Nontyphoidal Salmonella species are the leading bacterial cause of foodborne disease in the United States. Whole-genome sequences and paired antimicrobial susceptibility data are available for Salmonella strains because of surveillance efforts from public health agencies.
KEYWORDS: antimicrobial susceptibility testing, deep learning, diagnostics, genome sequencing, machine learning
ABSTRACT
Nontyphoidal Salmonella species are the leading bacterial cause of foodborne disease in the United States. Whole-genome sequences and paired antimicrobial susceptibility data are available for Salmonella strains because of surveillance efforts from public health agencies. In this study, a collection of 5,278 nontyphoidal Salmonella genomes, collected over 15 years in the United States, was used to generate extreme gradient boosting (XGBoost)-based machine learning models for predicting MICs for 15 antibiotics. The MIC prediction models had an overall average accuracy of 95% within ±1 2-fold dilution step (confidence interval, 95% to 95%), an average very major error rate of 2.7% (confidence interval, 2.4% to 3.0%), and an average major error rate of 0.1% (confidence interval, 0.1% to 0.2%). The model predicted MICs with no a priori information about the underlying gene content or resistance phenotypes of the strains. By selecting diverse genomes for the training sets, we show that highly accurate MIC prediction models can be generated with less than 500 genomes. We also show that our approach for predicting MICs is stable over time, despite annual fluctuations in antimicrobial resistance gene content in the sampled genomes. Finally, using feature selection, we explore the important genomic regions identified by the models for predicting MICs. To date, this is one of the largest MIC modeling studies to be published. Our strategy for developing whole-genome sequence-based models for surveillance and clinical diagnostics can be readily applied to other important human pathogens.
INTRODUCTION
Nontyphoidal Salmonella species are the leading bacterial cause of foodborne disease in the United States (1, 2), causing over 1 million illnesses per year (3) and an estimated 80 million illnesses annually worldwide (4). Nontyphoidal Salmonella causes acute gastroenteritis and is usually contracted via fecal contamination of food sources (5). Although these infections are usually self-limiting and typically do not require antibiotic treatment (6), severe infections can occur (7). Antimicrobial resistance (AMR) is prevalent in Salmonella isolates, and infections caused by highly antimicrobial-resistant Salmonella strains result in worse outcomes than infections caused by susceptible strains (8–11).
In 1996, the National Antimicrobial Resistance Monitoring System (NARMS) was established as a collaboration between the U.S. Centers for Disease Control and Prevention (CDC), the U.S. Food and Drug Administration (FDA), the U.S. Department of Agriculture (USDA), and state and local health departments. A primary goal of NARMS is to monitor antimicrobial resistance in Salmonella and other foodborne bacteria, including Campylobacter, Escherichia, and Enterococcus (12). The data collected by NARMS are used to inform public health decisions aimed at identifying contaminated food sources and reducing the spread of AMR through enhanced stewardship. In recent years, NARMS has adopted whole-genome sequencing (WGS) as a routine monitoring tool. The WGS data are used to determine the source of outbreak strains and the virulence factor and AMR genes carried by each strain. As a result, a large collection of bacterial whole-genome sequences with extensive metadata is available for downstream research efforts (13).
Whole-genome sequencing is now routinely used for public health surveillance and to guide diagnostic and patient care decisions (14–18). For routine surveillance, WGS provides the highest possible resolution for individuating traits in bacteria, assessing phylogenetic relationships, conducting outbreak investigations, and predicting virulence and epidemicity. From the clinical perspective, rapid diagnostics are key to improving patient care. For a conventional microbiology laboratory diagnosis, the total time for organism growth, isolation, taxonomic identification, and antimicrobial MIC determination may exceed 36 h for relatively fast-growing bacteria and several days for slower-growing organisms (19–21). Since reducing the time to optimal antimicrobial therapy significantly improves patient outcomes (22–24), rapid sequencing-based approaches that predict MICs may have clinical utility. The extensive WGS data sets generated by health agencies and the scientific community, such as those for nontyphoidal Salmonella strains, provide the necessary training sets required for building predictive models.
Several investigations have recently built models for predicting AMR phenotypes from WGS data. To date, the most common approach has relied on using a curated reference set of genes and polymorphisms that are implicated in AMR (25–33). This reference-guided approach best predicts susceptibility and resistance when organisms are well studied and the AMR mechanisms are known. As larger collections of genomes have become available, several studies have used machine learning algorithms to predict susceptible and resistant phenotypes (27, 29, 31, 34–38). By using WGS and AMR phenotype data to train a machine learning model, predictions without a priori information about the underlying gene content of the genome or molecular mechanism for resistance to each agent are possible. Although this reference-free approach requires many genomes, it is unbiased and can potentially be used to enable the discovery of new genomic features that are involved in AMR (36, 37). These two general approaches have also been used to predict MICs for Streptococcus, Neisseria, and Klebsiella (35, 38–40). When a curated reference collection of genes and single nucleotide polymorphisms (SNPs) is used for predicting MICs, a rules-based or machine learning algorithm is required for determining how much a given feature contributes to the MIC. Thus, for MIC prediction, both reference-guided and reference-free approaches are expected to have similar advantages and disadvantages if the collection of genes and SNPs used by the reference-guided method is sufficient for predicting all MICs, including those that are in the susceptible range. For example, in previous work, we built a machine learning model to predict MICs for a comprehensive population-based collection of 1,668 Klebsiella pneumoniae clinical isolates (38). For each genome, we used nucleotide 10-mers and the MICs for each antibiotic as features to train the model. Extreme gradient boosting (XGBoost) was chosen as the machine learning algorithm (41). The model could rapidly predict the MICs for 20 antibiotics with an average accuracy of 92%. This demonstrated that it is possible to successfully predict MICs without using a precompiled set of AMR genes or polymorphisms.
In this study, we build models that use whole-genome sequence data to predict MICs for nontyphoidal Salmonella based on the strains collected and sequenced by NARMS from 2002 to 2016. Our strategy can be used to guide responses to outbreaks and inform antibiotic stewardship decisions.
MATERIALS AND METHODS
Genomes and metadata.
A total of 5,278 nontyphoidal Salmonella genome sequences were used in this study. All strains were collected and sequenced as part of the NARMS program. The strains were recovered either from raw retail meat and poultry or directly from livestock animals at slaughter. Antimicrobial susceptibility testing was performed using broth microdilution on a Sensititre system (Thermo Scientific) for 15 antibiotics, ampicillin (AMP), amoxicillin-clavulanic acid (AUG), ceftriaxone (AXO), azithromycin (AZI), chloramphenicol (CHL), ciprofloxacin (CIP), trimethoprim-sulfamethoxazole (COT), sulfisoxazole (FIS), cefoxitin (FOX), gentamicin (GEN), kanamycin (KAN), nalidixic acid (NAL), streptomycin (STR), tetracycline (TET), and ceftiofur (TIO), at FDA and USDA NARMS laboratories (13). Clinical breakpoints are based on CLSI and FDA guidelines (42). Whole-genome sequencing was performed using the Illumina HiSeq and MiSeq platforms and standard methods (25). Accession numbers and MICs for each isolate are listed in Table S1 in the supplemental material. All non-AMR metadata, including the serotype, host, geographic location of isolation, and isolation year, were taken from the metadata associated with each NCBI Sequence Read Archive (SRA) entry.
Genomic analyses.
