The recent outbreaks of Zika virus (ZIKV) and associated birth defects in regions of dengue virus (DENV) endemicity emphasize the need for sensitive and specific serodiagnostic tests. We reported previously that enzyme-linked immunosorbent assays (ELISAs) based on the nonstructural protein 1 (NS1) of DENV serotype 1 (DENV1) and ZIKV can distinguish primary DENV1, secondary DENV, and ZIKV infections.
KEYWORDS: Zika virus, dengue virus, nonstructural protein 1, serodiagnosis
ABSTRACT
The recent outbreaks of Zika virus (ZIKV) and associated birth defects in regions of dengue virus (DENV) endemicity emphasize the need for sensitive and specific serodiagnostic tests. We reported previously that enzyme-linked immunosorbent assays (ELISAs) based on the nonstructural protein 1 (NS1) of DENV serotype 1 (DENV1) and ZIKV can distinguish primary DENV1, secondary DENV, and ZIKV infections. Whether ELISAs based on NS1 proteins of other DENV serotypes can discriminate various DENV and ZIKV infections remains unknown. We herein developed DENV2, DENV3, and DENV4 NS1 IgG ELISAs to test convalescent- and postconvalescent-phase samples from reverse transcription-PCR-confirmed cases, including 25 primary DENV1, 24 primary DENV2, 10 primary DENV3, 67 secondary DENV, 36 primary West Nile virus, 38 primary ZIKV, and 35 ZIKV with previous DENV infections as well as 55 flavivirus-naive samples. Each ELISA detected primary DENV infection with a sensitivity of 100% for the same serotype and 23.8% to 100% for different serotypes. IgG ELISA using a mixture of DENV1-4 NS1 proteins detected different primary and secondary DENV infections with a sensitivity of 95.6% and specificity of 89.5%. The ZIKV NS1 IgG ELISA detected ZIKV infection with a sensitivity of 100% and specificity of 82.9%. On the basis of the relative optical density ratio, the combination of DENV1-4 and ZIKV NS1 IgG ELISAs distinguished ZIKV with previous DENV and secondary DENV infections with a sensitivity of 91.7% to 94.1% and specificity of 87.0% to 95.0%. These findings have important applications to serodiagnosis, serosurveillance, and monitoring of both DENV and ZIKV infections in regions of endemicity.
INTRODUCTION
The rapid spread of ZIKV during the 2015 to 2016 outbreak and its association with fetal microcephaly and other birth defects, known as congenital Zika syndrome (CZS), highlight the need for sensitive and specific diagnostic tests, especially for pregnant women (1–4). According to the guidelines from the U.S. Centers for Disease Control and Prevention (CDC), the laboratory diagnosis of ZIKV infection includes a positive nucleic acid test as soon as possible post-symptom onset (PSO) to confirm ZIKV infection and a negative IgM test to exclude ZIKV infection (5, 6). As most (∼80%) ZIKV infections are asymptomatic and many individuals, including pregnant women, undergo ZIKV testing beyond the period when RNA is detectable, serological tests remain a critical component of ZIKV diagnosis (5, 6). Moreover, ZIKV is present in multiple body fluids and can be transmitted sexually or after asymptomatic infection (7–9), emphasizing the importance of serological tests.
ZIKV belongs to the genus Flavivirus of the family Flaviviridae, in which several vector-borne viruses cause medically important human diseases, including the four serotypes of dengue virus (DENV1 to DENV4), West Nile virus (WNV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV) (10). As the major target of antibody response after flaviviral infection, the envelope (E) protein has been the main antigen for serological tests, including the use of recombinant E protein, inactivated virions, or virus-like particles (10–13). However, due to the cross-reactivity of the anti-E antibody to ZIKV and other flaviviruses (13–17), positive or equivocal IgM results based on E protein require further testing with laborious plaque reduction neutralization tests (PRNT) (5, 6). PRNT can confirm individuals who had ZIKV as the first flaviviral infection, known as primary ZIKV (pZIKV) infection, but the results are more difficult to interpret for those who previously acquired DENV or other flaviviral infections, hampering the application of E-protein-based serological tests for ZIKV in regions of DENV endemicity.
Several recent studies have reported that DENV immune sera and monoclonal antibodies (MAbs) against DENV E protein enhance ZIKV replication in vitro and in vivo (18–22), so-called antibody-dependent enhancement (23), and raised the possibility that the risk and severity of CZS might be increased by previous DENV infection. Given the explosive spread of ZIKV and CZS during the 2015 to 2016 outbreak in regions of DENV endemicity, serological tests that can distinguish pZIKV infection from ZIKV infection with previous DENV (ZIKVwprDENV) infection are critical to further our understanding of the pathogenesis of ZIKV and CZS.
A study of human MAbs against nonstructural protein 1 (NS1) revealed that most anti-NS1 MAbs derived from patients with pZIKV infection were specific to ZIKV, and >50% of those from patients with ZIKVwprDENV infection reacted to DENV (21); other studies using ZIKV-specific NS1 MAbs in blockade-of-binding enzyme-linked immunosorbent assays (ELISAs) showed that ZIKV NS1 protein in serodiagnosis can distinguish ZIKV and DENV (24, 25). We reported previously that the combination of ZIKV and DENV1 NS1 ELISAs distinguishes pZIKV, primary DENV1 (pDENV1), secondary DENV (sDENV,) and ZIKVwprDENV infections (26). Whether an NS1 ELISA based on other DENV serotypes can detect different DENV infections and distinguish them with ZIKV infection remains unknown. In this study, we developed DENV2, DENV3, and DENV4 NS1 ELISAs to detect primary DENV infections of different serotypes and showed that a mixture of DENV1-4 NS1 proteins in an ELISA can detect various primary DENV infections, and its combination with a ZIKV NS1 ELISA can distinguish DENV and ZIKV infections.
MATERIALS AND METHODS
Clinical samples.
The study of coded serum or plasma samples was approved by the institutional review board (IRB) of the University of Hawaii (CHS 17568, CHS 23786). The numbers, serotypes, sampling time, and sources of different panels of samples are summarized in Table 1. Samples collected <3 months or ≥3 months PSO were designated convalescent- or postconvalescent-phase samples, respectively. Samples from reverse transcription-PCR (RT-PCR)-confirmed Zika cases that were DENV naive or previously DENV exposed, designated pZIKV and ZIKVwprDENV panels, respectively, were collected between July and March 2017 from the Pediatric Dengue Cohort Study (PDCS) and the pediatric Dengue Hospital-based Study in Managua, Nicaragua. The DENV immune status was based on anti-DENV antibody testing by an inhibition ELISA at entry and annually for the PDCS as described previously (26–28). These studies were approved by the IRBs of the University of California, Berkeley, and Nicaraguan Ministry of Health. Parents or legal guardians of all subjects provided written informed consent, and subjects 6 years old and older provided assent. Convalescent-phase samples from patients presenting with symptoms compatible with Zika and detectable anti-DENV IgG antibodies during the acute phase, designated the probable ZIKVwprDENV panel, were collected between November 2015 and May 2016 at the Complexo Hospital at Federal University of Bahia, Bahia, Brazil (26). Thirty-six plasma samples from blood donors who tested positive for WNV transcription-mediated amplification and IgM and IgG antibodies between 2006 and 2015, designated primary WNV (pWNV) infection, were provided by the American Red Cross at Gaithersburg, MD (21, 29). Convalescent- and postconvalescent-phase samples from RT–PCR-confirmed cases with different primary DENV infections (including pDENV1, primary DENV2 [pDENV2], and primary DENV3 [pDENV3], except primary DENV4 infection) or sDENV infection were from Taiwan, Hawaii, and Nicaragua prior to the 2015 to 2016 Zika outbreak; 55 flavivirus-naive samples from a seroprevalence study were included as the controls in this study (26, 30–33). Primary DENV or sDENV infection was determined by IgM/IgG ratio or focus reduction neutralization tests as described previously (26, 30–32).
