Abstract
Anaplastic large-cell lymphoma, a T-cell neoplasm, is primarily a pediatric disease. Seventy-five percent of pediatric anaplastic large-cell lymphoma cases harbor the chromosomal translocation t(2;5)(p23;q35) leading to the ectopic expression of NPM-ALK, a chimeric tyrosine kinase. NPM-ALK consists of an N-terminal nucleophosmin (NPM) domain fused to an anaplastic lymphoma kinase (ALK) cytoplasmic domain. Pediatric NPM-ALK+ anaplastic large-cell lymphoma is often a disseminated disease and young patients are prone to chemoresistance or relapse shortly after chemotherapeutic treatment. Furthermore, there is no gold standard protocol for the treatment of relapses. To the best of our knowledge, this is the first study on the potential role of the microRNA, miR-497, in NPM-ALK+ anaplastic large-cell lymphoma tumorigenesis. Our results show that miR-497 expression is repressed in NPM-ALK+ cell lines and patient samples through the hypermethylation of its promoter and the activity of NPM-ALK is responsible for this epigenetic repression. We demonstrate that overexpression of miR-497 in human NPM-ALK+ anaplastic large-cell lymphoma cells inhibits cellular growth and causes cell cycle arrest by targeting CDK6, E2F3 and CCNE1, the three regulators of the G1 phase of the cell cycle. Interestingly, we show that a scoring system based on CDK6, E2F3 and CCNE1 expression could help to identify relapsing pediatric patients. In addition, we demonstrate the sensitivity of NPM-ALK+ cells to CDK4/6 inhibition using for the first time a selective inhibitor, palbociclib. Together, our findings suggest that CDK6 could be a therapeutic target for the development of future treatments for NPM-ALK+ anaplastic large-cell lymphoma.
Introduction
Anaplastic large cell lymphoma (ALCL) is an aggressive form of T-cell non-Hodgkin lymphoma (NHL) with a constant membrane expression of the CD30 antigen, a cytokine receptor from the tumor necrosis factor receptor family. Four distinct entities of ALCL are currently recognized based on the 2016 revised World Health Organization (WHO) lymphoma classification: 1) anaplastic lymphoma kinase (ALK)-positive(+) ALCL; 2) ALK-negative (ALK−) ALCL; 3) primary cutaneous ALCL; and 4) breast implant-associated ALCL.1 ALCL accounts for approximately 10-15% of all NHL in children but is rare in adults (1-2% of adult NHL). Pediatricians have adopted ALCL99 chemotherapy including low cumulative doses of methotrexate, as standard therapy.2 Patients with pediatric ALCL have a favorable prognosis; however, systemic ALCL is often a disseminated disease and pediatric patients are prone to chemoresistance or to relapse after stopping the treatment. All relapses occur in patients with advanced disease and there is still no gold standard for treating relapses.
In children and adolescents, more than 90% of systemic ALCL cases harbor a chromosomal translocation t(2;5)(p23;q35) that leads to the expression of the NPM-ALK chimeric tyrosine kinase. This kinase consists of an N-terminal nucleophosmin (NPM) domain fused to the ALK cytoplasmic domain.3,4 ALK is a tyrosine kinase receptor whose expression is normally restricted to neural progenitor cells during development.5 At diagnosis, ALCL patients are grouped into risk groups based on the presence of NPM-ALK transcripts in peripheral blood and/or bone marrow6 and the quantification of anti-ALK antibody in peripheral blood,7 and chemotherapy treatment is adapted accordingly.
In ALCL, oncogenic NPM-ALK signaling is mediated by different pathways, such as RAS/ERK and JNK/STAT pathways, which play major roles in lymphomagenesis by controlling key cellular processes such as cell cycle progression.8,9 Studies showed increased proliferation of Rat1a fibroblasts upon ectopic expression of NPM-ALK as a consequence of accelerated entry into the S phase of the cell cycle. Furthermore, this was associated with a significant upregulation of cyclin A and cyclin D1 and increased levels of the mitogenic proto-oncogenes Jun, Fos, and Myc.10 Moreover, in 293T and Jurkat cells, it has been shown that forced expression of NPM-ALK induces JNK (JUN N-terminal kinase) and Jun phosphorylation, thereby up-regulating the activity of AP1 transcription factors. This results in uncontrolled cell cycle progression and cell growth due to the downregulation of p21 and the concomitant upregulation of cyclin A and cyclin D3.11 The ectopic expression of constitutively active NPM-ALK in Ba/F3 mouse cells leads to FOXO3A phosphorylation by Akt, upregulation of cyclin D2 expression and downregulation of p27 and Bim-1 expression to promote cell survival and cell cycle progression.12 In Ba/F3 mouse cells, NPM-ALK, via the PI3K/Akt pathway, also controls cell division cycle 25 A (Cdc25A), a key regulator of the G1 phase and the G1/S transition.13
Many microRNAs (miRNAs) modulate several major proliferation pathways by controlling critical regulators such as Cyclin-CDK complexes.14 miRNAs are single-stranded small non-coding RNAs that are pivotal in physiological and pathological processes such as development, cell proliferation and apoptosis. In general, by binding to specific targets with distinct degrees of complementarity, miRNAs exhibit a negative regulatory role at the post-transcriptional level through the inhibition of translation and/or degradation of their messenger RNA targets. There is growing evidence to show that differentially expressed miRNAs are associated with tumor types and cancer development.15 Indeed, several miRNAs display defective expression patterns in tumors, consequently altering oncogenic or tumor suppressive targets. miRNAs such as miR-16, miR-17-92, miR-21, miR-26a, miR-29a, miR-96, miR-101, miR-135b, miR-146a, miR-150, miR-155 and miR-219 are dysregulated and serve as oncogenes or tumor suppressors in NPM-ALK+ ALCL.16–20 Most of these miRNAs have been found to be down-regulated (miR-16, miR-21, miR-26a, miR-29a, miR-96, miR-101, miR-146a, miR-150, miR-155 et miR-219) in NPM-ALK+ ALCL. Our laboratory showed, for the first time, that NPM-ALK+ ALCL cell lines and primary tissues express low levels of several miRNAs mediated by the hypermethylation of their gene promoter.17,21 Both NPM-ALK and STAT3 activities contributed to epigenetic silencing in NPM-ALK+ ALCL cell lines and biopsy specimens by up-regulating and recruiting DNMT1 to the promoter of miR-29a, miR-125b and miR-150.17,19,21 The repressive methylation catalyzed by DNMT1 can be partially reversed by treatment with 5-aza-2’-deoxycytidine (5-aza-dC, decitabine, Dacogen,® SuperGen Inc., Dublin, CA, USA), a DNMT inhibitor. This DNA-demethylating agent has been shown to restore miR-497 expression, which is suppressed in HT29 colorectal cancer cells.22 In addition, miR-497 downregulation has been consistently demonstrated in a variety of solid tumor types such as hepatocellular carcinoma, ovarian cancer, colorectal adenomas, and in multiple myeloma cells.22,23 MiR-497, a highly conserved miRNA encoded by the first intron of the MIR497HG gene on human chromosome 17p13.110 belongs to the miR-15/16 family (miR-15a, miR-15b, miR-16-1/2, miR-195, miR-424 and miR-497) sharing the same seed sequence AGCAGCA.24 Downregulation of miR-497 controls cell cycle progression by regulating cell cycle regulators such as Cyclin A2, Cyclin D1, Cyclin D2, Cyclin D3 and Cdc25a. In a previous study, using microarray miRNA-expression profiling, we showed that miR-195 and miR-497 was differentially expressed in NPM-ALK+ ALCL lymph node primary tissues compared to reactive lymph nodes of healthy donors.21 As miR-195 and miR-497 are encoded as a cluster within the same host gene, MIR497HG (a highly conserved miRNA cluster),25 we sought to simultaneously study the roles of miR-195 and miR-497 in NPM-ALK+ ALCL tumorigenesis. Accordingly, we measured miR-195 and miR-497 expression in human NPM-ALK+ ALCL primary biopsies and cell lines. First, we studied the biological functions of these miRNAs in human NPM-ALK+ ALCL cells. We showed that overexpression of miR-497 inhibits cellular growth and causes cell cycle arrest. We identified cyclin E1, E2F3 and CDK6 as the main miR-497-targets responsible for the observed phenotype. Several CDK4/6 inhibitors have been developed [PD-0332991/palbociclib (Pfizer), LEE011/ribociclib (Novartis), and LY2835219/abemaciclib (Lilly)] and are currently being tested in clinical trials for patients with solid tumors and B lymphomas.26 Previous studies have demonstrated that palbociclib caused cycle arrest and apoptosis in T-cell leukemia in vitro and delayed disease progression in mouse models of T-cell acute lymphoblastic leukemia.27 However, this class of drugs has not been tested in T-cell lymphomas. Thus, we demonstrate, for the first time, the sensitivity of NPM-ALK+ cell lines to CDK4/6 inhibition using palbociclib. Furthermore, we show that a scoring system based on CDK6, E2F3 and CCNE1 expression could help identify relapsing patients with a very short survival. Altogether, our results indicate that miR-497 functions as a tumor suppressor and that some of the miR-497 functional downstream targets could be used as predictors of clinical outcomes. Amongst them, CDK6 could be a therapeutic target for the development of future treatments for NPM-ALK+ ALCL.
