Abstract
Primary testicular lymphoma is a rare lymphoid malignancy, most often, histologically, representing diffuse large B-cell lymphoma. The tumor microenvironment and limited immune surveillance have a major impact on diffuse large B-cell lymphoma pathogenesis and survival, but the impact on primary testicular lymphoma is unknown. Here, the purpose of the study was to characterize the tumor microenvironment in primary testicular lymphoma, and associate the findings with outcome. We profiled the expression of 730 immune response genes in 60 primary testicular lymphomas utilizing the Nanostring platform, and used multiplex immunohistochemistry to characterize the immune cell phenotypes in the tumor tissue. We identified a gene signature enriched for T-lymphocyte markers differentially expressed between the patients. Low expression of the signature predicted poor outcome independently of the International Prognostic Index (progression-free survival: HR=2.810, 95%CI: 1.228-6.431, P=0.014; overall survival: HR=3.267, 95%CI: 1.406-7.590, P=0.006). The T-lymphocyte signature was associated with outcome also in an independent diffuse large B-cell lymphoma cohort (n=96). Multiplex immunohistochemistry revealed that poor survival of primary testicular lymphoma patients correlated with low percentage of CD3+CD4+ and CD3+CD8+ tumor-infiltrating lymphocytes (P<0.001). Importantly, patients with a high T-cell inflamed tumor microenvironment had a better response to rituximab-based immunochemotherapy, as compared to other patients. Furthermore, loss of membrane-associated human-leukocyte antigen complexes was frequent and correlated with low T-cell infiltration. Our results demonstrate that a T-cell inflamed tumor microenvironment associates with favorable survival in primary testicular lymphoma. This further highlights the importance of immune escape as a mechanism of treatment failure.
Introduction
Primary testicular lymphoma (PTL) is a rare and aggressive lymphoid malignancy; the majority of the cases are diffuse large B-cell lymphomas (DLBCL).1 Patients with PTL have a substantial risk of recurrence and a tendency of lymphoma involvement in additional extranodal sites, such as contralateral testis and the central nervous system (CNS). Most cases are classified as non-germinal center (non-GC) cell-of-origin subtype, which may partially account for the aggressive nature of the disease. The current standard of care consists of rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) immunochemotherapy, CNS prophylaxis, and locoregional radiotherapy or surgery of the remaining contralateral testicle. While the addition of rituximab to systemic chemotherapy has improved the overall prognosis of DLBCL, treatment responses in PTL have not been equally evident.1,2
The tumor microenvironment (TME) and limited immune surveillance play important roles in the lymphoma pathogenesis and survival.3,4 The TME of B-cell lymphomas consists of immune cells [e.g. T cells, macrophages, natural killer (NK) cells], stromal cells, blood vessels, and extracellular matrix. The TME may confer effective tumor recognition and clearance, but on the other hand, can also provide a protective niche for the tumor cells, and facilitate cell proliferation and survival.5 The interactions between lymphoma cells and the TME contribute to the ability of tumor cells to escape host immune surveillance; these mechanisms include loss of expression of human leukocyte antigen (HLA) class I and II molecules that interferes with correct tumor cell recognition by the cellular immune system and may provide tumor cells with an escape mechanism.6 Several studies have reported loss of HLA I and II expression in B-cell lymphomas, including PTL.7–11 Another immune escape mechanism is the expression of immune checkpoint molecules, such as programmed cell death 1 (PD-1) and the cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4).6
In this study, we aimed to characterize cellular and molecular immunological profiles in PTL, and associate the findings with outcome. We identified a gene signature enriched for T-cell- and NK-cell-related genes, the low expression of which predicts high risk of recurrence and mortality in patients with PTL and DLBCL. Our results emphasize the key role of the TME and immune escape in regulating therapy resistance in aggressive B-cell lymphomas.