TABLE 1.
Sampling time, serotypes, and sources of different serum/plasma panels
| Panela | No. of subjects testing positive/total | Category (no. of samples) | Sampling time PSOb (mean [range]) | Source (n) of samples, yr(s) |
|---|---|---|---|---|
| pDENV1 | 21/25 | Convalescent (9) | 49 (19–89) days | Taiwan (8), 2006–2009 Hawaii (11), 2015 Nicaragua (2), 2006–2008 |
| Postconvalescent (16) | 6.7 (3–15) mo | |||
| pDENV2 | 18/24 | Convalescent (11) | 11 (6–20) days | Taiwan (16), 2001–2002 Nicaragua (2), 2006–2008 |
| Postconvalescent (13) | 19.4 (3–96) mo | |||
| pDENV3 | 4/10 | Postconvalescent (10) | 11 (3–19) mo | Nicaragua (2), 2006–2008 |
| pWNV | 36/36 | Early convalescent (36) | NAc | U.S. ARC, 2006–2015 |
| pZIKV | 20/38 | Convalescent (20) | 17 (14–24) days | Nicaragua, 2016 |
| Postconvalescent (18) | 6.9 (6–8) mo | |||
| ZIKVwprDENV | 20/35 | Convalescent (20) | 16 (14–19) days | Nicaragua, 2016 |
| Postconvalescent (15) | 7.0 (6–8) mo | |||
| Probable ZIKVwprDENV | 19/19 | Convalescent (19) | 10 (6–14) days | Brazil, 2015–2016 |
| sDENV | 59/63 | Convalescent (24) | 14 (8–35) days | Taiwan, 2001–2002 |
| Postconvalescent (20) | 9.4 (3–12) mo | Taiwan (2), 2006–2009 Nicaragua (18), 2006–2008 |
||
| Postconvalescent (14) | 19.7 (18–24) mo | Taiwan (10), 2006–2009 Nicaragua (4), 2006–2008 |
||
| Postconvalescent (5) | 71 (67–72) mo | Taiwan, 2006–2009 |
pDENV1, primary DENV1 infection; pDENV2, primary DENV2 infection; pDENV3, primary DENV3 infection; pWNV, primary WNV infection; pZIKV, primary ZIKV infection; sDENV, secondary DENV infection; ZIKVwprDENV, ZIKV infection with previous DENV infection.
PSO, post-symptom onset.
NA, not applicable. Index samples tested positive for WNV transcription-mediated amplification and IgM and IgG from blood donors at the American Red Cross (ARC).
Recombinant NS1 proteins.
The sequence for the gene encoding NS1 (residues 1 to 352) of ZIKV (HPF2013 strain) with a His tag at the C terminus was codon optimized (Integrated DNA Technologies, Skokie, IL) and cloned into the pMT-Bip vector (26). Drosophila S2 stable clones expressing ZIKV NS1 protein were established, followed by CuSO4 induction and purification by a fast purification chromatography system (AKTA Pure; GE Health Care Bio-Science, Pittsburgh, PA) (26). Purified DENV1 to DENV4 NS1 proteins were purchased from the Native Antigen Company (Oxford, UK).
ELISAs.
DENV1 and ZIKV NS1 IgG ELISAs have been described previously (26). DENV2, DENV3, DENV4, and DENV1-4 NS1 IgG ELISAs were technically developed and set up in the same way. Briefly, 96-well plates were coated with purified NS1 proteins (16 ng for individual NS1 proteins per well and 8/4/8/4 ng for mixed DENV1/2/3/4 NS proteins per well) at 4°C overnight, followed by blocking (StartingBlock blocking buffer; Thermo Scientific, Waltham, MA) at room temperature for 1 h, incubating with primary antibody (serum or plasma at 1:400 dilution) at 37°C for 2 h, washing with washing buffer (0.5% Tween 20 in 1× phosphate-buffered saline [PBS]) 4 times, incubating with secondary antibody (anti-human IgG conjugated to horseradish peroxidase [HRP] at 1:10,000 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA) at 37°C for 1 h, and washing with washing buffer 6 times (26, 31, 32). After adding tetramethylbenzidine substrate (Thermo Scientific, Waltham, MA) at room temperature for 15 min followed by stop solution, the optical density (OD) at 450 nm was read with a reference wavelength of 630 nm. Each ELISA plate included two positive controls (OD higher than 1; two confirmed Zika or confirmed dengue samples), four negative controls (flavivirus-naive sera or plasma), and test samples (all in duplicates). The OD values were divided by the mean OD value of one positive control (OD close to 1) in the same plate to calculate the relative OD (rOD) values for comparison between plates. The cutoff rOD was defined by the mean rOD value of negatives plus 12 standard deviations, which gave a confidence level of 99.9% from 4 negatives (34). The ratio of rOD of ZIKV NS1 IgG ELISA to that of DENV1-4 NS1 IgG ELISA was determined, and a cutoff rOD ratio at 0.24 was chosen as described previously (26). ZIKV, DENV1, DENV2, and DENV1-4 NS1 IgM ELISAs were performed similarly with some modifications to detect NS1-specfic IgM responses. Each sample was preincubated with an IgG absorbent, Gullsorb reagent (Meridian Bioscience, Cincinnati, OH), for 10 min and anti-human IgM conjugated with HRP (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as secondary antibody (26, 35). Each ELISA was performed twice (each in duplicates).
Statistical analysis.
Two-tailed Mann-Whitney tests were used to determine the P values between two groups (GraphPad Prism 6). The 95% confidence interval (CI) was calculated by Excel.
RESULTS
DENV1 to DENV4 NS1 IgG ELISAs.
