The objective of the present study was to evaluate the value of the PCR cycle threshold (CT) for predicting the recurrence/severity of infection compared to that of toxin detection plus clinical variables. First episodes of Clostridium difficile infection (CDI) diagnosed during 2015 at our institution were included.
KEYWORDS: Clostridium difficile, PCR CT, retrospective study, outcome, predictive model, recurrence
ABSTRACT
The objective of the present study was to evaluate the value of the PCR cycle threshold (CT) for predicting the recurrence/severity of infection compared to that of toxin detection plus clinical variables. First episodes of Clostridium difficile infection (CDI) diagnosed during 2015 at our institution were included. Samples were tested for glutamate dehydrogenase (GDH) and toxin A/B by use of a single enzyme immunoassay (EIA). The Xpert C. difficile PCR assay was performed on GDH-positive samples. Medical data were reviewed by investigators blinded to diagnostic results for comparison of patients with and without recurrence or a poor outcome (severe/severe-complicated CDI episodes and all-cause death). We generated two sets of predictive models by incorporating the presence of a positive toxin EIA (“EIA-including model”) or the optimal PCR CT cutoff value (“PCR-including model”) into the clinical variables. Among 227 episodes of CDI included in the study, the rates of recurrence and poor outcome were 15.8% and 30.8%, respectively. The mean PCR CT was lower for episodes with recurrence (24.00 ± 3.28 versus 26.02 ± 4.54; P = 0.002) or a poor outcome (24.9 ± 4.24 versus 26.05 ± 4.47; P = 0.07). The optimal cutoff value for recurrence was 25.65 (sensitivity, 77.8% [95% confidence interval {CI}, 60.9 to 89.9]; and specificity, 46.6% [95% CI, 39.4 to 53.9]). The area under the receiver operator characteristics curve (auROC) for the “PCR-including model” was similar to that for the “EIA-including model” (0.785 versus 0.775, respectively). The optimal PCR CT value for poor outcome was 27.55 (sensitivity, 78.6% [95% CI, 67.1 to 87.5]; and specificity, 35.7% [95% CI, 28.2 to 43.7]). The auROC of the “PCR-including model” was again similar to that of the “EIA-including model” (0.804 versus 0.801). Despite the inverse correlation between PCR CT and the risk of CDI recurrence/severity, this determination does not meaningfully increase the predictive value of clinical variables plus toxin EIA.
INTRODUCTION
The optimal diagnostic approach for Clostridium difficile infection (CDI) is still a controversial subject. The latest guidelines endorsed by the Infectious Diseases Society of America (IDSA) and the Society for Healthcare Epidemiology of America (SHEA) recommended a multistep algorithm (1) as the most effective strategy for diagnosing CDI and minimizing overdiagnosis among colonized individuals. The algorithm should start with a rapid and sensitive screening test with a high negative predictive value (NPV), such as a glutamate dehydrogenase (GDH) enzyme immunoassay (EIA) or nucleic acid amplification test (NAAT), and samples with a positive screening test should subsequently be retested with a toxin A/B EIA to identify patients infected with a toxigenic strain, who have the highest likelihood of clinically relevant CDI and need for specific treatment (2). A recent prospective study (3) concluded that patients with a positive molecular test but a negative toxin A/B EIA had outcomes comparable to those of patients with no evidence of CDI. On the basis of this and other studies (4), it may be concluded that half of patients with a positive C. difficile PCR test are likely overdiagnosed and exposed to unnecessary treatment. However, the results obtained by other groups support an approach based on the unique diagnostic performance of NAAT, since they suggest that the PCR cycle threshold (CT) may accurately predict the existence of free toxin (5, 6) or be used as a predictor of a poor outcome (7).
In a recently published retrospective cohort study (8), we found that both the occurrence of severe or complicated forms of CDI and recurrence were significantly more common among patients with a positive EIA for both GDH and toxin A/B than among those with GDH-positive, toxin-negative samples for whom the diagnosis of CDI was made by a positive PCR-based assay. In the present study, our aim was to evaluate if toxin B PCR CT adds something to the combination of clinical variables and free toxin detection by EIA in predicting recurrence or a poor CDI outcome.
