded with Fura-2 AM and Ca2+ measurements performed as reported previously (Moreno and Zhong 1996, Pace et al. 2014) (Fig. 2E). Addition of extracellular Ca2+ to tachyzoites previously suspended in a buffer with 100 μM EGTA led to an increase in cytosolic Ca2+ that we attribute to Ca2+ influx. This is a highly reproducible phenomenon (Fig. 2E, blue trace). The Δtg216620 parasites showed a lower increase in cytosolic Ca2+ as compared with control parasites, which we interpret as a defect in Ca2+ influx. This defect was also observed after pre-addition of thapsigargin, which increases cytosolic Ca2+ by leaking it from the ER (Fig. 2F) and this normally accelerates the rate of Ca2+ influx (Figs. 2E and F, compare the blue rate). The response to thapsigargin by the Δtg216620 tachyzoites, which is shown by an increase in cytosolic calcium due to leakage from the endoplasmic reticulum, was similar to the response obtained from the parental strain. This means that the free calcium in the ER has not been affected by the deletion of the plasma membrane protein.
Localization and characterization of TgGT1_280480
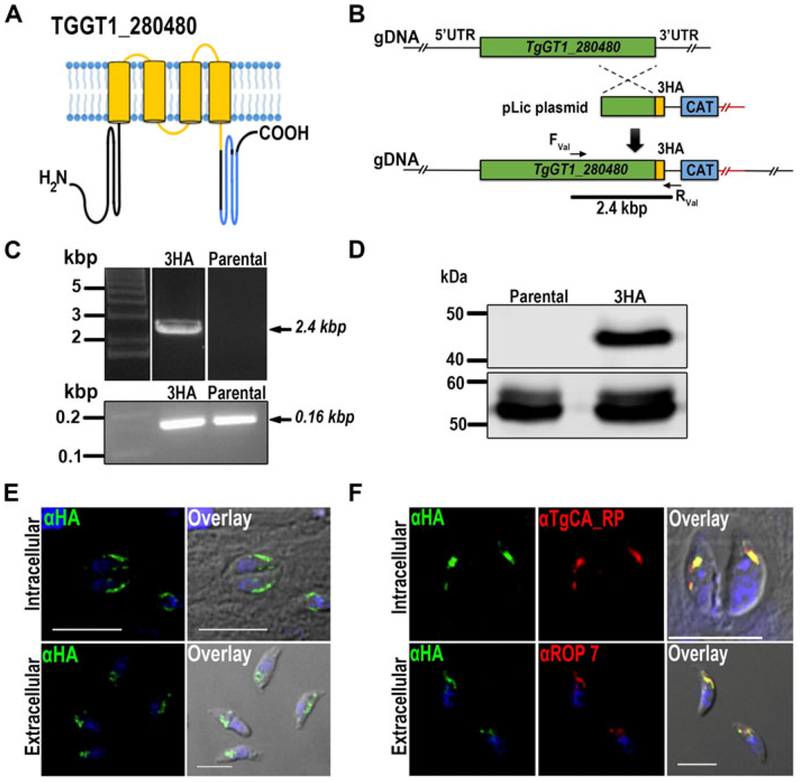
The TgGT1_280480 gene predicts a protein of 305 amino acids with an apparent molecular weight of 34.6 kDa, and two EF-hand-motifs located between amino acids 231-266 and 267-302 and one coat protein I (COPI) domain anchored to its transmembrane domains (amino acids 85-217) (Fig. 3A). Proteins carrying this domain are predicted to co-localize with COPI vesicle coat proteins that form part of the early secretory pathway (Beck et al. 2009). The predicted topology of the protein was generated with Protter (Omasits et al. 2014) and is shown in Fig. 3A.
Figure 3.
Tagging and localization of TgGT1_280480. (A) Model showing the predicted transmembrane topology of TgGT1_280480. The EF-hand Ca2+ binding domains are marked in blue, and the COPI associate domain marked in yellow. (B) Scheme showing the strategy used for C-terminal tagging of TgGT1_280480 through homologous recombination in RHTatIΔku80 strain. The genomic region and recombination fragment are shown in green, the C-terminal HA tag in yellow and the selection marker, chloramphenicol acetyl transferase (CAT), in blue. (C) Top panel, PCR validation of TgGT1_280480-3HA tagged strain (Table S2). A 2.4 kbp band confirmed correct HA integration as indicated by the bar in B. Bottom panel, PCR positive control of parental and 3HA tagged parasites. (D) Top panel, Western blot analysis of total lysates from TgGT1_280480-3HA and parental cells, showing a band at 37 kDa in the tagged line. Bottom panel, tubulin was used as loading control (α-Tubulin antibodies) (E) IFA analysis of intracellular and extracellular tachyzoites showing that TgGT1_280480-3HA localizes to the rhoptry bulb region. Scale bars = 5 μm (F) Top panel, IFA of extracellular tachyzoites showing co-localization of α-HA and α-ROP7, a rhoptry bulb marker. Bottom panel, IFA showing co-localizations of rhoptry marker α-TgCA_RP (carbonic anhydrase related protein) and α-HA antibodies in intracellular parasites. Scale bars = 5 μm.