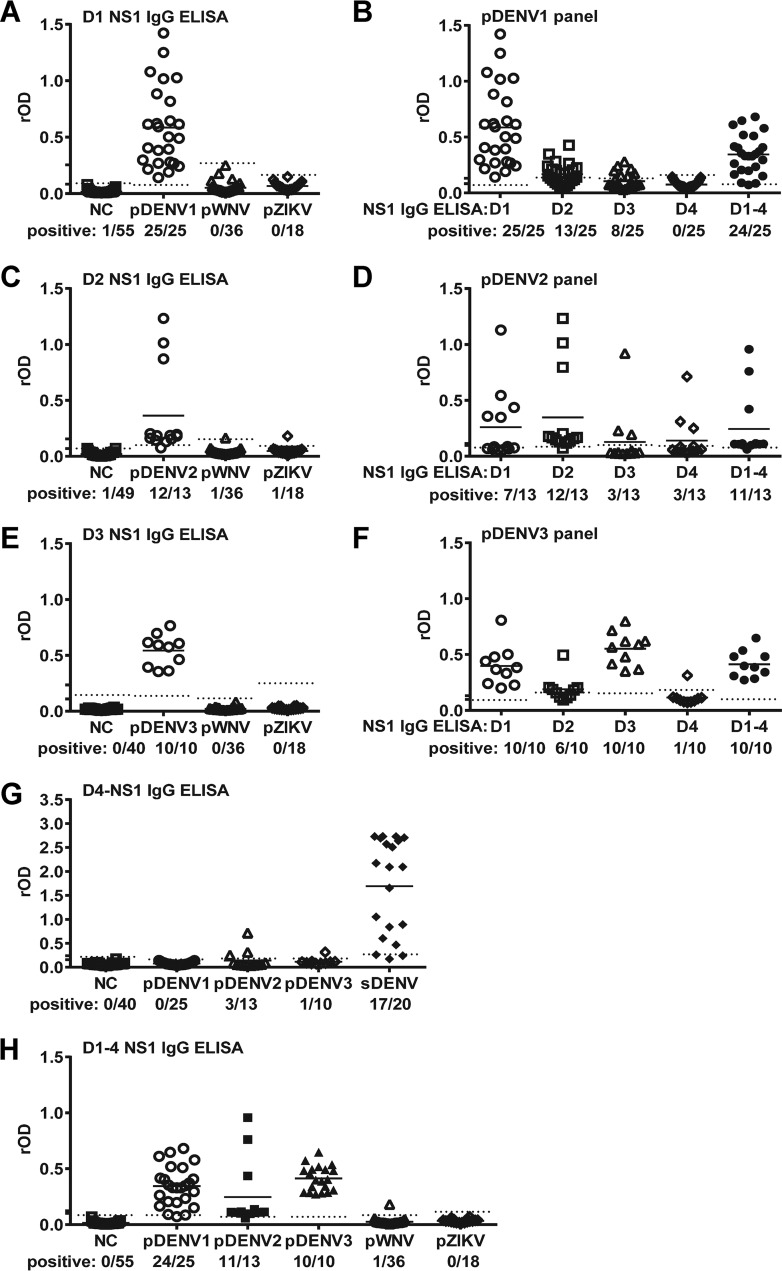
We first employed DENV1, DENV2, and DENV3 NS1 IgG ELISAs to test samples from primary DENV infection (pDENV1, pDENV2, and pDENV3) panels. As shown in Fig. 1A, the DENV1 NS1 IgG ELISA detected the pDENV1 panel (100%) but not the pZIKV, pWNV, or naive panel. Similarly, DENV2 and DENV3 NS1 IgG ELISAs detected pDENV2 (92.3%) and pDENV3 (100%) panels, respectively, but not the pZIKV, pWNV, or naive panel (Fig. 1C and E). We next examined if the pDENV1 panel was detected by DENV2, DENV3, or DENV4 NS1 IgG ELISA and found the detection rates were greatly reduced (52.0%, 32.0%, and 0%, respectively) (Fig. 1B). Similarly, the detection rates for pDENV2 and pDENV3 panels by ELISAs using heterologous NS1 protein were generally reduced, ranging from 23.1% to 53.8% for pDENV2 and 10.0% to 100% for pDENV3 panels compared with those using homologous NS1 protein (Fig. 1B, D, and F). We have also established a DENV4 NS1 IgG ELISA, which detected the sDENV panel (85.0%) and some primary DENV panels (10.0% to 23.1%) but not the pZIKV, pWNV, or naive panel (Fig. 1G).
FIG 1.
Results of DENV1, DENV2, DENV3, DENV4, and DENV1-4 NS1 IgG ELISAs tested with primary DENV infection panels. NS1 IgG ELISAs based on DENV1 (A), DENV2 (C), DENV3 (E), DENV4 (G), and DENV1-4 (H) were tested with convalescent-phase samples of different panels (negative control [NC], primary DENV, pZIKV, pWNV, or sDENV). Results of pDENV1 (B), pDENV2 (D), and pDENV3 (F) panels tested with different NS1 IgG ELISAs. Dotted lines indicate cutoff rODs. Data are the means from two experiments (each in duplicates); horizontal lines are the means from each panel. D1, DENV1; D2, DENV2; D3, DENV3; D4, DENV4; D1-4, DENV1-4.
Mixed DENV1-4 NS1 IgG ELISA.
To establish a single DENV-NS1 IgG ELISA that can detect primary DENV infections of different serotypes, we coated a ELISA plate with a mixture of DENV1-4 NS1 proteins. It had a detection rate of 96.0%, 84.6%, or 100% for the pDENV1, pDENV2, or pDENV3 panel, respectively, which was comparable to that using homologous NS1 protein (Fig. 1B, D, F, and H). We thus used the mixed DENV1-4 NS1 IgG ELISA to test other panels in this study. As expected, the DENV1-4 NS1 IgG ELISA did not detect samples from the naive, pZIKV, or pWNV panel except one in the pWNV panel (Fig. 1H).
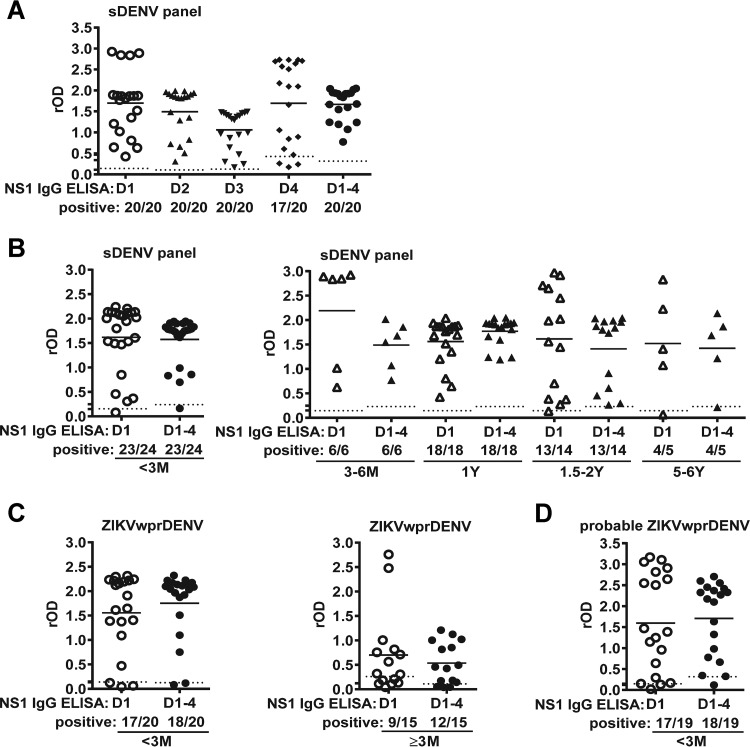
Previously, we reported that the DENV1 NS1 ELISA detects the sDENV panel with detection rates ranging from 95.0% to 100% (within 2 years PSO) to 80.0% (5 to 6 years PSO) (26). When testing with the sDENV panel (3 to 12 months PSO), the DENV1-4 NS1 IgG ELISA had a detection rate of 100%, comparable to those of the DENV1, DENV2, and DENV3 NS1 IgG ELISAs (Fig. 2A), suggesting that after secondary DENV infection, cross-reactive anti-NS1 antibodies can recognize NS1 protein of different serotypes well except from DENV4, which was more distantly related to DENV1-3 and less likely to be the exposed serotype of our study participants based on the epidemiological histories (27, 30). When testing the sDENV panels collected at different times, the detection rates ranged from 95.8% to 100% (<1 month to 3 to 12 months PSO) to 92.9% to 80% (1.5 to 6 years PSO), which were comparable to that of the DENV1 NS1 IgG ELISA reported previously (Fig. 2B) (26). When testing with the ZIKVwprDENV panel (<1 month and 6 to 8 months PSO) from Nicaragua and probable ZIKVwprDENV panel from Brazil (<1 month PSO), the DENV1-4 NS1 IgG ELISA had detection rates of 80.0% to 94.7% (Fig. 2C and D), which were higher than those of the DENV1 NS1 IgG ELISA (60.0% to 89.5%), probably due to the presence of serotypes other than DENV1 in these regions (26). Taken together, these findings suggest that the DENV1-4 NS1 IgG ELISA is a convenient and single DENV NS1 IgG ELISA that detects three serotypes of primary DENV infection as well as secondary DENV infection.
