Abstract
A high proportion of patients with acute myeloid leukemia who achieve minimal residual disease negative status ultimately relapse because a fraction of pathological clones remains undetected by standard methods. We designed and validated a high-throughput sequencing method for minimal residual disease assessment of cell clonotypes with mutations of NPM1, IDH1/2 and/or FLT3-single nucleotide variants. For clinical validation, 106 follow-up samples from 63 patients in complete remission were studied by sequencing, evaluating the level of mutations detected at diagnosis. The predictive value of minimal residual disease status by sequencing, multiparameter flow cytometry, or quantitative polymerase chain reaction analysis was determined by survival analysis. The sequencing method achieved a sensitivity of 10−4 for single nucleotide variants and 10−5 for insertions/deletions and could be used in acute myeloid leukemia patients who carry any mutation (86% in our diagnostic data set). Sequencing–determined minimal residual disease positive status was associated with lower disease-free survival (hazard ratio 3.4, P=0.005) and lower overall survival (hazard ratio 4.2, P<0.001). Multivariate analysis showed that minimal residual disease positive status determined by sequencing was an independent factor associated with risk of death (hazard ratio 4.54, P=0.005) and the only independent factor conferring risk of relapse (hazard ratio 3.76, P=0.012). This sequencing-based method simplifies and standardizes minimal residual disease evaluation, with high applicability in acute myeloid leukemia. It is also an improvement upon flow cytometry- and quantitative polymerase chain reaction-based prediction of outcomes of patients with acute myeloid leukemia and could be incorporated in clinical settings and clinical trials.
Introduction
Cytogenetic and molecular alterations at diagnosis and response to treatment are the most useful criteria for predicting the relative risk of relapse in acute myeloid leukemia (AML), and for guiding the choice between chemotherapy and hematopoietic stem cell transplantation in first complete remission.1 The definition of complete remission for AML includes criteria for the identification of patients with poor prognosis using cytomorphological methods,2 but these studies do not have a good predictive value because most of the patients in complete remission relapse within 3 years of diagnosis.3
Assessment of minimal residual disease (MRD) is critical in monitoring patients in morphological remission, to inform decisions about further therapy.1 Indeed, several studies have reported MRD status as a stronger predictor of relapse, because patients who are MRD negative have a better prognosis than those who are MRD positive.4,5 In support of this, recent non-randomized studies from prospective multicenter trials suggested better outcomes when leukemia therapy was selected based on the results of MRD assessment.6–8
AML is, nevertheless, a biologically complex and heterogeneous disease, which makes MRD testing more challenging in this condition than in other hematologic neoplasms such as acute lymphoblastic leukemia or multiple myeloma. The detection of very low levels of MRD by conventional methods such as quantitative (q) polymerase chain reaction (PCR) or multiparameter flow cytometry (MFC) provides powerful independent prognostic information. Unfortunately, as described for cytomorphological complete remission, many patients who achieve MRD negative status relapse as a result of the progression of undetected leukemic cells. The most common method for MRD detection is MFC, which has intermediate applicability (70–80%) and limited sensitivity.9,10 However, there is no consensus on multi-antibody panels with regards to inter-laboratory performance, and the technique requires a high level of expertise. The other principal method for monitoring MRD, qPCR, has good sensitivity (10−4-10−6), but its applicability is limited to the approximately 40% of patients who present with molecular alterations (RUNX1-RUNX1T1, CBFβ-MYH11 or NPM1) at diagnosis.11
For the above reasons, new methods with higher sensitivity, specificity, applicability and performance are needed for MRD assessment in AML. Against this background, next-generation sequencing (NGS) and digital PCR (dPCR) have recently emerged as potentially promising platforms for the assessment of MRD.12 Here, we optimized and clinically validated a new deep targeted NGS-based method, supported with dPCR technical validation, for the detection and quantification of MRD [both small insertion/deletions (indels) and single nucleotide variants (SNV)] in AML patients, in an attempt to improve and/or complement the current techniques for MRD evaluation, and to establish its potential as a predictor of patients’ outcome.
Methods
More detailed information can be found in the Online Supplementary Data (1–6).
Patients and samples
One hundred and ninety patients with de novo or secondary non–M3 AML were included in mutational profile screening at diagnosis. We performed a new selection for retrospective MRD assessment using the following criteria: presence of the NPM1 type A mutation, or SNV in FLT3, IDH1 and/or IDH2 at diagnosis, and availability of at least one follow-up genomic (g)-DNA sample.