Methods
Patients’ characteristics and samples
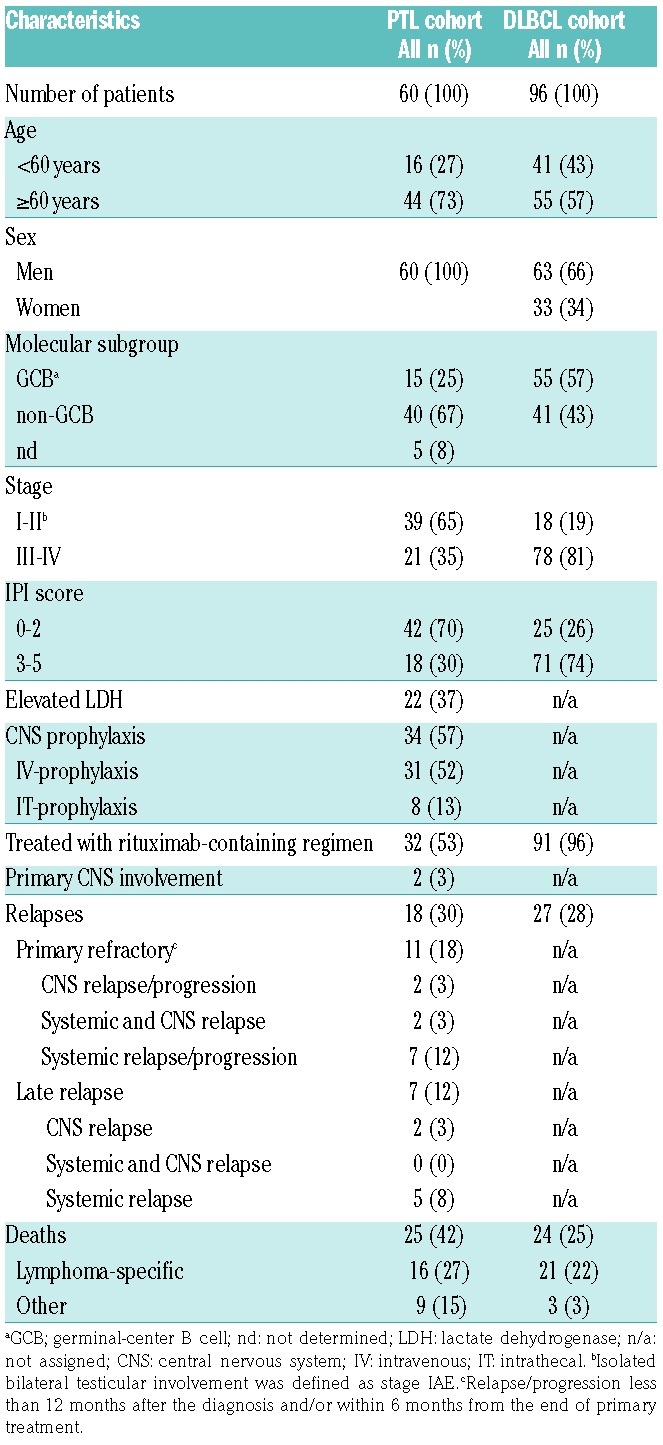
The study consisted of formalin-fixed paraffin-embedded (FFPE) samples from primary orchiectomy of 60 patients diagnosed with PTL in 1993-2013 (Table 1). Twenty-eight of the patients were diagnosed and treated in the pre-rituximab era with anthracyclin-based chemotherapy, whereas 32 of the patients were treated with rituximab-containing immunochemotherapy. In addition, 34 patients received CNS prophylaxis, consisting mainly of intravenously administered high-dose methotrexate and/or high-dose cytarabine. Nineteen patients received treatment of the contralateral testicle (radiotherapy or orchiectomy). The cell-of-origin (COO) was determined by immunohistochemistry using the Hans algorithm.12 The study was approved by the Ethics Committees in Helsinki and Tampere, Finland, and by the Finnish National Authority for Medicolegal Affairs, and by the Institutional Review Boards of the institutes involved in the study.
Table 1.
Patients’ characteristics of the primary testicular lymphoma (PTL) and diffuse large B-cell lymphomas (DLBCL).
The dataset of primary DLBCL from the Cancer Genome Characterization Initiative (CGCI) (the database of Genotypes and Phenotypes study accession: phs000532.v2.p1; n=96)13 was used for comparison (Table 1).
Gene expression analysis
Gene expression from the FFPE samples of 60 PTL patients was profiled with the Nanostring nCounter Human PanCancer Immunoprofiling Panel (XT-CSO-HIP1-12, NanoString Technologies, Seattle, WA, USA). The detailed protocol is provided in the Online Supplementary Methods.
Immunohistochemistry
Formalin-fixed paraffin-embedded tissue microarray (TMA) was utilized for the immunohistochemical (IHC) analyses. The protocol for HLA-ABC, HLA-DR, and β2 microglobulin staining is provided in the Online Supplementary Methods. Membranous staining in the majority (>90%) of tumor cells was scored as normal (highly positive). Cases with mixed cytoplasmic and membranous staining were scored as moderately positive. Cases with no membranous staining were scored as negative. When determining the triple-positive cases, the highly and moderately positive groups were merged. Scoring was performed independently by MA and SMa.
Multiplex immunohistochemistry (mIHC) using a panel with antibodies for CD3 (clone EP449E, Abcam), CD4 (clone EPR6855, Abcam), CD8 (clone C8/144B, Abcam), CD56 (clone MRQ-42, Cell Marque, Rocklin, CA, USA) was performed as previously described.14 Further details are provided in the Online Supplementary Methods. The digital image analysis platform CellProfiler 2.1.215 was used for cell segmentation, intensity measurements, and immune cell classification. Marker co-localization was computed with the single-cell analysis software FlowJo v.10 (FlowJo LLC.). Spots with less than 5000 cells were excluded from analysis and duplicate spots from the same patient merged.
Table 2.
Pathways enriched among T-lymphocyte signature genes.
Statistical analysis
Statistical analyses were performed with IBM SPSS v.24.0 (IBM, Armonk, NY, USA). The χ2 test was used to assess differences in categorical variables. Mann-Whitney U and Kruskal-Wallis tests were used to compare differences between two or more groups, respectively. The Kaplan-Meier method was used to estimate survival rates (log-rank test). Cox univariate regression analysis was performed to study the prognostic value of the factors. Multivariate analyses were performed according to the Cox proportional hazards regression model using categorical data. Overall survival (OS) and disease-specific survival (DSS) were determined as the interval from diagnosis to death from any cause or death due to lymphoma, respectively. Progression-free survival (PFS) was defined as the period between diagnosis and progression or death from any cause. Correlation analyses were performed with Spearman rank analysis. All comparisons were two-tailed, and P<0.05 was considered significant.