Methods
Human cell lines, tumoral and normal samples
The two human NPM-ALK+ ALCL cell lines, KARPAS-299 and SU-DHL1, were obtained from DSMZ (German Collection of Microorganisms and Cell Culture, Braunschweig, Germany). The COST cell line was established in our laboratory.28 CD4+ cells were stimulated for two days using CD3/CD28 antibodies coupled to magnetic beads (Dynabeads, Invitrogen, Waltham, USA) in RPMI-1640 with 20% fetal calf serum (FCS). All cells were cultured in RPMI-1640 supplemented with 10% FCS (KARPAS-299 and COST) or 15% FCS (SU-DHL1) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, and 100 U/mL penicillin/streptomycin (all from Invitrogen, Waltham, USA). Cells were cultured at 37°C with 5% CO2 and maintained in exponential growth phase. Tumor samples from NPM-ALK+ and NPM-ALK− ALCL diagnosed based on morphological and immunophenotypical criteria (according to the last WHO classification) were obtained from our tumor tissue bank. Only cases with at least 50% lymph node involvement (assessed by CD30 staining of frozen biopsies) and good RNA integrity were selected from our tumor bank. Studies were conducted in accordance with the Declaration of Helsinki, and with the approval of the relevant ethics committees. Lymph nodes collected from patients with reactive non-malignant disease, who were considered to be healthy donors, were retrieved from the CRB-Cancer du CHU de Bordeaux n. BRIF BB-0033-00036, a member of the “Cancéropôle Grand Sud-Ouest” network.
Cell treatment
Anaplastic large cell lymphoma cell lines were treated with 2 μM decitabine (Sigma-Aldrich, Saint Quentin Fallavier, France) for four days [fresh drug was added at 48 hours (h)] and NPM-ALK activity was inhibited using 500 nM crizotinib (@rtMolecule, Poitiers, France) for three days. ALCL cell lines were treated with 1 μM of PD-0332991 (Selleck Chemicals, Houston, TX, USA) during 24 h.
MiRNA and siRNA transfections
siRNAs (si-CDK6, si-CCNE1, si-CDC25A and si-STAT3) at 0.5 nmol, si-E2F3 (Online Supplementary Table S1) at 1 nmol or miRNA mimics (miR-497-5p: mirVana MC10490 and miR-CTL: mirVana AM17110, Invitrogen, Waltham, USA) were transfected by electroporation according to a previously described procedure.17 In the case of co-transfection of several siRNAs, 0.2 nmol of each si-CDK6 and si-CCNE1 and 1 nmol of si-E2F3 were used. The same amount of duplex siRNA (Eurogentec, Angers, France) was transfected and used as negative control.
Purification of biotinylated-miRNA containing complexes
Five million KARPAS-299, COST and SU-DHL1 cells were transfected with 0.25 nmol of biotinylated hsa-miR-497-5p (hsa-miR-497 mercury LNA miRNA mimics premium) or biotinylated-irrelevant miRNA (negative control) (Exiqon, Vedbaek, Denmark). Total cell lysates, 24 h post transfection, were prepared in IP-buffer: 25 mM Tris-HCl pH 7.5, 200 mM NaCl, 0.2% Triton-X100, 5 mM MgAc, 1 mM DTT, protease inhibitors (Roche France, Meylan, France) and 0.2 U/μL RNase OUT (Invitrogen, Waltham, USA). The lysates were incubated for 2 h with streptavidin coupled to magnetic beads (Thermofisher, Waltham, USA) previously saturated with 0.5 mg/mL BSA and 0.2 mg/mL yeast tRNA. Purified RNAs were recovered after 5 washes with IP buffer by Trizol reagent-mediated extraction (Invitrogen, Waltham, USA), according to the manufacturer’s instructions. RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen, Waltham, USA) and subjected to quantitative PCR. Biotin pull-down efficiency was expressed as a percentage of input. GAPDH amplification was used to remove pull-down background. The specificity of purification was evaluated for each putative target based on the enrichment in the Biotin-miR-497 condition relative to Biotin-CTL.
RNA extraction, reverse transcription and quantitative PCR
Total RNA from cell lines and CD4+ cells were extracted using the Trizol reagent (Invitrogen, Waltham, USA), according to the manufacturer’s instructions. MiRNAs were reverse transcribed using the Universal cDNA Synthesis Kit II (Exiqon, Vedbaek, Denmark). The expression of miR-195, miR-497, RNU1A1 and SNORD44 (used as reference genes) was measured by quantitative PCR (qPCR) using a specific Exiqon PCR primer set and SYBR qPCR Premix Ex Taq (Tli RNaseH Plus) Kit (Takara Bio Europe, Saint-Germain-en-Laye, France). Messenger RNAs (mRNAs) were reverse transcribed using Primescript RT reagent kit (Takara Bio Europe, Saint-Germain-en-Laye, France), according to the manufacturer’s recommendations. qPCR was performed using the primers listed in Online Supplementary Table S1.
Protein extraction and Western blotting analyses
Total cell lysates were prepared by incubating ALCL cells in Laemmli 1X buffer (Bio-Rad, Marnes-la-Coquette, France) for 10 minutes (min) on ice. Cells were then subjected to sonication using a Bioblock Vibra-cell 72446 apparatus at 35% of its power (Fisher Scientific, Illkirch, France). Proteins were separated by electrophoresis on SDS-polyacrylamide gels and electroblotted onto nitrocellulose membrane (GE Healthcare Europe, Velizy-Villacoublay, France). Proteins of interest were detected using primary antibodies [(CDC25A 1/500: sc-7389, CDK6 1/2000: CST#3136, CCNE1 1/2000: sc-247, E2F3 1/2000: GTX102302, CDK4 1/2000: sc-260, Rb 1/2000: OP66, phosphor-Rb (ser780) 1/2000: CST#9307)] and HRP -conjugated anti-mouse (Promega, Madison, USA) or anti-rabbit secondary antibodies (Cell Signaling Technology, Danvers, USA). Densitometric analysis was performed using ImageJ software.29
Cell proliferation assays and cell cycle analysis
Cell proliferation was evaluated using CellTiter 96AQueus One Solution (Promega, Madison, USA) and cell counting. For cell cycle analysis, NPM-ALK+ ALCL cells were washed with PBS and fixed overnight at 4°C in 70% ethanol. Cells were then washed with PBS supplemented with 0.1% BSA and incubated for 30 min with 10 μg/mL propidium iodide (Invitrogen, Waltham, USA) and 500 μg/mL of RNAse (Sigma-Aldrich, Saint Quentin Fallavier, France). Thereafter, cells were analyzed with a flow cytometer MACSQUANT VYB (Miltenyi, Paris, France).
Apoptosis luminescent assay
The Caspase-Glo® 3/7 Assay Kit (Promega, Madison, WI, USA) was used to assess cell apoptosis, according to the manufacturer’s instructions.
Bisulfite conversion and DNA methylation analysis
Genomic DNA was bisulfite converted using the MethylEdge Bisulfite Conversion System (Promega, Madison, USA) following the manufacturer’s protocol. The region of interest was amplified by PCR and methylation level was measured by pyrosequencing using PyromarkQ24 (Qiagen France, Courtaboeuf, France). (Primers are listed in Online Supplementary Table S1).
Xenograft tumor assay
Mice were housed under pathogen-free conditions in an animal room at constant temperature (20-22°C), with a 12 h/12 h light:dark cycle and free access to food and water. All animal procedures were performed following the principle guidelines of INSERM, and our protocol was approved by the Midi-Pyrénées Ethics Committee on Animal Experimentation. ALCL cell lines were transfected either with control (miR-CTL) or miR-497 miRNA mimics. Twenty-four hours later, a total of 3×106 COST or KARPAS-299 cells or 2×106 SU-DHL1 cells were injected subcutaneously into both flanks of 7-week old female non-obese diabetic/severe combined immunodeficient (NOD-SCID) or NOD-SCID Gamma (NSG) mice (Janvier Labs, Le Genest, St Isle, France). Mouse body weight and tumor volumes were measured three times a week with calipers, using the formula “length × width2 × π/6”. At the end of the experiment, mice (6 per group) were humanely sacrificed. Subcutaneous tumors were then excised and sections were fixed in 10% neutral buffered formalin for histochemical analysis.