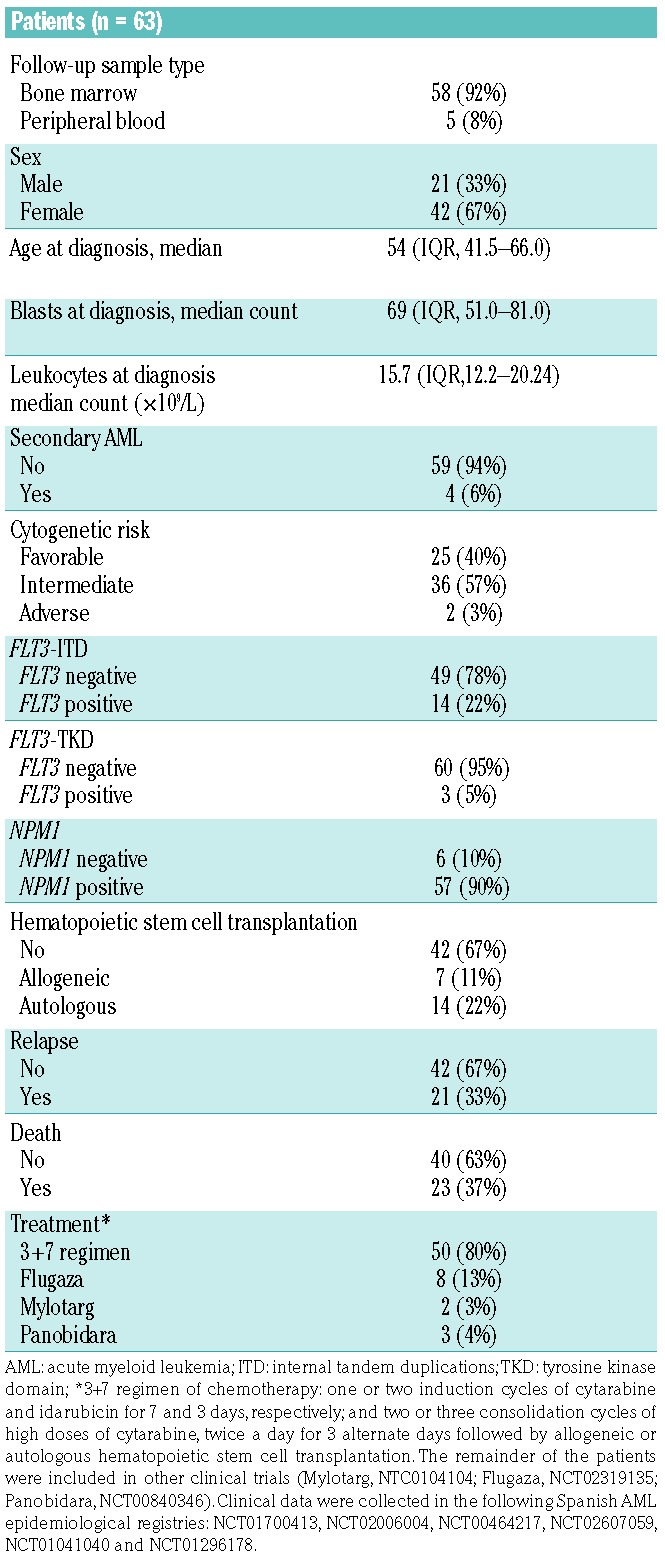
The MRD approach was applied to 51 (48%) follow-up samples taken after induction therapy and 55 (52%) taken after consolidation, corresponding to 63 patients diagnosed between 2006 and 2016 (for selection criteria see Online Supplement 6 and Supplementary Table S1). Patients were treated according to PETHEMA (Programa Español de Tratamientos en Hematología) or CETLAM (Grupo cooperativo de Estudio y Tratamiento de Leucemias Agudas y Mielodisplasias) protocols. The study was conducted according to Spanish law 14/2007 on biomedical research, and was approved by the Research Ethics Board of each participating institution. All patients provided informed consent. The main clinical characteristics of the patients are summarized in Table 1. All patients achieved complete remission according to cytomorphological criteria after induction therapy (<5% of bone marrow blasts).
Table 1.
Main characteristics of the patients with acute myeloid leukemia included in the minimal residual disease study.
To construct calibration curves, commercial (Horizon Discovery, UK) reference standard gDNA was used for somatic SNV in IDH1 (R132C) and IDH2 (R172K). As a further source of gDNA, we used the OCI-AML3 cell line (ACC 582, DSMZ, Germany) with the NPM1 type A mutation (c.863_864insCCTG) to examine indels. As OCI-AML3 cells also present a SNV in DNMT3A (R882C), this was included only for technical optimization.