Results
Unsupervised hierarchical clustering reveals genes differentially expressed between primary testicular lymphoma patients
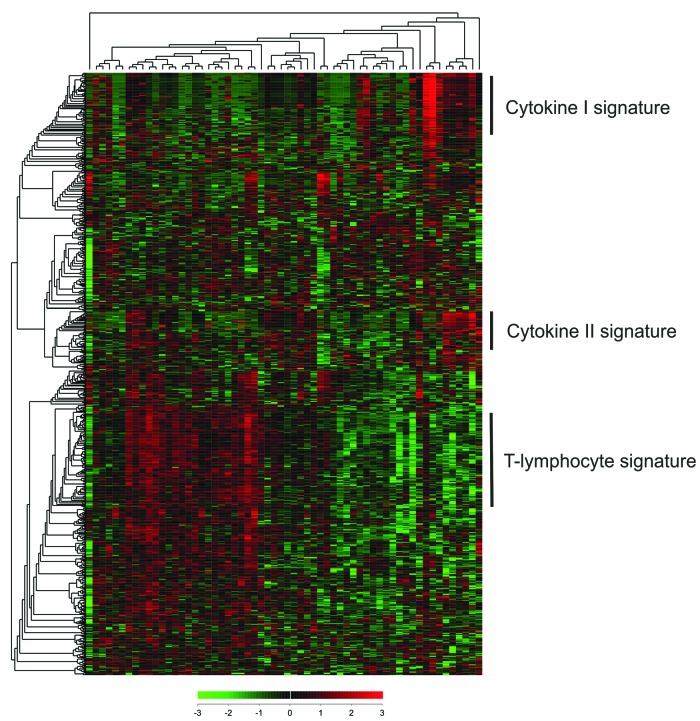
We profiled the expression of 730 immune-associated genes and 40 housekeeping genes from the FFPE samples of 60 primary testicular DLBCLs utilizing the Nanostring PanCancer Immune Profiling Panel. Unsupervised hierarchical clustering revealed three gene signatures differentially expressed between the patients (Figure 1). We identified a large cluster of genes, which clearly separated the patients. The core signature contained 121 genes (Online Supplementary Table S1), which were enriched for T-cell and NK-cell markers and signaling (e.g. CD3D/E/G, CD4, CD8A/B, ITGB2, PRF1, GZMA/B/H/M/K, KLRB1/G1/K1) (Online Supplementary Tables S1 and S2). This signature was named “T-lymphocyte signature”. In addition, two minor cytokine signatures were recognized. “Cytokine signature I” included 44 genes, enriched for cytokines (e.g. CSF3, CSF2, IL3, IFNA1, IL5, IL11, IL2) (Online Supplementary Table S2), whereas “Cytokine signature II” included 25 genes, enriched for both cytokines and cytokine receptors (e.g. IL4, IL17A, TNFRSF11A, IL17B, TNFSF11, PPBP, IL9, CXCR1, TNFSF18, CCL28) (Online Supplementary Table S3).
Figure 1.
Hierarchical clustering reveals gene signatures differentially expressed in primary testicular lymphoma (PTL) patients. The expression of PanCancer Immune profiling panel genes was assayed by Nanostring nCounter from 60 PTL samples. The data were z-score transformed and visualized by unsupervised hierarchical clustering using Euclidean distance. Relative levels of gene expression are depicted according to the color scale shown. Each row represents a different gene and each column a different sample. Three main gene signatures were discovered: T-lymphocyte signature, and Cytokine I and II signatures.
T-lymphocyte signature is associated with survival in patients with PTL
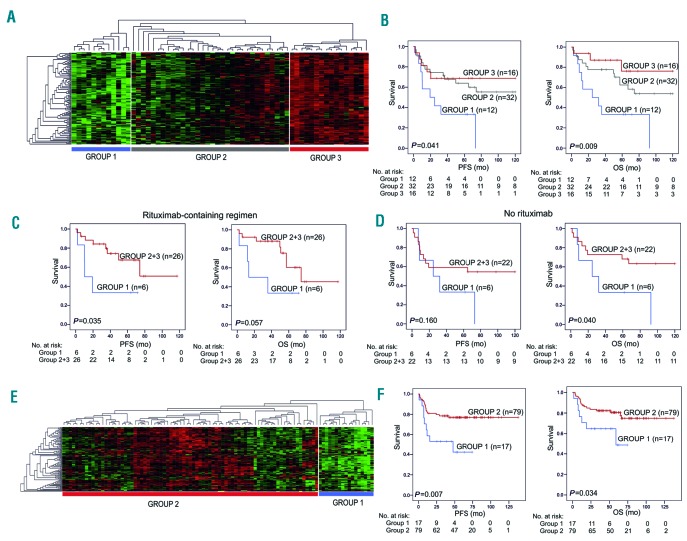
To study the association of the gene signatures with survival, we re-clustered samples according to the expression of the signature genes. Re-clustering the data based on the expression of the 121 genes of the T-lymphocyte signature separated the patients into three distinct groups with high, intermediate and low expression (Figure 2A and Online Supplementary Figure S1A). Interestingly, patients with low expression of the T-lymphocyte signature genes (n=12) had a significantly worse PFS, OS, and DSS in comparison to patients with intermediate (n=32) and high (n=16) gene expression (log-rank P=0.041, P=0.009, and P=0.033, respectively) (Figure 2B and Online Supplementary Figure S2A). In Cox multivariate analysis, low expression of T-lymphocyte signature had prognostic impact on survival independently of the International Prognostic Index (IPI) (PFS: HR=2.810, 95%CI: 0.228-6.431, P=0.014; OS: HR=3.267, 95%CI: 1.406-7.590, P=0.006; and DSS: HR=2.910, 95%CI: 1.004-8.436, P=0.049). This was evident for PFS and OS also with the individual IPI factors (Online Supplementary Table S4). Baseline characteristics, including age, molecular subgroup, stage, and IPI were equally distributed in the subgroups with higher and lower T-lymphocyte signature expression (Online Supplementary Table S5). When the survival analyses were adjusted according to treatment, adverse prognostic impact of the low expression of T-lymphocyte signature on outcome was particularly evident in the patients treated with the rituximab-containing regimen (Figure 2C and D and Online Supplementary Figure S2B).