The short-read sequence data for each strain were assembled with the PATRIC genome assembly service (43), using the full SPAdes pipeline, which uses the BayesHammer tool (44) for read correction and SPAdes software for assembly (45). All genomes were annotated using the PATRIC annotation service (43), which uses a variation of the RAST tool kit annotation pipeline (46). Annotated genomes are available on the PATRIC website (https://patricbrc.org). PATRIC genome identifiers are displayed in Table S1. Protein annotations, including those for proteins specifically asserted to be involved in AMR (47), were downloaded from the PATRIC work space and used for subsequent analyses. A phylogenetic tree was generated for the strains in the analysis by aligning the genes for the beta and beta prime subunits of the RNA polymerase using MAFFT software (48), concatenating the alignments, and computing a tree with the FastTree tool (49). The tree was rendered using the iTOL tool (50).
MIC prediction.
(i) Model generation. A model for predicting the MICs for the 15 antibiotics was built following the methods previously described by Nguyen and colleagues (38). Briefly, each genome was divided into the set of nonredundant overlapping nucleotide 10-mers using the k-mer counting program KMC (51). A matrix in which the k-mers, antibiotics, and MICs are treated as features for each genome was built. Each row in the matrix contains the k-mers for a genome as well as the MIC for a single antibiotic. The MIC prediction model was built using an XGBoost (41) regressor predicting linearized MICs. All model parameters were identical to those used by Nguyen et al. (38). Briefly, XGBoost is a computationally scalable method for generating gradient-boosted models. Gradient boosting is an ensemble method by which decision trees are generated to minimize an error function. We chose this method because of its scalability and its built-in ability to perform feature selection. The sensitivity and accuracy of the models generated in this study were tested by performing 10-fold cross validations. The cross validation partitions the matrix into 10 equal parts, such that each part has an equal (or nearly equal) number of antibiotic-MIC combinations. Ten rounds (folds) of training are performed: one part is used for testing, one part is used for validation, and eight parts are used for training. In this way, each part is used once for testing purposes, and any biases can be observed by tracking the average accuracy and confidence interval (CI) size over each fold. The validation set was used to monitor each model to prevent overfitting.
Unless otherwise stated, the accuracy of a model is reported as the ability to predict the correct MIC within ±1 2-fold dilution step of the laboratory-derived MIC. Defining an accuracy to be within ±1 2-fold dilution step is consistent with FDA requirements for automated MIC measuring device standards and is consistent with established clinical microbiology practices (20, 52–54). A comparison of raw accuracies and accuracies within ±1 2-fold dilution step is shown in Table S2. To assess the accuracy of a model over various metadata categories, including date, serotype source, and location, the training set genomes are used to make the model. The test set genomes are used to assess the model accuracy for a given fold. For models based on date ranges, all parameters are identical and the accuracy over the genomes is reported from the held-out dates.
(ii) Subsampling. All models were built and tested on a server with 4 Intel E5-4669v4 central processing units (Xeon at 2.2 GHz, 22 cores hyperthreaded to 44) with 1.585 terabytes (TB) of random-access memory (RAM). In order to perform the model building on this machine (machines with more memory are currently somewhat uncommon), we reduced the matrix size to sets of size n, where n is ≤250, 500, 1,000, 2,000, 3,000, 4,000, and 4,500 genomes. To create a diverse subset of size n, a hierarchical clustering method (55) was used to create n clusters by using the 10-mer distribution of each genome as the input features. To avoid the curse of dimensionality (56, 57), the taxicab/Manhattan distance (L1 norm) rather than the Euclidean distance (L2 norm) was used, since previous research has shown it to be both computationally fast and more accurate for high-dimensional data (58). From the resulting n clusters, one genome from each cluster was randomly selected from a uniform distribution to create the subset containing n genomes. For each subset of genomes, a matrix was generated, and models were generated as described above.
(iii) Feature identification. In order to unambiguously identify k-mers that are important to MIC prediction, we built separate models for each individual antibiotic using the method described above, except that we increased the k-mer length to 15 nucleotides in order to reduce the number of redundant k-mers within each genome and to enable analyses with the BLAST program (59). We also measured k-mer hits as presence versus absence, rather than counts, in order to simplify the analysis. Each model was built using the set of 1,000 diverse genomes from the subsampling experiment described above, and 10-fold cross validations were performed on each model.
The XGBoost feature importance score was computed using the internal XGBoost function xgboost.Booster.get_score(), using gain as the metric. This was computed for each fold within the 10-fold cross validation. This results in an importance score per feature (15-mer) from each fold. We summed the feature importance scores from each fold for the top 10 features in order to generate an overall importance score across all folds. This overall importance score captures both the importance of the 15-mer to a given fold and the number of times that the 15-mer was chosen as a top feature within each of the 10 folds.
XGBoost’s internal feature importance is unable to provide correlations between features and label values and thus does not provide an indication of whether a k-mer is related to antibiotic resistance or susceptibility. This is partially due to the fact that many nonlinear correlations that may use multiple features exist. In order to see if the high-scoring k-mers correlate with resistance or susceptibility, we computed the distribution of MICs for the genomes containing each high-scoring k-mer. For example, a k-mer conferring susceptibility should be found in more genomes with lower MICs, while a k-mer conferring resistance should exist in genomes with higher MICs. Each high-scoring k-mer was also compared to the set of protein-encoding genes within each Salmonella genome. If a k-mer was found within a known AMR gene, that gene is reported. Otherwise, we report the distance (in nucleotides) between the center of the k-mer and the center of nearby AMR genes. This search was scoped to 3 kb to reduce the search space.
To find k-mers that are being used by the individual antibiotic models to predict susceptible MICs, we computed the presence or absence of each k-mer with high XGBoost feature importance values (described above) for the entire data set of 5,278 genomes. The k-mers with the largest difference in occurrence between the susceptible and resistant genomes are the ones that are chosen by the models for predicting susceptible MICs. To determine if there were significant SNPs in these k-mers, we found the genomic features containing the k-mer—protein-encoding gene, RNA gene, or intergenic region—using the BLASTn program (59). The corresponding feature or region was then found for all genomes in the collection. The features were aligned using MAFFT (48) and manually curated using the Jalview program (60). Poor-quality sequence was removed, all duplicates and paralogs were removed, and the subalignment covering the k-mer was extracted. To prevent possible biases due to clonality that may exist in the full set of genomes, the analysis was repeated on the diverse subset of 1,000 genomes (described above). We report an SNP in a k-mer region as being significant if the susceptible and resistant sets were significantly different (P < 0.001) for a given nucleotide position, based on a chi-square test, for both the full set of 5,278 genomes and the set of 1,000 diverse genomes. Sequence logos for k-mers containing significant SNPs were generated using the WebLogo application (61). k-mers from the azithromycin and ciprofloxacin models were excluded from this analysis because they each had seven resistant genomes. Comparisons of codon usage versus the genome average, genome mode, and high-expression gene sets were computed as described previously (62, 63).
Software availability.
The Salmonella MIC prediction model based on 4,500 genomes—including the software and documentation for running the model—is available at our GitHub page (https://github.com/PATRIC3/mic_prediction).
Accession number(s).
Data are available under BioProject accession numbers PRJNA292661 and PRJNA292666. The SRA run accession number for each genome is displayed in Table S1 in the supplemental material.
RESULTS
Model construction.
For this study, we used a publicly available collection of 5,278 Salmonella whole-genome sequences generated by the NARMS project between 2002 and 2016. The strains were isolated from retail meat and food animal sources in the United States. The collection included 98 different serotypes, including Salmonella enterica serotypes Heidelberg (678 genomes), Kentucky (618 genomes), and Typhimurium var. 5− (588 genomes), from 41 states (see Table S1 in the supplemental material). Isolates were tested for resistance to up to 15 antimicrobial agents using the broth microdilution method. Many of the strains were randomly selected for sequencing as part of a compulsory nationwide collection program (Table 1).
TABLE 1.
