Abstract
Chronic lymphocytic leukemia (CLL) patients with differential somatic hypermutation status of the immunoglobulin heavy variable genes, namely mutated or unmutated, display fundamental clinico-biological differences. Considering this, we assessed prognosis separately within mutated (M-CLL) and unmutated (U-CLL) CLL in 3015 patients, hypothesizing that the relative significance of relevant indicators may differ between these two categories. Within Binet A M-CLL patients, besides TP53 abnormalities, trisomy 12 and stereotyped subset #2 membership were equivalently associated with the shortest time-to-first-treatment and a treatment probability at five and ten years after diagnosis of 40% and 55%, respectively; the remaining cases exhibited 5-year and 10-year treatment probability of 12% and 25%, respectively. Within Binet A U-CLL patients, besides TP53 abnormalities, del(11q) and/or SF3B1 mutations were associated with the shortest time-to-first-treatment (5- and 10-year treatment probability: 78% and 98%, respectively); in the remaining cases, males had a significantly worse prognosis than females. In conclusion, the relative weight of indicators that can accurately risk stratify early-stage CLL patients differs depending on the somatic hypermutation status of the immunoglobulin heavy variable genes of each patient. This finding highlights the fact that compartmentalized approaches based on immunogenetic features are necessary to refine and tailor prognostication in CLL.
Introduction
Despite mounting evidence for the existence of distinct biological variants of chronic lymphocytic leukemia (CLL), the 2016 update of the World Health Organization (WHO) classification still considers CLL as a single, homogeneous entity, in contrast to other hematologic malignancies (e.g. diffuse large B-cell lymphoma, DLBCL) which are segregated in different subgroups, based on the integration of genetic, morphological, immunophenotypic and clinical features.1
Since the introduction of the Rai and Binet clinical staging systems in the 1970s,2,3 it has become increasingly evident that the clinical heterogeneity in CLL is linked to and reflects the underlying biological heterogeneity. Hence, several initiatives have focused on identifying biomarkers that would refine prognostication, especially for cases who present with early stage disease, who nowadays represent the great majority of patients (80-85%).4–12 Consequently, numerous prognostic indices have been proposed; however, none has been adopted in every-day clinical practice.13 This is partly due to the fact that different variables have been assessed in each evaluated cohort while the actual routine diagnostic and monitoring practice varies between different institutions. Moreover, most reported cohorts were rather small, thus inherently limited in their capacity to both encompass the remarkable clinico-biological heterogeneity of CLL and reveal possible interactions and interdependencies among the evaluated prognosticators.
The clonotypic B-cell receptor immunoglobulin (BcR IG) is a unique molecular signature for every CLL clone, present from its genesis and remaining unaltered throughout the course of the disease, thus sharply contrasting other tumor-derived biomarkers.14–19 Seminal studies from the late 1990s have established that the somatic hypermutation (SHM) status of the immunoglobulin heavy variable (IGHV) gene expressed by the clonotypic BcR IG is a robust prognostic and predictive biomarker for CLL, stratifying patients into two non-interchangeable categories with different clinical behavior.20,21 More specifically, CLL with a significant SHM load (“mutated” CLL, M-CLL) generally follow an indolent clinical course, whereas CLL carrying no or few mutations (“unmutated” CLL, U-CLL) generally have an aggressive disease and an overall inferior response to chemoimmunotherapy.22–24
This subclassification into M-CLL and U-CLL reflects fundamental clinico-biological differences extending from the genomic and epigenomic to the transcriptomic and proteomic levels, alluding to distinct ontogeny and evolution patterns, including response to treatment, for the two patient categories.14,24–27 That said, within both M-CLL and U-CLL, a sizeable proportion of cases exhibit a clinico-biological behavior pattern that deviates from the expected, thus highlighting that the heterogeneity of CLL persists even within a given SHM category.28–31 The paradigmatic example is offered by CLL subset #2, defined by the expression of stereotyped IGHV3-21/IGLV3-21 BcR IG, within which M-CLL cases follow an aggressive clinical course similar to U-CLL.30,32,33
Notably, other established prognosticators such as cytogenetic aberrations or recurrent gene mutations are asymmetrically distributed within M-CLL or U-CLL.10,34–36 On these grounds, it is not unreasonable to think that definitive conclusions about the precise clinical implications of any given biomarker should be drawn only after considering the SHM status of the clonotypic BcR IG.
In this study, we followed a compartmentalized approach where we assessed the prognostic impact of traditional and novel prognostic parameters separately within M-CLL and U-CLL in a large multi-institutional cohort of well characterized CLL patients, based on the hypothesis that not all variables would carry equal weight within the two SHM categories. Considering that the key challenge at the time of diagnosis is determining if, and consequently when, early stage/asymptomatic patients will require treatment, we focused on identifying a robust prognostication scheme for time-to-first-treatment (TTFT) in these separate disease categories.
Methods
Patients’ characteristics
Overall, 2366 general practice patients with CLL diagnosed following the 2008 International Workshop on CLL (IWCLL) diagnostic criteria37 from 10 European institutions were included in this multicenter retrospective study (Online Supplementary Table S1). The external validation cohort comprised of 649 Binet A cases evaluated at the Munich Leukemia Laboratory (n=508) and from a Scandinavian population-based study (n=141) (Online Supplementary Table S2). Ethical approval was granted by the local review committees and informed consent was collected according to the Declaration of Helsinki.