Figure 2.
T-lymphocyte signature is associated with survival. (A) The gene expression data from 60 primary testicular lymphoma (PTL) patients were re-clustered according to the expression of the T-lymphocyte signature genes. The data clustered into three groups: Group 1: low expression; Group 2: intermediate expression; Group 3: high expression. (B) Kaplan-Meier (log-rank test) survival plots depict progression-free survival (PFS) and overall survival (OS) in the three patient groups. (C and D) Kaplan-Meier plots show the PFS and OS of PTL patients stratified for the treatment with rituximab-containing regimen (C) versus no rituximab (D). (E and F) RNA-seq data from the CGCI cohort with 96 de novo DLBCL cases was clustered based on the T-lymphocyte signature gene expression. This divided the patients into two groups with higher (Group 2) and lower (Group 1) expression (E). Kaplan-Meier plots depict survival differences between the two groups (F).
At the individual gene level, 72 genes from the T-lymphocyte signature were significantly (P<0.05) associated with survival in Cox univariate analysis with continuous variables and had lower expression in the poor prognosis patient group (Online Supplementary Table S6). These included, for example, T-cell surface markers (CD3D/E/G, CD4, CD8A/B), cytolytic factors (PRF1, GZMA/K/M), chemokines (CCL2/3/4/5), and killer-cell lectin-like receptors (KLRB1, KLRG1, KLRK1).
For the Cytokine I signature, no association with survival was found (data not shown). However, when patients were re-clustered according to the expression of the 25 genes of the Cytokine II signature, they could be separated into two groups with higher and lower expression. The group with higher expression of the Cytokine II signature had a shorter 5-year PFS as compared to those with low or no expression of the signature (36% vs. 66%; P=0.005) (Online Supplementary Figure S3). In Cox multivariate analysis with IPI, the Cytokine II signature was also found to be an independent predictor of PFS (HR=3.393, 95%CI: 1.531-7.521; P=0.003). Baseline characteristics, including age, molecular subgroup, stage and IPI were equally distributed in the subgroups with higher and lower expression of the Cytokine II signature (data not shown). In general, the absolute expression levels of the Cytokine II signature genes were low (Online Supplementary Figure S1B).
Low expression of the T-lymphocyte signature is associated with poor outcome in patients with de novo diffuse large B-cell lymphomas
Next, we tested whether the signatures could be identified also from other B-cell lymphomas. To this end, we used RNA-sequencing data from 96 primary DLBCL patients from the Cancer Genome Characterization Initiative (CGCI) cohort (Table 1). Following hierarchical clustering of the gene expression of the T-lymphocyte signature, a subgroup of patients with low expression of the signature was identified (Figure 2E and Online Supplementary Figure S1C). This group of patients had shorter survival as compared to patients with higher expression of the signature genes (PFS: P=0.007, OS: P=0.034) (Figure 2F). In a multivariate analysis with IPI, low T-lymphocyte signature remained an independent prognostic factor for PFS (HR=2.560, 95%CI: 1.151-5.695; P=0.021). The baseline characteristics were equally distributed between the patient groups (Online Supplementary Table S5). The results indicate that the T-lymphocyte signature identified from the PTL cohort has prognostic impact on survival also in patients with de novo DLBCL. On the contrary, genes from the cytokine signatures were neither differentially expressed between the patients or associated with survival in DLBCL (data not shown). As in PTL, the absolute expression levels of the cytokine signature genes were low (data not shown), suggesting that they are not clinically relevant.