We followed the strategy described above (Fig. 1B) for inserting a 3HA tag at the C-terminus of the genomic locus of TgGT1_280480 (Fig. 3B). Selection and subcloning followed, and a clonal cell line TgGT1_280480-3HA was produced. Validation of the correct insertion was done by PCR and sequencing. A fragment of 2.4 kbp (Fig. 3C) was amplified from DNA isolated from TgGT1_280480-3HA parasites with primers complementary to the upstream part of the homology region and the 3’UTR of the 3HA plasmid (Fig. 3C and Table S2).
Expression of the tagged protein was confirmed by western blot analysis as shown in Fig. 3D. Immunoblots of total TgGT1_280480-3HA parasite lysates with an anti-HA antibody showed a band of 37 kDa, while no bands were detected in lysates of control parasites (Fig. 3D). IFAs of TgGT1_280480-3HA parasites showed that the protein localized to the rhoptry blub in extracellular and intracellular parasites (Fig. 3E, F). Co-localization studies with anti-TgCA_RP antibodies (red) in intracellular parasites and with anti-ROP7 antibodies (red) in extracellular tachyzoites confirmed rhoptry localization (Fig. 3F).
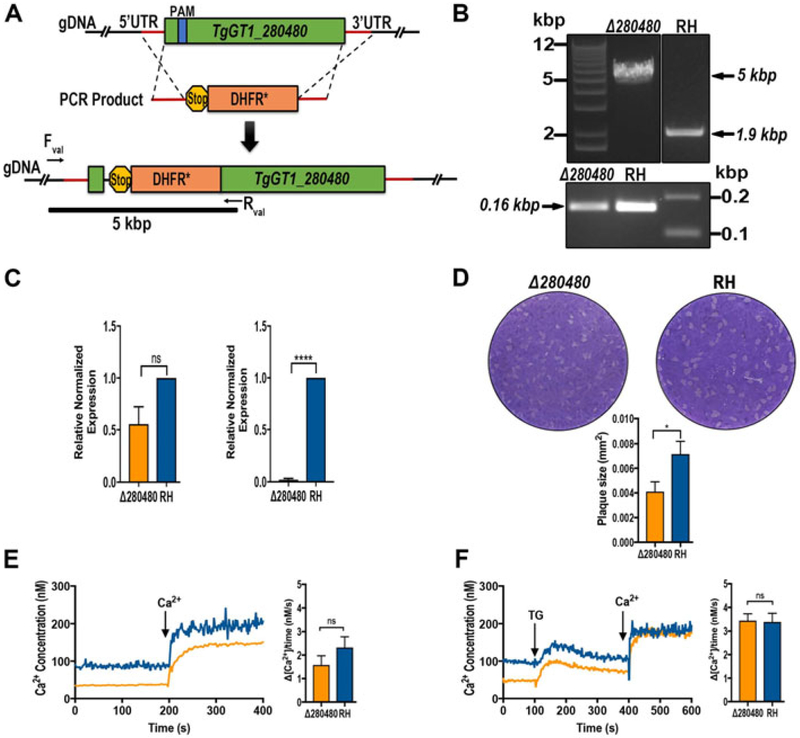
Using CRISPR/Cas9 a copy of the DHFR resistant gene was introduced into the TgGT1_280480 gene locus (Fig. 4A, top) with the aim of disrupting gene transcription (Fig. 4A, bottom). Proper integration into the locus was validated by PCR. A band of 5 kbp corresponding to the size of DHFR cassette and flanking homology regions was amplified (Fig. 4B). Quantitative RT-PCR analysis showed a reduction of 30% in the levels of TgGT1_280480 mRNA in Δtg280480 parasites when using primers targeting downstream the drug cassette insertion (Fig. 4C, left), and a decrease of ~95% in mRNA levels using primers located upstream and downstream of the drug cassette (Fig. 4C, right). This result indicates that it is very unlikely that full transcripts of TgGT1_280480 are being made in the knockout strain line.
Figure 4.