Deep targeted sequencing workflow
The sequencing workflow included a first study at diagnosis and a second study at follow-up. Mutational profile screening at diagnosis was done with a customized NGS myeloid panel of 32 genes frequently mutated in myeloid diseases,13 (Online Supplementary Table S2) and NPM1 analysis was carried out with qPCR.14
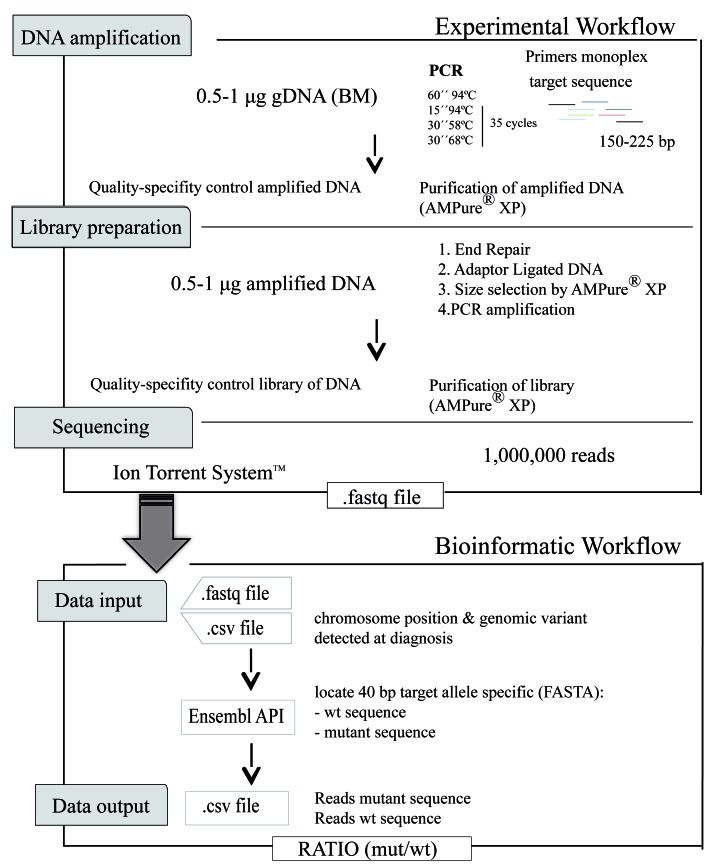
The specific mutations detected at diagnosis were studied at follow-up. We first tested a variety of experimental steps to define optimal conditions (Online Supplement 1). We established an optimal protocol (Figure 1) that included DNA amplification, library preparation and sequencing as experimental steps (Online Supplement 2).
Figure 1.
Workflow of the next-geneartion sequencing – minimal residual disease method. DNA amplification, library preparation and sequencing experimental workflow. Genomic DNA (gDNA) is amplified by quantitative (q) polymerase chain reaction (PCR) using specific primers. Libraries are prepared in four steps: end repair, adaptor ligation, size selection, and PCR amplification. The libraries are then sequenced. A customized bioinformatic pipeline analyzes the sequences obtained. The results are expressed as a ratio of mutated sequences (mut) among wild-type (wt) sequences.
Libraries were sequenced on the Ion Proton System platform (Life Technologies, Thermo Fisher Scientific Inc.) with an estimated depth ≥1,000,000 of reads, generating .fastq files. These files were analyzed using a customized bioinformatic pipeline; which leads from the .fastq file and a .csv file that contains information about name identifier, run and barcode identifier, chromosomal position and the variant detected in the diagnosis to be evaluated in the follow-up sample. Through Ensembl Perl API,15 the aligned mutated sequence and the aligned wild-type (wt) sequence are presented in FASTA format (sequences of 40 bp). Finally, we obtained a .csv file containing the name identifier, run and barcode identifier, chromosomal position, the variant, the specific target sequence in FASTA format (mutated forward, mutated reverse, wt forward and wt reverse), the counts of each and the ratio (mutated/wt) in absolute values.
Results
A high percentage of acute myeloid leukemia patients could benefit from deep sequencing minimal residual disease assessment
In total, 211 (80%) SNV and 46 (20%) indels were detected in the 190 patients analyzed at diagnosis using the customized NGS panel. We detected one variant (SNV or indel) in 48 (25%) cases, two or more variants in 116 (61%) cases and no variants in 26 (14%) cases. In addition, we detected the NPM1 type A mutation in 53 (28%) patients by qPCR. Genes (TET2, ASXL1, or DNMT3A) with evidence of an association with clonal hematopoiesis of indeterminate potential (CHIP) were excluded from the analysis.11 Consequently, 82% of patients in our cohort could benefit from this approach.
Based on those genes reported as potential markers for monitoring MRD,16 and also the availability of follow-up samples, we focused on IDH1/2 and FLT3-SNV. We identified at diagnosis IDH1 mutations in 13 patients (7%), IDH2 mutations in 27 patients (14%) and FLT3-SNV mutations (18%) in 34 patients.
Deep sequencing minimal residual disease assessment has a sensitivity of 10−4 for single nucleotide variants and 10−5 for insertions-deletions
To establish the limit of quantification (LOQ) of the method, we used 10-fold serial dilutions of mixed mutated and control DNA. To study prototype indels, we used gDNA from OCI-AML3 cells (NPM1 type A) and to study prototype SNV, we used both gDNA from OCI-AML3 cells (DNMT3A) and commercial reference gDNA (IDH1/IDH2). As a control, we used a pool of gDNA from ten individuals without somatic mutations in these chromosomal regions. In all cases, initial allele frequency was 50% and a total of six dilutions were tested to construct a calibration curve, covering a theoretical dynamic range from 10−1 to 10−7.