Low amount of tumor-infiltrating T cells is associated with poor outcome in patients with PTL
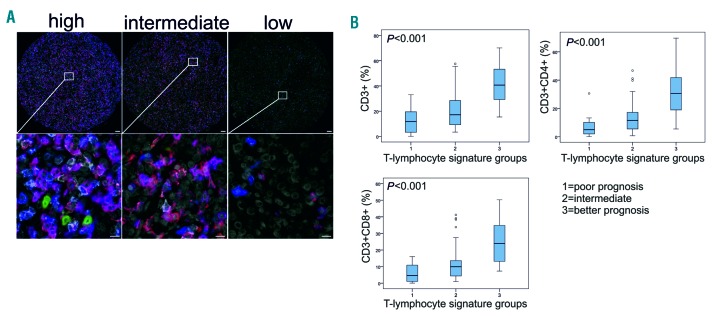
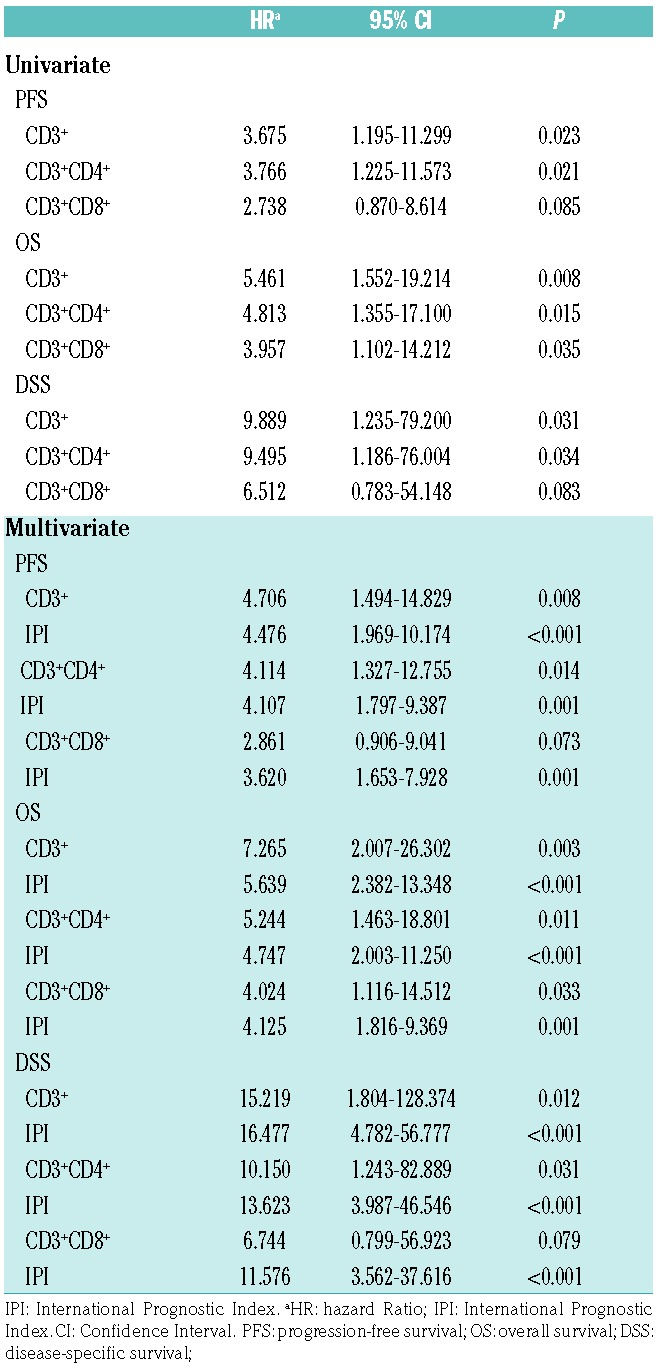
To study the presence of different T-cell and NK-cell subtypes in the tumor milieu, we performed mIHC with CD3, CD4, CD8, and CD56 cell surface markers on a TMA of 60 PTL patients (Figure 3A and Online Supplementary Figure S4). The relative levels of tumor-infiltrating CD3−CD56+ cells were very low (median 0.3%, range 0.0-52.7%), and there were no significant differences in the amount of CD56+ cells between the good and poor prognosis groups (data not shown). However, mIHC analysis confirmed significantly (P<0.001) lower proportions of CD3+, CD3+CD4+, and CD3+CD8+ lymphocytes in patients with a poor prognosis in comparison to patients with a favorable outcome (Figure 3B). Expression of CD3, CD4, and CD8 at the protein level correlated with the corresponding gene expressions (Spearman P=0.779, P<0.001; P=0.645, P<0.001; and P=0.768, P<0.001, respectively). Furthermore, low amounts of tumor-infiltrating CD3+, CD3+CD4+, and CD3+CD8+ lymphocytes were associated with a poor outcome (Online Supplementary Figure S5), which was independent of IPI (Table 3). Consistent with the findings on the gene expression level, an adverse prognostic impact of the low tumor-infiltrating CD4+ and CD8+ T-cell counts was particularly evident in patients treated with the rituximab-containing regimen (Online Supplementary Figure S6).
Figure 3.
Lower number of tumor-infiltrating T cells is associated with poor survival in primary testicular lymphoma (PTL). (A) Representative images (high, intermediate, low) from mIHC analysis of PTL tumor-associate macrophages probed with a 4-plex panel of T-cell markers. Blue: CD3; red: CD8; white: CD4; green: CD56; gray: DAPI. Scale bars 50 μm (upper panel) and 20 μm (lower panel). Images from individual channels are presented in Online Supplementary Figure S4. (B) Boxplots visualizing the expression of CD3+, CD3+CD4+, and CD3+CD8+ lymphocytes in the three groups based on the T-lymphocyte signature (1: poor prognosis, 2: intermediate, and 3: better prognosis). Statistical significance was determined by Kruskall-Wallis test.
Table 3.
Univariate and multivariate Cox regression survival analysis of multiplex immunohistochemistry data.
Loss of membrane-associated HLA class I and II expression is frequent in PTL
Loss or aberrant expression of HLA molecules may cause tumor cells to escape from immunosurveillance. Here, the expression of HLA I and HLA II genes was lower in the poor prognosis group of PTL patients (P<0.05) (Online Supplementary Figure S7A). This was evident also in the de novo DLBCL cohort (Online Supplementary Figure S7B).