Methodologies
Detailed information about the methodologies used to analyze the prognostic markers and definitions about stereotyped subsets and the criteria for subset assignment are provided in the Online Supplementary Appendix. Briefly: i) mutational screening was performed for the following genes: NOTCH1, entire exon 34 or targeted analysis for del7544-45/p.P2514Rfs*4; TP53, exons 4–8 but also exons 9–10 for some centers; SF3B1, exons 14–16; BIRC3, exons 6–9 and MYD88, exons 3 and 5 or targeted analysis for p.L265P. The great majority (80%) of cases included in this study were screened for the aforementioned recurrent mutations by Sanger sequencing. In the remaining cases (20%), next generation sequencing (NGS) was applied and only those clones with higher than 10% variant allele frequency (VAF >10%) were considered; ii) fluorescence in situ hybridization (FISH) was performed in 1825 (77%) cases using probes for the 13q14, 11q22, 17p13 regions and trisomy 12 (cut off: 5%) while results were interpreted following Döhner’s hierarchical model;38 and iii) sequence analysis and interpretation of IGHV-IGHD–IGHJ rearrangements (including BcR IG stereotypy) was performed as described previously.39
Statistical analysis
We assessed the prognostic impact on TTFT of the following variables: age at the time of diagnosis; sex; CD38 expression; cytogenetic aberrations [del(17p), del(11q), del(13q) and trisomy 12 (+12)]; mutations within the TP53, SF3B1, NOTCH1, BIRC3 and MYD88 genes; and immunogenetic features, including borderline germline identity (GI: 97-97.99%) and assignment to stereotyped subsets #1, #2 and #4.
The following methodology was applied separately for M-CLL and U-CLL patients. The χ2 test was used to assess the homogeneity of all categorical prognostic variables within the different Binet stages (A, B and C). In order to evaluate homogeneity and detect significant differences within each categorical variable, the Bonferroni correction was applied to adjust for multiple comparison error and the significance level was set to P<0.017. TTFT was evaluated from the diagnostic date until the date of initial treatment; untreated cases were censored at the time of last follow up. Survival curves were constructed using the Kaplan-Meier method, and the log-rank test was applied to determine statistically significant differences between survival proportions.
The univariable Cox regression model was used to determine the most important prognostic factors. Multivariable Cox regression analysis was subsequently applied to evaluate the simultaneous effect of the important variables on TTFT. For the multivariable Cox analysis, we considered only those cases with available data for all the factors included in the model (n=918 for M-CLL and n=384 for U-CLL) on the grounds that imputing the values of the biomarkers could introduce substantial bias. The same rule was applied for the binary recursive partitioning. That said, no major differences were observed between the entire cohort and the proportion of cases included in the multivariable Cox analysis (Online Supplementary Table S3); this suggests that the patients included in the multivariable analysis were representative of the entire cohort.
The proportional hazard assumption was assessed for both the univariable and multivariable analysis. The stability of the multivariable Cox model was internally validated using a bootstrapping procedure. Harrell’s C index was also calculated to further assess the discriminatory power of the multivariable analysis when: a) all important prognostic variables were included in the model; and b) the resultant prognostic index was included in the model as the sole prognostic variable (C=1 indicates perfect discrimination; C=0.5 equates to chance).40
Binary recursive partitioning, based on the development of conditional inference trees, was used to further validate the results of Cox regression analysis.41 The patients were initially divided in subgroups (terminal nodes) with different survival behavior. An amalgamation algorithm was subsequently applied to merge the terminal nodes that did not exhibit a significant difference in survival. All tests were two-sided and P<0.05 was considered statistically significant. Statistical analyses were performed using the Statistica Software 10.0 (StatSoftInc, Tulsa, OK, USA) and R-3.2.2. Details are provided in the Online Supplementary Appendix.
Results
Prognostic impact of clinical staging within M-CLL and U-CLL
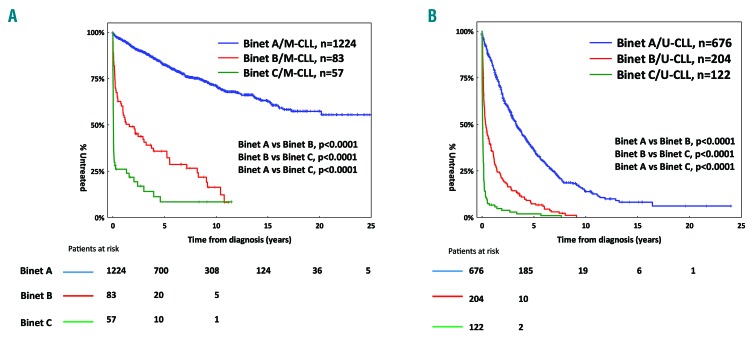
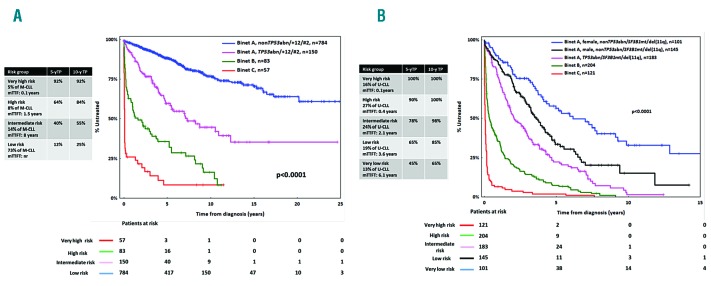
The median follow-up time for the entire cohort was 7.1 years (range, 0.1-33.1) with a median TTFT of 6.4 years (95%CI: 0.01-20.2 years) (Online Supplementary Figure S1). As expected, in both M-CLL and U-CLL, Binet A patients exhibited significantly longer TTFT compared to Binet B and C cases (Figure 1). Most likely, this reflects the sharp clinico-biological differences between Binet A and Binet B/C patients while also underscoring the current indications for treatment that broadly overlap with the criteria for Binet stage B and C assignment (Online Supplementary Tables S4 and S5). In keeping with previous reports, M-CLL cases (n=1364, 58%) exhibited significantly (P<0.0001) longer TTFT compared to U-CLL cases (n=1002, 42%) (Online Supplementary Figure S2A). Significant differences (P<0.0001) between M-CLL versus U-CLL remained when the analysis was restricted to Binet A cases (Online Supplementary Figure S2B).