Characterization of TgGT1_280480. (A) Scheme showing the strategy used for TgGT1_280480 gene knockout. CRISPR/Cas9 gene knockout strategy was done in the RH strain. Genomic region (green) and recombination fragment, protospacer (blue), and selection marker (orange; DHFR) are shown. (B) Top panel, insertion of the drug cassette disrupts the TgGT1_280480 gene. PCR product using the primers indicated in the cartoon in A produce a product of 5 kbp validating the insertion in Δtg280480 (fragment is marked in the scheme in A) while a fragment of 1.9 kbp is obtained from the PCR reaction of the parental strain. Bottom panel, PCR positive control from parental and Δtg280480. (C) qPCR analysis of total RNA harvested from Δtg280480 mutant strain and parental strain RH. Left, expression level obtained with primers downstream of the drug cassette insertion site. Right, mRNA levels using primers upstream and downstream of the drug cassette insertion site. (D) Plaque assays showing growth of plaques formed by Δtg280480 mutant and RH parasites. Plaque size measurements and statistic analysis n = 3, P<0.05. (E) Changes in cytosolic Ca2+ levels of Fura-2 AM loaded tachyzoites [Δtg280480 mutants and RH). Parasites were in suspension as described in Material and Methods and Ca2+ (2 mM) was added at the time indicated. The bar graph to the right shows the quantification and statistical analysis from three biological experiments, each in duplicate. (F) Changes in cytosolic Ca2+ levels of Fura-2 AM loaded tachyzoites [Δtg280480 mutants and RH). 1 μM Thapsigargin and 2 mM Calcium (Ca2+) were added at the times indicated. The bar graph to the right shows the quantification and statistical analysis of three biological experiments, each in duplicate. P<0.05.
Plaque assays of the ΔTgGT1_280480 parasites showed that they grew at a slower rate than the parental cell line (Fig 4D).
Because of the presence of Ca2+-binding domains we analyzed these mutant parasites for any potential defect in cytosolic Ca2+ homeostasis or influx. We loaded these parasites with the calcium indicator Fura-2 AM and performed measurements following published protocols (Moreno and Zhong 1996). Cytosolic Ca2+ levels of these mutants were similar to those of control parasites and they appeared to respond to thapsigargin similarly to control parasites, suggesting that TgGT1_280480 does not impact Ca2+ leakage from the ER (Fig. 4F). Ca2+ influx was not affected in the TgGT1_280480 mutants (Fig. 4E).
DISCUSSION
Calcium ions impact nearly every aspect of cellular life. Ca2+ binding proteins (CBPs) with a wide range of affinities for Ca2+ play a central role by orchestrating the localization and specificity of Ca2+ signals. CBPs bind Ca2+ at specific domains like the EF-hand-motifs. EF-hand-motif containing proteins in mammalian cells have been divided in two groups: Ca2+ sensors and Ca2+ buffers (Ikura 1996). In Toxoplasma, a cytosolic Ca2+ increase precedes essential virulence traits that form part of its lytic cycle (Arrizabalaga and Boothroyd 2004, Lourido and Moreno 2015). The molecular players and the detailed mechanisms involved are not completely known. However, it is very likely that CBPs, through EF-hand-motifs, play a role in the orchestration of a variety of biological responses important for the infection cycle of the parasite. The genome of T. gondii contains a large number of genes predicted to encode proteins possessing at least one EF-hand-motif. Most of these genes have unknown function and only a handful have been characterized (Seeber et al. 1999, Lourido et al. 2010, McCoy et al. 2012, Treeck et al. 2014, Wang et al. 2016, Long et al. 2017). In this work we report the localization and preliminary characterization of two EF-hand-motif-containing proteins TgGT1_216620 and TgGT1_280480.
The TgGT1_216620 gene predicts the expression of a protein with seven transmembrane domains and four EF-hand motifs predicted to face toward the extracellular side of the membrane. The gene does not show apparent homology with previously described proteins apart from the presence of the EF-Hand-motifs and a permease of the drug/metabolite transporter (DMT) superfamily domain (RhaT domain) (amino acids 2008 to 2189). The TgGT1_216620 amino acid sequence shows 65% and 41% identity with orthologs from Eimeria tenella and Cryptosporidium parvum annotated as hypothetical EF-hand-motif containing proteins.
According to the immunofluorescence assay (IFA) results the TgGT1_216620-3HA protein is predominantly expressed at the plasma membrane of intracellular and extracellular tachyzoites. Gene disruption of TgGT1_216620 in the RH strain provoked defects in parasite growth, as seen in plaque assays, and deficient Ca2+ influx. The response to the SERCA-Ca2+-ATPase inhibitor, thapsigargin was not affected indicating that the function of the ER Calcium store is likely not influenced by this protein. The presence of seven transmembrane domains, its plasma membrane localization, the presence of potential Ca2+ binding domains and the Ca2+ influx phenotype of mutant clones supported a potential role in Ca2+ influx and downstream signaling.