As shown in Figure 2A,B, MRD NGS testing of NPM1 (indel) could quantify one mutated cell in the order of 10−5 normal ones and in the case of SNV (IDH1, IDH2 and DNMT3A) the LOQ was 10−4, which was reproducible for all SNV tested.
Figure 2.
Calibration curve of minimal residual disease in serial dilutions. (A,B) Ten-fold dilution curves for the assessment of the sensitivity of next-generation sequencing (NGS) in (A) insertions-deletions (InDel), using OCI-AML3 gDNA with 50% NPM1 type A mutation (R2 = 0.98); and (B) single nucleotide variabts (SNV), using OCI-AML3 gDNA with 50% mutated DNMT3A (R2 = 0.98), and gDNA with 50% mutated IDH1 or IDH2 from a commercial standard (R2 = 0.91 and R2 = 0.98, respectively). (C,D) The same 10-fold dilution curves for the assessment of sensitivity of digital polymerase chain reaction (dPCR) in (C) InDel (R2 = 0.98); and (D) SNV (R2 = 0.91 for IDH1 and R2 = 0.98 for IDH2). The vertical red bars indicate the limit of quantification (LOQ) according to the sample. Clone frequency is expressed as target concentration as mutated copies/μL in wild-type copies/μL. Negative controls are included in the calibration curves and had levels below the corresponding LOQ values.
Next-generation sequencing is more sensitive than digital polymerase chain reaction analysis for minimal residual disease assessment
We compared the sensitivity of the sequencing method with that of dPCR using the same LOQ dilution protocol. Clone frequency expressed as target concentration (mutated copies/μL in wt copies/μL) gradually decreased with each dilution, reaching a LOQ of 10−3 for NPM1, IDH1 and IDH2 (Figure 2C,D). While both methods showed similar detection limits and good linearity, the LOQ for the sequencing method was one order of magnitude higher than that for dPCR (IDH1 and IDH2), and two orders of magnitude higher for indels (NMP1).
Minimal residual disease status assessed by sequencing has prognosis value in acute myeloid leukemia
The median depth coverage was 401,300 aligned reads (interquartile range 195,100–825,700) for the 88 NPM1 and 18 SNV (9 IDH1, 7 IDH2, and 2 FLT3) follow-up samples evaluated. We detected no mutated sequence in 13 (12%) samples, one to five mutated sequences in 19 (18%) samples, and more than ten in 74 (70%) samples. The ratio of mutated sequences to wt sequences defined MRD levels. Considering MRD levels from the 106 samples evaluated we established the optimal cutoff to classify MRD status (positive versus negative) by receiver operating characteristic curves (Online Supplementary Figure S1) at each check-point of MRD evaluation [post-induction (n=51), post-consolidation (n=55), or both together (n=106)].
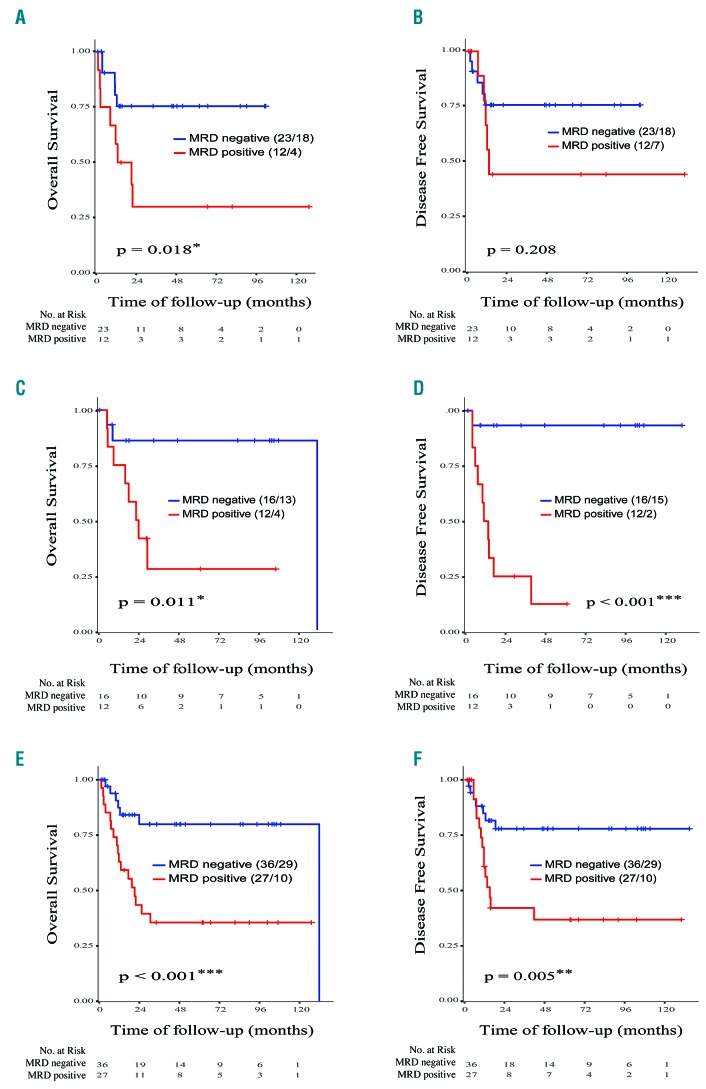
