Chronic lymphocytic leukemia (CLL) derives from the expansion of clonal and antigen-experienced mature B lymphocytes, whose accumulation results from a dynamic imbalance between cell death and proliferation. The former is impaired by the overexpression of the anti-apoptotic protein BCL2 by CLL cells and the latter is mainly driven by the B-cell receptor (BCR), the key molecule to elucidate the pathogenic and evolution mechanisms of the disease.
Chronic lymphocytic leukemia is a highly heterogeneous disease both in terms of biological landscape and clinical course, including the interval from diagnosis to first progression requiring treatment (time-to-first-treatment, TTFT), the degree and duration of response to therapy, overall survival (OS), and risk of transformation into an aggressive lymphoma (Richter’s syndrome). The prognosis of CLL patients can be accurately defined by combining clinical and biological parameters that include BCR features, cytogenetic lesions, immunophenotypic markers, and gene mutations. Some biomarkers are also useful predictors of response to therapy.
Mutations of the genes codifying for the immunoglobulin heavy chain variable region (IGHV) of the BCR represent one of the most robust prognostic biomarkers, and, indeed, was one of the first to be identified. IGHV mutations never change over time, and thus represent the fingerprint of the disease. Back in 1999, it was reported that CLL patients with mutated IGHV genes (M-CLL) (i.e. <98% cut-off of IGHV identity to the germline counterpart) display a longer TTFT and a longer survival than CLL with unmutated IGHV genes (U-CLL) (≥98%).1,2 The subsequent identification in about 30% of CLL of stereotyped BCRs was even more intriguing.3,4 Stereotyped BCRs (namely those with a nearly identical length of the HCDR3 region, shared amino acids in key positions and the non-stochastic pairing of IGHV and light chain genes) identify subgroups defined “subsets”. More frequent in U-CLL (40%) than in M-CLL (10%) in Caucasians, CLL subsets display distinctive clinicobiological associations: subset #4, mostly M-CLL, is associated to a young age at diagnosis and an indolent disease; subset #1, U-CLL, to a very aggressive clinical course; subset #8, U-CLL, to a higher risk of developing Richter’s syndrome; subset #2 to a poor prognosis regardless of the percentage of IGHV mutations.5 Although the IGHV gene usage and the frequency of BCR subsets can vary across populations with a different incidence of CLL (i.e. Caucasian vs. Chinese), it is interesting that these clinicobiological associations hold true across all ethnic groups.6
In 2015, the value of the IGHV status in predicting the outcome after chemoimmunotherapy also emerged, since M-CLL patients have a significantly longer progression-free survival (PFS), particularly when devoid of poor-risk fluorescence in situ hybridization (FISH) lesions.7 In contrast, it became apparent that the IGHV status does not influence the efficacy of the BTK inhibitor ibrutinib.8,9 Thus, it has been suggested that both the IGHV status and TP53 deletions/mutations should be investigated at the time of disease progression in order to guide the first-line therapeutic choice between chemoimmunotherapy and novel agents.10 Given the clinical implications, the European Research Initiative on CLL (ERIC) group has conducted an international harmonization process across labs for the analysis and reporting of IGHV and TP53 genes in CLL, and this has led to the recently up-dated recommendations.11,12
Although the pathogenic mechanisms operational in CLL are far from being fully elucidated, the oncogenic function of the BCR is indirectly demonstrated by the high anti-leukemic efficacy of kinase inhibitors that block BCR signaling (i.e. ibrutinib, idelalisib, acalabrutinib, duvelisib). On the one hand, in CLL, unlike other lymphoproliferative diseases, the BCR is capable of generating a cell-autonomous signaling driven by the interactions between HCDR3 of near BCR (BCR-BCR) on the cell surface.13 On the other hand, the quality of BCR signaling is heterogeneous: U-CLL are more responsive in vitro to IgM ligation in terms of modulation of the gene expression profile, advance in the cell cycle, and increase in proliferation compared to M-CLL.14 As for a commonly accepted model, U-CLL show a weak autonomous BCR-BCR signaling, a low affinity binding to auto-antigens, an increased BCR responsiveness, and an aggressive clinical course, while M-CLL patients show a strong autonomous BCR-BCR signaling that leads to an anergic state, a lower proliferative response after BCR stimulus, and an overall indolent course.15,16 This model conciliates a shared pathogenic mechanism with the biological and clinical heterogeneity of CLL. In addition, BCR stereotyping likely supports the role of an antigenic pressure in the selection of the leukemic clone.3–5 Among the various factors that contribute to modulate the BCR responsiveness, the microenvironment certainly has a relevant role, since the CLL cells within the lymph node show an upregulation of genes involved in BCR signaling and NfKB activation, at variance from the circulating CLL cells from the same individual.17
Although the mechanisms driving the heterogeneity of CLL genetics are currently unknown, since leukemia kinetics and genetic complexity are usually closely related, it would seem that the BCR can play a role in maintaining genetic stability or in acquiring genetic instability in CLL. Indeed, U-CLL and M-CLL display a variable proportion of the different genetic lesions, as well as of the BCR subsets; U-CLL is enriched with biomarkers with an adverse prognostic significance, although not exclusively.