TgGT1_280480 has specific subcellular localization in both intracellular and extracellular tachyzoites. Localization of TgGT1_280480-3HA to the rhoptry bulb was confirmed by IFAs of intracellular and extracellular tachyzoites with the rhoptry markers ROP7 and TgCA_RP. TgGT1_280480 does not appear to be essential for parasite survival and Ca2+ influx was not affected in the Δtg280480 parasites. TgGT1_280480 is predicted to possess two EF-hand motifs and a COPI-associated protein domain. COPI domains are usually found in proteins that colocalize with COPI coated vesicles that mediate trafficking from Golgi to the ER (Adisa et al. 2002). We did not observe a Golgi localization of TgGT1_280480, but is known that secretory proteins (MIC and ROP) traffic from the ER and Golgi during tachyzoite division (Venugopal et al. 2017).
qPCR data showed that transcripts levels for the TgGT1_216620 and TgGT1_280480 genes were reduced in the ΔTgGT1_216620 and ΔTgGT1_280480 mutants. Considering that the CRISPR reaction only resulted in insertion of the selection marker gene it is possible that transcripts are still being made although it is very unlikely that they produce active protein.
In summary, we identified two membrane proteins containing EF-hand motifs, TgGT1_216620 and TgGT1_280480. These two proteins may be part of a more complex signaling network and their localization may assist in the characterization of the Toxoplasma Ca2+ signaling toolkit Especially intriguing is the localization of TgGT1_216620 to the plasma membrane and the prediction for 7 transmembrane domains giving the protein the possibility of potentially transducing extracellular signals to a specific intracellular signaling network. TgGT1_280480 localization to the rhoptries may indicate a need for the rhoptries for calcium for the diverse maturation functions going on in the organelle. Further studies are needed to understand the TgGT1_216620 and TgGT1_280480 detailed function in the parasite.
Supplementary Material
List of EF-hand domain containing proteins with transmembrane domains
Primers used in this study
ACKNOWLEDGMENTS
Beejan Asady and Melissa Storey provided technical assistance. We thank Dr. Muthugapatti K. Kandasamy and the Biomedical Microscopy Core (BMC), Coverdell, UGA for the use of the microscopes and also for training and advice. We thank John Boothroyd for the anti-SAG1 antibody and Peter Bradley for the anti-ROP7 antibody. This work was supported by U.S. National Institutes of Health grants AI128356 and AI110027 to SNJM. LC was supported by the China Scholarship Council (CSC) as a Visiting scholar for two years at the University of Georgia.
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/jeu.12675
LITERATURE CITED
- Adisa A, Rug M, Foley M & Tilley L 2002. Characterisation of a delta-COP homologue in the malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol, 123:11–21. [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G & Boothroyd JC 2004. Role of calcium during Toxoplasma gondii invasion and egress. Int J Parasitol, 34:361–8. [DOI] [PubMed] [Google Scholar]
- Baltgalvis KA, Jaeger MA, Fitzsimons DP, Thayer SA, Lowe DA & Ervasti JM 2011. Transgenic overexpression of gamma-cytoplasmic actin protects against eccentric contraction-induced force loss in mdx mice. Skelet Muscle, 1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Rawet M, Wieland FT & Cassel D 2009. The COPI system: molecular mechanisms and function. FEBS Lett, 583:2701–9. [DOI] [PubMed] [Google Scholar]
- Blader IJ, Coleman BI, Chen CT & Gubbels MJ 2015. Lytic Cycle of Toxoplasma gondii: 15 Years Later. Annu Rev Microbiol, 69:463–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasen NM, Asady B, Lemgruber L, Vommaro RC, Kissinger JC, Coppens I & Moreno SNJ 2017. A Glycosylphosphatidylinositol-Anchored Carbonic Anhydrase-Related Protein of Toxoplasma gondii Is Important for Rhoptry Biogenesis and Virulence. Msphere, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D & Means AR 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol, 10:322–8. [DOI] [PubMed] [Google Scholar]
- Clapham DE 2007. Calcium signaling. Cell, 131:1047–58. [DOI] [PubMed] [Google Scholar]
- Fang J, Marchesini N & Moreno SN 2006. A Toxoplasma gondii phosphoinositide phospholipase C (TgPI-PLC) with high affinity for phosphatidylinositol. Biochem J, 394:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenal K, Dubremetz JF, Lebrun M & Soldati-Favre D 2017. Gliding motility powers invasion and egress in Apicomplexa. Nat Rev Microbiol, 15:645–660. [DOI] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ, Wang H & Brunk BP 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Research, 36:D553–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarek Z 2006. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol, 359:509–25. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M & Tsien RY 1985. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem, 260:3440–50. [PubMed] [Google Scholar]