RNA sequencing
RNA sequencing was performed at the LISBP, Toulouse, France, on an Ion Proton (Thermofischer, Waltham, MA, USA) as single-end oriented 95pb-long reads. RNAs co-purified with biotinylated hsa-miR-497-5p or with a biotin-negative control mimic (biotin-miR-CTL) were used for library preparation using the Ion Total RNA-seq Kit V2 and Ion PI HiQ OT2 Kit (Thermofischer, Waltham, MA, USA). TopHat2 (v.2.1.0) was used for sequence alignment on the human genome GRCH38.83 and counting reads performed using HTSeq-Count (v.0.6.1P1).
Statistical analysis
Results are presented as mean values±Standard Error of Mean (SEM) from at least 3 independent experiments unless otherwise indicated. Differences between groups were examined using the unpaired two-tailed Student’s t-test, unpaired two-tailed Student’s t-test with Welch’s correction or two-way ANOVA with Bonferroni post-test using GraphPad Prism software version 6.00 for Windows (GraphPad software) (La Jolla, CA, USA). For all tests, P<0.05 (*), P<0.01 (**) or P<0.001 (***) were considered statistically significant. Event-free survival was analyzed using a Receiver Operating Characteristics (ROC) method. A cut-off value was then chosen which would assume a minimum error rate and a limited number of misclassifications within the cohort.
Results
The miR-497∼195 cluster is down-regulated in human NPM-ALK+ primary lymphoma samples and cell lines
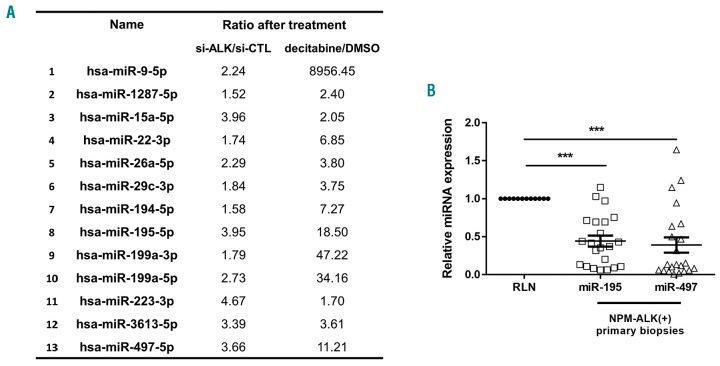
To determine the profile of methylated miRNA genes driven by NPM-ALK oncogene in ALCL cells, we performed a high throughput analysis of miRNA expression (small RNA sequencing) from NPM-ALK+ KARPAS-299 cells following decitabine treatment or transfection with a siRNA targeting ALK mRNA (si-ALK). A set of 13 miRNAs (miR-9-5p, miR-1287-5p, miR-15a-5p, miR-22-3p, miR-26a-5p, miR-29c-3p, miR-194-5p, miR-195-5p, miR-199a-3p, miR-199a-5p, miR-223-3p, miR-3613-5p and miR-497-5p) was found to be up-regulated >1.5-fold in treated cells in both conditions (si-ALK and decitabine) compared either to negative control siRNA (si-CTL) or drug vehicle alone (DMSO) (Figure 1A). Amongst these 13 miRNAs, using microarray data previously published of miRNA-expression of NPM-ALK+ (n=52) and NPM-ALK− (n=5) ALCL patients,21 we observed that miR-195 and miR-497 were down-regulated only in NPM-ALK+ ALCL patients compared to normal reactive lymph node (RLNs, n=3) (Online Supplementary Table S2). To corroborate the miRNA array data, we performed qPCR analysis for miR-195 and miR-497 on 23 samples from NPM-ALK+ by selecting cases with at least 50% lymph node involvement compared to 12 RLNs (Figure 1B). Significant downregulation of both miR-195 and miR-497 was confirmed in primary biopsies despite the presence of a mixed population of neoplastic and normal cells (Figure 1B).
Figure 1.
MiR-195 and miR-497 are down-regulated in human NPM-anaplastic lymphoma kinase (ALK)-positive(+) primary lymphoma samples and cell lines. (A) RNA sequencing from NPM-ALK+ KARPAS-299 cells following decitabine or DMSO treatment or transfection with a control siRNA (si-CTL) or siRNA targeting ALK mRNA (si-ALK). A set of 13 miRNAs was found to be up-deregulated >1.5 fold in treated cells in both conditions (si-ALK and decitabine) compared to either negative control siRNA (si-CTL) or drug vehicle alone (DMSO). (B) Quantitative real-time PCR (qRT-PCR) analysis of miR-195 and miR-497 in NPM-ALK+ anaplastic large cell lymphoma patients (n=52). Data were normalized against equivalent miRNA levels from reactive lymph node (RLN) tissue samples (n=19). Data represent means±Standard Error of Mean (bars), ***P<0.0001; unpaired two-tailed Student’s t-test with Welch’s correction.
NPM-ALK activity and DNA methylation mediate silencing of miR-195 and miR-497 in lymphoma cells
The downregulation of miR-195 and miR-497 observed in NPM-ALK+ cells suggests that the NPM-ALK+ protein itself might drive this phenomenon. This hypothesis is corroborated by the fact that, in our initial screen, miR-195 and miR-497 were up-regulated following NPM-ALK knockdown in KARPAS-299 cells (Figure 1A). To verify whether NPM-ALK is involved in miR-195 and miR-497 silencing, NPM-ALK expression was knocked down in 2 other human NPM-ALK+ ALCL cell lines (COST and SU-DHL1) using siRNA directed against ALK mRNA (Figure 2A and D). This was followed by reducing NPM-ALK activity with crizotinib treatment, a pharmacological inhibitor of ALK tyrosine kinase activity (Figure 2B and E). Western blotting analysis (Online Supplementary Figure S1A and B) showed that NPM-ALK was down-regulated and inhibited. Both inhibition of ALK gene expression and its activity induce a significant increase in both miR-195 and miR-497 expression in NPM-ALK+ cells (Figure 2A–E). Next, we measured the levels of both miR-195 and miR-497 after decitabine treatment in order to confirm that the observed miR-195 and miR-497 downregulation in NPM-ALK+ lymphoma cells was, indeed, due to DNA methylation (Figure 2C and F). We detected a significant increase in miR-195 and miR-497 expression in DNMT inhibitor-treated cells compared to the drug vehicle alone (DMSO) in all NPM-ALK+ cell lines (Figure 2C and F). STAT3 appears to be implicated in this mechanism (data not shown). To determine the DNA methylation status of the MIR497HG promoter, the miR-195 and miR-497 host gene on chromosome 17, we carried out bisulfite sequencing analysis of genomic DNA isolated from the three NPM-ALK+ cell lines: KARPAS-299, SU-DHL1 and COST and normal CD4 T lymphocytes expressing CD30 antigen activated using CD3/CD28 antibodies (n=3) (Online Supplementary Figure S2A). The results showed significantly high levels of methylation of all CpGs located in the region upstream of miR-195 and miR-497 promoters in the KARPAS-299 and SU-DHL1, and a few CpGs (CpG1, CpG2 and CpG3) in COST tumor cells. We observed lower levels of CpG methylation in the region upstream of miR-195 and miR-497 promoters in normal cells (Online Supplementary Figure S2B). Decitabine significantly decreased methylation levels of CpGs in NPM-ALK+ cancer cells (Online Supplementary Figure S2C). Collectively, these data suggest that the silencing of both miR-195 and miR-497 is due, at least in part, to NPM-ALK expression and aberrant MIR497HG promoter methylation in ALCL cells.
Figure 2.
NPM-anaplastic lymphoma kinase (ALK) activity and DNA methylation mediate downregulation of miR-195 and miR-497. Quantitative RT-PCR analysis of miR-195 and miR-497 expression in NPM-ALK-positive lymphoma cell lines, COST, KARPAS-299 (KARPAS) and SU-DHL1, transfected with either an irrelevant siRNA as the negative control (si-CTL) or a siRNA targeting ALK mRNA (si-ALK) (A and D) or treated for 72 hours (h) or not (PBS) with crizotinib (B and E) or treated for 96 h or not (PBS) with decitabine (C and F). SNORD44 was used as an internal control and the relative ratio of miR-195 and miR-497 expression was expressed as 2−ΔΔCt relative to untreated cells. Data represent means±Standard Error of Mean (bars) from 3 independent experiments, **P<0.01; ***P<0.0001; unpaired two-tailed Student’s t-test with Welch’s correction.