Survival analysis revealed that positive MRD status (MRD levels >0.1%) after induction (n=35) was associated with a significantly lower rate of overall survival [33% versus 78%; hazard ratio (HR): 3.5; 95% confidence interval (CI): 1.1–10.7; P=0.019], but a non-significant lower rate of disease-free survival (58% versus 78%; HR: 2.18; 95% CI: 0.63–7.5; P=0.208) (Figure 3A,B). In post-consolidation samples (n=28), MRD positive status (MRD levels >0.025%) was associated with both significantly shorter overall survival (33% versus 81%; HR: 6.0; 95% CI: 1.3–28.7; P<0.001) and significantly shorter disease-free survival (17% versus 94%; HR: 19.6; 95% CI: 2.5–155.6; P<0.001) (Figure 3C,D). Survival outcomes were also analyzed combining post-induction and post-consolidation (n=63) tests, in order to compare survival analysis with MFC and qPCR data sets. We observed that positive MRD status (MRD levels >0.035%) was associated with a higher risk of relapse (48% versus 81%; HR: 3.4; 95% CI: 1.4–8.5; P=0.005) and death (37% versus 81%; HR: 4.2; 95% CI: 1.6–10.7; P<0.001) (Figure 3E,F). In order to test the power of NPM1 and SNV as independent predictive markers, we performed the analysis separately. Evaluating NPM1 as an MRD marker (n=54), we found that MRD positive status was associated with both significantly shorter overall survival (43% versus 78%; HR: 3.3; 95% CI: 1.2–8.8; P=0.011), and shorter disease-free survival (57% versus 85%; HR: 2.9; 95% CI: 0.9–7.6; P=0.052). Similar results were found when we evaluated IDH1, IDH2 or FLT3-SNV as MRD markers (n=11). Accordingly, MRD positive status was associated with both significantly shorter overall survival (17% versus 100%; HR: not applicable; P=0.041), and shorter disease-free survival (17% versus 75%; HR: 6.3; 95% CI:0.7–54; P=0.058).
Figure 3.
Analysis of overall survival and disease-free survival in patients with acute myeloid leukemia stratified according to minimal residual disease levels determined by sequencing. Analysis of overall survival for (A) the induction data set, (C) the consolidation data set, and (C) both together. Analysis of disease-free survival for (B) the induction data set, (D) the consolidation data set, and (F) both together. The cutoff used for overall and disease-free survival was 0.001 at the post–induction check-point (n=35), 0.00026 at the post-consolidation check–point (n=28) and 0.00035 for both check-points (all data set) (n=63). The numbers of censored patients with respect to the stratified groups and the numbers at risk are indicated. Statistically significant values: *P<0.05, **P<0.01.
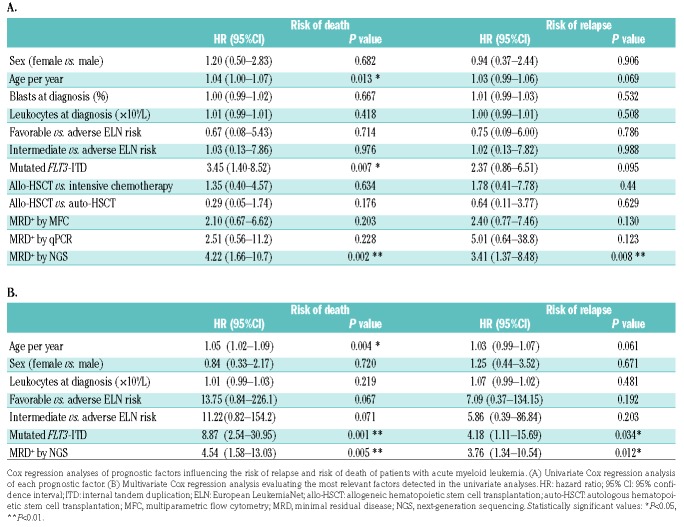
In univariate Cox analysis (Table 2A), the risk of death was significantly higher with increasing age (HR: 1.04; P=0.013), in patients with FLT3-ITD (HR: 3.45; P=0.007), and in those with MRD positive status as determined by NGS (HR: 4.22; P=0.002). The risk of relapse was significantly higher only in those patients with MRD positive status determined by NGS (HR: 3.4; P=0.008). In multivariate analysis (Table 2B), the risk of death was significantly higher with increasing age (HR: 1.05; P=0.004), in patients with mutated FLT3-ITD (HR: 8.87; P=0.001), and in those with MRD positive status determined by NGS (HR: 4.54; P=0.005). The risk of relapse was higher only in patients who were MRD positive as determined by NGS (HR: 3.76; P=0.012).
Table 2.
Cox regression analyses.