Figure 1.
Kaplan-Meier curves for time-to-first-treatment (TTFT). (A) In Binet A, B and C mutated chronic lymphocytic leukemia (M-CLL) patients and (B) Binet A, B and C unmutated chronic lymphocytic leukemia (U-CLL) patients.
Prompted by these findings, and also considering that Binet A cases predominated in both M-CLL and U-CLL (90% and 67%, respectively), we focused our attention on 1900 Binet A cases (M-CLL: n=1224; U-CLL: n=676). We refrained from investigating the impact of biomarkers within Binet B/C cases since the limited number of cases within each subgroup would not allow firm conclusions to be drawn.
Analysis for time-to-first-treatment within early stage mutated chronic lymphocytic leukemia
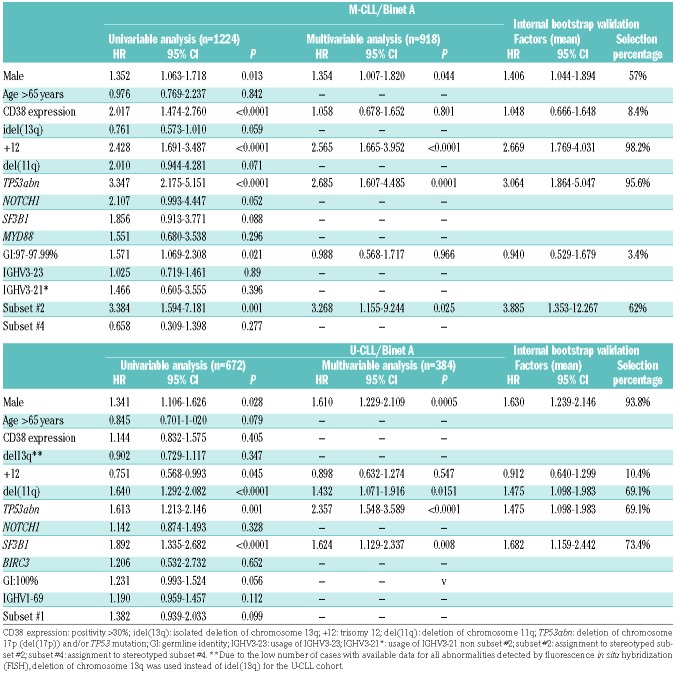
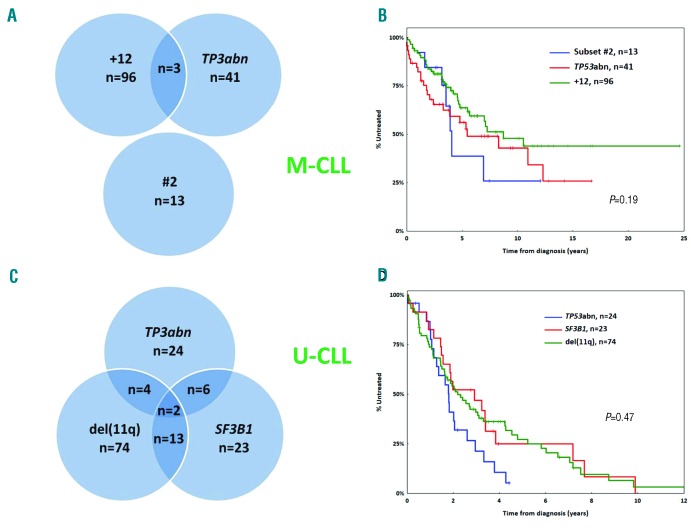
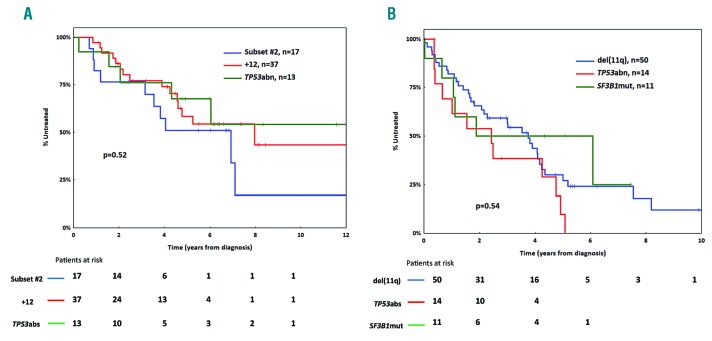
Univariable Cox regression analysis within M-CLL Binet A cases (n=1224) revealed that male sex, CD38 positivity, +12, subset #2, TP53 abnormalities (TP53abn) and borderline IGHV gene germline identity (GI: 97-97.9%) were associated with significantly shorter TTFT (Table 1). Bordeline GI remained significant in the univariable analysis even when subset #2 cases were excluded. TP53abn, +12, subset #2 membership and male sex retained independent adverse prognostic significance in the multivariable analysis (n=918) (Table 1). Of note, TP53abn, +12 and subset #2 membership identified 3 groups of almost mutually exclusive cases (altogether: n=153, 16% of all Binet A M-CLL) (Figure 2A) with no statistical differences in TTFT (median TTFT for TP53abn, +12, subset #2: 5.4, 7.1 and 4.1 years respectively; P=0.19) (Figure 2B).
Table 1.
Univariable and multivariable analysis for time-to-first-treatment within Binet A mutated chronic lymphocytic leukemia (M-CLL) and unmutated chronic lymphocytic leukemia (U-CLL). Harrell’s C-index (standard error in parenthesis) was calculated as 0.745 (se=0.013) and 0.753 (se=0.013) for the M-CLL and the U-CLL, respectively. Internal bootstrap validation for the multivariable analysis. Means of the Hazard Ratio (HR), 95% Confidence Interval (CI) and percentage of selection within 1000 bootstrap samples are displayed per factor.
Figure 2.