In the present issue of Haematologica, on behalf of the ERIC group, out of a large cohort of 2366 CLL cases, Baliakas et al.18 focused on 1900 stage A CLLs that were divided into the two main CLL immunogenetic subgroups: U-CLL and M-CLL. Considering each of them separately, they analyzed the relative weight of different prognostic markers in determining the TTFT.18 These prognostic markers included: age, sex, CD38, FISH lesions, TP53, SF3B1, NOTCH1, BIRC3, MYD88 gene mutations, and the major BCR subsets #1, #2 and #4.
This IGHV-based prognostic approach suggests that, among stage A M-CLL, cases with trisomy 12 and stereotyped subset #2 show a short TTFT at five and ten years after diagnosis, similar to those with TP53 abnormalities; within stage A U-CLL, cases with del(11q) and/or SF3B1 mutations experience a TTFT as short as cases with TP53 abnormalities. These markers are almost entirely mutually exclusive. The validity of the model is confirmed also in an external validation cohort of 649 Binet A CLL. Interestingly, male sex comes out as a determinant of disease course within U-CLL, a recurrent observation that has never been truly addressed.19
Overall, considering patients in all stages, within IGHV M-CLL, the lowest risk category (stage A/non-TP53 abnormalities/+12/subset #2), representing 73% of all M-CLL, shows a TTFT of 12% and 25% at five and ten years, respectively; this means that only 1 out of 4 patients has required treatment after ten years from diagnosis. The intermediate-risk category (stage A/TP53 abnormalities/+12/subset #2), representing 14% of all M-CLL, shows a TTFT of 40% and 55% at five and ten years, implying that 1 out of 2 patients still remains untreated after ten years from diagnosis.
On the other hand, within IGHV U-CLL, the very low-risk category (stage A/female/non-SF3B1 mutation/del11q), representing 13% of all U-CLL, shows a TTFT of 45% and 65% at five and ten years, respectively, much shorter (median TTFT 6.1 years) than low-risk M-CLL patients (median TTFT not reached). The U-CLL low-risk (stage A/male/non-SF3B1 mutation/del11q) and intermediate-risk (stage A/SF3B1 mutation/del11q) patients, representing 19% and 24% of all U-CLL, respectively, show a median TTFT of 3.6 and 2.1 years.
Although the retrospective nature of the study and the heterogeneity of the treatments administered prevent the authors from assessing OS in this series, these data identify important differences between the two main CLL immunogenetic subgroups and may help in building a novel CLL patient risk stratification where BCR plays a major role as the core of the prognostic model. For example, trisomy 12, associated to an intermediate prognosis in CLL analyzed as a whole, now has a better definition, having a detrimental prognostic impact on TTFT within M-CLL but not within U-CLL. Moreover, subset #2 should be identified and reported since it has an independent impact on TTFT within M-CLL.
The CLL prognostic algorithms proposed so far have always included IGHV status. However, in six different scoring systems (CLL-IPI, MDACC 2007, MDACC 2011, GCLLSG, Barcelona-Brno, O-CLL1), the outcome prediction fails in a proportion ranging from 22% to 35% of early phase CLL.20 Especially within low-risk CLL, there is still a sizable proportion of patients experiencing “early” progression. A prognostic algorithm built within the U-CLL and M-CLL IGHV categories may overcome this limitation. This has clinical implications for patients’ counseling, follow-up planning, and potential enrollment in trials exploring early treatment approaches for stage A CLL.
It is clear that, in an era of new drugs, the value of all the algorithms proposed so far in terms of prediction of OS will need to be re-evaluated, since the BCR/BCL2 inhibitors have the power to overcome the prognostic impact of the IGHV mutational status. For the time being, besides TP53 deletions/mutations, it is advisable to consider IGHV gene sequencing in all CLL patients requiring first-line treatment as a guide to choose between conventional and innovative approaches. In this respect, the ERIC initiative on the IGHV and TP53 harmonization project and the certification system, with external quality controls for the accreditation of the laboratories performing IGHV/TP53 analysis, is a very timely effort.
Supplementary Material
Acknowledgments
The authors wish to thank Associazione Italiana per la Ricerca sul Cancro (AIRC) 5 × 1000, Milan, Italy (Special Program Molecular Clinical Oncology MCO-10007; Metastases Special Program, n. 21198). They thank Dr. Sara Raponi and Dr. Luca Vincenzo Cappelli for their contribution in the design of Figure 1.