Minimal residual disease assessment by sequencing predicts overall survival and disease-free survival better than multiparameter flow cytometry or quantitative polymerase chain reaction analysis
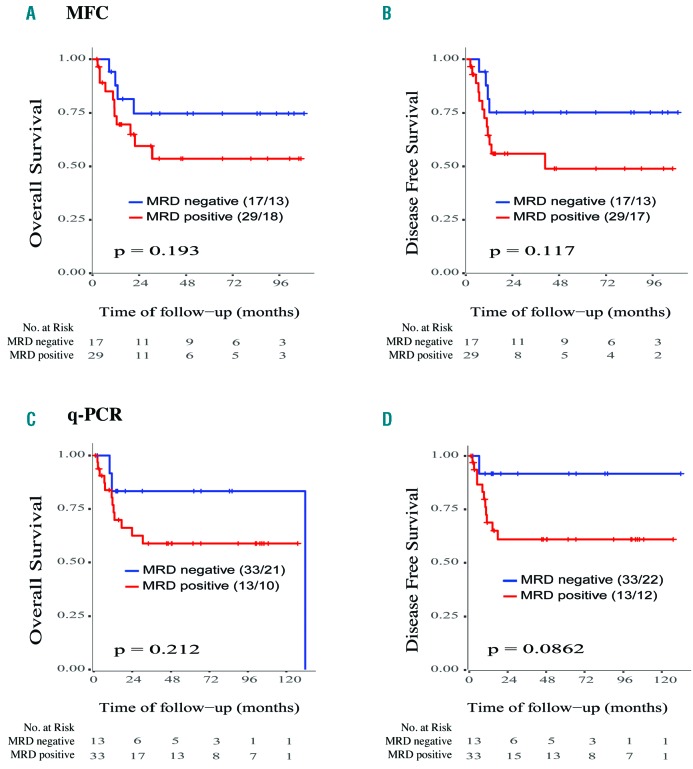
A positive correlation was found when comparing MRD assessment by NGS versus MFC (r=0.47, P=0.005, n=75), and NGS versus qPCR (r=0.62, P<0.001, n=80) (Online Supplementary Figure S2). There were differences between positive MRD and negative MRD groups of patients tested by MFC, but they were not significant for either overall survival (P=0.193) or disease-free survival (P=0.117) (n=46) (Figure 4A). Similarly, differences were observed between positive MRD and negative MRD groups defined by qPCR of NPM1, although statistical significance was not reached for either overall survival (P=0.212) or disease-free survival (P=0.086) (n=46) (Figure 4B).
Figure 4.
Analysis of overall survival and disease-free survival in patients with acute myeloid leukemia stratified according to minimal residual disease levels determined by conventional methods. Kaplan-Meier plots of (A) overall survival and (B) disease-free survival according to minimal residual disease (MRD) assessment by multiparametric flow cytometry (MRC) and (C) overall survival and (D) disease-free survival according to MRD assessment by quantitative polymerase chain reaction (qPCR) analysis. The numbers of censored patients with respect to each stratified group and numbers at risk are indicated. Statistically significant values: *P<0.05, **P<0.01.
Discussion
We have optimized and validated a high sensitivity NGS method for the detection and quantification of NPM1, IDH1, IDH2 and FLT3-SNV mutated sequences at very low allele frequency in follow-up gDNA samples. NGS has demonstrated prognostic value for pre-treatment status in patients with AML,17 and may also be a useful tool for detecting MRD.18,19 We first studied the mutational profile of patients with AML using a customized NGS panel to ensure a high applicability (82% of patients). This approach is also a useful screening method for detecting all potential MRD markers and choosing those most relevant. The combination of several markers is possible and recommended to overcome limitations of MRD assessment due to sub-clonal heterogeneity of AML and CHIP.11 Accordingly, our method has the capacity to evaluate multiple markers simultaneously and, considering that 61% of patients in our cohort had two or more genetic alterations, this approach is sufficiently robust to monitor MRD even in patients with clonal evolution.
Reported variants associated with CHIP are frequently located in DNMT3A, TET2 or ASXL1 genes, and are detected in the preleukemic phase and during complete AML remission.20–23 Indeed, any gene could carry both CHIP and non-CHIP variants, and these should be evaluated for each patient. Moreover, studies have shown that genes related to CHIP (IDH1/2) are useful for predicting prognosis because in these cases the genetic alterations have been acquired in the leukemic clone and not before.24
The sensitivity of this method equates to one mutated cell per 100,000 cells (LOQ 10−5) for NPM1 and one mutated cell per 10,000 cells (LOQ 10−4) for IDH1, IDH2 and FLT3-SNV. This difference in sensitivity is related to the fact that the NPM1 type A mutation (insCCTG) is rarely generated erroneously by NGS, and the quantification is precise.