Subgroups of patients with similar prognosis within mutated chronic lymphocytic leukemia (M-CLL) and unmutated chronic lymphocytic leukemia (U-CLL). (A) TP53abn, trisomy 12 (+12) and stereotyped subset #2 assignment define three almost non-overlapping groups within early stage M-CLL. (B) Binet A M-CLL cases with TP53abn, +12 or assignment to stereotyped subset #2 display similar time-to-first-treatment (TTFT.) (C) Distribution of TP53abn, SF3B1 mutations and del(11q) in Binet A U-CLL. (D) Binet A U-CLL cases carrying TP53abn, SF3B1 mutations or del(11q) exhibit similar TTFT. N: number of patients.
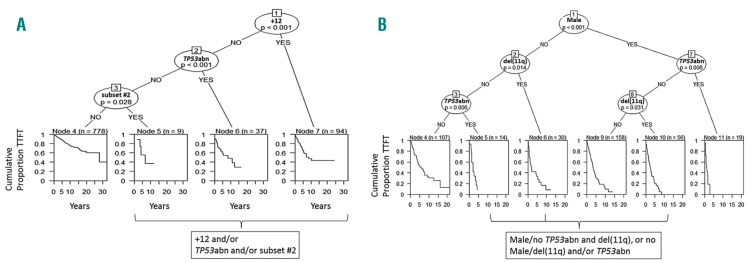
To further validate the results of the Cox regression analysis, we applied recursive partitioning models within a conditional inference framework. TP53abn, +12 and subset #2 membership were again identified as the most important predictors since the 3 groups defined by these predictors displayed similar TTFT that was significantly shorter from the remaining Binet A M-CLL (Figure 3A).
Figure 3.
Application of binary recursive partitioning in mutated chronic lymphocytic leukemia (M-CLL) and unmutated chronic lymphocytic leukemia (U-CLL). (A) Decision tree for Binet A M-CLL based on binary recursive partitioning and the subsequent application of an amalgamation algorithm. Trisomy 12 (+12), TP53abn and subset #2 membership were found to be the most significant factors as determined by the partitioning algorithm. The Binet A population is split in 4 terminal nodes (4, 5, 6 and 7). The amalgamation algorithm applied subsequently merged 3 of them in a larger terminal node. In particular, +12 was considered as the covariate with the strongest association to time-to-first-treatment (TTFT). Amongst patients lacking +12, TP53abn was the co-variate with the strongest association to TTFT and so on. After applying the amalgamation algorithm, patients with +12 and/or TP53abn and/or assignment to subset #2 were grouped into a larger node, resulting in 2 terminal nodes. The splitting is performed from right to left, following the criterion of strongest factor association with TTFT. The right branch represents the presence of a particular factor and the left branch the absence of that factor. P-value corresponds to a log-rank scores based test. The Kaplan-Meier curves estimate the TTFT of patients within each terminal node and n represents the number of patients per node. (B) Decision tree for Binet A U-CLL based on binary recursive partitioning and the subsequent application of an amalgamation algorithm. Male sex, TP53abn and del(11q) were the most significant factors as determined by the partitioning algorithm. The Binet A population was split into 6 terminal nodes (4, 5, 6, 9, 10 and 11). The amalgamation algorithm applied merged 3 of the terminal nodes into a larger terminal node. Sex was deemed to be the co-variate with the strongest association to TTFT. Amongst male patients, TP53abn was the co-variate with strongest association to TTFT. Amongst female patients, del(11q) was the co-variate with strongest association to TTFT and so on. After applying the amalgamation algorithm, male patients without TP53abn and with del(11q), and female patients with del(11q) and/or TP53abn were grouped into a larger node. The final number of terminal nodes was 4. The splitting is performed from top to bottom, following the criterion of strongest factor association with TTFT. The right branch represents the presence of a particular factor and the left one the absence of that factor. P-value corresponds to a log-rank scores-based test. The Kaplan-Meier curves estimate the TTFT of patients within each terminal node and n represents the number of patients per node.
Analysis for time-to-first-treatment within early stage unmutated chronic lymphocytic leukemia
Univariable Cox regression analysis revealed that TP53abn, SF3B1 mutations, del(11q) and male sex had a shorter TTFT within Binet-A U-CLL (n=676), while +12 had a longer TTFT (Table 1). TP53abn, SF3B1 mutations, del(11q) and male sex retained significance in the multivariable analysis (n=384) (Table 1). TP53abn and/or SF3B1 mutations and/or del(11q) [TP53abn/SF3B1mut/del(11q)] were positive in 146 cases (42% of the evaluated cohort with available data for all 3 parameters) (Figure 2C) and exhibited a similar TTFT (median TTFT for TP53abn, SF3B1mut, del(11q): 1.8, 2.8 and 2.1 years, respectively; P=0.47) (Figure 2D). The co-occurrence of these aberrations was not found to aggravate the prognosis when compared to single aberrations alone (Online Supplementary Figure S3). Male sex was associated with a shorter TTFT (median TTFT: 3.9 years, 95%CI: 0.1-5.9 years) amongst non-TP53abn/SF3B1mut/del(11q) U-CLL cases (Online Supplementary Figure S4).
Similar to M-CLL, we applied recursive partitioning (Figure 3B) which highlighted TP53abn, del(11q) and male sex as the most important variables within Binet A U-CLL. SF3B1 mutations did not reach significance, potentially due to the low number of cases with isolated SF3B1 mutations at the stage of the final nodes. In short, the evaluated U-CLL Binet A cohort was allocated to subgroups with differing TTFT, from shorter to longer, as follows: males with TP53abn; males with del(11q)/females with TP53abn and/or del(11q); males without TP53abn/del(11q); and, females without TP53abn/del(11q).