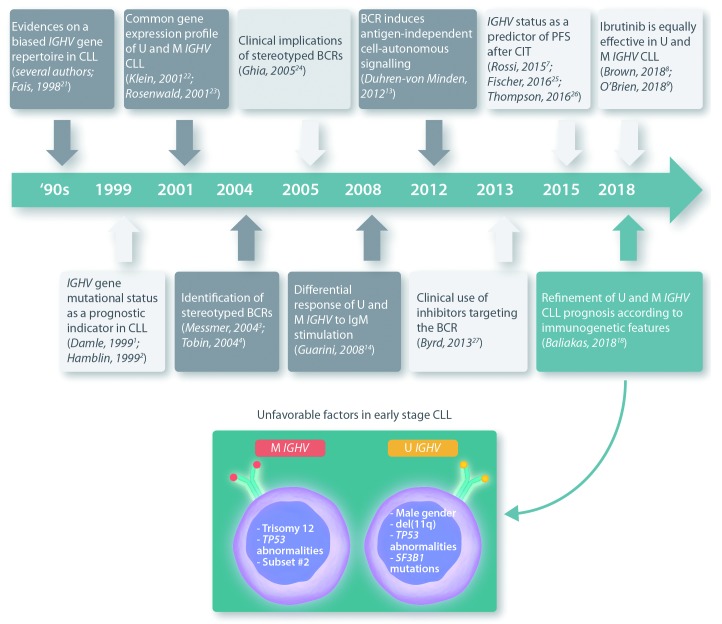
Figure 1.
The 20-year history of IGHV mutations in chronic lymphocytic leukemia (CLL). IGHV: immunoglobulin heavy chain variable region; BCR: B-cell receptor; M: mutated; U: unmutated; CIT: chemoimmunotherapy; PFS: progression-free survival.
References
- 1.Damle RN, Wasil T, Fais F, et al. IgV gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 2.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 3.Messmer BT, Albesiano E, Efremov DG, et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200(4):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tobin G, Thunberg U, Karlsson K, et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104(9):2879–2885. [DOI] [PubMed] [Google Scholar]
- 5.Stamatopoulos K, Agathangelidis A, Rosenquist R, Ghia P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia. 2017;31(2):282–291. [DOI] [PubMed] [Google Scholar]
- 6.Marinelli M, Ilari C, Xia Y, et al. Immunoglobulin gene rearrangements in Chinese and Italian patients with chronic lymphocytic leukemia. Oncotarget. 2016;7(15):20520–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi D, Terzi-di-Bergamo L, De Paoli L, et al. Molecular prediction of durable remission after first-line fludarabine-cyclophosphamide-rituximab in chronic lymphocytic leukemia. Blood. 2015;126(16):1921–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JR, Hillmen P, O’Brien S, et al. Extended follow-up and impact of high-risk prognostic factors from the phase 3 RESONATE study in patients with previously treated CLL/SLL. Leukemia. 2018;32(1):83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien S, Furman RR, Coutre S, et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood. 2018;131(17):1910–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745–2760. [DOI] [PubMed] [Google Scholar]
- 11.Rosenquist R, Ghia P, Hadzidimitriou A, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia. 2017;31(7):1477–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malcikova J, Tausch E, Rossi D, et al. ; European Research Initiative on Chronic Lymphocytic Leukemia (ERIC) - TP53 network. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia. 2018;32(5):1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dühren-von Minden M, Übelhart R, Schneider D, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489(7415):309–312. [DOI] [PubMed] [Google Scholar]
- 14.Guarini A, Chiaretti S, Tavolaro S, et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood. 2008;112(3):782–792. [DOI] [PubMed] [Google Scholar]
- 15.Iacovelli S, Hug E, Bennardo S, et al. Two types of BCR interactions are positively selected during leukemia development in the Eμ-TCL1 transgenic mouse model of CLL. Blood. 2015;125(10):1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minici C, Gounari M, Übelhart R, et al. Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia. Nat Commun. 2017;8:15746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herishanu Y, Perez-Galan P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baliakas P, Moysiadis T, Hadzidimitriou A, et al. Tailored approaches grounded on immunogenetic features for refined prognostication in chronic lymphocytic leukemia. Haematologica. 2018. September 27 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catovsky D, Wade R, Else M. The clinical significance of patients’ sex in chronic lymphocytic leukemia. Haematologica. 2014;99(6):1088–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molica S, Giannarelli D, Mirabelli R, et al. Reliability of six prognostic models to predict time-to-first-treatment in patients with chronic lymphocytic leukaemia in early phase. Am J Hematol. 2017;92(6):E91–E93. [DOI] [PubMed] [Google Scholar]
- 21.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102(8):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghia P, Stamatopoulos K, Belessi C, et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood. 2005;105(4):1678–1685. [DOI] [PubMed] [Google Scholar]
- 25.Fischer K, Bahlo J, Fink AM, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127(2):208–215. [DOI] [PubMed] [Google Scholar]
- 26.Thompson PA, Tam CS, O’Brien SM, et al. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127(3):303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.