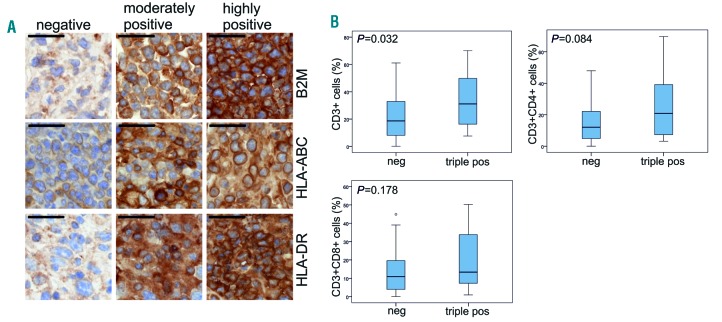
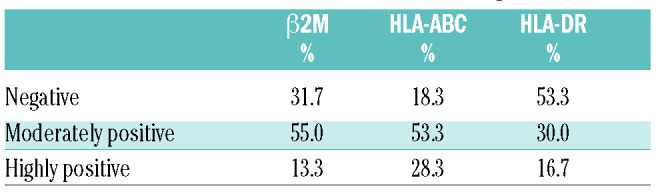
To study the subcellular localization of HLA class I and II, we analyzed HLA-DR, HLA-ABC and β2M, a component of HLA I, immunohistochemically (Figure 4A). A minority of the PTL cases showed highly positive membrane staining for HLA-DR, HLA-ABC, and β2M (17%, 28%, and 13%, respectively) (Table 4). Interestingly, triple-positive membrane staining of HLA I, HLA II and β2M was associated with a higher number of tumor-infiltrating CD3+, CD3+CD4+, and CD3+CD8+ lymphocytes (Figure 4B).
Figure 4.
Loss of membrane-associated HLA class I and II expression is frequent in primary testicular lymphoma (PTL) and correlates with low T-cell infiltration. (A) Representative images of HLA-ABC, HLA-DR, and β2 microglobulin antibody of PTL tissue sections. The samples were scored as negative (neg), moderately positive (pos), and highly positive according to the membrane staining. Scale bar 50 μm. (B) Boxplots visualizing the correlation of HLA-ABC, HLA-DR and β2M triple-positive membrane staining (either moderate or high membrane staining) with T-cell numbers in PTL (n=14 and n=46 in positive and negative groups, respectively). P-values were determined by Mann-Whitney test.
Table 4.
HLA and β2M immunohistochemical staining results.
Discussion
The TME plays an important role in lymphoma pathogenesis and patient prognosis.3 In this study, we aimed to characterize the immunological profiles and their association with outcome in patients with PTL. We found that a gene signature enriched for T-cell genes was associated with outcome in patients treated with chemotherapy and immunochemotherapy. We could further demonstrate that lower amounts of tumor-infiltrating CD4+ and CD8+ T cells was associated with a poor prognosis in patients with PTL. The impact was particularly evident among patients treated with rituximab-containing immunochemotherapy. The limitation of our study is the lack of a proper validation cohort due to the rare nature of PTL. However, the T-lymphocyte signature had a prognostic impact in an independent cohort of de novo DLBCL patients treated with immunochemotherapy, demonstrating the importance of the signature genes also in other aggressive B-cell lymphomas. Our data extend previous findings on DLBCL patients treated with CHOP and R-CHOP-like regimens.16–18 Together, the results emphasize the important role of the T-cell inflamed TME in regulating therapy resistance in PTL.
T lymphocytes, mostly comprising CD4+ and CD8+ T cells, play a major role in cell-mediated immunity. Lymphoma cells have been shown to escape immunosurveillance due to loss of expression or mislocalization of HLA I and II molecules.7–11,19 We found that reduced membranous staining of HLA I and II molecules and β2M correlated with lower T-cell infiltration, implying that defects in HLA complexes may impair the recruitment of the tumor-infiltrating T-cell subsets.
Indeed, our data suggest that immune escape does not only provide a mechanism for lymphoma pathogenesis, but also plays a role in promoting resistance to immunochemotherapy. We propose that lymphomas with inflammatory profile characterized by high content of tumor-infiltrating CD4+ and CD8+ T cells, “the hot tumors”, display pre-existing antitumor immune response. In response to therapy, and particularly rituximab-containing regimen, tumor-infiltrating T cells are stimulated further to participate in immune response against lymphoma cells. In contrast, lymphomas that lack T-cell infiltration, “the cold tumors”, reflect the absence of pre-existing anti-tumor immunity and have a lower likelihood of having an optimal response to therapy. Consistent with our hypothesis, it has been shown that many chemotherapeutic drugs, including cyclophosphamide and doxorubicin, which are the main components in the CHOP regimen, can activate anti-tumor immune response by increasing immunogenicity of malignant cells as well as by directly relieving immunosuppressive networks.20 Rituximab and other therapeutic CD20 antibodies can, in turn, further promote a long-term anti-tumor immune response, called the “vaccinal” effect, which is dependent on the presence of both CD4+ and CD8+ lymphocytes.21–23 Further studies should aim to characterize in more detail the underlying mechanisms for the loss of T-cell trafficking and infiltration. For example, differences in the mutational density between the T-cell inflamed “hot” and non-inflamed “cold” tumors might explain the loss of T cells in a subset of tumors. Additional gene expression profiling studies could provide information as to which genes and molecular pathways are differentially expressed or activated in the T-cell inflamed and non-inflamed tumors, and thus might mediate T-cell exclusion from the TME. For example, in melanoma and bladder cancer, the Wnt/β-catenin pathway has been shown to be causal in preventing T-cell activation and trafficking into the tumor microenvironment.24,25 In addition, it would be interesting to examine the distribution of other cell lineages and their phenotypes, and determine whether PTLs express other immunoregulatory molecules, including PD-L1, LAG-3 or IDO-1 and IDO-2, which can be targeted by novel therapies.