Number of genomes susceptible, intermediate, or resistant to the 15 antibiotics for the 5,278 Salmonella genomes used in this study
| Antibiotic | No. of genomes: |
||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| AMP | 3,682 | 2 | 1,593 |
| AUG | 4,145 | 355 | 778 |
| AXO | 4,508 | 1 | 769 |
| AZI | 2,409 | 0 | 7 |
| CHL | 5,026 | 87 | 164 |
| CIP | 5,217 | 53 | 7 |
| COT | 5,219 | 0 | 58 |
| FIS | 3,356 | 0 | 1,573 |
| FOX | 4,501 | 98 | 679 |
| GEN | 4,577 | 68 | 633 |
| KAN | 837 | 3 | 84 |
| NAL | 5,233 | 0 | 45 |
| STR | 872 | 0 | 1,919 |
| TET | 2,364 | 28 | 2,885 |
| TIO | 4,517 | 8 | 753 |
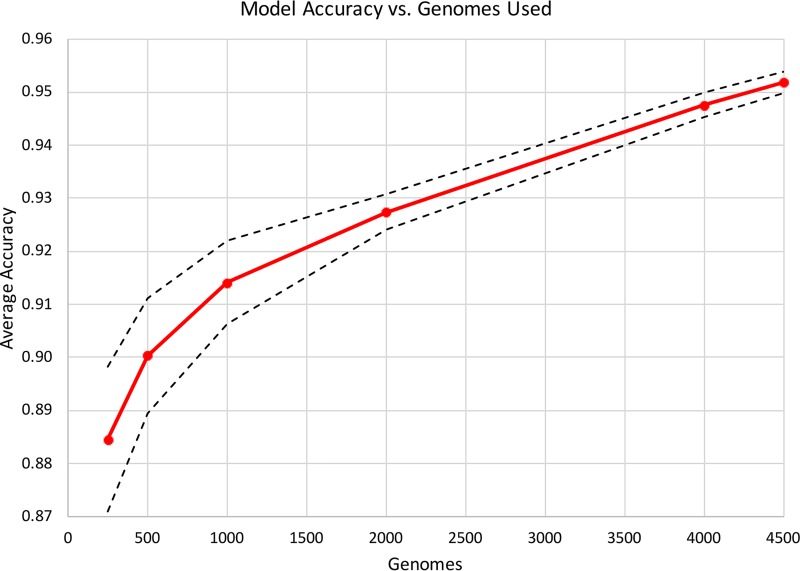
The nontyphoidal Salmonella MIC prediction model was built by a strategy similar to our previously described strategy used to predict MICs for K. pneumoniae clinical isolates (38). Since the Salmonella data set has many more genomes and greater sampling in the range of susceptible MICs, it provides a critical test case for determining if the approach remains robust for other pathogens. In the Klebsiella study, we built individual models for each antibiotic, as well as a single large integrated model by combining the data from all antibiotics. We found that the combined model achieved slightly higher overall accuracies (by ∼1% to 2%); however, the matrix that was necessary to train this model had a large memory footprint. Indeed, if we were to build a similar matrix for the current Salmonella data set using all 5,278 genomes, the model training would exceed 1.5 TB of RAM. RAM usage and training times are displayed in Fig. S1. Therefore, we first built models for all antibiotics using subsets of the genomes ranging in size from 250 to 4,500 genomes, which were rationally selected to maximize genetic diversity (Fig. 1). A matrix built from 4,500 genomes is the largest that we can train on a 1.5-TB machine using this protocol. As the training set size increased from 250 to 1,000 genomes, the accuracy increased from 88.5% to 91.4%. Then, as the training set increased beyond 1,000 genomes, the accuracy continued to improve more modestly, with the 4,500-genome model having an average accuracy of 95.2%. The results indicated that the overall MIC prediction approach which was developed previously for Klebsiella pneumoniae also works for Salmonella, despite the differences in sampling, genetic diversity, and MICs. Also, we discovered that a smaller number of well-chosen diverse genomes can serve as a useful proxy for the entire set, since models built from ≥500 genomes have accuracies exceeding 90%.
FIG 1.
MIC prediction model accuracy for subsamples of genomes. Diverse subsamples of genomes were chosen, and the model accuracy within ±1 2-fold dilution step based on a 10-fold cross validation is shown with the red plot line. The dashed lines represent the high and low values for the 95% confidence interval for the average accuracy at each given plot point.
Model accuracy.
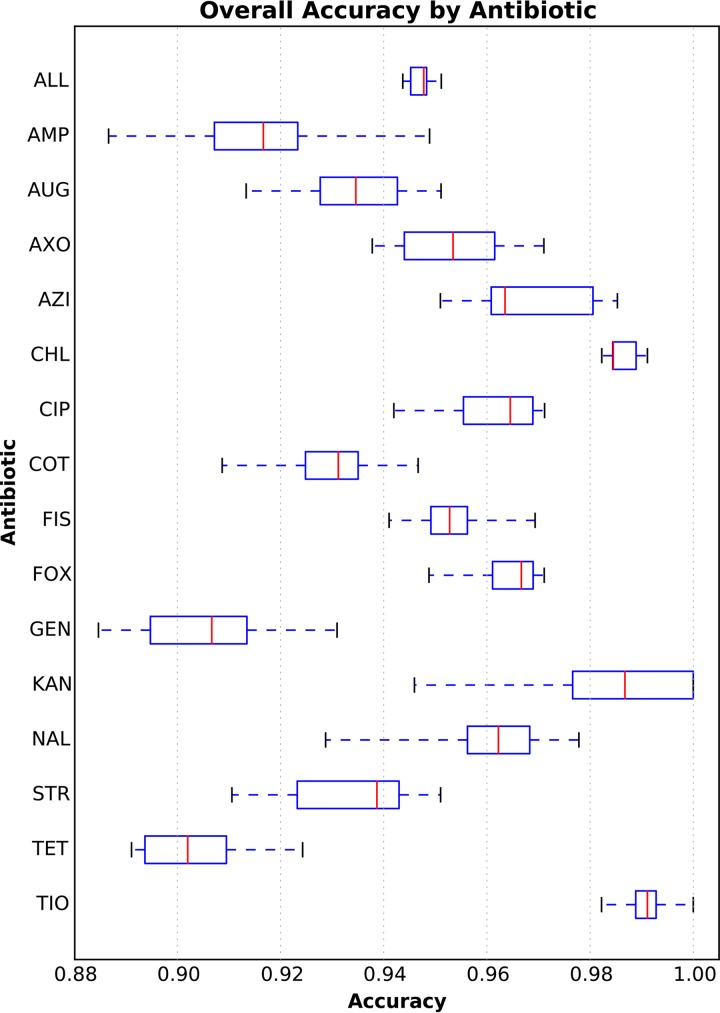
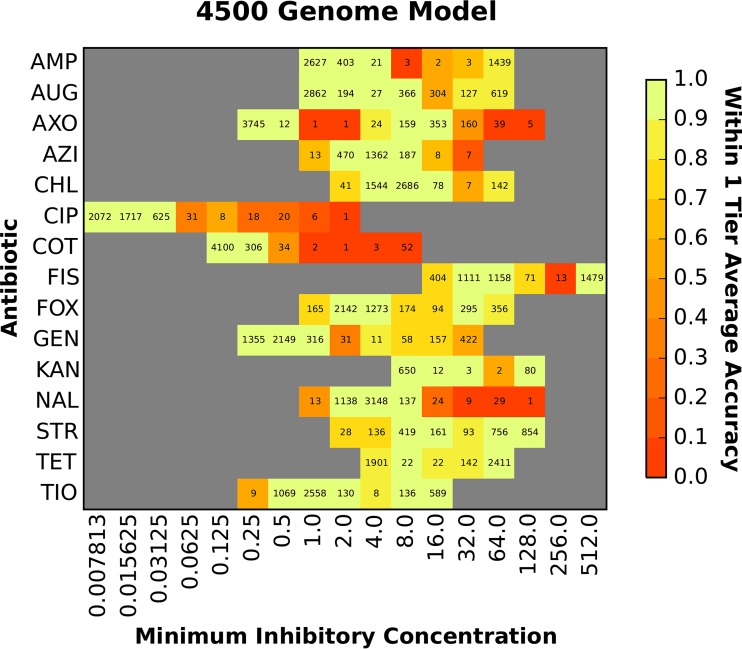
We computed the overall accuracy for each antibiotic using the model that was based on 4,500 genomes. For this model, all 15 antibiotics had average accuracies of ≥90%, with their first quartile being bound ≥89% (Fig. 2). Chloramphenicol and ceftiofur had the highest accuracies (99%), and gentamicin and tetracycline had the lowest accuracies (91% and 90%, respectively) (Table S2). Since the model is robust to the various mechanisms of resistance for the 15 antibiotics, it is possible that the slightly lower accuracies for gentamicin and tetracycline could be due to the distribution of multiple AMR genes/mechanisms across the population of strains with resistant genomes (which will be analyzed in more detail below). Figure 3 depicts the accuracy of the 4,500-genome model for each MIC. Overall, the model was robust for both the resistant and susceptible MICs, and it tended to be more accurate when a MIC was represented by many genomes. The model tended to have lower accuracies for the highest and lowest MICs, perhaps because of underlying genetic differences between strains that have been reported with values greater than or equal to or less than or equal to a MIC, which represents a range of MICs rather than a discrete value.