FIG 2.
Results of DENV1-4 NS1 IgG ELISA tested with secondary DENV infection panels. (A) DENV1-4 NS1 IgG ELISA was tested with postconvalescent-phase samples of the sDENV panel. Comparison of DENV1 and DENV1-4 NS1 IgG ELISAs tested with convalescent- or postconvalescent-phase samples of sDENV (B), ZIKVwprDENV (C), and probable ZIKVwprDENV (D) panels. Dotted lines indicate cutoff rOD values. Data are the means from two experiments (each in duplicates); horizontal lines are the means from each panel. D1, DENV1; D2, DENV2; D3, DENV3; D4, DENV4; and D1-4, DENV1-4.
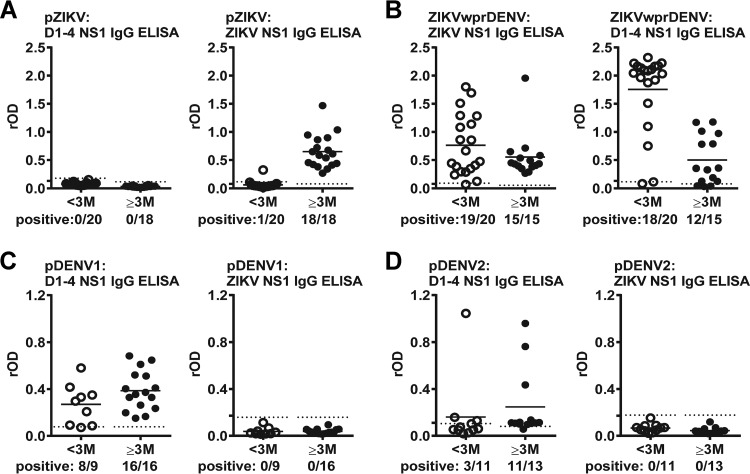
Comparison of convalescent- and postconvalescent-phase samples.
Since several of our panels included both convalescent-phase (<1 to 3 months PSO) and postconvalescent-phase (≥3 months PSO) samples, we next compared the results of DENV1-4 and ZIKV NS1 IgG ELISAs with different sampling times. For the pZIKV panel, we observed increased detection rates (5.0% versus 100%) and rOD values of ZIKV NS1 IgG ELISA when comparing convalescent- with postconvalescent-phase samples (Fig. 3A). This was different from the ZIKVwprDENV panel, which showed high detection rates (95.0% to 100%) for both convalescent- and postconvalescent-phase samples (Fig. 3B). A trend of increased detection rates from convalescent to postconvalescent phases was also observed for pDENV1 and pDENV2 panels (Fig. 3C and D), which was further supported by sequential samples from patients with primary DENV1 or DENV2 infection (see Fig. S1 in the supplemental material).
FIG 3.
Results of DENV1-4 and ZIKV NS1 IgG ELISAs for different panels over time. DENV1-4 and ZIKV NS1 IgG ELISAs were tested for convalescent- and postconvalescent-phase samples of pZIKV (A), ZIKVwprDENV (B), pDENV1 (C), and pDENV2 (D) panels. Dotted lines indicate cutoff rOD values. Data are the means from two experiments (each in duplicates); horizontal lines are the means from each panel. D1-4, DENV1-4.
The results of all samples tested with DENV1, DENV2, DENV3, DENV4, DENV1-4, and ZIKV NS1 IgG ELISAs are summarized in Table 2. For the statistical analysis comparing different panels, only one sample from each participant was included. The overall sensitivity of the DENV1-4 NS1 IgG ELISA was comparable to that of the DENV1 NS1 IgG ELISA (95.6% versus 94.5%, respectively) but higher than those of DENV2 and DENV3 NS1 IgG ELISAs (Table 3). Similarly, the overall specificity of the DENV1-4 NS1 IgG ELISA was comparable to that of the DENV1 NS1 IgG ELISA (89.5% versus 91.9%, respectively) but higher than that of the DENV2 NS1 IgG ELISA. For the ZIKV NS1 IgG ELISA, the sensitivity was 100% and specificity was 82.9%, mainly due to the cross-reactivity from the sDENV panel.
TABLE 2.
Results of NS1 IgG ELISAs in different serum/plasma panels
| NS1 IgG ELISA | No. of IgG+/total samples (%) in different serum/plasma panelsa |
|||||||
|---|---|---|---|---|---|---|---|---|
| Naive | pWNV | pDENV1 | pDENV2 | pDENV3 | pZIKV | sDENV | ZIKVwprDENV | |
| DENV1 | 1/55 (1.8) | 0/36 (0) | 25/25 (100) | 9/24 (37.5) | 10/10 (100) | 0/38 (0) | 64/67 (95.5) | 26/35 (74.3) |
| DENV2 | 1/49 (2.0) | 1/36 (2.7) | 13/25 (52) | 16/24b (66.7) | 6/10 (60) | 1/18 (5.6) | 20/20 (100) | 12/15 (80) |
| DENV3 | 0/40 (0) | 0/36 (0) | 8/25 (32) | 4/24 (16.7) | 10/10 (100) | 0/18 (0) | 20/20 (100) | 8/15 (53.3) |
| DENV4 | 0/40 (0) | 0/12 (0) | 0/25 (0) | 4/24 (16.7) | 1/10 (10) | 0/18 (0) | 17/20 (85) | 3/15 (20) |
| DENV1-4 | 0/55 (0) | 1/36 (2.7) | 24/25 (96) | 14/24 (58.3) | 10/10 (100) | 0/38 (0) | 64/67 (95.5) | 30/35 (85.7) |
| ZIKV | 0/48 (0) | 0/36 (0) | 0/25 (0) | 0/24 (0) | 0/10 (0) | 19/38c (50) | 32/67 (47.8) | 34/35 (97.1) |
pWNV, primary WNV infection; pDENV1, primary DENV1 infection; pDENV2, primary DENV2 infection; pDENV3, primary DENV3 infection; pZIKV, primary ZIKV infection; sDENV, secondary DENV infection; ZIKVwprDENV, ZIKV infection with previous DENV infection.
IgG+ rate equals 4/11 (36.4%) and 12/13 (92.3%) for convalescent and postconvalescent-phase samples, respectively.
IgG+ rate equals 1/20 (5%) and 18/18 (100%) for convalescent and postconvalescent-phase samples, respectively.