- Hill DE, Chirukandoth S & Dubey JP 2005. Biology and epidemiology of Toxoplasma gondii in man and animals. Anim Health Res Rev, 6:41–61. [DOI] [PubMed] [Google Scholar]
- Huynh MH & Carruthers VB 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell, 8:530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M 1996. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci, 21:14–7. [PubMed] [Google Scholar]
- Lewit-Bentley A & Rety S 2000. EF-hand calcium-binding proteins. Curr Opin Struct Biol, 10:637–43. [DOI] [PubMed] [Google Scholar]
- Liu J, Pace D, Dou ZC, King TP, Guidot D, Li ZH, Carruthers VB & Moreno SNJ 2014. A vacuolar-H+-pyrophosphatase (TgVPl) is required for microneme secretion, host cell invasion, and extracellular survival of Toxoplasma gondii. Molecular Microbiology, 93:698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Marmaro J & Todd JA 1995. Towards fully automated genome-wide polymorphism screening. Nat Genet, 9:341–2. [DOI] [PubMed] [Google Scholar]
- Long S, Brown KM, Drewry LL, Anthony B, Phan IQH & Sibley LD 2017. Calmodulin-like proteins localized to the conoid regulate motility and cell invasion by Toxoplasma gondii. PLoS Pathog, 13:el006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido S & Moreno SN 2015. The calcium signaling toolkit of the Apicomplexan parasites Toxoplasma gondii and Plasmodium spp. Cell Calcium, 57:186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido S, Shuman J, Zhang C, Shokat KM, Hui R & Sibley LD 2010. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature, 465:359–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy JM, Whitehead L, van Dooren GG & Tonkin CJ 2012. TgCDPK3 regulates calcium- dependent egress of Toxoplasma gondii from host cells. PLoS Pathog, 8:el003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SN & Zhong L 1996. Acidocalcisomes in Toxoplasma gondii tachyzoites. Biochem J, 313 ( Pt 2):655–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S & Wollscheid B 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics, 30:884–6. [DOI] [PubMed] [Google Scholar]
- Pace DA, McKnight CA, Liu J, Jimenez V & Moreno SN 2014. Calcium entry in Toxoplasma gondii and its enhancing effect of invasion-linked traits. J Biol Chem, 289:19637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P & Cardona A 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods, 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber E, Beuerle B & Schmidt ΗH 1999. Cloning and functional expression of the calmodulin gene from Toxoplasma gondii. Mol Biochem Parasitol, 99:295–9. [DOI] [PubMed] [Google Scholar]
- Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O., Behnke MS., White MW. & Striepen B. 2011. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog, 7:el002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD & Sibley LD 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio, 5:e01114–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Hackett CG, Tran F, Westwood NJ & Lourido S 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One, 9:el00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, Thiru R, Saeij JPJ., Carruthers VB., Niles JC. & Lourido S. 2016. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell, 166:1423–1435 el2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenter AM, Heckeroth AR & Weiss LM 2000. Toxoplasma gondii: from animals to humans. Int J Parasitol, 30:1217–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treeck M, Sanders JL, Gaji RY, LaFavers KA, Child MA, Arrizabalaga G, Elias JE & Boothroyd JC 2014. The calcium-dependent protein kinase 3 of toxoplasma influences basal calcium levels and functions beyond egress as revealed by quantitative phosphoproteome analysis. PLoS Pathog, 10:el004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal K, Werkmeister E, Barois N, Saliou JM, Poncet A, Huot L, Sindikubwabo F, Hakimi MA, Langsley G, Lafont F & Marion S 2017. Dual role of the Toxoplasma gondii clathrin adaptor AP1 in the sorting of rhoptry and microneme proteins and in parasite division. PLoS Pathog, 13:el006331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Huang SY, Li TT, Chen K, Ning HR & Zhu XQ 2016. Evaluation of the basic functions of six calcium-dependent protein kinases in Toxoplasma gondii using CRISPR- Cas9 system. Parasitol Res, 115:697–702. [DOI] [PubMed] [Google Scholar]
- Yanez M, Gil-Longo J & Campos-Toimil M 2012. Calcium binding proteins. Adv Exp Med Biol, 740:461–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of EF-hand domain containing proteins with transmembrane domains
Primers used in this study