Forced expression of miR-497 suppresses cell cycle progression and prevents the growth of NPM-ALK+ ALCL cells
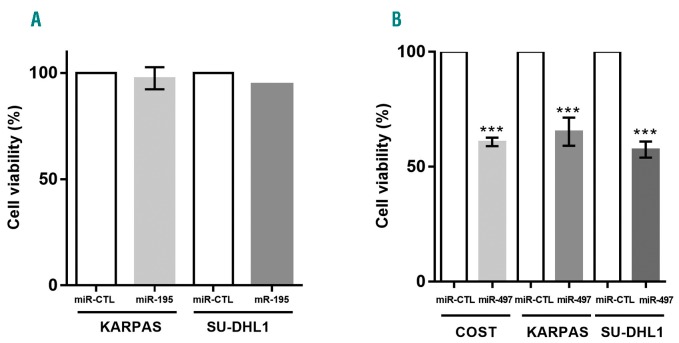
To investigate possible biological functions (potential effects on cell proliferation) of miR-195 and miR-497 in NPM-ALK+ cells, the two miRNAs were ectopically expressed by transfecting synthetic mimics into KARPAS-299, SU-DHL1 and COST cell lines. We observed that upregulation of miR-195 by transfection with miR-195 mimic does not impair cell viability of NPM-ALK+ cells (Figure 3A). By contrast, upregulation of miR-497 significantly affects cell viability of the three cell lines (Figure 3B). The successful overexpression of miR-497 and of miR-195 were confirmed by qPCR (Online Supplementary Figure S3). Taken together, these data suggest that only miR-497 in the miR-497/miR-195 cluster plays a role in cell proliferation of NPM-ALK+ cells.
Figure 3.
Overexpression of miR-497 reduces in vitro viability and growth of NPM-anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells. Cell viability determined by MTS assay of NPM-ALK+ lymphoma COST, KARPAS-299 (KARPAS) and SU-DHL1 cells 72 hours (h) after transfection with mimic miR-195 (miR-195) (A) or mimic miR-497 (miR-497) (B). Data represent means±Standard Error of Mean (bars) from 3 independent biological replicates, ***P<0.0001; unpaired two-tailed Student’s t-test with Welch’s correction.
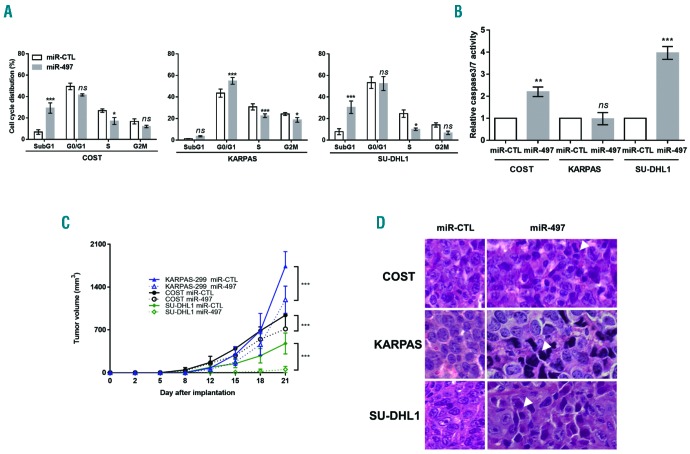
Since cell proliferation is closely associated with cell cycle control and progression, we performed flow cytometric analysis of cell cycle with propidium iodide DNA staining to examine the effects of miR-497 on cell cycle distribution in all three NPM-ALK+ cell lines. Accumulation in the G1 phase of the cell cycle was significant in KARPAS-299 cells, 48 h post transfection with miR-497 compared with those transfected with miR-CTL (Figure 4A). In addition, in the same condition, ectopic expression of miR-497 in COST and SU-DHL1 cells led to a significant accumulation of cells in a sub-G1 phase of cell cycle suggesting cell death (Figure 4A). To test whether apoptosis is involved in the growth inhibition caused by miR-497 overexpression, we measured caspase 3 and 7 activity. As shown in Figure 4B, compared to that of miR-CTL, miR-497 overexpression induced activation of apoptosis pathways in COST and SU-DHL1 cells (2.2- and 4-fold, respectively) but not in KARPAS-299 cells. These findings suggest that miR-497 overexpression induces growth-inhibitory effects on cancer cells by impeding cell cycle progression and leads to programmed cell death. We next investigated the effect of miR-497 transfection on tumor growth of NPM-ALK+ cell lines KARPAS-299, SU-DHL1 and COST in vivo. MiR-CTL or miR-497-transfected NPM-ALK+ cells were inoculated subcutaneously into the left or right flanks of each immunodeficient mouse, respectively (n = 6 for each condition). MiR-497-transfected NPM-ALK+ cells resulted in significant reduction of tumor growth (Figure 4C) when compared to those transfected with miR-CTL. Morphological analyses showed that forced expression of miR-497 in NPM-ALK+ cells caused phenotypic hallmarks of cellular degeneration (uncommon chromatin condensation, nuclear piknosis, decreased cellular volume and disruption of the nuclear envelope) (Figure 4D). These results show that miR-497 overexpression drastically reduces the growth of NPM-ALK+ cells in vivo. Taken together, these experiments suggest a tumor-suppressive function of miR-497 in NPM-ALK+ cells.
Figure 4.
Ectopic expression of miR-497 induces cell cycle arrest and reduces growth in vivo of NPM-anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells. NPM-ALK+ ALCL COST, KARPAS-299 (KARPAS) and SU-DHL1 cells were transfected with negative control miRNA (miR-CTL) or mimic miR-497 (miR-497). Cell cycle was analyzed (A) and caspase 3/7 activity was measured (B). Data were normalized against the miR-CTL condition. Each experiment was repeated 3 times. (C) NPM-ALK+ COST, KARPAS-299 (KARPAS) and SU-DHL1 cells transfected either with miR-CTL or miR-497 were injected subcutaneously in the left or right flank of immunodeficient mice, respectively (n=6). Tumor volume was evaluated over time by caliper measurements. Data represent mean±Standard Error of Mean, *P<0.05; **P<0.001; ***P<0.0001; ns: not significant; unpaired two-tailed Student’s t-test with Welch’s correction. (D) Micrographs of hematoxylin & eosin staining of excised miR-CTL or miR-497 tumors (original magnification ×40). Arrows indicate cells with phenotypical hallmarks of cellular degeneration.
Effect of miR-497 on the expression of cell cycle regulatory genes in NPM-ALK+ lymphoma cells
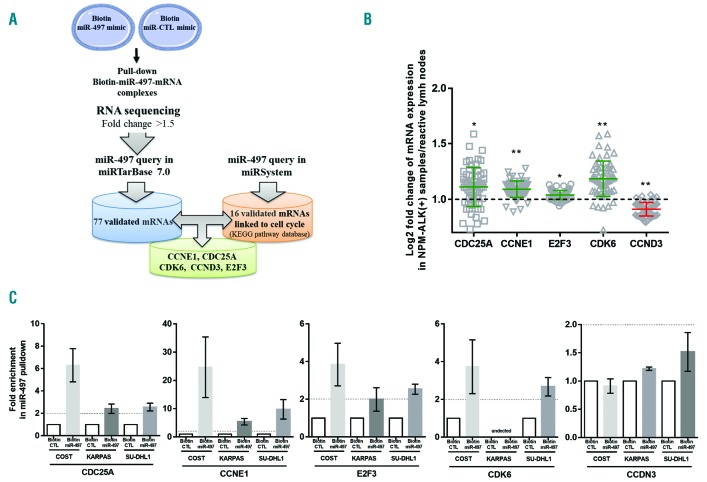
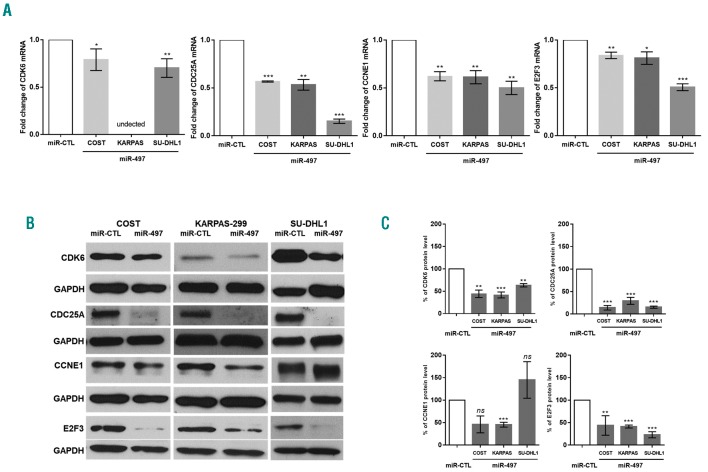
To determine the mechanism(s) by which miR-497 regulates tumor growth and progression of NPM-ALK+ cancer cells, we searched for mRNA targets of miR-497. To capture miR-497 associated mRNAs, KARPAS-299 cells were transfected with biotinylated human miR-497. We isolated and sequenced RNAs that co-purified with biotin-miR-497 mimic to map miR-497-mRNA interaction. As a negative control, we performed the same experiment with a biotin-negative control mimic (biotin-miR-CTL). A large set of mRNAs is regulated by miR-497 (enrichment ratio >1.5). In this data set, 77 validated candidate targets of miR-497 were collected from the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/, Release 7.0) (Figure 5A). In addition, using the miRSystem database (http://mirsystem.cgm.ntu.edu.tw/), we identified 16 miR-497 mRNA targets, experimentally validated and associated with cell cycle regulation (KEGG pathway database) (Figure 5A). We identified 5 mRNA gene targets that were common to both databases: CCNE1, CDC25A, CDK6, CCND3 and E2F3 (Figure 5A). Next, we used gene expression array data (Daugrois et al., 2018, submitted publication) to identify the veracity of these 5 targets captured by biotin-miR-497. Despite the presence of a mixed population of neoplastic and normal cells, CCNE1, CDC25A, CDK6 and E2F3 mRNAs were found to be significantly over-expressed in NPM-ALK+ ALCL patients (n=55) compared to reactive lymph node controls (RLN) (Figure 5B). As previously reported by Thompson et al., CCND3 was not over-expressed in NPM-ALK+ ALCL patients.30 Finally, to validate the specificity of the biotin pulldown and applicability to other cell lines, biotinylated forms of miR-497 or negative miRNA-control were used to co-purify CCNE1, CCDN3, CDC25A, CDK6 and E2F3 mRNAs in three KARPAS-299, SU-DHL1 and COST cell lines. Enrichment of target mRNAs by streptavidin-mediated purification (enrichment ratio >2) was measured by qPCR and normalized to miR-CTL enrichment. In the biotin-miR-497 pulldown of COST, SU-DHL1 and KARPAS-299 cells, CDC25A, CCNE1 and E2F3 transcripts but not CCDN3 were captured 24 h after transfection (Figure 5C). Moreover, biotin pulldown enriches CDK6 mRNA in COST and SU-DHL1 cells but not in KARPAS-299 cells (Figure 5C). This result was in accordance with data published by Nagen et al.,31 who reported very weak CDK6 expression in KARPAS-299 cells compared to SU-DHL1 cells (Online Supplementary Figure S5). Moreover, for the first time, we observed that CDK6 protein is over-expressed in COST cells (similar to SU-DHL1 cells) in comparison to KARPAS-299 cells (Online Supplementary Figure S4). To corroborate the results of miR-497–bound mRNAs capture and transcriptomic analyses, the effect of miR-497 on the endogenous expression of CCNE1, CDC25A, CDK6 and E2F3 was subsequently examined using qPCR and Western blotting. Transfection of miR-497 mimic induced a significant decrease of all four mRNAs in all three cell lines compared to cells transfected with miR-CTL (Figure 6A). Overexpression of miR-497 significantly down-regulated the protein level of CDK6, CDC25A and E2F3 in all three cell lines (Figure 6B). In addition, CCNE1 expression was down-regulated in KARPAS-299 cells but not in COST or SU-DHL1 cells (Figure 6B). Densitometric analysis of the mean relative intensity for each target protein of miR-497 supports the Western blotting results (Figure 6C). Taken together, these results strongly suggest that miR-497 regulates the expression of CCNE1, CDC25A, CDK6 and E2F3 in NPM-ALK+ cells.