TABLE 3.
Sensitivity and specificity of different NS1 IgG ELISAsa
| Panelb | DENV1 NS1 ELISA |
DENV2 NS1 ELISA |
DENV3 NS1 ELISA |
DENV1-4 NS1 ELISA |
ZIKV NS1 ELISA |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| % Sens (95% CI) | % Spec (95% CI) | % Sens (95% CI) | % Spec (95% CI) | % Sens (95% CI) | % Spec (95% CI) | % Sens (95% CI) | % Spec (95% CI) | % Sens (95% CI) | % Spec (95% CI) | |
| Overall | 94.5 (89.8–96.9) | 91.9 (87.1–94.4) | 78.9 (67.8–84.5) | 87.3 (81.3–90.4) | 61.5 (48.3–68.3) | 92.7 (87.8–95.2) | 95.6 (91.4–97.8) | 89.5 (84.1–92.3) | 100 (67.8–84.5) | 82.9 (77.3–85.7) |
| pDENV1 | 100 (100–100) | NAc | 47.6 (26.3–58.5) | NA | 23.8 (5.6–33.1) | NA | 95.2 (86.1–99.9) | NA | NA | 100 (100–100) |
| pDENV2d | 71.4 (38.0–88.5) | NA | 100 (100–100) | NA | 42.9 (6.2–61.6) | NA | 100 (100–100) | NA | NA | 100 (100–100) |
| pDENV3 | 100 (100–100) | NA | 75 (32.6–96.7) | NA | 100 (100–100) | NA | 100 (100–100) | NA | NA | 100 (100–100) |
| sDENV | 94.9 (89.3–97.8) | NA | 100 (100–100) | NA | 100 (100–100) | NA | 94.9 (89.3–97.8) | NA | NA | 49.2 (36.4–55.7) |
| Naive | NA | 98.2 (94.7–100) | NA | 98.0 94.0–100) | NA | 100 (100–100) | NA | 100 (100–100) | NA | 100 (100–100) |
| pWNV | NA | 100 (100–100) | NA | 97.2 (91.9–100) | NA | 100 (100–100) | NA | 97.2 (91.9–100) | NA | 100 (100–100) |
| pZIKVd | NA | 100 (100–100) | NA | 94.4 (83.9–99.8) | NA | 100 (100–100) | NA | 100 (100–100) | 100 (100–100) | NA |
| ZIKVwprDENVd | NA | 40 (15.2–52.7) | NA | 20 (0–30.3) | NA | 46.7 (21.4–59.6) | NA | 20 (0–30.3) | 100 (100–100) | NA |
ELISAs, enzyme-linked immunosorbent assays; Sens, sensitivity; Spec, specificity; CI, confidence interval. For those with repeated samples, only one sample from each subject was included.
pDENV1, primary DENV1 infection; pDENV2, primary DENV2 infection; pDENV3, primary DENV3 infection; pWNV, primary WNV infection; pZIKV, primary ZIKV infection; sDENV, secondary DENV infection; ZIKVwprDENV, ZIKV infection with previous DENV infection.
NA, not applicable.
Samples at postconvalescent phase (≥3 months post-symptom onset) are presented.
Distinction between sDENV and ZIKVwprDENV panels.
Previously, we reported that the convalescent-phase sDENV panel not only recognized DENV1 NS1 protein but also ZIKV NS1 protein (95.8% and 66.7%, respectively, in the IgG ELISA); similarly, the ZIKVwprDENV panel recognized both ZIKV and DENV1 NS1 proteins (95.0% and 85.0%, respectively) (26). Using the rOD ratio of ZIKV NS1 to DENV1 NS1 with a cutoff at 0.24, we can distinguish ZIKVwprDENV and sDENV panels. We next tested the same panels (<3 months PSO) using the mixed DENV1-4 and ZIKV NS1 IgG ELISAs and found that the rOD ratio with a cutoff at 0.24 discriminated the ZIKVwprDENV and sDENV panels with a sensitivity of 94.1% and a specificity of 87.0% (Fig. 4A to C). We further tested the postconvalescent-phase (3 to 12 months PSO) sDENV and ZIKVwprDENV panels and found that a cutoff rOD ratio at 0.24 distinguished these two panels with a sensitivity of 91.7% and a specificity of 95.0% (Fig. 4D to F). Consistent with the high specificity, the rOD ratio was <0.24 in 17 of 18 samples of the sDENV panel at later time points (1.5 to 6 years PSO) (see Fig. S2 in the supplemental material).
FIG 4.
Results of DENV1-4 and ZIKV NS1 IgG ELISAs for sDENV and ZIKVwprDENV panels. DENV1-4 NS1 (A, D) and ZIKV NS1 (B, E) IgG ELISAs were tested for convalescent-phase (<3 month PSO [A to C]) and postconvalescent-phase (3 to 12 months PSO [D to F]) samples of sDENV and ZIKVwprDENV panels; the rOD ratio of ZIKV to DENV1-4 NS1 (C, F) is shown. Dotted lines indicate cutoff rOD values; dashed lines are the cutoff rOD ratio (0.24). Data are the means from two experiments (each in duplicates); horizontal lines are the means from each panel. The two-tailed Mann-Whitney test was used to compare two groups. D1-4, DENV1-4.
NS1 IgM ELISA.
NS1 IgM ELISAs were also established to test samples of <3 months PSO. The detection rate of the ZIKV NS1 IgM ELISA was 90%, whereas those of DENV1, DENV2, and DENV1-4 NS1 IgM ELISAs ranged from 44.4% to 55.6% (Table 4). Of the sDENV panel, there was less cross-reactivity to the ZIKV NS1 IgM ELISA (4%) than to the ZIKV NS1 IgG ELISA (66.7%) (Fig. 4B). The ZIKVwprDENV panel showed 55% positivity in the ZIKV NS1 IgM ELISA, whereas the sDENV panel showed 16.7% positivity in the DENV1 NS1 IgM ELISA.
TABLE 4.
Results of NS1 IgM ELISAs in different serum/plasma panels
| NS1 IgM ELISAa | No. of IgM+/total samples (%) in different serum/plasma panelsb |
|||||
|---|---|---|---|---|---|---|
| Naive | pDENV1 | pDENV2 | pZIKV | sDENV | ZIKVwprDENV | |
| ZIKV | 0/8 (0) | 0/5 (0) | 18/20 (90) | 1/24 (4) | 11/20 (55) | |
| DENV1 | 0/16 (0) | 5/9 (55.6)c | 0/20 (0) | 4/24 (16.7) | 3/20 (15) | |
| DENV2 | 0/4 (0) | 5/9 (55.6) | ||||
| DENV1-4 | 0/4 (0) | 4/9 (44.4)c | 5/9 (55.6) | |||
ELISA, enzyme-linked immunosorbent assay; pDENV1, primary DENV1 infection; pDENV2, primary DENV2 infection; pZIKV, primary ZIKV infection; sDENV, secondary DENV infection; ZIKVwprDENV, ZIKV infection with previous DENV infection.
Only samples collected <3 months post-symptom onset were tested for IgM.
IgM+ rate was 3/3 (100%) for samples collected <1 month.