FIG 2.
Box plot of the overall accuracies within ±1 2-fold dilution step for each antibiotic in the 4,500-genome model. The y axis depicts each antibiotic (abbreviations are defined in Materials and Methods). The x axis depicts the accuracy. Each vertical red line represents the median accuracy over the holdout sets for each fold in the 10-fold cross validation. The blue box encompasses the data for the first and third quartiles. The dashed blue horizontal lines bounded by black vertical lines (or whiskers) depict the entire distribution of accuracies for each fold and antibiotic. The accuracy of the entire 4,500-genome model over all antibiotics and folds is depicted in the row marked “ALL.”
FIG 3.
Accuracy of the MIC prediction model based on 4,500 diverse genomes. The heat map depicts the accuracy within ±1 2-fold dilution step of the laboratory-derived MIC. The x axis shows the MIC (in micrograms per milliliter), and each antibiotic is shown on the y axis. The accuracy for each antibiotic-MIC combination is depicted by color, with bright yellow/green being the most accurate and red being the least accurate. The values shown in each cell are the number of genomes with that MIC for a given antibiotic.
The utility of AMR diagnostic devices is often described in terms of error rate. Major errors (MEs) are defined as susceptible genomes that have been incorrectly assigned resistant MICs by the model. Very major errors (VMEs) are defined as resistant genomes that have been incorrectly assigned susceptible MICs by the model. FDA standards for automated systems recommend a major error rate of ≤3% (54). All antibiotics used in the model had ME rates within this range (Table 2). The FDA standards for VME rates indicate that the lower 95% confidence limit should be ≤1.5% and that the upper limit should be ≤7.5% (54). Seven of the 15 antibiotics—amoxicillin-clavulanic acid, ceftriaxone, chloramphenicol, cefoxitin, streptomycin, tetracycline, and ceftiofur—had acceptable VME rates by this measure. Ampicillin and sulfisoxazole had VME rates with 95% confidence intervals approaching the ranges of 0.022 to 0.033 and 0.026 to 0.053, respectively, failing only on the lower bound. The VME rates for some of the remaining antibiotics were higher because there were fewer resistant genomes. As more resistant genomes are collected and the data set becomes more balanced, we expect VME rates to be reduced.
TABLE 2.
VME and ME rates for the 4,500-genome modela
| Antibiotic | VME rate |
ME rate |
No. of samples: |
|||
|---|---|---|---|---|---|---|
| Avg | 95% CI | Avg | 95% CI | Resistant | Susceptible | |
| All | 0.027 | 0.024 to 0.030 | 0.001 | 0.001 to 0.002 | 10,979 | 47,366 |
| AMP | 0.028 | 0.022 to 0.033 | 0.000 | 0.000 to 0.001 | 1,442 | 3,054 |
| AUG | 0.012 | 0.000 to 0.025 | 0.000 | 0.000 to 0.000 | 746 | 3,449 |
| AXO | 0.022 | 0.011 to 0.032 | 0.000 | 0.000 to 0.001 | 740 | 3,758 |
| AZI | 0.857 | 0.508 to 1.207 | 0.000 | 0.000 to 0.000 | 7 | 2,040 |
| CHL | 0.000 | 0.000 to 0.000 | 0.000 | 0.000 to 0.001 | 149 | 4,271 |
| CIP | 0.417 | 0.099 to 0.933 | 0.000 | 0.000 to 0.000 | 7 | 4,445 |
| COT | 0.670 | 0.515 to 0.825 | 0.000 | 0.000 to 0.001 | 55 | 4,443 |
| FIS | 0.039 | 0.026 to 0.053 | 0.000 | 0.000 to 0.000 | 1,479 | 2,757 |
| FOX | 0.009 | 0.001 to 0.020 | 0.000 | 0.000 to 0.000 | 651 | 3,754 |
| GEN | 0.090 | 0.066 to 0.113 | 0.000 | 0.000 to 0.000 | 579 | 3,862 |
| KAN | 0.074 | 0.012 to 0.136 | 0.000 | 0.000 to 0.000 | 82 | 662 |
| NAL | 0.917 | 0.819 to 1.014 | 0.000 | 0.000 to 0.001 | 39 | 4,460 |
| STR | 0.014 | 0.008 to 0.020 | 0.027 | 0.013 to 0.040 | 1,703 | 744 |
| TET | 0.000 | 0.000 to 0.000 | 0.018 | 0.012 to 0.025 | 2,575 | 1,901 |
| TIO | 0.004 | 0.001 to 0.009 | 0.000 | 0.000 to 0.000 | 725 | 3,766 |
The rates of very major errors (VME), defined as resistant genomes predicted to be susceptible, and major errors (ME), defined as susceptible genomes predicted to be resistant, are reported within ±1 twofold dilution step. CI, 95% confidence interval.