Figure 5.
MiR-497 targets G1/S regulators in anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells. (A) mRNA targets of miR-497 identification workflow. NPM-ALK+ KARPAS-299 cells transfected with biotinylated forms of human miR-497 (biotin-miR-497) or an irrelevant biotin-miRNA as negative control (biotin-miR-CTL) were used to co-purify associated mRNAs (enrichment ratio >1.5). Validated candidate targets of miR-497 were collected from miRTarBase (n=77) and experimentally validated miR-497 associated with the regulation of cell cycle from miRSystem database (n=16). CCNE1, CDC25A, CDK6, CCND3 and E2F3 were the only genes identified in the two computational analyses. (B) Microarray analysis of CCNE1, CDC25A, CDK6, CCND3 and E2F3 mRNA expression in human NPM-ALK+ (n=56; Daugrois et al., submitted publication). Data were normalized against equivalent miRNA levels from reactive lymph (RLN) node tissue samples (n=3). Data represent means±Standard Error of Mean (bars), *P<0.05; **P<0.001; ***P<0.0001; unpaired two-tailed Student’s t-test with Welch’s correction. (C) Quantitative real-time PCR (qRT-PCR) analysis of CCNE1, CDC25A, CDK6, CCND3 and E2F3 mRNA was performed after pulldown of the biotinylated miRNAs using streptavidin beads. The results are presented as the relative enrichment in miR-497 pulldown compared to miRNA control pulldown (enrichment ratio >2).
Figure 6.
CDK6, CDC25A, CCNE1 and E2F3 are target genes of miR-497 in anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells. (A) Quantitative real-time (RT)-PCR analysis of CDK6, CDC25A, CCNE1 and E2F3 mRNA expression in NPM-ALK+ lymphoma COST, KARPAS-299 (KARPAS) and SU-DHL1 cells transfected with negative control miRNA (miR-CTL) or a mimic miR-497 (miR-497). GAPDH was used as internal control and the relative ratios of mRNA expression was expressed as 2−ΔΔCt relative to those in cells transfected with miR-CTL. Data represent means±Standard Deviation (bars), **P<0.001; ***P<0.0001; unpaired two-tailed Student’s t-test with Welch’s correction. (B) Western blotting analysis of CDK6, CDC25A, CCNE1 and E2F3 expression in NPM-ALK+ COST, KARPAS-299 (KARPAS) and SU-DHL1 cells transfected by mir-CTL or mir-497. The GAPDH protein served as an internal control to ensure equal loading. Results from one representative experiment are shown. (C) Densitometric analysis was performed using ImageJ software from Wayne Rasband (NIH). The relative levels of the proteins of interest were normalized to GAPDH levels. Data represent means±Standard Error of Mean (bars) from 3 independent experiments, **P<0.001; ***P<0.0001; ns: not significant; unpaired two-tailed Student’s t-test with Welch’s correction.
Significance of identified miR-497 target genes in NPM-ALK+ lymphoma cells
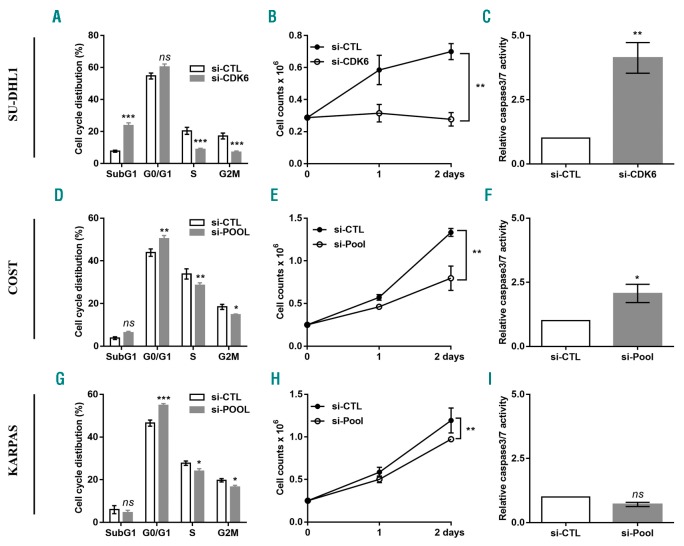
To evaluate whether CCNE1, CDC25A, CDK6 and E2F3 may be functional downstream targets of miR-497 and involved in regulation of NPM-ALK+ cell proliferation, we individually knocked down the expression of these genes using specific siRNAs. In order to check that the knockdown of gene expression had been efficiently achieved, we performed qPCR and Western blotting to detect the mRNA (Online Supplementary Figure S5A) and protein (Online Supplementary Figure S5B) levels, respectively. Densitometric analysis of the mean relative intensity for each target protein of miR-497 supports the Western blot results (Online Supplementary Figure S5C). We observed that, in SU-DHL1 cells, silencing of CDK6 mRNA simultaneously induced a decrease of cell viability and an increase of both subG1 fraction and caspase 3/7 activity, thereby inhibiting cell growth (Figure 7A–C). Hence, CDK6 silencing alone is sufficient to recapitulate the phenotype observed upon miR-497 ectopic expression in SU-DHL1 cells. These results are similar to those published by Nagen et al.31 showing an addiction to CDK6 expression for proper cell cycle progression in this cell line. In contrast, even though siRNA directed against CDK6 led to a significant accumulation of COST and KARPAS-299 cells in G0/G1 phase (Online Supplementary Figure S6B and E), it is not sufficient to fully recapitulate miR-497-induced blockage of cell cycle. The same observation was made after E2F3 and CCNE1 mRNA downregulation (Online Supplementary Figure S6A and D, and C and F, respectively). Finally, we observed that CDC25A knockdown did not modify the cell cycle profiles of COST and KARPAS-299 cells (data not shown). We also tested the effect of each siRNA on cellular apoptosis in COST and KARPAS-299 cells. Compared to the control condition (si-CTL), CDK6 and CCNE1 silencing induced apoptosis in COST cells but, as expected, not in KARPAS-299 cells (Online Supplementary Figure S7). In addition, the silencing of E2F3 mRNA did not affect caspase activity in both NPM-ALK+ COST and KARPAS-299 cells (Online Supplementary Figure S7). Moreover, regardless of the siRNA transfected, cell viability following RNAi delivery was never significantly modified in NPM-ALK+ cells (data not shown). Thus, individual silencing of three miR-497-targets, CDK6, CCNE1 and E2F3, can reproduce (only to a lesser extent) the miR-497-induced phenotype (Figure 4A and B). Consequently, in order to improve the effects observed in COST and KARPAS-299 cells, we decided to pool siRNAs targeting CCNE1, CDK6 and E2F3. We transfected KARPAS-299 or COST cells with siRNAs (pools of three siRNAs/targets) targeting each of the miR-497-down-regulated transcripts. In order to check whether the knockdown of gene expression had been efficiently achieved, we performed qPCR to detect the mRNA (Online Supplementary Figure S8). We then analyzed transfected cells for their cell cycle distributions. Similar to transfection of miR-497 mimics, siRNA pools induced significant accumulation of KARPAS-299 and COST cells in G0/G1 fraction (Figure 7D and G). Of note, the same result was observed in COST and SU-DHL1 cells, 24 h post transfection with the miR-497 mimic (Online Supplementary Figure S9A and B). The reduction in cell proliferation in the knockdown NPM-ALK+ cells was observed by cell counting (Figure 7E and H). In addition, compared to the control condition (si-CTL), CCNE1, CDK6, E2F3 silencing induced apoptosis in COST cells thereby inhibiting cell growth (Figure 7G and K) but, as expected, not in KARPAS-299 cells (Figure 7K and H). Taken together, these findings demonstrate that miR-497 controls multiple targets that collaborate to regulate cell cycle progression and cell growth.