(This study was partially presented at the 28th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], Madrid, Spain, 21 to 24 April 2018.)
MATERIALS AND METHODS
Study population and setting.
This retrospective cohort study was performed at the 12 de Octubre University Hospital (Madrid, Spain), a 1,360-bed tertiary care hospital. Incident episodes of CDI were identified by reviewing all GDH-positive stool samples sent to the microbiology laboratory from 1 January to 31 December 2015. We included cases diagnosed in adult patients (≥18 years), and in cases of recurrence, only the first episode of CDI was considered. Episodes of CDI without adequate information were excluded, as well as those without a traceable 8-week follow-up after the end of treatment and those for which the CT result for the tcdB PCR was not available. More details about the cohort on which the present study is based are described elsewhere (8). The local clinical ethics committee approved the study protocol. The need for specific informed consent was waived owing to its retrospective nature.
Study definitions.
“CDI” was defined as the occurrence of diarrhea in the presence of a positive stool test for toxigenic C. difficile. “Mild or moderate CDI” was defined as diarrhea without systemic symptoms, leukocytosis, or significant renal failure (9). “Severe CDI” was defined by the presence of systemic symptoms of infection and/or leukocytosis (white blood cell count [WBC] of ≥15,000 cells/ml) or significant acute renal failure (≥1.5-fold increase of serum creatinine from the premorbid level) (9), only if these features were deemed to be attributable to CDI. “Severe-complicated CDI” was defined by the presence of severe disease accompanied by life-threatening conditions, such as ileus, toxic megacolon, refractory hypotension, and/or multiorgan failure attributable to CDI (9). “Recurrent CDI” was defined as the recurrence of CDI symptoms within the first 8 weeks following the completion of an effective course of therapy (with complete resolution of symptoms) in the presence of a positive laboratory test for C. difficile (9). “Poor outcome” was defined as the occurrence of a severe or severe-complicated first CDI episode and/or all-cause death within the first 8 weeks after the end of treatment. Other study definitions are detailed elsewhere (8).
Study design and outcomes.
We compared epidemiological variables, clinical characteristics, anti-C. difficile therapies, and diagnostic test results between patients with and without recurrence and with and without poor outcomes. Clinical data were retrospectively reviewed through a standardized case report form by two independent investigators with long-term clinical experience with CDI. In cases of discrepancy, a third expert was consulted. All of these investigators remained blinded to the toxin A/B EIA and CT results. Criteria used for CDI evaluation were consistent across all investigators. Investigators evaluated whether CDI-related symptoms were the main reason for consultation, the severity of the CDI episode, and, in cases of concurrent infection, if the development of complications or death could be attributable to CDI.
Microbiological methods.
Unformed stools (taking the shape of the container) were processed immediately or, if that was logistically unfeasible, kept at 4°C for 24 to 48 h until processing. Samples were simultaneously tested for GDH and toxin A/B by use of a single enzyme immunoassay (TechLab C. diff Quik Chek Complete; Inverness Medical Innovations, Princeton, NJ, USA). For samples with discordant results (GDH positive but toxin A/B negative), toxigenicity was confirmed by use of the Xpert C. difficile PCR assay (Cepheid, Sunnyvale, CA, USA), a real-time PCR assay targeting the tcdB gene of C. difficile. For investigational purposes, the Xpert C. difficile PCR assay was also performed on samples with a toxin A/B EIA-positive result. The quantitative CT result was recorded from the assay software.
Statistical analysis.