DISCUSSION
In this study, we developed DENV2, DENV3, and DENV4 NS1 IgG ELISAs to detect convalescent- or postconvalescent-phase samples from RT–PCR-confirmed cases with DENV and ZIKV infections. Our findings that DENV1-4 NS1 IgG ELISAs detect different primary and secondary DENV infections and the combination with ZIKV NS1 IgG ELISAs distinguishes sDENV and ZIKVwprDENV panels with high sensitivity and specificity have important application to serodiagnosis and serosurveillance of DENV and ZIKV infections in regions where both viruses cocirculate.
We found that DENV1, DENV2, and DENV3 NS1 IgG ELISAs detected primary DENV infection of homologous serotype with a sensitivity (100%) higher than that for heterologous serotypes (23.8% to 100%) (Table 3). In contrast, DENV1, DENV2, DENV3, and DENV4 NS1 IgG ELISAs detected secondary DENV infection with a sensitivity of 94.9% to 100%. This was in agreement with our previous study showing that anti-NS1 antibodies following primary DENV infection recognized mainly the NS1 protein of the infecting serotype, whereas anti-NS1 antibodies after secondary DENV infection recognized those of multiple serotypes (13). This suggests the difficulty of using single NS1 IgG ELISAs to identify the infecting DENV serotype due to variable degrees of cross-reactivity after primary DENV infection and extensive cross-reactivity after secondary DENV infection. Notably, the mixed DENV1-4 NS1 IgG ELISA detected, though did not distinguish, different primary and secondary DENV infections with a sensitivity of 95.6% and a specificity of 89.5%, suggesting the convenience and feasibility of using mixed DENV1-4 NS1 IgG ELISAs alone to detect different DENV infections (within the DENV serocomplex) rather than distinguishing different serotypes.
A recent study reported an increased risk of severe disease accompanying breakthrough DENV infection among dengue-naive individuals receiving Dengvaxia, the currently licensed chimeric yellow fever dengue tetravalent dengue vaccine, and highlighted the importance of determining the DENV immune status prior to administering Dengvaxia and identifying DENV-naive individuals among Dengvaxia recipients as a risk group in regions of endemicity (36–39). Another study using a mixed DENV1-4 NS1 IgG ELISA to determine DENV immune status among Dengvaxia recipients reported high sensitivity and specificity. However, limited numbers of other flavivirus controls (4 ZIKV and 3 WNV samples) showed positivity (40), raising concerns about its practical application in regions of endemicity where multiple flaviviruses are prevalent. With >124 control samples, including naive, pZIKV, and pWNV panels, our DENV1-4 NS1 IgG ELISA showed specificity at levels to suggest it might be a useful tool to determine DENV immune status in regions of endemicity.
In addition, we observed a trend of increased detection rates for the NS1 IgG ELISA of convalescent to postconvalescent phases from primary infection panels (pZIKV, pDENV1, and pDENV2) (Fig. 3A, C, and D) as opposed to the high detection rates of both convalescent and postconvalescent phases from repeated infection panels (sDENV and ZIKVwprDENV) (Fig. 2B and 3B). This was in agreement with a recent study of a blockade-of-binding ZIKV NS1 ELISA (25), in which the detection rate increased (77.4% to 98.3%) from early convalescent to late convalescent phases for the pZIKV panel and remained high (92.3% to 94.4%) for the ZIKVwprDENV panel. Our ZIKV NS1 IgG ELISA had an overall sensitivity of 100%, which is higher or compatible with those reported recently (79% to 100%) using the Euroimmun ZIKV NS1 IgG ELISA kit (41–44).
Consistent with our previous report that sDENV and ZIKVwprDENV panels recognize both DENV1 and ZIKV NS1 proteins, we found that these two panels recognize both DENV1-4 and ZIKV NS1 proteins in IgG ELISAs (Fig. 4; Table 3). Moreover, we found that the rOD ratio of DENV1-4 NS1 to ZIKV NS1 distinguished these two panels with a sensitivity of 94.1% to 91.7% and a specificity of 87.0% to 95.0% (Fig. 4).
According to the current CDC guidelines, positive or equivocal IgM tests based on E protein require PRNT (5, 6), which can confirm pZIKV infection but not in those with previous flaviviral infections, including secondary DENV and ZIKVwprDENV infections. Although a recent study reported reduced cross-neutralization against ZIKV among those with secondary DENV infection more than 6 months PSO, 23% still cross-neutralize ZIKV (45). Another study of antibody cross-neutralization patterns showed that ZIKV lies outside the DENV serocomplex, suggesting the neutralizing antibody titers distinguish ZIKV and DENV infections (46). However, the sensitivity and specificity of neutralizing antibody titers to differentiate different DENV and ZIKV infections, especially sDENV and ZIKVwprDENV panels, were not reported. Moreover, the feasibility of using time-consuming neutralization tests for serodiagnosis and serosurveillance in the field remains a challenge.
Our combined DENV1-4 and ZIKV NS1 IgG ELISAs could be applied to routine serological tests for detecting ZIKV infection both in regions of DENV endemicity and in regions where it is nonendemic and to serosurveillance and Zika pregnancy studies to understand the epidemiology, transmission, and complication of ZIKV infection during pregnancy (47, 48). Given that the CZS may affect both microcephalic and normocephalic infants during their growth and development (48, 49), our IgG assays could be used in retrospective studies to understand the contribution of pZIKV infection alone or ZIKVwprDENV infection to the full spectrum of CZS. Additionally, these ELISAs are simple, inexpensive, and readily applicable to field sites in developing countries. They can be further developed into rapid tests or high-throughput formats for various clinical or field studies.
There are several limitations. First, the sample size in each panel with well-documented infections is small; future studies involving a larger sample size, including more sequential samples, are needed to validate these observations. Second, as limited samples of PSO of <3 months were available from patients with primary DENV infection (Table 1), the study focused on NS1-based IgG ELISAs rather than IgM ELISAs. The detection rates of DENV1, DENV2, and DENV1-4 NS1 IgM ELISAs were low (44.4% to 55.6%), whereas that of the ZIKV-NS1 IgM ELISA (90%) was higher than those reported previously (41–43). Future studies with a larger size of samples with PSO of <3 months are warranted. Third, other control panels including samples from uninfected pregnant women and individuals with IgG antibodies to chikungunya virus, yellow fever virus, Plasmodium species, and Japanese encephalitis virus should be included in future studies to validate the specificity of our assays. Fourth, since ZIKV has spread to regions where multiple flaviviruses cocirculate, serological tests that can distinguish ZIKV from other medically important flaviviruses, such as JEV, WNV, YFV, and TBEV, remain to be explored (50, 51). Additionally, given the success of several flaviviral vaccines and ongoing vaccine trials in regions of flavivirus endemicity, the development serological tests to distinguish ZIKV infection from immunizations with different flaviviral vaccines, including DENV, JEV, YFV, and TBEV vaccines, is warranted (50, 51).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants R01AI110769-01 (W.-K.W.), R21AI135292-01A1 (W.-K.W.), R01 AI099631 (A.B.), U54 AI065359 (A.B.), P01 AI106695 (E.H.), and U19 AI118610 (E.H.) from the National Institute of Allergy and Infectious Diseases, NIH, P30GM114737 from the National Institute of General Medical Sciences, NIH, MOHW107-TDU-B-212-123006 (J.-J.-T.) from the Ministry of Health and Welfare, Taiwan, NHRI-MR-107-PP-38 (J.-J.-T.) from the National Health Research Institute, Taiwan, and in part by a gift from the Outpatient Parenteral Antimicrobial Therapy Outcomes Registry in honor of the late Alan Tice. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01464-18.