Figure 7.
Silencing of CDK6, CCNE1 and E2F3 genes reduces cell proliferation in anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells. NPM-ALK+ ALCL SU-DHL1 (A-C), COST (D-F) and KARPAS-299 (KARPAS) (G-I) cells were transfected with either an irrelevant siRNA as the negative control (si-CTL) or a siRNA targeting CDK6 mRNA (si-CDK6) or CDK6, CCNE1 and E2F3 mRNA together (si-Pool). Cell cycle distribution (A, D and G), cell count at three different time points (0, 1, 2 days) (B, E and H) and caspase 3/7 activity (C, F and I) were measured. Data represent means±Standard Error of Mean (bars) from 3 independent experiments, *P<0.05; **P<0.001; ***P<0.0001; ns: not significant; unpaired two-tailed Student’s t-test with Welch’s correction.
ALK-positive cells are responsive to CDK4/6 inhibitors
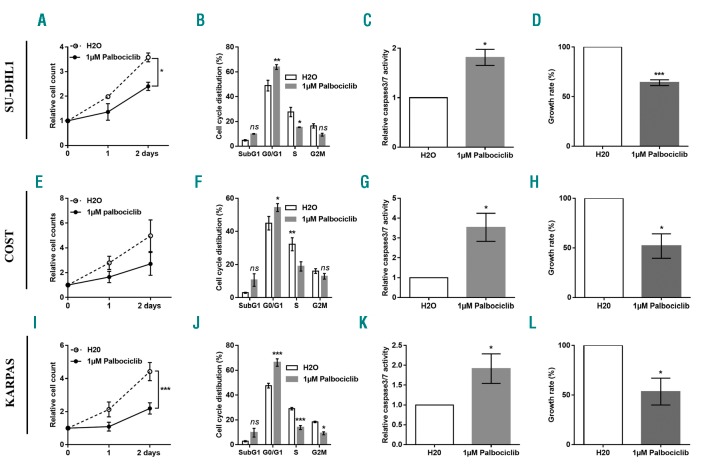
The new generation of selective CDK4/6 inhibitors, with anti-proliferative activity in Rb-proficient cells, target tumor types in which CDK4/6 has a pivotal role in the G1-to-S-phase cell cycle transition with improved effectiveness and fewer adverse effects. Palbociclib is an orally bioavailable CDK4/6 inhibitor recently approved by the US Food and Drug Administration for the treatment of metastatic breast cancer.32 As all three NPM-ALK+ cell lines, SU-DHL1, COST and KARPAS-299 expressed CDK4, CDK6 and Rb (Online Supplementary Figure S4), we evaluated the effect of palbociclib treatment. We observed that all NPM-ALK+ cell lines are sensitive to this molecule (Figure 8A, E and I). In addition, we showed that palbociclib markedly inhibited the proliferation of NPM-ALK+ cells through the induction of G0/G1 cell-cycle arrest (Figure 8B, F and J), increased apoptosis (Figure 8C, G and K) and reduced cell growth (Figure 8D, H and L). As a control of palbociclib efficacy, we observed a decrease in Cdk4/6-specific phosphorylation of the Rb protein as shown by Western blotting (Online Supplementary Figure S10A). Densitometric analysis of the mean relative intensity for pRB and RB corroborate Western blotting results (Online Supplementary Figure S10B). As palbociclib is a selective cyclin D kinase 4/6 inhibitor, NPM-ALK+ ALCL SU-DHL1, COST and KARPAS-299 cells were transfected with either an irrelevant siRNA as the negative control (si-CTL) or a siRNA targeting CDK4 mRNA. In order to check whether the knockdown of gene expression had been efficiently achieved, we performed qPCR to detect the CDK4 mRNA (Online Supplementary Figure S11C, F and I). As silencing of CDK4 does not reduce cell proliferation in ALK+ lymphoma cells (Online Supplementary Figure S11A, D and G), nor induce apoptosis (Online Supplementary Figure S11B, E and H), we suggest that the action of palbiciclib is only CDK6-dependent.
Figure 8.
Anaplastic lymphoma kinase (ALK)-positive(+) lymphoma cells are sensitive to CDK6 inhibitor. NPM-ALK+ ALCL SU-DHL1 (A-D), COST (E-H) and KARPAS-299 (KARPAS) (I-L) cells were treated or not (H2O) with palbociclib, an CDK4/6 inhibitor. Cell count at three different time points (0, 1, 2 days) (A, E and I), cell cycle distribution (B, F and J), caspase 3/7 activity (C, G and K) and cell proliferation rate (D, H and L) were measured. Data represent means±SEM (bars) from 3 independent experiments. *P<0.05; **P<0.001; ***P<0.0001; ns: not significant; unpaired two-tailed Student’s t-test with Welch’s correction. Data represent means±Standard Error of Mean (bars) from 3 independent experiments, *P<0.05; **P<0.001; unpaired two-tailed Student’s t-test with Welch’s correction.
Based on our findings that CDK6, CCNE1 and E2F3 are important for efficient proliferation of NPM-ALK+ ALCL cells, we hypothesized that the highest level of expression of these factors could be the most favorable for tumor development and possibly affect chemotherapy effectiveness. To test this hypothesis, we used qPCR to measure the expression of CDK6, CCNE1 and E2F3 in a cohort of NPM-ALK+ primary ALCL samples (n=55) obtained from chemotherapy-naive patients at diagnosis. Compared to normal reactive lymph nodes (RLNs) (n=19) and despite the presence of a mixed population of neoplastic and normal cells, we observed a significant CDK6, CCNE1 and E2F3 gene overexpression in NPM-ALK+ samples (Online Supplementary Figure S12A). In order to evaluate the clinical significance of CCNE1, CDK6 and E2F3 overexpression, first using linear regression, we derived a model based on microarray data on expression levels of CCNE1, CDK6, E2F3 genes in NPM-ALK+ samples (n=55). The samples included NPM-ALK+ primary biopsies of patients who had experienced early relapse and those without relapse after three years of minimal follow up after chemotherapy cure. By applying the formulae obtained from linear regression, we derived a ‘score’ for each patient: score = (−0.08378 × CDK6) + (−0.41269 × CCNE1) + (0.37893 × E2F3). Secondly, a ROC curve was generated from CCNE1, CDK6, E2F3 genes-based score from 44 NPM-ALK+ ALCL samples, including 8 adults and 36 children, for whom event-free survival is known (Online Supplementary Figure S12B). The cut-off value and Air Under Curve (AUC) values were determined as −0.038 and 0.65, respectively. Based on the optimal cut-off points from the ROC curve, patients were significantly categorized into two groups, high (n=25) and low (n=14) CCNE1, CDK6, E2F3 expression (Online Supplementary Figure S12C and D). NPM-ALK+ ALCL patients with a low expression of CCNE1, CDK6, E2F3 had a poor prognosis (P<0.046) (Online Supplementary Figure S12C). Moreover, this trend can be observed for high (n=14) and low (n=22) expression in NPM-ALK+ pediatric ALCL (Online Supplementary Figure S12D). In addition, for the same pediatric cases, we noted that CCNE1, CDK6, E2F3 gene-based score was increased in relapsing patients (n=23 vs. without relapse n=13) (Online Supplementary Figure S12E). As all patients were treated with chemotherapy following diagnosis, our data suggest that CDK4/6 inhibitors could benefit ALK+ patients harboring resistant tumors.