Two somatic hypermutation categories of chronic lymphocytic leukemia patients, two prognostic indices for time-to-first-treatment
Based on the above, we developed two prognostic indices for assessing TTFT, tailored to each SHM category. Within M-CLL, we defined 4 groups based on their clinico-biological profiles at the time of diagnosis: i) very high risk: Binet C with identical 5- and 10-year treatment-probability (TP) of 92%; ii) high risk: Binet B, 5y-TP and 10y-TP: 64% and 84%, respectively; iii) intermediate risk: Binet A with one of the following: TP53abn and/or +12 and/or subset #2 membership, 5y-TP and 10y-TP: 40% and 55%, respectively. [Of note, among 18 non-censored cases with no treatment indication for more than 10 years after diagnosis, 5 (30%) carried TP53abn]; and iv) low risk: non-TP53abn/+12/subset #2 Binet A, 5y-TP and 10y-TP: 12% and 25%, respectively (Figure 4A). Harrell’s C Index was calculated for the multivariable Cox model with the prognostic index above being the sole predictor and was found to equal 0.745 (se = 0.013).
Figure 4.
Prognostic index for time-to-first-treatment (TTFT) for mutated chronic lymphocytic leukemia (M-CLL) and unmutated chronic lymphocytic leukemia (UCLL). (A) Prognostic index for TTFT within M-CLL: i) very high risk: Binet C; ii) high risk: Binet B; iii) intermediate risk: Binet A with at least one of the following: TP53abn or +12 or assignment to subset #2 and; iv) low risk: non-TP53abn/+12/#2 Binet A. (B) Prognostic index for TTFT within U-CLL: i) very high risk: Binet C; ii) high risk: Binet B; iii) intermediate risk: Binet A with at least one of the following: TP53abn or +SF3B1 mutations or del(11q) membership; iv) low risk: non-TP53abn/SF3B1mut/del(11q) male Binet A and; v) very low risk: non-TP53abn/SF3B1mut/del(11q) female Binet A.
In U-CLL, we could define 5 groups: i) very high risk: Binet C with 5- and 10-year TP of 100%; ii) high risk: Binet B, 5y-TP and 10y-TP: 90% and 100%, respectively; iii) intermediate risk: Binet A with one of the following: TP53abn and/or SF3B1mut and/or del(11q), 5y-TP and 10y-TP: 78% and 98%, respectively; v) low risk: non-TP53abn/SF3B1mut/del(11q) male Binet A, 5y-TP and 10y-TP: 65% and 85%, respectively; and iv) very low risk: non-TP53abn/SF3B1mut/del(11q) female Binet A, 5y-TP and 10y-TP: 45% and 65%, respectively (Figure 4B). Harrell’s C Index was calculated for the multivariable Cox model with the prognostic index being the sole predictor and was found to equal 0.753 (se = 0.013).
Internal validation
In order to validate the results mentioned above, a bootstrapping procedure was performed. In early stage M-CLL patients, this showed that the average number of predictors included in the multivariable Cox model was 3.2 with three variables exhibiting selection percentages greater than 60%, i.e. TP53abn, +12 and subset #2 (Table 1). In early stage U-CLL patients, bootstrapping showed that the average number of predictors considered significant in the multivariable Cox model was 3.5. Four variables exhibited selection percentages greater than 60%, i.e. TP53abn, SF3B1mut, del(11q) and male sex.
External validation
Application of the above mentioned indices to the validation cohort (n=649) led to the following observations: i) in M-CLL, TP53abn, +12 and subset #2 membership (intermediate risk for M-CLL) exhibited similar TTFT, constituting a group (16% of all Binet cases also in the validation cohort) with almost identical 5y-TP (43%) and 10y-TP (60%) to those observed in the training cohort (40% and 55%, respectively) (Figure 5A); ii) similarly, in U-CLL, no difference was observed in TTFT for Binet A cases carrying TP53abn and/or SF3B1mut and/or del(11q) (intermediate risk for U-CLL) who exhibited similar 5y-TP and 10y-TP to that of the training cohort (74% vs. 78% and 92% vs. 98%, respectively; P-values: non significant) (Figure 5B); iii) amongst the remaining non-TP53abn/SF3B1mut/del(11q) U-CLL Binet A cases, the difference between male and female patients did not reach statistical significance (P=0.2) (Online Supplementary Figure S5A and B).
Figure 5.
Kaplan-Meier curves for time-to-first-treatment (TTFT) in the validation cohort. (A) Within mutated chronic lymphocytic leukemia (M-CLL), Binet A cases positive for TP53abn, trisomy 12 (+12) and stereotyped subset #2 assignment display similar TTFT. (B) No difference regarding TTFT among Binet A and unmutated chronic lymphocytic leukemia (U-CLL) cases carrying TP53abn, SF3B1 mutations or del(11q) in the validation cohort.
Discussion
Chronic lymphocytic leukemia constitutes a rather unique case amongst cancers in that the great majority of patients are asymptomatic at diagnosis and classified as early stage, and thus do not require immediate treatment.42 However, most patients will progress and meet the criteria for treatment initiation albeit at a variable time from diagnosis.37,42 Therefore, accurate prediction of the TTFT is of major importance for both patients and physicians having to address the challenge of living with and managing this (largely) invisible and incurable disease. In support of this argument, solid prognostication is increasingly recognized as both a priority and an unmet need by CLL patients themselves, who would benefit from accurate prognostic information in order to make educated life choices and, perhaps, also participate more actively in their care.
Several prognostic models have been developed for CLL based on combinations of biomarkers and host-derived features. However, none has been adopted in everyday clinical practice,13 e.g. to help decide about the follow-up strategy amongst untreated patients. These indices were mainly based on Cox regression models, where each respective training/validation cohort was considered as a single group. Herein, we followed a novel approach evaluating prognosis separately within M-CLL and U-CLL focusing on TTFT in the largest ever series studied to this purpose. We report that within early stage M-CLL, TP53abn, +12 and assignment to stereotyped subset #2 identified a subgroup of patients with uniformly shorter TTFT compared to the remaining early Binet A M-CLL. Similarly, within U-CLL the presence of TP53abn, del(11q) and SF3B1 mutations was found to be associated with the shortest TTFT, whereas the remaining U-CLL female patients had a significantly longer TTFT compared to males.