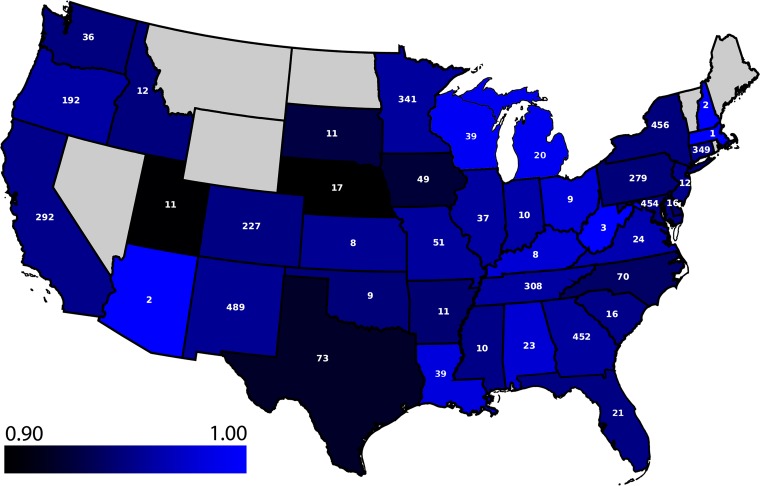
In addition to the extensive MIC data, NARMS reports rich metadata, including isolation date, food or animal source, collection year, geographic location, and serotype. We computed the accuracy of the model over each available metadata category to determine if the model is robust to these differences and to ensure that no subset biased the model. The period of collection of the genomes spanned 15 years, with all the years except 2002 (the oldest) and 2016 (the most recent) having over 100 isolates. The model accuracy ranged from 94% to 97% over each collection year (Table 3). That is, the genetic factors that contribute to the MICs either remained stable over the 15-year period or were learned as the model was trained. Although the data set was mostly comprised of poultry meat or live animal isolates, the accuracy ranged from 94% to 96% over the four contamination sources: turkey, beef, pork, and chicken (Table 4). No obvious biases in the accuracies were detected on the basis of the state of isolation (an average of 95% accuracy over 41 states with a 95% CI equal to 0.95 to 0.96) (Fig. 4) or the serovar of each isolate (94% accuracy over 97 serovars with a 95% CI equal to 0.94 to 0.96) (Table S3). Since the traditional Salmonella serotyping scheme is based on the lipopolysaccharide O and flagellar H antigens, which are encoded by genes that influence the cell surface (64), we also constructed a phylogenetic tree for Salmonella genomes to observe the model accuracy over the various clades. Overall, no phylogenetic bias in the model accuracy was detected (Fig. S2).
TABLE 3.
Model accuracy for the genomes from each sample collection year
| Collection date (yr) | Accuracy | No. of genomes | No. of binsa |
|---|---|---|---|
| 2002 | 0.97 | 55 | 624 |
| 2003 | 0.95 | 159 | 1,809 |
| 2004 | 0.96 | 235 | 2,850 |
| 2005 | 0.95 | 274 | 3,384 |
| 2006 | 0.95 | 313 | 3,880 |
| 2007 | 0.94 | 258 | 3,192 |
| 2008 | 0.95 | 388 | 4,821 |
| 2009 | 0.95 | 436 | 5,367 |
| 2010 | 0.94 | 230 | 2,820 |
| 2011 | 0.95 | 214 | 2,968 |
| 2012 | 0.96 | 257 | 3,694 |
| 2013 | 0.97 | 265 | 3,793 |
| 2014 | 0.95 | 506 | 7,100 |
| 2015 | 0.95 | 689 | 9,646 |
| 2016 | 0.96 | 83 | 1,161 |
The total number of MICs available for the genomes isolated in that year.
TABLE 4.
Model accuracy for the genomes isolated from various sources
| Source | Accuracy | No. of genomes | No. of binsa |
|---|---|---|---|
| Chicken | 0.96 | 1,981 | 25,869 |
| Cow/beef | 0.94 | 419 | 5,688 |
| Pig/pork | 0.95 | 448 | 6,144 |
| Turkey | 0.94 | 1,651 | 21,260 |
The total number of MICs available for the genomes of each category.
FIG 4.
Average accuracy of the model based on 4,500 diverse genomes for predicting MICs for the Salmonella genomes from each state. Light blue is the most accurate, and dark blue/black is the least accurate. Note that the scale starts at an accuracy of 0.90. Each state is labeled with the number of genomes collected from that state. States without a label contain no samples and are colored in gray; no genomes from Alaska and Hawaii exist in the collection.
One concern of using a model that is trained on the data from previous years and in some cases over 15 years old is that the training set is not representative of currently circulating strains. That is, the model may be inaccurate for predicting MICs for the genomes of strains that are currently circulating or will emerge in the future. For example, shifts in clonal groups, the evolution of AMR-associated genes, or the introduction of AMR genes by horizontal gene transfer is possible (65, 66). We evaluated this possibility by building models from subsets of the whole-genome sequence data using strains collected in earlier years and measuring the accuracy of the models on genomes collected in later years. Models were built for years prior to 2009 through 2014 and tested on the remaining genomes (Table 5). These models had accuracies ranging from 86% to 92%. As the number of years used for building the models increased, the number of genomes available for testing decreased, so we also tested each model on only the 462 genomes from 2015 and 2016. Similarly, the accuracy of each model on the genomes from 2015 and 2016 ranged from 87% to 90% (Table S4). The results indicate that within this data set, the models generated from genomes collected at earlier dates yielded stable MIC predictions for genomes collected at later dates. This finding is consistent with the pattern of AMR genes that was observed within the data set. Although the AMR gene content may vary from year to year, we did not observe any major sweeps or fixation events that drastically altered the AMR gene content of the collection between years, which would cause the MIC predictions to fail for a large fraction of the genomes (Table S5). Taken together, these data suggest that the MIC prediction models generated in this study are likely to be sustainable over time.
TABLE 5.
Ability of models trained on genomes from prior years to predict MICs for genomes collected in later years
| Training set yr | Test set yr | Accuracy | 95% CI for accuracy | No. of training binsa | No. of testing binsa | No. of training genomes | No. of testing genomes |
|---|---|---|---|---|---|---|---|
| 2002–2008 | 2009–2016 | 0.88 | 0.88–0.89 | 36,563 | 22,412 | 1,819 | 2,681 |
| 2002–2009 | 2010–2016 | 0.88 | 0.88–0.89 | 31,196 | 27,779 | 2,255 | 2,245 |
| 2002–2010 | 2011–2016 | 0.88 | 0.88–0.88 | 28,376 | 30,599 | 2,485 | 2,015 |
| 2002–2011 | 2012–2016 | 0.88 | 0.88–0.89 | 25,408 | 33,567 | 2,699 | 1,801 |
| 2002–2012 | 2013–2016 | 0.88 | 0.87–0.88 | 21,714 | 37,261 | 2,956 | 1,544 |
| 2002–2013 | 2014–2016 | 0.86 | 0.86–0.87 | 17,921 | 41,054 | 3,221 | 1,279 |
| 2002–2014 | 2015–2016 | 0.92 | 0.92–0.92 | 10,807 | 48,168 | 3,728 | 772 |
The total number of genome/antibiotic combinations.
Genomic regions important for MIC prediction.
The 4,500-genome model described above contained data from all antibiotics and MICs, making feature extraction to determine which k-mers contribute to the MIC predictions for each antibiotic difficult. To address this limitation, we modified the protocol by building separate models for each antibiotic. Instead of using 10-mers, we increased the k-mer length to 15 nucleotides to reduce redundancy and make them identifiable using BLAST analysis (59). We also searched for the presence or absence of k-mers, rather than using k-mer counts, to simplify the analysis of the XGBoost decision trees. Since a 15-mer matrix can be 45 times larger than a 10-mer matrix, we used ≤1,000 diverse genomes to reduce the memory footprint during training. Overall, the average accuracy for the individual models was nearly identical to the average accuracy for the combined 4,500-genome model (96% versus 95%, respectively), and in nearly all cases, the 95% confidence intervals overlapped between the combined and single antibiotic models (Table S6). Thus, for this data set, single antibiotic models with fewer genomes and larger k-mers performed as well as a combined model (Fig. S3).