REFERENCES
- 1.Petersen LR, Jamieson DJ, Powers AM, Honein MA. 2016. Zika virus. N Engl J Med 374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 2.Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, Carcelen AC, Ott CT, Sheffield JS, Ferguson NM, Cummings DA, Metcalf CJ, Rodriguez-Barraquer I. 2016. Assessing the global threat from Zika virus. Science 353:aaf8160. doi: 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauer F, Riesen M, Reveiz L, Oladapo OT, Martínez-Vega R, Porgo TV, Haefliger A, Broutet NJ, Low N, WHO Zika Causality Working Group. 2017. Zika virus infection as a cause of congenital brain abnormalities and Guillain-Barré syndrome: systematic review. PLoS Med 14:e1002203. doi: 10.1371/journal.pmed.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. 2017. An update on Zika virus infection. Lancet 390:2099–2109. doi: 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 5.CDC. 2017. Guidance for U.S. laboratories testing for Zika virus infection. CDC, Atlanta, GA. http://www.cdc.gov/zika/laboratories/lab-guidance.html.
- 6.Rabe IB, Staples JE, Villanueva J, Hummel KB, Johnson JA, Rose L, Hills S, Wasley A, Fischer M, Powers AM. 2016. Interim guidance for interpretation of Zika virus antibody test results. MMWR Morb Mortal Wkly Rep 65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 7.Paz-Bailey G, Rosenberg ES, Doyle K, Munoz-Jordan J, Santiago GA, Klein L, Perez-Padilla J, Medina FA, Waterman SH, Gubern CG, Alvarado LI, Sharp TM. 2018. Persistence of Zika virus in body fluids - final report. N Engl J Med 379:1234–1243. doi: 10.1056/NEJMoa1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. 2011. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks RB, Carlos MP, Myers RA, White MG, Bobo-Lenoci T, Aplan D, Blythe D, Feldman KA. 2016. Likely sexual transmission of Zika virus from a man with no symptoms of infection - Maryland, 2016. MMWR Morb Mortal Wkly Rep 65:915–916. doi: 10.15585/mmwr.mm6534e2. [DOI] [PubMed] [Google Scholar]
- 10.Pierson TC, Diamond MS. 2013. Flaviviruses, p 747–794. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott William & Wilkins, Philadelphia, PA. [Google Scholar]
- 11.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. 2000. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol 38:1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson AJ, Martin DA, Karabatsos N, Roehrig JT. 2000. Detection of anti-arboviral immunoglobulin G by using a monoclonal antibody-based capture enzyme-linked immunosorbent assay. J Clin Microbiol 38:1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C-Y, Tsai W-Y, Lin S-R, Kao C-L, Hu H-P, King C-C, Wu H-C, Chang G-J, Wang W-K. 2008. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol 82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson BW, Kosoy O, Martin DA, Noga AJ, Russell BJ, Johnson AA, Petersen LR. 2005. West Nile virus infection and serologic response among persons previously vaccinated against yellow fever and Japanese encephalitis viruses. Vector Borne Zoonotic Dis 5:137–145. doi: 10.1089/vbz.2005.5.137. [DOI] [PubMed] [Google Scholar]
- 16.van Meer MPA, Mögling R, Klaasse J, Chandler FD, Pas SD, van der Eijk AA, Koopmans MPG, Reusken CBEM, GeurtsvanKessel CH. 2017. Re-evaluation of routine dengue virus serology in travelers in the era of Zika virus emergence. J Clin Virol 92:25–31. doi: 10.1016/j.jcv.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Felix AC, Souza NCS, Figueiredo WM, Costa AA, Inenami M, da Silva RMG, Levi JE, Pannuti CS, Romano CM. 2017. Cross reactivity of commercial anti-dengue immunoassays in patients with acute Zika virus infection. J Med Virol 89:1477–1479. doi: 10.1002/jmv.24789. [DOI] [PubMed] [Google Scholar]
- 18.Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, Mongkolsapaya J, Screaton GR. 2016. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol 17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. 2016. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanha PMS, Nascimento EJM, Braga C, Cordeiro MT, de Carvalho OV, de Mendonça LR, Azevedo EAN, França RFO, Dhalia R, Marques ETA. 2017. Dengue virus (DENV)-specific antibodies enhance Brazilian Zika virus (ZIKV) infection. J Infect Dis 215:781–785. doi: 10.1093/infdis/jiw638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, García-Sastre A, Krammer F, Lim JK. 2017. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons CP, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. 2016. Specificity, cross-reactivity and function of antibodies elicited by Zika virus infection. Science 353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 23.Halstead SB, O'Rourke EJ. 1977. Antibody-enhanced dengue virus infection in primate leukocytes. Nature 265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- 24.Balmaseda A, Stettler K, Carrera RM, Collado D, Jin X, Zambrana JV, Jaconi S, Cameroni E, Saborio S, Rovida F, Percivalle E, Ijaz S, Dicks S, Ushiro-Lumb I, Barzon L, Siqueira P, Brown DWG, Baldanti F, Tedder R, Zambon M, Bispo de Filippis AM, Harris E, Corti D, 2017. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A 114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balmaseda A, Zambrana JV, Collado D, García N, Saborío S, Elizondo D, Mercado JC, Gonzalez K, Cerpas C, Nuñez A, Corti D, Waggoner JJ, Kuan G, Burger-Calderon R, Harris E. 2018. Comparison of four serological methods and two reverse transcription-PCR assays for diagnosis and surveillance of Zika virus infection. J Clin Microbiol 56:e01785-17. doi: 10.1128/JCM.01785-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai WY, Youn HH, Brites C, Tsai JJ, Tyson J, Pedroso C, Drexler JF, Stone M, Simmons G, Busch MP, Lanteri M, Stramer SL, Balmaseda A, Harris E, Wang WK. 2017. Distinguishing secondary dengue virus infection from Zika virus infection with previous dengue by combination of three simple serological tests. Clin Infect Dis 65:1829–1836. doi: 10.1093/cid/cix672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuan G, Gordon A, Avilés W, Ortega O, Hammond SN, Elizondo D, Nuñez A, Coloma J, Balmaseda A, Harris E. 2009. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narvaez F, Gutierrez G, Perez MA, Elizondo D, Nuñez A, Balmaseda A, Harris E. 2011. Evaluation of the Traditional and revised WHO classifications of dengue disease severity. PLoS Negl Trop Dis 5:e1397. doi: 10.1371/journal.pntd.0001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stramer SL, Fang CT, Foster GA, Wagner AG, Brodsky JP, Dodd RY. 2005. West Nile virus among blood donors in the United States, 2003 and 2004. N Engl J Med 353:451–459. doi: 10.1056/NEJMoa044333. [DOI] [PubMed] [Google Scholar]