Discussion
MiR-195 and miR-497 are important members of the miR-15/16 family and are aberrantly expressed in multiple diseases, such as solid cancers.23,33 Based on the expression of miRNAs, Steinhilber et al. have identified a gene signature including reduced expression of some miR-15/16 family members, such as miR-16, miR-195 and miR-497 that distinguish ALK+ from ALK− ALCL.34 Except miR-16,19 the functional characterization of miR-195 and miR-497, which are clustered on chromosome 17p13.1, has never been undertaken in ALK+ lymphoma cells.
MiR-195 and miR-497 downregulation has been consistently demonstrated in a variety of solid tumor types. miRNA-silencing in cancer is frequently related to abnormal promoter methylation.35 Accordingly, the downregulation of miR-195 and miR-497 has been reported to be linked to promoter-associated hypermethylation in solid cancers; demethylation following 5-aza-CdR treatment resulted in the re-expression of these two miRNAs.36,37 In the present work, we demonstrate for the first time that miR-195 and miR-497 are down-regulated in human NPM-ALK+ ALCL tissues and cell lines through a mechanism involving NPM-ALK and DNA hypermethylation. These results are highly reminiscent of what we previously observed for three other miRNA genes, MIR29A, MIR125B and MIR150, the levels of which were also reduced in NPM-ALK+ ALCL as a consequence of the promoter hypermethylation.17,20,21 In addition, we observed that only ectopic expression of miR-497 markedly attenuates in vitro cell proliferation of NPM-ALK+ cells.
It is estimated that more than half of human genes are targeted by at least one miRNA. One miRNA may have multiple different mRNA targets and at the same time, one mRNA might be targeted by multiple miRNAs. miR-497 regulates the expression of a plethora of targets, which are involved in cell cycle, apoptosis, proliferation, etc.23 Recent studies have revealed that miR-497 can directly reduce the CCNE1 protein level to suppress tumor growth by inducing G1 arrest in breast cancer38 and hepatocellular carcinoma.39 Indeed, CCNE1 protein binds to and activates cyclin-dependent kinase 2 (CDK2) to promote a cascade of events required for cell-cycle progression from G1 to S phase. Several other genes encoding cell-cycle regulators, including Cyclin D1/CCND1 and Cyclin D3/CCND3, Cyclin-dependent kinase 4 (CDK4) and CDK6, cell-cycle division factors CDC25A and/or E2F3 transcription factor, are also known targets of miR-497.23,40 The role of miR-497 in human cancer is not yet clear. Several studies have reported that increased miR-497 expression via miR-497 mimic transfection suppressed proliferation and increased apoptosis in various solid cancers, such as adrenocortical carcinoma41 and pancreatic cancer.42 Accordingly, miR-497 overexpression was found to block G0/G1 phase transition in breast cancer cells43 and induce G1/S arrest in gastric cancer cells.37 In our study, we revealed for the first time that enhanced miR-497 expression in human NPM-ALK+ lymphoma cells could suppress cell proliferation, by inducing cell cycle arrest, and inhibit tumor growth in vivo. These biological effects are the consequence of simultaneous downregulation of three mir-497 mRNA targets: CCNE1, E2F3 and CDK6, and their subsequent decreased protein level. Contribution of CCNE1 and E2F3 in uncontrolled cell-cycle progression and oncogenesis have never been studied in human NPM-ALK+ ALCL models. Of note, only two publications report an abnormal CDK6 expression in human NPM-ALK+ lymphoma cells. The amplification of the CDK6 locus in NPM-ALK+ SU-DHL1 cells and co-expression of high levels of CDK6 and CD31 (angio-genic marker) in NPM-ALK+ ALCL samples have been reported.31,44
NPM-ALK+ ALCL is a rare disease which accounts for about 1-2% of adult non-Hodgkin lymphomas (NHL) and 10-15% of childhood NHL. The prognosis of NPM-ALK+ ALCL is better than that of other T-cell lymphomas. Overall, patients with NPM-ALK+ ALCL who are resistant to primary chemotherapy or who relapse early have the worst prognosis.4,45 We checked the CCNE1, E2F3 and CDK6 overexpression in a cohort of NPM-ALK+ primary ALCL samples obtained from chemotherapy-naive patients at diagnosis. As aberrant growth control is a hallmark of cancer cells, we hypothesized that altered CCNE1, E2F3 and CDK6 expression in NPM-ALK+ patients could affect their response to treatment. Using a NPM-ALK+ cohort, including patients who had experienced early relapse after chemotherapy, we show that the three G1-S regulators together could predict treatment outcome in pediatric patients.
CDK6 protein is a member of the cyclin-dependent kinase family, which includes CDK4, with a 71% shared amino acid identity. These homologs are ubiquitously expressed and have largely overlapping functions. In mammalian cells, cell cycle is regulated by CDK4 and CDK6 in early G1 phase through interactions with cyclins D1, D2 and D3. Cyclin D/CDK4 and cyclin D/CDK6 complexes induce phosphorylation of the retinoblastoma protein (Rb) and the release of E2F3 transcription factor from RB-E2F3 complexes. Cell-cycle components, such as CDK4 and CDK6, are frequently altered in human cancer, reflecting the deregulated growth of transformed cells. In B-cell lymphoid malignancies, DNA hypermethylation-mediated epigenetic silencing of the tumor suppressors miR-124a and miR-29 results in an upregulation of their target CDK6.46 Downregulation of CDK6 was observed following introduction of miR-29a mimics into T-cell acute lymphoblastic leukemia Jurkat cells.47 CDK6 has also been implicated in thymic lymphoma formation in a transgenic mouse model.48 Moreover, Kollmann et al. have observed that NPM-ALK− transgenic mice lacking CDK6 expression developed disease with a significantly prolonged latency.44 At the same time, in a NOD/SCID xenograft human multiple myeloma model, palbociclib, a newly developed small-molecule inhibitor specific to CDK4 and CDK6, can enhance the killing of myeloma cells by dexamethasone.49 In addition, palbociclib inhibits CDK4/6 activity according to the activation status of cycling primary bone marrow myeloma cells freshly isolated from both new and relapsed patients.49 A phase II study has shown that progression-free survival was doubled with palbociclib plus standard hormone therapy in advanced hormone receptor-positive, HER2-negative breast cancer.32 Taken together, these findings provide the grounds for new therapeutic strategies in NPM-ALK+ ALCL either targeting the epigenetic regulation of miRNAs and/or directly targeting the CDK6 pathway. We have previously reported that hypomethylating drugs may benefit NPM-ALK+ patients.17 Bonvini et al. previously described the effect of flavopiridol, a pan-CDK inhibitor, on NPM-ALK+ ALCL cells.50 In the present study, as a proof of concept, we have presented in vitro data showing that palbociclib can be the first promising and specific inhibitor for therapeutic targeting of Cdk4/6 in NPM-ALK+ lymphoma cells. Our work also provides a rationale for targeting ALK+ ALCL progression with palbociclib. Indeed, it could be possible to achieve greater synergy by combining palbociclib with other cytotoxic agents or anti-ALK tyrosine kinase inhibitor such as crizotinib, recently approved for the treatment of metastatic and late-stage ALK-rearranged non-small-cell lung cancers (NSCLC). In summary, these findings suggest that CDK4/CDK6 inhibitors could be novel candidates for mechanism-defined combination therapy in ALK+ lymphomas and possibly other ALK+-cell cancers including a proportion of NSCLC.
Supplementary Material
Acknowledgments
This work is dedicated to CHA: «A winner is a dreamer who never gives up (Nelson Mandela)». English proofreading was performed by EnPro Language Solutions (www.enprols.fr) and Greenland scientific proofreading.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/2/347
Funding
This work was supported by grants from the “Ligue contre le Cancer”, Fondation ARC pour la Recherche sur le Cancer” (FM), and INCa (projet PAIR Lymphomes) (LL). CHA and AC were supported by a fellowship from the “Labex TOUCAN / Laboratoire d’excellence Toulouse Cancer”. For their technical assistance, the authors thank M. Tosolini (Pôle Technologique du CRCT – Plateau Bioinformatique INSERM-UMR1037); F. Capilla and C. Salon at the histology facility and the staff of the zootechnie Langlade, INSERM/UPS-US006/CREFRE (Toulouse, France).