During model training, XGBoost assigns an importance value to each k-mer used in a decision tree. When the model is used to predict the MICs for a new genome, the k-mers with the highest importance values are the most informative for the MIC prediction. Thus, by analyzing the feature importance values of each k-mer, we can use the models as a tool for understanding the genomic regions that differentiate MICs. For each antibiotic-specific model, we parsed the XGBoost decision trees from each fold of the 10-fold cross validation to extract the importance values for each k-mer. To understand the relationship between known AMR genes and the important k-mers that were chosen by each model, we then searched for k-mers that had high importance values within AMR genes or that occurred in close proximity to an AMR gene. Table 6 lists the highest-ranking k-mers from each model that occurred within or in close proximity to an AMR gene. In most cases, the k-mers corresponded to known AMR genes, including class A and C beta-lactamases for the beta-lactam antibiotics, aminoglycoside nucleotidyl- and acetyltransferases for the aminoglycosides, DNA gyrase and QnrB for the fluoroquinolones, TetA and TetR for tetracycline, and dihydrofolate reductase and dihydropteroate synthase for co-trimoxazole and sulfisoxazole. In the case of azithromycin, the collection mostly contained susceptible genomes (Table 1), so the first macrolide resistance gene observed corresponds to the eighth-ranking k-mer. The top 10 k-mers with the highest feature importance values from each of the 10 folds used in model training are listed in Tables S7 to S21. In addition to the top AMR k-mers displayed in Table 6, these tables show other highly ranking k-mers from the same AMR genes as well as k-mers from related genes that are known to confer resistance to the given antibiotic. In some cases, k-mers matching regions or genes from unrelated AMR mechanisms have high importance values, suggesting a pattern of co-occurrence on horizontally transferred genetic elements.
TABLE 6.
Highest-ranking AMR-related protein function (or genomic region) with a matching k-mer from the XGBoost models
| Antibiotic | k-mer rank | Distance between k-mer and AMR genea | k-mer sequence | PATRIC annotation(s) |
|---|---|---|---|---|
| AMP | 1 | Direct match | CTTAATCAGTGAGGC | Class A beta-lactamase (EC 3.5.2.6) => TEM family |
| AUG | 1 | Direct match | AAACGTCTTACTAAC | Class C beta-lactamase (EC 3.5.2.6) => CMY/CMY-2/CFE/LAT family |
| AXOb | 1 | 566.0 ± 39.7 | AAAGAGAAAGAAAGG | Class C beta-lactamase (EC 3.5.2.6) => CMY/CMY-2/CFE/LAT family |
| AZI | 8 | Direct match | CCCATTTCCGCCGCC | Macrolide 2′-phosphotransferase => Mph(A) family |
| CHLb | 1 | 611.8 ± 5.1 | AGACAAGTAAGCCGC | Chloramphenicol/florfenicol resistance, MFS efflux pump => FloR family |
| CIP | 1 | 313.5 ± 70.5 | ACAGTCCATCCAGGA | Pentapeptide repeat protein QnrB family => Quinolone resistance protein QnrB10 |
| COTb | 1 | 248.4 ± 7.0 | AAAAACGATAGCTGC | Dihydrofolate reductase (EC 1.5.1.3) |
| FISb | 1 | 408.9 ± 11.3 | CGCAACGGCTCAAGC | Dihydropteroate synthase type-2 (EC 2.5.1.15) @ Sulfonamide resistance protein |
| FOX | 1 | Direct match | AAAAAAACCTTGGCA | Class C beta-lactamase (EC 3.5.2.6) => CMY/CMY-2/CFE/LAT family |
| GEN | 1 | 439.0 ± 70.4, 451.2 ± 7.0 | AGTTAAGCCGCGCCG | Aminoglycoside 3ʺ-nucleotidyltransferase (EC 2.7.7.−) => APH(3ʺ)-Ia (AadA family); Aminoglycoside N(3)-acetyltransferase (EC 2.3.1.81) => AAC(3)-II,III,IV,VI,VIII,IX,X |
| KAN | 1 | Direct match | AAAAAGCCGTTTCTG | Aminoglycoside 3′-phosphotransferase (EC 2.7.1.95) => APH(3′)-I |
| NAL | 1 | Direct match | ATTCCGCAGTGTATG | DNA gyrase subunit A (EC 5.99.1.3) |
| STR | 1 | Direct match | ATTTGTACGGCTCCG | Aminoglycoside 3ʺ-nucleotidyltransferase (EC 2.7.7.−) => APH(3ʺ)-Ia (AadA family) |
| TET | 1 | 335.6 ± 12.6, 596.5 ± 22.5 | CGTTCTGCCTTGCGC | Tetracycline resistance regulatory protein TetR; Tetracycline resistance, MFS efflux pump => Tet(A) |
| TIOb | 1 | 566 ± 39.7 | AAAGAGAAAGAAAGG | Class C beta-lactamase (EC 3.5.2.6) => CMY/CMY-2/CFE/LAT family |
A direct match means that the k-mer is an exact match to the protein-encoding gene. Average distances with standard deviations, measured as the number of nucleotides from the center of the k-mer to the center of the AMR gene, are shown for the set of 1,000 diverse genomes.
TIO has other AMR genes within 3 kb listed in Tables S7 to S21.
Since each model predicts the entire range of MICs, some of the highly ranking k-mers may be used to predict susceptible MICs. To assess this, we computed the fraction of susceptible and resistant genomes with each k-mer from Tables S7 to S21. The set of k-mers that were most enriched in the susceptible genomes is shown in Table 7. Overall, 7 of the top 10 k-mers represented significantly different SNPs (P < 0.001) both in the complete set of 5,278 genomes and in the set of 1,000 diverse genomes used to build the models (Fig. S4). The top k-mer associated with susceptibility was from the nalidixic acid model and occurred in the DNA gyrase gyrA gene. This is also the top k-mer that was found in an AMR gene for nalidixic acid from Table 6. In this case, the model relied more heavily on the wild-type version of the k-mer rather than any of the resistant versions (the remaining k-mers from Table 6 occurred almost exclusively in resistant genomes). The same gyrA k-mer was also found as a highly ranking k-mer in the case of ciprofloxacin (Table S12). Two significant gyrA SNPs were captured by this k-mer (Fig. S4). These are missense mutations in the resistant genomes occurring at Ser83 and Asp87, and changes at these positions have been shown to confer quinolone resistance in Escherichia coli (67, 68). The remaining significant mutations from Fig. S4 that occurred in the protein-encoding genes were the same sense (not amino acid-changing) mutations. In the cases of eptA (Ser, TCG to TCA), oadA (Ala, GCC to GCA), the AraJ precursor gene (Leu, CTG to CTA), and the second gcd mutation (Thr, ACG to ACA), the codon changed from a commonly used codon in the susceptible genomes to the least preferred codon in the resistant genomes. In the cases of the nrfE and nrfF mutations (Asn, AAT to AAC) and the first gcd mutation (Asp., GAC to GAT), the resistant genomes had the preferred codon of the pair. Whether these SNPs have a modulating effect on protein translation or contribute to the fitness of the resistant organisms requires further analysis.
TABLE 7.