Quantitative data are shown as means ± standard deviations (SD) or medians with interquartile ranges (IQR). Qualitative variables are given as absolute and relative frequencies. Categorical variables were compared using the chi-square test or Fisher’s exact test, as appropriate. Student’s t test or the Mann-Whitney U test was applied for continuous variables. Episodes of CDI without adequate information were excluded, as well as those without traceable follow-up. Other missing data were excluded. Tukey box plots were used to depict the differences in mean tcdB gene CT values between patients with and without recurrence or a poor outcome. Optimal tcdB gene CT cutoff values were calculated by using the Youden index (J = sensitivity + specificity − 1) for the two study outcomes. CT cutoffs for both outcomes were also determined by fixing the sensitivity to 95% (see the supplemental material). Next, we explored the potential gain in the capacity to predict these outcomes that might result from incorporating data from tcdB gene CT values into the clinical prediction process. Backward stepwise logistic regression analysis was used to construct two sets of predictive models for recurrence and poor outcome; one of them was based exclusively on clinical variables ascertained at the time of symptom onset and showing P values of <0.05 at the univariate level (i.e., “clinical model”), whereas the second set incorporated the result of the toxin A/B EIA into the model (i.e, “EIA-including model”). Finally, we forced the tcdB gene CT value into each of these models dichotomized according to the previously established optimal cutoff point (i.e., “PCR-including model”). The goodness-of-fit and discriminative capacity values for the resulting models were assessed by means of the Hosmer-Lemeshow test and the area under the receiver operator characteristics curve (auROC), respectively. Obtained auROCs were compared to assess the incremental discriminative capacity that resulted from incorporating information derived from the PCR assay. In addition, the sensitivities, specificities, and likelihood ratios of these models were calculated for different thresholds. Because the Youden index has some limitations, we additionally performed two sets of predictive models based on different PCR CT cutoffs: a prespecified high-sensitivity threshold of >95% for each outcome and the CT cutoff of 26.35 reported by Senchyna et al. for predicting free-toxin status (5).
Associations were expressed by adjusted odds ratios (aORs) and 95% confidence intervals (CIs). All the significance tests were two-tailed. Statistical analysis was performed using SPSS, version 22.0 (IBM Corp., Armonk, NY, USA), and graphics were generated with Prism, version 6.0 (GraphPad Software Inc., La Jolla, CA).