Our method, as with any NGS method, has an intrinsic error rate that limits its sensitivity for most SNV to 1–2% of all reads. This limitation can nevertheless be overcome by virtue of the scalable nature of NGS.16 Thus, we boosted NGS sensitivity by increasing the amount of DNA by PCR prior to sequencing, which increased the depth of coverage to one million reads. By also optimizing the bioinformatic analysis, we focused the search for the precise variant in order to eliminate random sequencing errors, enhancing the specificity of the technique and reducing the computational time. To the best of knowledge, our NGS method presents possibly the highest sensitivity reported for NGS in AML.18,19,24–27
dPCR is a relatively novel technique for precise and absolute quantification of nucleic acids, which is based on limiting partitions of the PCR volume and Poisson statistics.28 It is also an extremely sensitive technique, with a high specificity due to the detection of mutant alleles.29 However, when we compared the same standard dilutions in NGS and dPCR, NGS afforded a 2-log increment in LOQ for indels (NPM1) and a 1-log increment for SNV (IDH1/2), with the sensitivity of dPCR for indels being similar to that reported in a previously published study (10−2).30 Compared with NGS, dPCR is a faster measurement technique but, as it is focused, it requires allele-specific primers that can complicate the experimental procedure, and a high number of parallel experiments are needed to raise the sensitivity, which increases the cost of the assay. Additionally, although it is possible to multiplex dPCR, unfortunately only a few targets can be monitored simultaneously within each sample.29 Another advantage of NGS technology is that it does not require calibration curves in each assay, and the results are reported in absolute values, facilitating its standardization.
The NGS method described here showed comparable sensitivities (10−4 for SNV and 10−5 for indels) to those of MFC methods in those cases with immunophenotyphically aberrant populations.10,31 Although our method had a similar sensitivity to that of qPCR, it does not require oligonucleotides that hybridize specifically to a particular sequence, so all nucleotides in the amplified region can be studied. Consequently, the NGS test is capable of detecting all NPM1 subtype mutations in the same assay.
We found positive correlations when MRD levels were evaluated by NGS versus MFC and versus qPCR, but not with the expected results. In the case of MFC, this could be explained, in part, by the fact that NPM1 mutations are usually associated with monocytic subtype-AML, which frequently presents more difficulties for identifying MRD by MFC. Indeed, Salipante et al.27 described that the level of success of MFC depends greatly on the immunophenotype of the abnormal blasts and how to discriminate them from background regenerative blasts. Moreover, due to the lack of standardization, MFC shows substantial variability across laboratories, including that of sample processing, instrument configuration, number of events, and training of pathologists.32 The lack of a strong correlation between NGS and qPCR could be explained by the nature of the sample (sequencing uses gDNA whereas qPCR uses cDNA). Although RNA overexpression allows a higher sensitivity of detection, RNA levels do not correlate with the number of tumor cells, in contrast to mutated DNA. Accordingly, mutated DNA is more representative of the tumor burden than is overexpression of mutated RNA.35 It should be noted that the prediction of survival and progression of AML using MRD NGS was better than that of the other methodologies employed, at least in the cohorts evaluated.
Finally, survival analysis showed that MRD positive status determined by NGS was associated with a higher risk of relapse and death and that MRD negative status in post-consolidation ssmples was associated with longer overall and disease-free survival, in accordance with recently published studies.23 Supporting these findings, previous studies reported that an MRD check-point after consolidation could be the best moment for analysis because it afforded better prediction.8,34–37 Cox regression multivariate analyses confirmed that MRD positive status determined by sequencing was the only statistically significant predictor of the risk of relapse (P=0.012).
In conclusion, we have optimized a new targeted sequencing method with high sensitivity for MRD evaluation with applicability in a high percentage of AML patients, improving the capacity, over MFC or qPCR, to predict the survival outcomes of the AML patients in our cohort.