Important k-mers used by the individual antibiotic models for predicting susceptible MICsa
| Antibiotic | k-mer sequence | Sus | Res | Frac sus | Frac res | Genomic region | PATRIC annotation or genomic region |
|---|---|---|---|---|---|---|---|
| NAL | ATTCCGCAGTGTATG | 5,233 | 45 | 1.00 | 0.38 | PEG | DNA gyrase subunit A (EC 5.99.1.3) |
| AXO | TGGTATTCGCATCAA | 4,508 | 769 | 0.78 | 0.48 | PEG | Phosphoethanolamine transferase EptA |
| KAN | CTGGCTTTTTTTTTT | 837 | 84 | 0.30 | 0.00 | RNA | RyhB RNA |
| STR | CCCTTATCCAACACG | 872 | 1,919 | 0.85 | 0.55 | PEG | Respiratory nitrate reductase delta chain (EC 1.7.99.4) |
| AXO | CAGAACCAGAATTTG | 4,508 | 769 | 0.74 | 0.46 | PEGs | Formate-dependent nitrite reductase complex subunit NrfF, and Cytochrome c-type heme lyase subunit NrfE, nitrite reductase complex assembly |
| TIO | AGAGAAGCCTGCCGC | 4,517 | 753 | 0.68 | 0.40 | PEG | Oxaloacetate decarboxylase alpha chain (EC 4.1.1.3) |
| AXO | ATCCCCGCCATTACA | 4,508 | 769 | 0.73 | 0.46 | PEG | Tagatose-1,6-bisphosphate aldolase GatY (EC 4.1.2.40) |
| AXO | TGCTGCAAAACGCCA | 4,508 | 769 | 0.69 | 0.45 | PEG | Protein AraJ precursor |
| AXO | GAAAACAGGGTGTAG | 4,508 | 769 | 0.47 | 0.23 | INT | Upstream of IlvGMEDA operon leader peptide |
| FOX | GGATACCACGCCGGG | 4,501 | 679 | 0.58 | 0.35 | PEGs | Glucose dehydrogenase, PQQ-dependent (EC 1.1.5.2), and IncF plasmid conjugative transfer protein TraP |
Sus and Res, total number of susceptible and resistant genomes in the entire collection, respectively; Frac sus and Frac res, fraction of susceptible and resistant genomes with the k-mer in the entire collection, respectively; PEG, protein-encoding gene; RNA, RNA gene; INT, intergenic region.
DISCUSSION
In this study, we built, using XGBoost (41), machine learning-based MIC prediction models for nontyphoidal Salmonella genomes that achieved overall accuracies of 95% to 96% within a ±1 2-fold dilution factor. To our knowledge, this is one of the largest and most accurate MIC prediction models to be published to date. Importantly, it provides a model strategy for performing MIC prediction directly from genome sequence data that could be applied to other human or veterinary pathogens.
The success of our MIC prediction model was dependent on the large, publicly available, population-based collection of genomes with associated metadata. Since researchers often focus on collecting highly resistant or otherwise unusual strains, the opportunities to generate balanced models are rare. We demonstrate the many benefits from comprehensive sampling for the entire range of possible MICs. First, the use of diverse and balanced data sets improved model accuracies because there was better sampling across all MIC dilutions. Second, having balanced data enabled us to achieve acceptable ME and VME rates for 7 of the 15 antibiotics used in the study. Third, compared with our recent model for Klebsiella pneumoniae, the larger and more balanced data set for nontyphoidal Salmonella enabled us to build models for individual antibiotics that had accuracies similar to those of the combined model. This enabled us to begin to disambiguate the important genomic regions relating to resistant and susceptible MICs. Finally, we show that MICs in the susceptible range can be accurately predicted with the algorithm using all genomic data rather than scoping it to known AMR genes or gene polymorphisms. This contrasts with prior work correlating MICs to known resistance mechanisms in Salmonella (69). In future studies, our strategy could be used as a starting point for identifying the subtle genomic changes that result in different MICs.
For each single-antibiotic model, we analyzed the k-mers that had high feature importance values and were important to the models for predicting MICs. The highly ranking k-mers that were enriched in the resistant genomes mainly occurred within or in close proximity to well-known AMR genes. With the exception of the gyrA k-mer, the highly ranking k-mers that were enriched in the susceptible genomes were significant in several cases but were more difficult to interpret. Some of these susceptibility k-mers hint at a possible relationship between AMR and oxidative stress or electron transport, such as the k-mers matching components of the nitrate and nitrite reductases and pqq-dependent glucose dehydrogenase, which is consistent with the known link between antibiotics and oxidative stress (70, 71). Although XGBoost would be expected to yield similar results with other ensemble machine learning methods, it may be possible to generate a more interpretable set of features using support vector machines or a single perceptron. However, these methods are not ideal, given the size of the input data. Future machine learning work that eliminates the effect of the resistance k-mers and bench work specifically examining these mutations may shed light on the subtle effects that result in the gradient of susceptible MICs.
The genomes used in this study were collected over a 15-year period from 41 U.S. states. By building models encompassing genomes collected over ranges of earlier dates, we demonstrated stable and accurate MIC prediction for genomes collected at later dates. Presently, we are not aware of any large publicly available collections of Salmonella genomes with MIC data from other countries. Since the AMR gene content may vary across pathogen populations, validation of the Salmonella models using strains from other countries is important to its application in global health. Nevertheless, the present analysis clearly demonstrates that the current model provides accurate MIC predictions for United States isolates. Similarly, an analysis of this model on Salmonella enterica serotype Typhi strains would provide information about the utility of the model over broader phylogenetic distances.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Service (contract no. HHSN272201400027C).
We thank Emily Dietrich for her helpful edits.
M.N. designed the study, performed experiments, generated data, and prepared the manuscript, S.W.L. designed the study, P.F.M. designed the study and generated data, R.J.O. designed the study, R.O. performed software engineering, R.L.S. designed the study, G.H.T. designed the study and generated data, S.Z. designed the study and generated data, and J.J.D. designed the study, generated data, and prepared the manuscript.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the U.S. Department of Health and Human Services, the U.S. Food and Drug Administration, the Centers for Disease Control and Prevention, or the U.S. government. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or the U.S. Food and Drug Administration.
We claim no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01260-18.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2017. Surveillance for foodborne disease outbreaks, United States, 2015. Annual report. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/foodsafety/pdfs/2015FoodBorneOutbreaks_508.pdf. [Google Scholar]
- 2.Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K. 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2006-2014. MMWR Morb Mortal Wkly Rep 64:495–499. [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States. Emerg Infect Dis 17:7–40. doi: 10.3201/eid1701.091101p1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2015. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Andino A, Hanning I. 2015. Salmonella enterica: survival, colonization, and virulence differences among serovars. ScientificWorldJournal 2015:520179. doi: 10.1155/2015/520179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aserkoff B, Bennett JV. 1969. Effect of antibiotic therapy in acute salmonellosis on the fecal excretion of salmonellae. N Engl J Med 281:636–640. doi: 10.1056/NEJM196909182811202. [DOI] [PubMed] [Google Scholar]
- 7.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varma JK, Mølbak K, Barrett TJ, Beebe JL, Jones TF, Rabatsky-Ehr T, Smith KE, Vugia DJ, Chang HH, Angulo FJ. 2005. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis 191:554–561. doi: 10.1086/427263. [DOI] [PubMed] [Google Scholar]
- 9.Varma JK, Greene KD, Ovitt J, Barrett TJ, Medalla F, Angulo FJ. 2005. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg Infect Dis 11:943. doi: 10.3201/eid1106.041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger AL, Greene SA, Barzilay EJ, Henao O, Vugia D, Hanna S, Meyer S, Smith K, Pecic G, Hoefer D, Griffin PM. 2014. Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal Salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog Dis 11:335–341. doi: 10.1089/fpd.2013.1642. [DOI] [PubMed] [Google Scholar]
- 11.Angulo FJ, Mølbak K. 2005. Human health consequences of antimicrobial drug—resistant Salmonella and other foodborne pathogens. Clin Infect Dis 41:1613–1620. doi: 10.1086/497599. [DOI] [PubMed] [Google Scholar]
- 12.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott PF. 2017. National Antimicrobial Resistance Monitoring System: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557. doi: 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.U.S. Food and Drug Administration (FDA). 2018. NARMS now. U.S. Food and Drug Administration, Rockville, MD: Updated 20 July 2018 https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm. [Google Scholar]