RESULTS
Study population.
Overall, 3,846 stool specimens were sent to the microbiology laboratory for C. difficile detection during the study period. A total of 231 episodes of CDI were identified, of which 227 (98.3%) had the tcdB gene CT value available and were therefore included in the present analysis (see Fig. S1 in the supplemental material). There were 8 episodes that had been preceded by a previous CDI diagnosis, but they were included because the time intervals between both diagnoses were longer than 8 months (median of 20 [IQR, 5.5 to 88] months).
Predictive models for recurrence.
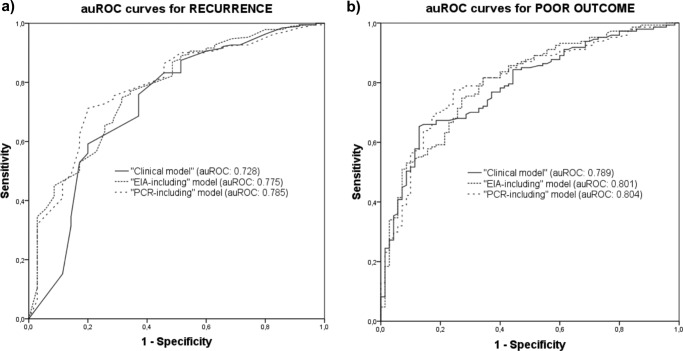
In our cohort, the rate of recurrence within the first 8 weeks after the completion of an effective course of therapy for CDI was 15.8% (36/227 episodes). The univariate comparison between patients with and without recurrence is depicted in Table 1. The mean tcdB gene CT was lower for episodes with recurrence (24.00 ± 3.28 versus 26.02 ± 4.54; P = 0.002) (Fig. 1a). Various predictive models were constructed. The “clinical model” was based on the following variables: presence of chronic renal failure (aOR, 2.78; 95% CI, 1.10 to 6.99; P = 0.03), number of hospital admissions in the previous 6 months (aOR [per unitary increment], 1.39; 95% CI, 1.06 to 1.84; P = 0.017), fulfillment of the diagnostic criteria for severe CDI in the initial episode (aOR, 3.79; 95% CI, 1.59 to 9.08; P = 0.003), and CDI-attributable symptoms as the main reason for consultation (aOR, 2.43; 95% CI, 1.08 to 5.46; P = 0.031) (Table 2). The optimal tcdB gene CT cutoff value for recurrence was set at 25.65, yielding a sensitivity of 77.8% (95% CI, 60.9 to 89.9%), a specificity of 46.6% (95% CI, 39.4 to 53.9%), a positive predictive value (PPV) of 21.5% (95% CI, 18.1 to 25.5%), and a negative predictive value (NPV) of 91.8% (95% CI, 85.6 to 95.4%). Next, we added to this model the presence of a positive result for A/B toxin EIA (aOR, 3.52; 95% CI, 1.49 to 8.31; P = 0.004) and a tcdB gene CT value of <25.65 (aOR, 3.41; 95% CI, 1.35 to 8.61; P = 0.009) to obtain the “EIA-including model” and the “PCR-including model,” respectively (Table 2). The auROC for each of these models is depicted in Fig. 2a. The auROC for the “PCR-including model” was only slightly superior to that resulting from the “EIA-including model” (0.785 versus 0.775, respectively), suggesting a low incremental predictive value.
TABLE 1.
Univariate analysis of risk factors predicting recurrence of CDIh
| Factor | Value |
Univariate analysis result |
||
|---|---|---|---|---|
| No recurrence (n = 191) | Recurrence (n = 36) | OR (95% CI) | P value | |