Recently, we showed that tumor-associated macrophages (TAMs) play a major role in PTL.26 High infiltration of PD-L1+CD68+ TAMs was associated with favorable survival and correlated with CD4+ and CD8+ T cells positive for PD1. In addition, both PD-L1+ TAMs and PD-1+ T cells emerged as independent indicators of survival for the patients with PTL. The interaction of PD-L1+ TAMs and PD-1+ T cells may modify the TME in PTL, or otherwise promote an anti-tumor immune response following immunochemotherapy.
The poor outcome associated with a low tumor-infiltrating T-cell content might provide the objective for therapeutic interventions. Cancer immunotherapy, especially protocols aiming to activate T-cell-mediated anti-tumor responses, and T-cell trafficking into the non-inflamed tumors has indeed gained a lot of attention recently. Antibodies targeting inhibitory molecules, such as PD-1, PD-L1, and CTLA-4 that regulate T-cell cytotoxicity have achieved impressive clinical responses.27–29 However, despite clear clinical responses, only a fraction of patients respond to treatment. Based on the findings from solid tumors, it has been suggested that the status of the HLA machinery affects the success of immunotherapy. For example, inactivating mutations in the β2M and HLA I complex in lung cancer caused a lack of response to immunotherapy because the cytotoxic cells were unable to find and attack the tumor cells.30 It was suggested that the recurrent inactivation of HLA I is an acquired mechanism for avoiding tumor immune recognition. In lymphomas, novel immunotherapies, including bispecific antibodies, cancer vaccines, and chimeric antigen receptor (CAR)-T cells may help T cells to identify and attack cancer cells.31–36 Another therapeutic implication that could be tested is to recuperate the loss of HLA II expression and the subsequent inability of T cells to recognize lymphoma cells by using histone deacetylase inhibitors.37,38
In conclusion, we have combined immune gene signatures and mIHC data with clinical information in a cohort of patients with PTL. We found three immune signatures, of which the T-lymphocyte signature was associated with survival both in PTL and DLBCL. The patients with T-cell-inflamed TME had a significantly increased risk of progression and death independently of IPI. Furthermore, reduced membranous staining of HLA I and II correlated with low T-cell infiltration. Taken together, the results presented herein are novel and emphasize the importance of immune escape as a mechanism regulating therapy resistance in patients with PTL.
Supplementary Material
Acknowledgments
We thank Dr. Petri Auvinen and Lars Paulin (Institute of Biotechnology, University of Helsinki, Finland) for the Nanostring analyses. Anne Aarnio and Marika Tuukkanen are acknowledged for technical assistance. The DLBCL dataset is part of the Cancer Genomics Characterization Initiative (CGCI), supported by NCI contract N01-C0-12400 (http://cgap.nci.nih.gov/cgci.html).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/2/338
Funding
This work was supported by grants from the Academy of Finland (to SL); Finnish Cancer Foundation (to SL and SMu); Finnish Cancer Institute (to SMu); Sigrid Juselius Foundation (to SL and SMu); Gyllenberg Foundation (to SMu); University of Helsinki (to SL); Helsinki University Hospital (to SL and SMu); Pirkanmaa Cancer Society (to MP); Seppo Nieminen Foundation (to MP); Eino Saarinen Foundation (to MP); and Finnish Oncology Association (to MP).