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugieres L, Le Deley MC, Rosolen A, et al. Impact of the methotrexate administration dose on the need for intrathecal treatment in children and adolescents with anaplastic large-cell lymphoma: results of a randomized trial of the EICNHL Group. J Clin Oncol. 2009;27(6):897–903. [DOI] [PubMed] [Google Scholar]
- 3.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263(5151):1281–1284. [DOI] [PubMed] [Google Scholar]
- 4.Delsol G, Falini B, Mûller-hermelink HK, Campo E, et al. Anaplastic large cell lymphoma, ALK-positive. Lyon: IARC Press, 2008. [Google Scholar]
- 5.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14(4):439–449. [DOI] [PubMed] [Google Scholar]
- 6.Damm-Welk C, Busch K, Burkhardt B, et al. Prognostic significance of circulating tumor cells in bone marrow or peripheral blood as detected by qualitative and quantitative PCR in pediatric NPM-ALK-positive anaplastic large-cell lymphoma. Blood. 2007;110(2):670–677. [DOI] [PubMed] [Google Scholar]
- 7.Ait-Tahar K, Damm-Welk C, Burkhardt B, et al. Correlation of the autoantibody response to the ALK oncoantigen in pediatric anaplastic lymphoma kinase-positive anaplastic large cell lymphoma with tumor dissemination and relapse risk. Blood. 2010;115(16):3314–3319. [DOI] [PubMed] [Google Scholar]
- 8.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8(1):11–23. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell SR, Hwang SR, Basrur V, et al. NPM-ALK signals through glycogen syn-thase kinase 3beta to promote oncogenesis. Oncogene. 2012;31(32):3733–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellmann A, Doseeva V, Butscher W, et al. The activated anaplastic lymphoma kinase increases cellular proliferation and oncogene up-regulation in rat 1a fibroblasts. FASEB J. 1997;11(12):965–972. [DOI] [PubMed] [Google Scholar]
- 11.Leventaki V, Drakos E, Medeiros LJ, et al. NPM-ALK oncogenic kinase promotes cell- cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood. 2007;110(5):1621–1630. [DOI] [PubMed] [Google Scholar]
- 12.Gu TL, Tothova Z, Scheijen B, Griffin JD, Gilliland DG, Sternberg DW. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signaling through modulation of FOXO3a. Blood. 2004;103(12):4622–4629. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Vidal A, Mazars A, Gautier EF, Prevost G, Payrastre B, Manenti S. Upregulation of the CDC25A phosphatase down-stream of the NPM/ALK oncogene participates to anaplastic large cell lymphoma enhanced proliferation. Cell Cycle. 2009;8(9):1373–1379. [DOI] [PubMed] [Google Scholar]
- 14.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812(5):592–601. [DOI] [PubMed] [Google Scholar]
- 15.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. [DOI] [PubMed] [Google Scholar]
- 16.Hoareau-Aveilla C, Merkel O, Meggetto F. MicroRNA and ALK-positive anaplastic large cell lymphoma. Front Biosci (Schol Ed). 2015;7:217–225. [DOI] [PubMed] [Google Scholar]
- 17.Hoareau-Aveilla C, Valentin T, Daugrois C, et al. Reversal of microRNA-150 silencing disadvantages crizotinib-resistant NPM-ALK(+) cell growth. J Clin Invest. 2015;125(9):3505–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkel O, Robert G, Hamacher F, Grabner L, Greil R, Kenner L. Subtype specific expression of immnune-modulating miR-155 and miR-146a in anaplastic large cell lymphoma. Cancer Res. 2009;72(8): supplemental 1. [Google Scholar]
- 19.Dejean E, Renalier MH, Foisseau M, et al. Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia. 2011;25(12):1882–1890. [DOI] [PubMed] [Google Scholar]
- 20.Desjobert C, Renalier MH, Bergalet J, et al. MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood. 2011;117(24):6627–6637. [DOI] [PubMed] [Google Scholar]
- 21.Congras A, Caillet N, Torossian N, et al. Doxorubicin-induced loss of DNA topoisomerase II and DNMT1- dependent suppression of MiR-125b induces chemoresistance in ALK-positive cells. Oncotarget. 2018;9(18):14539–14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He XX, Kuang SZ, Liao JZ, et al. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol Biosyst. 2015;11(2):532–539. [DOI] [PubMed] [Google Scholar]
- 23.Yang G, Xiong G, Cao Z, et al. miR-497 expression, function and clinical application in cancer. Oncotarget. 2016;7(34):55900–55911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Casalini P, Tagliabue E, Menard S, Croce CM. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44(18):2753–2759. [DOI] [PubMed] [Google Scholar]
- 26.Pikman Y, Alexe G, Roti G, et al. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clin Cancer Res. 2017;23(4):1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawai CM, Freund J, Oh P, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell. 2012;22(4):452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamant L, Espinos E, Duplantier M, et al. Establishment of a novel anaplastic large-cell lymphoma-cell line (COST) from a ’small-cell variant’ of ALCL. Leukemia. 2004;18(10):1693–1698. [DOI] [PubMed] [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson MA, Stumph J, Henrickson SE, et al. Differential gene expression in anaplastic lymphoma kinase-positive and anaplastic lymphoma kinase-negative anaplastic large cell lymphomas. Hum Pathol. 2005;36(5):494–504. [DOI] [PubMed] [Google Scholar]
- 31.Nagel S, Leich E, Quentmeier H, et al. Amplification at 7q22 targets cyclin-dependent kinase 6 in T-cell lymphoma. Leukemia. 2008;22(2):387–392. [DOI] [PubMed] [Google Scholar]
- 32.Degenhardt T, Wuerstlein R, Eggersmann T, Harbeck N. The safety of palbociclib for the treatment of advanced breast cancer. Expert Opin Drug Saf. 2018;28:1–6. [DOI] [PubMed] [Google Scholar]
- 33.He JF, Luo YM, Wan XH, Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol. 2011;25(6):404–408. [DOI] [PubMed] [Google Scholar]
- 34.Steinhilber J, Bonin M, Walter M, Fend F, Bonzheim I, Quintanilla-Martinez L. Next-generation sequencing identifies deregulation of microRNAs involved in both innate and adaptive immune response in ALK+ ALCL. PLoS One. 2015;10(2):e0117780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Chen X, Yu X, et al. Regulation of microRNAs by epigenetics and their interplay involved in cancer. J Exp Clin Cancer Res. 2013;32:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng H, Guo Y, Song H, et al. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene. 2013;518(2):351–359. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Jin X, Deng X, Zhang G, Zhang B, Ma L. The putative tumor suppressor microRNA-497 modulates gastric cancer cell proliferation and invasion by repressing eIF4E. Biochem Biophys Res Commun. 2014;449(2):235–240. [DOI] [PubMed] [Google Scholar]
- 38.Luo Q, Li X, Gao Y, et al. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furuta M, Kozaki K, Tanimoto K, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8(3):e60155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Zhang Z, Li Z, et al. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep. 2016;36(3):1293–1300. [DOI] [PubMed] [Google Scholar]
- 41.Ozata DM, Caramuta S, Velazquez-Fernandez D, et al. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18(6):643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu JW, Wang TX, You L, et al. Insulin-like growth factor 1 receptor (IGF-1R) as a target of MiR-497 and plasma IGF-1R levels associated with TNM stage of pancreatic cancer. PLoS One. 2014;9(3):e92847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen L, Li J, Xu L, et al. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3(3):475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kollmann K, Heller G, Schneckenleithner C, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angio-genesis. Cancer Cell. 2013;24(2):167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreri AJ, Govi S, Pileri SA, Savage KJ. Anaplastic large cell lymphoma, ALK-positive. Crit Rev Oncol Hematol. 2012;83(2):293–302. [DOI] [PubMed] [Google Scholar]
- 46.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69(10): 4443–4453. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira LH, Schiavinato JL, Fraguas MS, et al. Potential roles of microRNA-29a in the molecular pathophysiology of T-cell acute lymphoblastic leukemia. Cancer Sci. 2015;106(10):1264–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu MG, Deshpande A, Enos M, et al. A requirement for cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res. 2009;69(3):810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baughn LB, Di Liberto M, Wu K, et al. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66(15):7661–7667. [DOI] [PubMed] [Google Scholar]
- 50.Bonvini P, Zorzi E, Mussolin L, et al. The effect of the cyclin-dependent kinase inhibitor flavopiridol on anaplastic large cell lymphoma cells and relationship with NPM-ALK kinase expression and activity. Haematologica. 2009;94(7):944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.