| Male gender (n [%]) | 94 (49.2) | 13 (36.1) | 1.71 (0.82–3.58) | 0.14 |
| Age (yr) (mean ± SD) | 63.32 ± 19.48 | 68.84 ± 17.78 | 3.49 (−12.4–1.36) | 0.11 |
| CCI (median [IQR]) | 4 (2–7) | 5 (4–6) | 0.59 | |
| Diabetes mellitus (n [%]) | 27 (14.1) | 8 (22.2) | 1.73 (0.71–4.20) | 0.21 |
| Active malignancya (n [%]) | 33 (17.3) | 7 (19.4) | 1.15 (0.46–2.86) | 0.75 |
| Hematological diseasea (n [%]) | 12 (6.3) | 1 (2.8) | 0.42 (0.05–3.38) | 0.69 |
| Chemotherapya (n [%]) | 23 (12.0) | 4 (11.1) | 0.91 (0.29–2.82) | 1.00 |
| Hematopoietic stem cell transplantationa (n [%]) | 2 (1.0) | 1 (2.8) | 2.70 (0.24–30.59) | 0.40 |
| Solid organ transplantation (n [%]) | 15 (7.9) | 6 (16.7) | 2.34 (0.84–6.52) | 0.11 |
| Chronic renal failure (n [%]) | 29 (15.2) | 10 (27.8) | 2.15 (0.94–4.92) | 0.06 |
| Cirrhosis (n [%]) | 15 (7.9) | 0 (0.0) | 0.83 (0.78–0.88) | 0.13 |
| Concurrent corticosteroid therapy (any dose) (n [%]) | 33 (17.3) | 8 (22.2) | 1.37 (0.57–3.27) | 0.48 |
| Concurrent corticosteroid therapy (high dose)b (n [%]) | 9 (4.7) | 3 (8.3) | 1.84 (0.47–7.15) | 0.41 |
| Other immunosuppression (n [%]) | 36 (18.8) | 7 (19.4) | 1.04 (0.42–2.56) | 0.93 |
| Inflammatory bowel disease (n [%]) | 12 (6.3) | 4 (11.1) | 1.86 (0.56–6.14) | 0.29 |
| Cognitive impairment (n [%]) | 12 (6.3) | 1 (2.8) | 0.42 (0.05–3.38) | 0.70 |
| Admission to long-term care facility (n [%]) | 13 (6.8) | 0 (0.0) | 0.83 (0.78–0.88) | 0.23 |
| PPI therapyc (n [%]) | 115 (60.2) | 26 (72.2) | 1.72 (0.78–3.76) | 0.17 |
| H2 blocker therapyc (n [%]) | 9 (4.7) | 2 (5.6) | 1.19 (0.24–5.75) | 0.68 |
| Prior hospital admissiond (n [%]) | 94 (49.2) | 23 (65.7) | 1.98 (0.93–4.20) | 0.07 |
| No. of admissionsd (median [IQR]) | 1 (0–1) | 1 (0–2) | 0.024 | |
| Prior antibiotic therapy (n [%]) | ||||
| Within 4 weeks prior to diagnosis | 147 (77.0) | 29 (80.6) | 1.24 (0.51–3.02) | 0.63 |
| Within 12 weeks prior to diagnosis | 167 (87.4) | 33 (91.7) | 1.58 (0.45–5.55) | 0.58 |
| CDI symptoms as the main reason for consultatione (n [%]) | 84 (44.0) | 23 (63.9) | 2.25 (1.08–4.71) | 0.028 |
| Leukocytosisf (n [%]) | 41/171 (24.0) | 13/36 (36.1) | 1.79 (0.83–3.85) | 0.13 |
| White blood cell count (median [IQR]) | 8,900 (6,600–14,800) | 11,550 (6,775–16,800) | 0.22 | |
| Fever (n [%]) | 68/191 (35.6) | 15/36 (41.7) | 1.29 (0.62–2.67) | 0.48 |
| Acute renal failureg (n [%]) | 25/181 (13.8) | 9/36 (25.0) | 2.1 (0.87–4.94) | 0.09 |
| Maximum no. of daily bowel movements (median [IQR]) | 5 (3–7) | 7 (5–9) | 0.003 | |
| Severity of symptoms at presentation of CDI (n [%]) | ||||
| Mild or moderate CDI | 147 (77.0) | 22 (61.1) | 1 | |
| Severe CDI | 27 (14.1) | 13 (36.1) | 3.43 (1.55–7.58) | 0.002 |