- 14.Abrams AJ, Trees DL. 2017. Genomic sequencing of Neisseria gonorrhoeae to respond to the urgent threat of antimicrobial-resistant gonorrhea. Pathog Dis 75:ftx041. doi: 10.1093/femspd/ftx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. 2015. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio 6(6):e01888-15. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown EW, Gonzalez-Escalona N, Stones R, Timme R, Allard MW. 2017. The rise of genomics and the promise of whole genome sequencing for understanding microbial foodborne pathogens, p 333–351. In Gurtler JB, Doyle MP, Kornacki JL (ed), Foodborne pathogens: virulence factors and host susceptibility. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 18.McArthur AG, Tsang KK. 2017. Antimicrobial resistance surveillance in the genomic age. Ann N Y Acad Sci 1388:78–91. doi: 10.1111/nyas.13289. [DOI] [PubMed] [Google Scholar]
- 19.Opota O, Croxatto A, Prod'hom G, Greub G. 2015. Blood culture-based diagnosis of bacteraemia: state of the art. Clin Microbiol Infect 21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Reller LB, Weinstein M, Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 21.Saha SK, Darmstadt GL, Baqui AH, Hanif M, Ruhulamin M, Santosham M, Nagatake T, Black RE. 2001. Rapid identification and antibiotic susceptibility testing of Salmonella enterica serovar Typhi isolated from blood: implications for therapy. J Clin Microbiol 39:3583–3585. doi: 10.1128/JCM.39.10.3583-3585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llor C, Bjerrum L. 2014. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf 5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 24.Palmer H, Palavecino E, Johnson J, Ohl C, Williamson J. 2013. Clinical and microbiological implications of time-to-positivity of blood cultures in patients with Gram-negative bacilli bacteremia. Eur J Clin Microbiol Infect Dis 32:955–959. doi: 10.1007/s10096-013-1833-9. [DOI] [PubMed] [Google Scholar]
- 25.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, Harris SR. 2017. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehaus KE, Walker TM, Crook DW, Peto TE, Clifton DA. 2014. Machine learning for the prediction of antibacterial susceptibility in Mycobacterium tuberculosis, p 618–621. Abstr 2014 IEEE-EMBS Int Conf Biomed Health Informatics. [Google Scholar]
- 28.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TEA, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesesky MW, Hussain T, Wallace M, Patel S, Andleeb S, Burnham C-AD, Dantas G. 2016. Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in Gram-negative bacilli from whole genome sequence data. Front Microbiol 7:1887. doi: 10.3389/fmicb.2016.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipworth SIW, Hough N, Leach L, Morgan M, Jeffrey K, Andersson M, Robinson E, Smith G, Crook D, Peto T. 2018. Whole genome sequencing for predicting Mycobacterium abscessus drug susceptibility. bioRxiv 251918 10.1101/251918. [DOI] [PMC free article] [PubMed]
- 31.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, De Cesare M. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison OB, Clemence M, Dillard JP, Tang CM, Trees D, Grad YH, Maiden MCJ. 2016. Genomic analyses of Neisseria gonorrhoeae reveal an association of the gonococcal genetic island with antimicrobial resistance. J Infect 73:578–587. doi: 10.1016/j.jinf.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, Bentley SD, Trees D, Lipsitch M. 2016. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis 214:1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho JR, Carriço JA, Knight D, Martínez J-L, Morrissey I, Oggioni MR, Freitas AT. 2013. The use of machine learning methodologies to analyse antibiotic and biocide susceptibility in Staphylococcus aureus. PLoS One 8:e55582. doi: 10.1371/journal.pone.0055582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyre DW, De Silva D, Cole K, Peters J, Cole MJ, Grad YH, Demczuk W, Martin I, Mulvey MR, Crook DW, Walker AS, Peto TEA, Paul J. 2017. WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J Antimicrob Chemother 72:1937–1947. doi: 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drouin A, Giguère S, Déraspe M, Marchand M, Tyers M, Loo VG, Bourgault A-M, Laviolette F, Corbeil J. 2016. Predictive computational phenotyping and biomarker discovery using reference-free genome comparisons. BMC Genomics 17:754. doi: 10.1186/s12864-016-2889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. 2016. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 6:27930. doi: 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H. 2018. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 8:421. doi: 10.1038/s41598-017-18972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metcalf BJ, Chochua S, Gertz R, Li Z, Walker H, Tran T, Hawkins PA, Glennen A, Lynfield R, Li Y. 2016. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect 22:1002.e1–1002.e8. doi: 10.1016/j.cmi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Walker H, Hawkins PA, Tran T, Whitney CG, McGee L. 2016. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. mBio 7(3):e00756-16. doi: 10.1128/mBio.00756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T, Guestrin C. 2016. Xgboost: a scalable tree boosting system, p 785–794. Proc 22nd ACM SIGKDD Int Conf Knowledge Discovery Data Mining ACM. [Google Scholar]
- 42.U.S. Food and Drug Administration. 2011. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): executive report. U.S. Department of Health and Human Services. Food and Drug Administration, Rockville, MD. [Google Scholar]
- 43.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolenko SI, Korobeynikov AI, Alekseyev MA. 2013. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14:S7. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonopoulos DA, Assaf R, Aziz RK, Brettin T, Bun C, Conrad N, Davis JJ, Dietrich EM, Disz T, Gerdes S, Kenyon RW, Machi D, Mao C, Murphy-Olson DE, Nordberg EK, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Santerre J, Shukla M, Stevens RL, VanOeffelen M, Vonstein V, Warren AS, Wattam AR, Xia F, Yoo H. 31 July 2017. PATRIC as a unique resource for studying antimicrobial resistance. Brief Bioinform doi: 10.1093/bib/bbx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I, Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 51.Deorowicz S, Kokot M, Grabowski S, Debudaj-Grabysz A. 2015. KMC 2: fast and resource-frugal k-mer counting. Bioinformatics 31:1569–1576. doi: 10.1093/bioinformatics/btv022. [DOI] [PubMed] [Google Scholar]
- 52.U.S. Food and Drug Administration. 2009. Guidance for industry and FDA. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. Center for Devices and Radiological Health, U.S. Food and Drug Administration, U.S. Department of Health and Human Services, Silver Spring, MD. [Google Scholar]
- 53.Jorgensen JH. 1993. Selection criteria for an antimicrobial susceptibility testing system. J Clin Microbiol 31:2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.U.S. Food and Drug Administration. 2009. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. U.S. Food and Drug Administration, Rockville, MD. [Google Scholar]
- 55.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel OBM, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Machine Learning Res 12:2825–2830. [Google Scholar]
- 56.Bellman R. 2013. Dynamic programming. Courier Corporation, New York, NY. [Google Scholar]
- 57.Shalev-Shwartz S, Ben-David S. 2014. Understanding machine learning: from theory to algorithms. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 58.Aggarwal CC, Hinneburg A, Keim DA (ed). 2001. On the surprising behavior of distance metrics in high dimensional space In International conference on database theory. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 59.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis JJ, Olsen GJ. 2010. Modal codon usage: assessing the typical codon usage of a genome. Mol Biol Evol 27:800–810. doi: 10.1093/molbev/msp281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis JJ, Olsen GJ. 2011. Characterizing the native codon usages of a genome: an axis projection approach. Mol Biol Evol 28:211–221. doi: 10.1093/molbev/msq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ranieri ML, Shi C, Switt AIM, Den Bakker HC, Wiedmann M. 2013. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J Clin Microbiol 51:1786–1797. doi: 10.1128/JCM.03201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida H, Kojima T, Yamagishi J-I, Nakamura S. 1988. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet 211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida H, Bogaki M, Nakamura M, Nakamura S. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother 34:1271–1272. doi: 10.1128/AAC.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyson GH, Zhao S, Li C, Ayers S, Sabo JL, Lam C, Miller RA, McDermott PF. 2017. Establishing genotypic cutoff values to measure antimicrobial resistance in Salmonella. Antimicrob Agents Chemother 61:e02140-16. doi: 10.1128/AAC.02140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 71.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. 2012. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.