Supplementary Material
Acknowledgments
This study was supported by the Subdirección General de Investigación Sanitaria (Instituto de Salud Carlos III, Spain) grants PI13/02387 and PI16/01530, and the CRIS against Cancer foundation grant 2014/0120. ML holds a postdoctoral fellowship of the Spanish Ministry of Economy and Competitiveness (FPDI-2013-16409). PRP holds a postdoctoral fellowship of the Spanish Instituto de Salud Carlos III: Contrato Predoctoral de Formación en Investigación en Salud i-PFIS (IFI 14/00008).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/2/288
References
- 1.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–4649. [DOI] [PubMed] [Google Scholar]
- 3.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 4.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374(5):422–433. [DOI] [PubMed] [Google Scholar]
- 5.Kronke J, Schlenk RF, Jensen KO, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian Acute Myeloid Leukemia Study Group. J Clin Oncol. 2011;29(19):2709–2716. [DOI] [PubMed] [Google Scholar]
- 6.Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6): 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balsat M, Renneville A, Thomas X, et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: a study by the Acute Leukemia French Association Group. J Clin Oncol. 2017;35(2):185–193. [DOI] [PubMed] [Google Scholar]
- 8.Zhu HH, Zhang XH, Qin YZ, et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood. 2013;121(20):4056–4062. [DOI] [PubMed] [Google Scholar]
- 9.Freeman SD, Virgo P, Couzens S, et al. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol. 2013;31(32):4123–4131. [DOI] [PubMed] [Google Scholar]
- 10.Terwijn M, van Putten WL, Kelder A, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31(31):3889–3897. [DOI] [PubMed] [Google Scholar]
- 11.Schuurhuis GJ, Heuser M, Freeman S, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131(12):1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “prime time”¿ Hematology Am Soc Hematol Educ Program. 2014;2014(1):222–233. [DOI] [PubMed] [Google Scholar]
- 13.Cedena MT, Rapado I, Santos-Lozano A, et al. Mutations in the DNA methylation pathway and number of driver mutations predict response to azacitidine in myelodysplastic syndromes. Oncotarget. 2017;8(63):106948–106961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorello P, Cazzaniga G, Alberti F, et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia. 2006;20(6):1103–1108. [DOI] [PubMed] [Google Scholar]
- 15.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson B, Lazarus HM. Enhancing acute myeloid leukemia therapy - monitoring response using residual disease testing as a guide to therapeutic decision-making. Expert Rev Hematol. 2017;10(6):563–574. [DOI] [PubMed] [Google Scholar]
- 17.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thol F, Kolking B, Damm F, et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer. 2012;51(7):689–695. [DOI] [PubMed] [Google Scholar]
- 19.Kohlmann A, Nadarajah N, Alpermann T, et al. Monitoring of residual disease by next-generation deep-sequencing of RUNX1 mutations can identify acute myeloid leukemia patients with resistant disease. Leukemia. 2014;28(1):129–137. [DOI] [PubMed] [Google Scholar]
- 20.Metzeler KH, Herold T, Rothenberg-Thurley M, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(5):686–698. [DOI] [PubMed] [Google Scholar]
- 21.Bullinger L, Dohner K, Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35(9):934–946. [DOI] [PubMed] [Google Scholar]
- 22.Koeffler HP, Leong G. Preleukemia: one name, many meanings. Leukemia. 2017;31(3):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378(13):1189–1199. [DOI] [PubMed] [Google Scholar]
- 24.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314(8):811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Getta BM, Devlin SM, Levine RL, et al. Multicolor flow cytometry and multigene next-generation sequencing are complementary and highly predictive for relapse in acute myeloid leukemia after allogeneic transplantation. Biol Blood Marrow Transplant. 2017;23(7):1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debarri H, Lebon D, Roumier C, et al. IDH1/2 but not DNMT3A mutations are suitable targets for minimal residual disease monitoring in acute myeloid leukemia patients: a study by the Acute Leukemia French Association. Oncotarget. 2015;6(39): 42345–42353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salipante SJ, Fromm JR, Shendure J, et al. Detection of minimal residual disease in NPM1-mutated acute myeloid leukemia by next-generation sequencing. Mod Pathol. 2014;27(11):1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunetti C, Anelli L, Zagaria A, et al. Droplet digital PCR is a reliable tool for monitoring minimal residual disease in acute promyelocytic lseukemia. J Mol Diagn. 2017;19(3): 437–444. [DOI] [PubMed] [Google Scholar]
- 29.Roloff GW, Lai C, Hourigan CS, et al. Technical advances in the measurement of residual disease in acute myeloid leukemia. J Clin Med. 2017;6(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minervini A, Francesco Minervini C, Anelli L, et al. Droplet digital PCR analysis of NOTCH1 gene mutations in chronic lymphocytic leukemia. Oncotarget. 2016;7(52): 86469–86479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ossenkoppele G, Schuurhuis GJ. MRD in AML: does it already guide therapy decision-making¿ Hematology Am Soc Hematol Educ Program. 2016;2016(1):356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz NM, Mencia-Trinchant N, Hassane DC, et al. Minimal residual disease in acute myelogenous leukemia. Int J Lab Hematol. 2017;39(Suppl 1):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duployez N, Nibourel O, Marceau-Renaut A, et al. Minimal residual disease monitoring in t(8;21) acute myeloid leukemia based on RUNX1-RUNX1T1 fusion quantification on genomic DNA. Am J Hematol. 2014;89(6): 610–615. [DOI] [PubMed] [Google Scholar]
- 34.Jacobsohn DA, Tse WT, Chaleff S, et al. High WT1 gene expression before haematopoietic stem cell transplant in children with acute myeloid leukaemia predicts poor event-free survival. Br J Haematol. 2009;146(6):669–674. [DOI] [PubMed] [Google Scholar]
- 35.Hourigan CS, Gale RP, Gormley NJ, et al. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31(7): 1482–1490. [DOI] [PubMed] [Google Scholar]
- 36.Rossi G, Carella AM, Minervini MM, et al. Optimal time-points for minimal residual disease monitoring change on the basis of the method used in patients with acute myeloid leukemia who underwent allogeneic stem cell transplantation: a comparison between multiparameter flow cytometry and Wilms’ tumor 1 expression. Leuk Res. 2015;39(2):138–143. [DOI] [PubMed] [Google Scholar]
- 37.Jourdan E, Boissel N, Chevret S, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121(12): 2213–2223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.