| Severe-complicated CDI | 17 (8.9) | 1 (2.7) | 0.29 (0.04–2.27) | 0.32 |
| Positive binary toxin (n [%]) | 34 (17.8) | 7 (19.4) | 1.15 (0.45–2.75) | 0.81 |
| Positive EIA result for A/B toxin (n [%]) | 77 (40.3) | 27 (75.0) | 4.44 (1.98–9.96) | 0.000 |
| Toxin B CT value (mean ± SD) | 26.02 ± 4.54 | 24.00 ± 3.28 | 0.79 (0.45–3.58) | 0.002 |
| Concomitant antibiotic during CDI-specific treatment (n [%]) | 134 (70.2) | 24 (66.7) | 0.85 (0.40–1.81) | 0.67 |
| Delay between symptom onset and start of treatment (days) (median [IQR]) | 4.0 (1.0–8.0) | 4.0 (1.0–13.0) | 0.74 | |
| Type of therapy and delay from sample submission to lab | ||||
| No treatment (n [%]) | 35 (18.3) | 1 (2.8) | 7.85 (1.04–59.27) | 0.02 |
| Empirical treatment (n [%]) | 27 (14.1) | 7 (19.4) | 1.46 (0.58–3.68) | 0.41 |
| Advance treatment (days) (median ± SD) | 2.22 ± 1.45 | 2.57 ± 2.63 | 0.73 (−1.15–1.84) | 0.64 |
| Targeted treatment (n [%]) | 126 (81.8) | 28 (77.8) | 1.8 (0.78–4.18) | 0.18 |
| Delay (days) (median ± SD) | 2.19 ± 3 − 04 | 1.07 ± 1.69 | 0.59 (−0.59–2.29) | 0.01 |
| Unknown (n [%]) | 3 (1.6) | 0 (0.0) | 0.84 (0.79–0.89) | 1.00 |
| Treatment (n [%]) | ||||
| Oral metronidazole | 105 (54.9) | 19 (52.7) | 1.41 (0.59–3.38) | 0.43 |
| Intravenous (i.v.) metronidazole | 42 (21.9) | 11 (30.5) | 1.62 (0.74–3.59) | 0.22 |
| Oral vancomycin | 60 (31.4) | 17 (47.2) | 2.06 (0.99–4.28) | 0.049 |
| Enemas of vancomycin | 1 (0.52) | 0 (0.0) | 0.84 (0.79–0.89) | 1.00 |
| Fidaxomicin | 0 (0.0) | 0 (0.0) | ||
| Rifaximin | 7 (3.6) | 0 (0.0) | 0.83 (0.79–0.88) | 0.60 |
| Probiotics | 15 (7.8) | 2 (5.5) | 0.69 (0.15–3.16) | 1.00 |
| Polyclonal gamma globulin | 0 (0.0) | 1 (2.7) | 0.15 (0.11–0.21) | 0.16 |
| i.v. tigecycline | 8 (4.2) | 1 (2.7) | 0.65 (0.08–5.39) | 1.00 |
| Surgical treatment | 0 (0.0) | 0 (0.0) | ||
Within the 6 months prior to the diagnosis of CDI.
Daily dose of >20 mg of prednisone or equivalent for more than 3 weeks at diagnosis.
Within the 4 weeks prior to the diagnosis of CDI.
Within the 12 weeks prior to the admission in which diagnosis of CDI was made.
To the primary care physician, emergency department, or outpatient facility.
White blood cell count of ≥15,000 cells/ml.
Increase of serum creatinine of ≥1.5-fold compared to the premorbid level.
CCI, age-adjusted Charlson comorbidity index; CDI, Clostridium difficile infection; CI, confidence interval; CT, threshold cycle; EIA, enzyme immunoassay; IQR, interquartile range; OR, odds ratio; PPI, proton pump inhibitors; SD, standard deviation. Statistically significant results are shown in bold.
FIG 1.
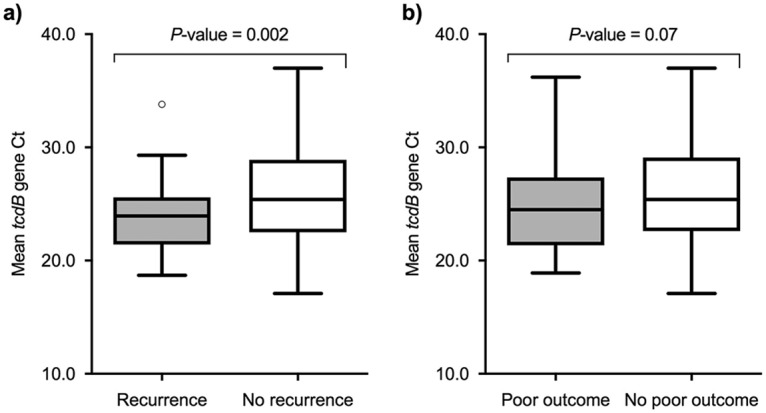
Box plots of mean tcdB gene CT values among patients with and without recurrence (a) and with and without a poor outcome (b).
TABLE 2.