Classification of CLL patients based on the SHM status of the clonotypic BcR IG into M-CLL and U-CLL categories offers robust prognostic information, differing from other prognostic markers (e.g. genetic aberrations) that may evolve over time.15,17,18,43 Of note, studies by us and others have documented an asymmetric distribution of certain genetic aberrations in patients with distinct immunogenetic features extending from the M-CLL or U-CLL categorization to different stereotyped subsets.32,44,45 This has prompted speculation that particular modes of immune signaling initiated by specific BcR IG may trigger different pathways of clonal evolution leading to the emergence of distinct disease variants.
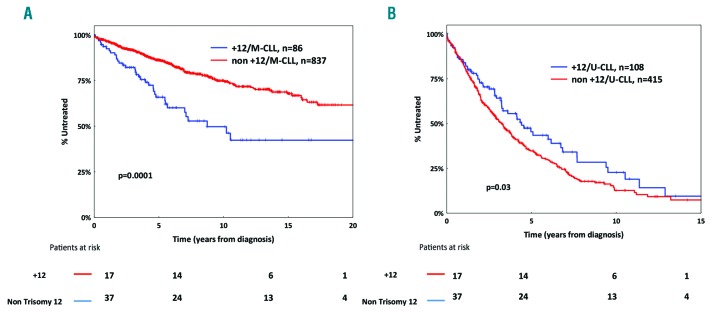
For these reasons, the BcR IG appears an obvious starting point for developing a biologically-orientated prognostication scheme for CLL, as in our present study, offering the possibility to dissect the precise impact of a given biomarker within a particular immunogenetic category (e.g. M-CLL or U-CLL). For example, within M-CLL, +12 was found to define a subgroup of patients with a TTFT similar to that of patients harboring TP53abn, whereas, in contrast, +12 was associated with favorable outcome within U-CLL (Figure 6A and B). Of note, mutations within the NOTCH1 gene had no impact on survival among U-CLL cases with +12 (Online Supplementary Figure S6). These findings may explain: i) why +12 is considered as an intermediate-risk aberration in prognostic indices where CLL is evaluated as one group regardless of the SHM status;38 and, ii) the contradictory results reported in different cohorts with different relative proportion of M-CLL and U-CLL patients, regarding the significance of a given indicator that can show an asymmetric distribution within each SHM group.
Figure 6.
Kaplan-Meier curves for time-to-first-treatment (TTFT) for mutated chronic lymphocytic leukemia (M-CLL) and unmutated chronic lymphocytic leukemia (U-CLL) cases carrying trisomy 12 (+12). (A) +12 is an unfavorable prognosticator in M-CLL. (B) +12 is associated with a more indolent clinical course in U-CLL.
Our initial results based on Cox regression analysis were validated internally, being highly reproducible in an independent validation cohort. In particular, the median TTFT for the subgroups of patients with the shortest TTFT in both SHM categories, namely TP53abn/+12/#2 and TP53abn/SF3B1mut/del(11q) for M-CLL and U-CLL respectively, was almost identical between the validation and the training cohort. Interestingly, the latter exhibited significant differences in terms of the biological background compared to the training cohort (Online Supplementary Table S2); this may be taken as further evidence for the robustness of our approach, since similar results were obtained across cohorts with differing patient composition. Cox regression results were further confirmed through the application of an alternative statistical approach, namely binary recursive partitioning, which offers a different framework, thereby conveying a hierarchical order of importance and classification for the evaluated prognostic factors.
Admittedly, despite providing a robust risk stratification scheme, the prognostic indices proposed here will not solve the problem of outliers, while they also overlook the potential effect of other variables with proven prognostic significance, e.g. cytogenetic complexity or methylation signatures. Thus, they cannot be considered the last word in biomarker-orientated risk stratification for TTFT. They do, however, highlight the need for further studies, while providing the conceptual frame of compartimentalization. It should be further emphasized that our approach is built on the pivotal role of IGHV SHM status in the prognostic setting, which appears to be less important in the era of novel agents, namely BTK and BCL2 inhibitors, where response rates seem to be similar between M-CLL and U-CLL.46,47
Recently, the International Prognostic Index for patients with CLL (CLL-IPI) was developed for assessing overall survival (OS).7 This has provided a robust prognostic classification scheme as it includes well-characterized patients followed in the context of clinical trials. A caveat of CLL-IPI concerns the fact that the evaluated patients had been treated with various regimens in the context of different clinical trials. Moreover, CLL-IPI does not allow the identification of distinct groups within each SHM category. For example, following the CLL-IPI score, a young (<65 years), early-stage patient, negative for TP53abn and belonging to M-CLL would never be characterized as very high risk. It is important to note, therefore, that the clinically aggressive stereotyped subset #2 would be overlooked by the CLL-IPI as it largely falls into the category of M-CLL cases lacking TP53abn, with 60% of patients also under 65 years of age. This is not a trivial issue, considering that subset #2 accounts for up to 5% of all CLL requiring treatment and, therefore, is equal in size to the CLL-IPI very high risk group with very limited if any overlap.30 A final, more general concern is that CLL-IPI was developed based on the analysis of cases treated prior to the introduction of novel therapies, which are likely to change the treatment expectations and OS in CLL, thus eventually creating the need for new predictive schemes.48
Our study identified subset #2 membership and SF3B1 mutations as prognostically important biomarkers for early-stage M-CLL and U-CLL, respectively. In contrast, other recurrent gene mutations such as NOTCH1 or BIRC3 failed to reach significance even in univariable analysis. Interestingly, SF3B1 mutations are remarkably enriched within subset #2 (approx. 50% of cases harbor a mutation), however, their impact within this very aggressive subset remains equivocal.30,32 Overall, these results emphasize the value of investigating IG sequences for stereotyped subset #2 membership (easily determined through the use of a free online tool available at: http://bat.infspire.org/arrest/assignsubsets/49) and searching for SF3B1 mutations in routine clinical practice as this would enable a more accurate assessment of prognosis (TTFT) at diagnosis of CLL.