References
- 1.Cheah CY, Wirth A, Seymour JF. Primary testicular lymphoma. Blood. 2014;123(4):486–493. [DOI] [PubMed] [Google Scholar]
- 2.Deng L, Xu-Monette ZY, Loghavi S, et al. Primary testicular diffuse large B-cell lymphoma displays distinct clinical and biological features for treatment failure in rituximab era: a report from the International PTL Consortium. Leukemia. 2016;30(2):361–372. [DOI] [PubMed] [Google Scholar]
- 3.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517–534. [DOI] [PubMed] [Google Scholar]
- 4.Shain KH, Dalton WS, Tao J. The tumor microenvironment shapes hallmarks of mature B-cell malignancies. Oncogene. 2015;34(36):4673–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholas NS, Apollonio B, Ramsay AG. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim Biophys Acta. 2016;1863(3):471–482. [DOI] [PubMed] [Google Scholar]
- 7.Booman M, Douwes J, Glas AM, et al. Mechanisms and effects of loss of human leukocyte antigen class II expression in immune-privileged site-associated B-cell lymphoma. Clin Cancer Res. 2006;12(9):2698–2705. [DOI] [PubMed] [Google Scholar]
- 8.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of beta2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20(6):728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijland M, Veenstra RN, Visser L, et al. HLA dependent immune escape mechanisms in B-cell lymphomas: Implications for immune checkpoint inhibitor therapy¿ Oncoimmunology. 2017;6(4):e1295202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riemersma SA, Jordanova ES, Schop RF, et al. Extensive genetic alterations of the HLA region, including homozygous deletions of HLA class II genes in B-cell lymphomas arising in immune-privileged sites. Blood. 2000;96(10):3569–3577. [PubMed] [Google Scholar]
- 11.Riemersma SA, Oudejans JJ, Vonk MJ, et al. High numbers of tumour-infiltrating activated cytotoxic T lymphocytes, and frequent loss of HLA class I and II expression, are features of aggressive B cell lymphomas of the brain and testis. J Pathol. 2005;206(3):328–336. [DOI] [PubMed] [Google Scholar]
- 12.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. [DOI] [PubMed] [Google Scholar]
- 13.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blom S, Paavolainen L, Bychkov D, et al. Systems pathology by multiplexed immunohistochemistry and whole-slide digital image analysis. Sci Rep. 2017;7(1):15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansell SM, Stenson M, Habermann TM, Jelinek DF, Witzig TE. Cd4+ T-cell immune response to large B-cell non-Hodgkin’s lymphoma predicts patient outcome. J Clin Oncol. 2001;19(3):720–726. [DOI] [PubMed] [Google Scholar]
- 17.Keane C, Gill D, Vari F, Cross D, Griffiths L, Gandhi M. CD4(+) tumor infiltrating lymphocytes are prognostic and independent of R-IPI in patients with DLBCL receiving R-CHOP chemo-immunotherapy. Am J Hematol. 2013;88(4):273–276. [DOI] [PubMed] [Google Scholar]
- 18.Keane C, Vari F, Hertzberg M, et al. Ratios of T-cell immune effectors and checkpoint molecules as prognostic biomarkers in diffuse large B-cell lymphoma: a population-based study. Lancet Haematol. 2015;2(10):e445–455. [DOI] [PubMed] [Google Scholar]
- 19.Rimsza LM, Roberts RA, Miller TP, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004;103(11):4251–4258. [DOI] [PubMed] [Google Scholar]
- 20.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28(6):690–714. [DOI] [PubMed] [Google Scholar]
- 21.Abes R, Gelize E, Fridman WH, Teillaud JL. Long-lasting antitumor protection by anti-CD20 antibody through cellular immune response. Blood. 2010;116(6):926–934. [DOI] [PubMed] [Google Scholar]
- 22.Battella S, Cox MC, La Scaleia R, et al. Peripheral blood T cell alterations in newly diagnosed diffuse large B cell lymphoma patients and their long-term dynamics upon rituximab-based chemoimmunotherapy. Cancer Immunol Immunother. 2017;66(10):1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilchey SP, Hyrien O, Mosmann TR, et al. Rituximab immunotherapy results in the induction of a lymphoma idiotype-specific T-cell response in patients with follicular lymphoma: support for a “vaccinal effect” of rituximab. Blood. 2009;113(16):3809–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. [DOI] [PubMed] [Google Scholar]
- 25.Sweis RF, Spranger S, Bao R, et al. Molecular Drivers of the Non-T-cell-Inflamed Tumor Microenvironment in Urothelial Bladder Cancer. Cancer Immunol Res. 2016;4(7):563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollari M, Bruck O, Pellinen T, et al. PD-L1+ tumor-associated macrophages and PD-1+ tumor infiltrating lymphocytes predict survival in primary testicular lymphoma. Haematologica. 2018;103(11):1908–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Onc. 2017;14(4):203–220. [DOI] [PubMed] [Google Scholar]
- 28.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. New Engl J Med. 2015;372(26):2521–2532. [DOI] [PubMed] [Google Scholar]
- 29.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. New Engl J Med. 2017;377(14):1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira C, Gimenez-Xavier P, Pros E, et al. Genomic profiling of patient-derived xenografts for lung cancer identifies B2M inactivation impairing immunorecognition. Clin Cancer Res. 2017;23(12):3203–3213. [DOI] [PubMed] [Google Scholar]
- 31.Allegra A, Russo S, Gerace D, et al. Vaccination strategies in lymphoproliferative disorders: Failures and successes. Leuk Res. 2015;39(10):1006–1019. [DOI] [PubMed] [Google Scholar]
- 32.Brudno JN, Kochenderfer JN. Chimeric antigen receptor T-cell therapies for lymphoma. Nat Rev Clin Oncol. 2018;15(1):31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goebeler ME, Knop S, Viardot A, et al. Bispecific T-Cell Engager (BiTE) Antibody Construct Blinatumomab for the Treatment of Patients With Relapsed/Refractory Non-Hodgkin Lymphoma: Final Results From a Phase I Study. J Clin Oncol. 2016;34(10): 1104–1111. [DOI] [PubMed] [Google Scholar]
- 34.Pishko A, Nasta SD. The role of novel immunotherapies in non-Hodgkin lymphoma. Transl Cancer Res. 2017;6(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. New Engl J Med. 2017;377(26):2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viardot A, Goebeler ME, Hess G, et al. Phase 2 study of the bispecific T-cell engager (BiTE) antibody blinatumomab in relapsed/refractory diffuse large B-cell lymphoma. Blood. 2016;127(11):1410–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cycon KA, Mulvaney K, Rimsza LM, Persky D, Murphy SP. Histone deacetylase inhibitors activate CIITA and MHC class II antigen expression in diffuse large B-cell lymphoma. Immunology. 2013;140(2):259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Ortega-Molina A, Geng H, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discov. 2017;7(1):38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.