Multivariate models for predicting recurrence of CDIc
| Factor | Value for clinical model |
Statistical analysis result |
||||||
|---|---|---|---|---|---|---|---|---|
| Clinical model |
Clinical model + positive toxin EIA |
Clinical model + toxin B PCR CT |
||||||
| No recurrence (n = 191) | Recurrence (n = 36) | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Chronic renal failure (n [%]) | 29 (15.2) | 10 (27.8) | 2.78 (1.1–6.99) | 0.03 | 3.00 (1.16–7.77) | 0.024 | 3.03 (1.17–7.84) | 0.022 |
| No. of admissionsa (median [IQR]) | 1 (0–1) | 1 (0–2) | 1.39 (1.06–1.84) | 0.017 | 1.37 (1.03–1.83) | 0.03 | 1.37 (1.04–1.80) | 0.024 |
| Severe CDI symptoms at presentation (n [%]) | 27 (14.1) | 13 (36.1) | 3.79 (1.59–9.08) | 0.003 | 3.15 (1.27–7.75) | 0.013 | 3.66 (1.51–8.86) | 0.004 |
| CDI symptoms as the main reason for consultationb (n [%]) | 84 (44.0) | 23 (63.9) | 2.43 (1.08–5.46) | 0.031 | 2.11 (0.92–4.83) | 0.077 | 2.20 (0.97–5.02) | 0.059 |
| Positive EIA result for A/B toxin (n [%]) | 77 (40.3) | 27 (75.0) | 3.52 (1.49–8.31) | 0.004 | ||||
| Toxin B CT of <25.65 (n [%]) | 102 (53.4) | 28 (77.8) | 3.41 (1.35–8.61) | 0.009 | ||||
Within the 6 months prior to the diagnosis of CDI.
To the primary care physician, emergency department, or outpatient facility.
CDI, Clostridium difficile infection; CI, confidence interval; CT, threshold cycle; EIA, enzyme immunoassay; IQR, interquartile range; OR, odds ratio; SD, standard deviation. Statistically significant results are shown in bold.
FIG 2.
ROC curves for recurrence (a) and poor outcome (b) according to the different predictive models.
Predictive models for poor outcome.
Overall, 70 first episodes of CDI had an unfavorable outcome (40 episodes of severe CDI, 18 episodes of severe-complicated CDI, and 27 deaths) within the first 8 weeks following the completion of therapy. Table 3 details the univariate comparison between patients suffering and not suffering from such an outcome. The mean tcdB gene CT was lower for episodes with a poor outcome (24.9 ± 4.24 versus 26.05 ± 4.47; P = 0.07) (Fig. 1b). Previous diagnosis of inflammatory bowel disease (aOR, 0.08; 95% CI, 0.01 to 0.7; P = 0.02) and solid organ transplantation (aOR, 0.18; 95% CI, 0.04 to 0.92; P = 0.04) were independent protective factors against a poor outcome. CDI-attributable symptoms as the main reason for consultation (aOR, 3.16; 95% CI, 1.55 to 6.46; P = 0.002) and the concurrent receipt of antibiotic therapy during the course of CDI-specific treatment (aOR, 4.54; 95% CI, 1.88 to 10.9; P = 0.001) were found to be independent predictors of poor outcome and were therefore included in the “clinical model” (Table 4). The optimal cutoff for the tcdB gene CT value was established at 27.55, yielding a sensitivity of 78.6% (95% CI, 67.1 to 87.5), a specificity of 35.7% (95% CI, 28.2 to 43.7), a PPV of 35.3% (95% CI, 31.5 to 39.2), and an NPV of 78.9% (95% CI, 69.5 to 85.9). Again, we generated the “EIA-including” and “PCR-including” models by incorporating the presence of a positive A/B toxin EIA result (aOR, 2.51; 95% CI, 1.29 to 4.91; P = 0.007) and a tcdB gene CT value of <27.55 (aOR, 2.55; 95% CI, 1.18 to 5.49; P = 0.017), respectively (Table 4). The auROC for each model is depicted in Fig. 2b. Similar to what happened in the models for recurrent CDI, the auROC of the “PCR-including model” was not meaningfully superior to that of the “EIA-including model” (0.804 versus 0.801), respectively).
TABLE 3.