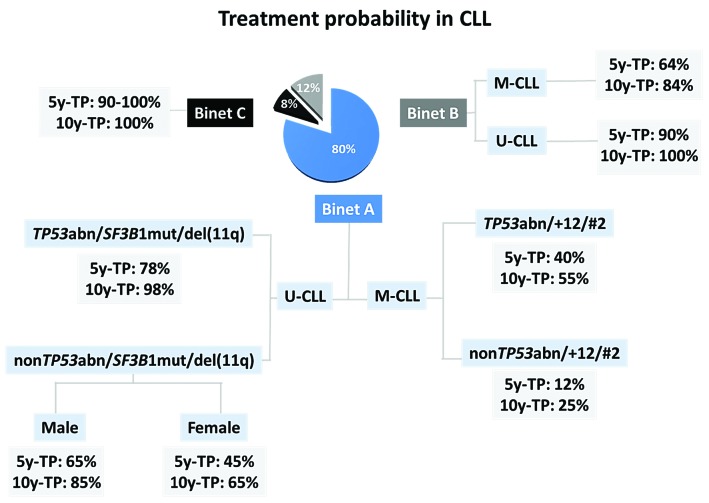
In conclusion, we propose a novel approach to prognostic assessment in CLL grounded on the fact that not all CLL are equal, but instead that M-CLL and U-CLL categories are fundamentally different regarding their ontogeny and molecular landscape, at least for early-stage patients (Figure 7). Our results support that compartmentalizing CLL with the BcR IG as the starting point allows accurate prognostication in early-stage CLL. This further shows that the relative weight of well established prognostic indicators differs based on the immunogenetic features of each individual case. From a broader perspective, such compartmentalized approaches might prove relevant in other B-cell lymphomas as well e.g. diffuse large B-cell lymphoma where different biomarkers are emerging as prognostically relevant for the activated B-cell or the germinal center subtype, respectively,50 that display distinct immunogenetic features and signaling signatures.50
Figure 7.
Prognostic algorithm regarding treatment probability for chronic lymphocytic leukemia (CLL). 5y-TP: treatment probability five years from diagnosis; 10y-TP: treatment probability ten years from diagnosis; U-CLL: CLL with unmutated IGHV genes; M-CLL: CLL with mutated IGHV genes; TP53abn: deletion of chromosome 17p (del(17p)) and/or TP53 mutation; +12:trisomy 12, del(11q): deletion of chromosome 11q; SF3B1mut: SF3B1 mutation; #2: assignment to stereotyped subset #2. The pie chart refers to the entire cohort with each slice indicating the proportion of patients according to Binet clinical staging.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/2/360
Funding
This work was supported in part by the Swedish Cancer Society, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, Karolinska Institutet, Stockholm, the Lion’s Cancer Research Foundation, Uppsala, the Marcus Borgström Foundation and Selander’s Foundation, Uppsala; H2020 “AEGLE, An analytics framework for integrated and personalized healthcare services in Europe” by the EU; H2020 “MEDGENET, Medical Genomics and Epigenomics Network” (No.692298) by the EU; H2020 “CLLassify, Innovative risk assessment for individualizing treatment in chronic lymphocytic leukemia” (No.702714) by the EU; Associazione Italiana per la Ricerca sul Cancro AIRC Investigator grants #20246, and Special Program Molecular Clinical Oncology AIRC 5 per mille #9965; Progetti di Rilevante Interesse Nazionale (PRIN) #2015ZMRFEA, MIUR, Rome, Italy; TRANSCAN-179 NOVEL JTC 2016; project CEITEC 2020 (LQ1601) by MEYS-CZ, project AZV-MH-CZ 15-30015A-4/2015; JCS was funded by Bloodwise (11052, 12036), the Kay Kendall Leukaemia Fund (873), Cancer Research UK (C34999/A18087, ECMC C24563/A15581), Wessex Medical Research and the Bournemouth Leukaemia Fund; Special Program Molecular Clinical Oncology 5 × 1000 No. 10007, Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy; Progetto Ricerca Finalizzata RF-2011-02349712, Ministero della Salute, Rome, Italy.
TM is recipient of a Marie Sklodowska-Curie individual fellowship (grant agreement No. 702714), funded by the EU H2020 research and innovation programme.