- 30.Wang WK, Chen HL, Yang CF, Hsieh SC, Juan CC, Chang SM, Yu CC, Lin LH, Huang JH, King CC. 2006. Slower rates of clearance of viral load and virus-containing immune complexes in patients with dengue hemorrhagic fever. Clin Infect Dis 43:1023–1030. doi: 10.1086/507635. [DOI] [PubMed] [Google Scholar]
- 31.Tsai W-Y, Durbin A, Tsai J-J, Hsieh S-C, Whitehead S, Wang W-K. 2015. Complexity of neutralization antibodies against multiple dengue viral serotypes after heterotypic immunization and secondary infection revealed by in-depth analysis of cross-reactive antibodies. J Virol 89:7348–7362. doi: 10.1128/JVI.00273-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai CY, Williams KL, Wu YC, Knight S, Balmaseda A, Harris E, Wang WK. 2013. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl Trop Dis 7:e2451. doi: 10.1371/journal.pntd.0002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai JJ, Liu CK, Tsai WY, Liu LT, Tyson J, Tsai CY, Lin PC, Wang WK. 2018. Seroprevalence of dengue in two districts of Kaohsiung city after the largest dengue outbreak in Taiwan since World War II. PLoS Negl Trop Dis 12:e0006879. doi: 10.1371/journal.pntd.0006879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frey A, Di Canzio J, Zurakowski D. 1998. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods 221:35–41. doi: 10.1016/S0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
- 35.Paldanius M, Bloigu A, Leinonen M, Saikku P. 2003. Measurement of Chlamydia pneumoniae-specific immunoglobulin A (IgA) antibodies by the microimmunofluorescence (MIF) method: comparison of seven fluorescein-labeled anti-human IgA conjugates in an in-house MIF test using one commercial MIF and one enzyme immunoassay kit. Clin Diagn Lab Immunol 10:8–12. doi: 10.1128/CDLI.10.1.8-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, Moodie Z, Westling T, Mascareñas C, Frago C, Cortés M, Chansinghakul D, Noriega F, Bouckenooghe A, Chen J, Ng SP, Gilbert PB, Gurunathan S, DiazGranados CA. 2018. Effect of Dengue serostatus on dengue vaccine safety and efficacy. N Engl J Med 379:327–340. doi: 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
- 37.Halstead SB. 2017. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 35:6355–6358. doi: 10.1016/j.vaccine.2017.09.089. [DOI] [PubMed] [Google Scholar]
- 38.Iacobucci G. 2018. WHO recommends additional tests for Sanofi's dengue vaccine after safety concerns. BMJ 361:k1765. doi: 10.1136/bmj.k1765. [DOI] [PubMed] [Google Scholar]
- 39.Normile D. 2017. Safety concerns derail dengue vaccination program. Science 358:1514–1515. doi: 10.1126/science.358.6370.1514. [DOI] [PubMed] [Google Scholar]
- 40.Nascimento EJM, George JK, Velasco M, Bonaparte MI, Zheng L, DiazGranados CA, Marques ETA, Huleatt JW. 2018. Development of an anti-dengue NS1 IgG ELISA to evaluate exposure to dengue virus. J Virol Methods 257:48–57. doi: 10.1016/j.jviromet.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Steinhagen K, Probst C, Radzimski C, Schmidt-Chanasit J, Emmerich P, van Esbroeck M, Schinkel J, Grobusch MP, Goorhuis A, Warnecke JM, Lattwein E, Komorowski L, Deerberg A, Saschenbrecker S, Stöcker W, Schlumberger W. 2016. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: a multicohort study of assay performance, 2015 to 2016. Euro Surveill 21:30426. doi: 10.2807/1560-7917.ES.2016.21.50.30426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lustig Y, Zelena H, Venturi G, Van Esbroeck M, Rothe C, Perret C, Koren R, Katz-Likvornik S, Mendelson E, Schwartz E. 2017. Sensitivity and kinetics of an NS1-based Zika virus enzyme-linked immunosorbent assay in Zika virus-infected travelers from Israel, the Czech Republic, Italy, Belgium, Germany, and Chile. J Clin Microbiol 55:1894–1901. doi: 10.1128/JCM.00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasquier C, Joguet G, Mengelle C, Chapuy-Regaud S, Pavili L, Prisant N, Izopet J, Bujan L, Mansuy JM. 2018. Kinetics of anti-ZIKV antibodies after Zika infection using two commercial enzyme-linked immunoassays. Diagn Microbiol Infect Dis 90:26–30. doi: 10.1016/j.diagmicrobio.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Safronetz D, Sloan A, Stein DR, Mendoza E, Barairo N, Ranadheera C, Scharikow L, Holloway K, Robinson A, Traykova-Andonova M, Makowski K, Dimitrova K, Giles E, Hiebert J, Mogk R, Beddome S, Drebot M. 2017. Evaluation of 5 commercially available Zika virus immunoassays. Emerg Infect Dis 23:1577–1580. doi: 10.3201/eid2309.162043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins MH, McGowan E, Jadi R, Young E, Lopez CA, Baric RS, Lazear HM, de Silva AM. 2017. Lack of durable cross-neutralizing antibodies against Zika virus from dengue virus infection. Emerg Infect Dis 23:773–781. doi: 10.3201/eid2305.161630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montoya M, Collins M, Dejnirattisai W, Katzelnick LC, Puerta-Guardo H, Jadi R, Schildhauer S, Supasa P, Vasanawathana S, Malasit P, Mongkolsapaya J, de Silva AD, Tissera H, Balmaseda A, Screaton G, de Silva AM, Harris E. 2018. Longitudinal analysis of antibody cross-neutralization following Zika virus and dengue virus infection in Asia and the Americas. J Infect Dis 218:536–545. doi: 10.1093/infdis/jiy164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, Ellington SR, Shapiro-Mendoza CK, Oduyebo T, Fine AD, Brown CM, Sommer JN, Gupta J, Cavicchia P, Slavinski S, White JL, Owen SM, Petersen LR, Boyle C, Meaney-Delman D, Jamieson DJ, U.S. Zika Pregnancy Registry Collaboration. 2017. Birth defects among fetuses and infants of U.S. women with evidence of possible Zika virus infection during pregnancy. JAMA 317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 48.Moore CA, Staples JE, Dobyns WB, Pessoa A, Ventura CV, Fonseca EB, Ribeiro EM, Ventura LO, Neto NN, Arena JF, Rasmussen SA. 2017. Characterizing the pattern of anomalies in congenital Zika syndrome for pediatric clinicians. JAMA Pediatr 171:288–295. doi: 10.1001/jamapediatrics.2016.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Linden V, Pessoa A, Dobyns W, Barkovich AJ, Júnior HV, Filho EL, Ribeiro EM, Leal MC, Coimbra PP, Aragão MF, Verçosa I, Ventura C, Ramos RC, Cruz DD, Cordeiro MT, Mota VM, Dott M, Hillard C, Moore CA. 2016. Description of 13 infants born during October 2015–January 2016 with congenital Zika virus infection without microcephaly at birth–Brazil. MMWR Morb Mortal Wkly Rep 65:1343–1348. doi: 10.15585/mmwr.mm6547e2. [DOI] [PubMed] [Google Scholar]
- 50.Munoz-Jordan JL. 2017. Diagnosis of Zika Virus infections: challenges and opportunities. J Infect Dis 216:S951–S956. doi: 10.1093/infdis/jix502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons G, Stone M, Busch MP. 2018. Arbovirus diagnostics: from bad to worse due to expanding dengue virus vaccination and Zika virus epidemics. Clin Infect Dis 66:1181–1183. doi: 10.1093/cid/cix972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.