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48(1):198–206. [DOI] [PubMed] [Google Scholar]
- 3.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46(2):219–234. [DOI] [PubMed] [Google Scholar]
- 4.Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia-what do we need to know¿ Nat Rev Clin Oncol. 2011;8(1):38–47. [DOI] [PubMed] [Google Scholar]
- 5.Gentile M, Mauro FR, Rossi D, et al. Italian external and multicentric validation of the MD Anderson Cancer Center nomogram and prognostic index for chronic lymphocytic leukaemia patients: analysis of 1502 cases. Br J Haematol. 2014;167(2):224–232. [DOI] [PubMed] [Google Scholar]
- 6.Haferlach C, Dicker F, Weiss T, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49(9):851–859. [DOI] [PubMed] [Google Scholar]
- 7.International CLLIPIwg. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17(6):779–790. [DOI] [PubMed] [Google Scholar]
- 8.Morabito F, Cutrona G, Gentile M, et al. Definition of progression risk based on combinations of cellular and molecular markers in patients with Binet stage A chronic lymphocytic leukaemia. Br J Haematol. 2009;146(1):44–53. [DOI] [PubMed] [Google Scholar]
- 9.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100(4):1177–1184. [PubMed] [Google Scholar]
- 10.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanafelt TD, Jenkins G, Call TG, et al. Validation of a new prognostic index for patients with chronic lymphocytic leukemia. Cancer. 2009;115(2):363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wierda WG, O’Brien S, Wang X, et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood. 2007;109(11):4679–4685. [DOI] [PubMed] [Google Scholar]
- 13.Baliakas P, Mattsson M, Stamatopoulos K, Rosenquist R. Prognostic indices in chronic lymphocytic leukaemia: where do we stand how do we proceed¿ J Intern Med. 2016;279(4):347–357. [DOI] [PubMed] [Google Scholar]
- 14.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3): 145–162. [DOI] [PubMed] [Google Scholar]
- 15.Sutton LA, Rosenquist R. Deciphering the molecular landscape in chronic lymphocytic leukemia: time frame of disease evolution. Haematologica. 2015;100(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutton LA, Rosenquist R. The complex interplay between cell-intrinsic and cell-extrinsic factors driving the evolution of chronic lymphocytic leukemia. Semin Cancer Biol. 2015;34:22–35. [DOI] [PubMed] [Google Scholar]
- 17.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson FK, Caligaris-Cappio F. Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood. 2004;103(12):4389–4395. [DOI] [PubMed] [Google Scholar]
- 20.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 21.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 22.Fischer K, Bahlo J, Fink AM, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127(2):208–215. [DOI] [PubMed] [Google Scholar]
- 23.Rossi D, Terzi-di-Bergamo L, De Paoli L, et al. Molecular prediction of durable remission after first-line fludarabine-cyclophosphamide-rituximab in chronic lymphocytic leukemia. Blood. 2015;126(16):1921–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson PA, Tam CS, O’Brien SM, et al. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127(3):303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236–1242. [DOI] [PubMed] [Google Scholar]
- 26.Marincevic M, Cahill N, Gunnarsson R, et al. High-density screening reveals a different spectrum of genomic aberrations in chronic lymphocytic leukemia patients with ‘stereotyped’ IGHV3-21 and IGHV4-34 B-cell receptors. Haematologica. 2010;95(9):1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ntoufa S, Vardi A, Papakonstantinou N, et al. Distinct innate immunity pathways to activation and tolerance in subgroups of chronic lymphocytic leukemia with distinct immunoglobulin receptors. Mol Med. 2012;18:1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multi-centre study. Lancet Haematology. 2014;1(2):74–84. [DOI] [PubMed] [Google Scholar]
- 29.Agathangelidis A, Vardi A, Baliakas P, Stamatopoulos K. Stereotyped B-cell receptors in chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55(10):2252–2261. [DOI] [PubMed] [Google Scholar]
- 30.B aliakas P, Agathangelidis A, Hadzidimitriou A, et al. Not all IGHV3-21 chronic lymphocytic leukemias are equal: prognostic considerations. Blood. 2015;125(5):856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xochelli A, Baliakas P, Kavakiotis I, et al. Chronic Lymphocytic Leukemia with Mutated IGHV4-34 Receptors: Shared and Distinct Immunogenetic Features and Clinical Outcomes. Clin Cancer Res. 2017;23(17):5292–5301. [DOI] [PubMed] [Google Scholar]
- 32.Jeromin S, Haferlach C, Dicker F, Alpermann T, Haferlach T, Kern W. Differences in prognosis of stereotyped IGHV3-21 chronic lymphocytic leukaemia according to additional molecular and cytogenetic aberrations. Leukemia. 2016;30(11):2251–2253. [DOI] [PubMed] [Google Scholar]
- 33.Baliakas P, Mattsson M, Hadzidimitriou A, et al. No improvement in long-term survival over time for chronic lymphocytic leukemia patients in stereotyped subsets #1 and #2 treated with chemo(immuno)therapy. Haematologica. 2018;103(4):e158–e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329–336. [DOI] [PubMed] [Google Scholar]
- 35.Jeromin S, Weissmann S, Haferlach C, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28(1):108–117. [DOI] [PubMed] [Google Scholar]
- 36.Baliakas P, Hadzidimitriou A, Agathangelidis A, et al. Prognostic relevance of MYD88 mutations in CLL: the jury is still out. Blood. 2015;126(8):1043–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 39.Agathangelidis A, Darzentas N, Hadzidimitriou A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119(19):4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. [DOI] [PubMed] [Google Scholar]
- 41.Hothorn T, Hornik K, Zeileis A. Unbaised Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical Statistics. 2006;15(3):651–674. [Google Scholar]
- 42.Hallek M. Chronic lymphocytic leukemia: 2015 Update on diagnosis, risk stratification, and treatment. Am J Hematol. 2015;90(5):446–460. [DOI] [PubMed] [Google Scholar]
- 43.Ljungstrom V, Cortese D, Young E, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2016;127(8):1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi D, Spina V, Bomben R, et al. Association between molecular lesions and specific B-cell receptor subsets in chronic lymphocytic leukemia. Blood. 2013;121(24):4902–4905. [DOI] [PubMed] [Google Scholar]
- 45.Sutton LA, Young E, Baliakas P, et al. Different spectra of recurrent gene mutations in subsets of chronic lymphocytic leukemia harboring stereotyped B-cell receptors. Haematologica. 2016;101(8):959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrd JC, Furman RR, Coutre SE, et al. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125(16):2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2016;374(4):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamanna N, O’Brien S. Novel agents in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2016;2016(1):137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bystry V, Agathangelidis A, Bikos V, et al. ARResT/AssignSubsets: a novel application for robust subclassification of chronic lymphocytic leukemia based on B cell receptor IG stereotypy. Bioinformatics. 2015;31(23):3844–3846. [DOI] [PubMed] [Google Scholar]
- 50.Karube K, Enjuanes A, Dlouhy I, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018;32(3):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.