Univariate analysis of risk factors predicting a poor outcomeh
| Factor | Value |
Univariate analysis result |
||
|---|---|---|---|---|
| Not poor outcome (n = 157) | Poor outcome (n = 70) | OR (95% CI) | P value | |
| Male gender (n [%]) | 81 (51.6) | 26 (37.1) | 1.8 (1.01–3.21) | 0.04 |
| Age (yr) (mean ± SD) | 62.8 ± 19.4 | 67.2 ± 18.8 | 2.76 (−9.7–1.16) | 0.12 |
| CCI (median [IQR]) | 4.0 (1.0–6.0) | 5.5 (4.0–7.0) | 0.005 | |
| Diabetes mellitus (n [%]) | 23 (14.6) | 12 (17.1) | 1.2 (0.56–2.58) | 0.63 |
| Active malignancya (n [%]) | 24 (15.3) | 16 (22.99 | 1.64 (0.81–3.33) | 0.17 |
| Hematological diseasea (n [%]) | 9 (5.7) | 4 85.7) | 0.99 (0.29–3.35) | 1.00 |
| Chemotherapya (n [%]) | 12 (7.6) | 15 (21.4) | 3.29 (1.45–7.48) | 0.003 |
| Hematopoietic stem cell transplantationa (n [%]) | 1 (0.6) | 2 (2.9) | 4.58 (0.41–51.4) | 0.22 |
| Solid organ transplantation (n [%]) | 19 (12.1) | 2 (2.9) | 0.21 (0.05–9.4) | 0.02 |
| Chronic renal failure (n [%]) | 29 (18.5) | 10 (14.3) | 0.73 (0.33–1.61) | 0.44 |
| Cirrhosis (n [%]) | 9 (5.7) | 6 (8.6) | 1.54 (0.52–4.51) | 0.43 |
| Concurrent corticosteroid therapy (any dose) (n [%]) | 32 (20.4) | 9 (12.9) | 0.57 (0.26–1.28) | 0.17 |
| Concurrent corticosteroid therapy (high doses)b (n [%]) | 7 (4.5) | 5 (7.1) | 1.65 (0.5–5.38) | 0.52 |
| Any immunosuppression (n [%]) | 45 (28.7) | 24 (34.3) | 1.30 (0.71–2.37) | 0.39 |
| Inflammatory bowel disease (n [%]) | 15 (9.6) | 1 (1.4) | 0.14 (0.02–1.06) | 0.02 |
| Cognitive impairment (n [%]) | 8 (5.1) | 5 (7.1) | 1.43 (0.45–4.54) | 0.54 |
| Admission to long-term care facility (n [%]) | 6 (3.8) | 7 (10.0) | 2.79 (0.90–8.65) | 0.12 |
| PPI therapyc (n [%]) | 94 (59.9) | 47 (67.1) | 1.37 (0.76–2.47) | 0.29 |
| H2 blocker therapyc (n [%]) | 7 (4.5) | 4 (5.7) | 1.30 (0.37–4.58) | 0.74 |
| Prior hospital admissiond (n [%]) | 85 (54.5) | 32 (45.7) | 0.70 (0.39–1.24) | 0.22 |
| No. of admissionsd (median [IQR]) | 1.0 (0.0–1.0) | 0.5 (0.0–1.0) | 0.43 | |
| Prior antibiotic therapy (n [%]) | ||||
| Within 4 weeks prior to diagnosis | 120 (76.4) | 56 (80.0) | 1.23 (0.62–2.46) | 0.55 |
| Within 12 weeks prior to diagnosis | 136 (86.6) | 64 (91.4) | 1.65 (0.63–4.28) | 0.30 |
| CDI symptoms as the main reason for consultatione (n [%]) | 68 (43.3) | 39 (55.7) | 1.64 (0.93–2.9) | 0.08 |
| Leukocytosisf (n [%]) | 13 (9.4) | 41 (60.3) | 14.72 (6.95–31.15) | 0.000 |
| White blood cell count [median (IQR)] | 8,300 (6,300–11,600) | 16,200 (8,575–21,925) | 0.000 | |
| Fever (n [%]) | 33 (21.0) | 50 (71.4) | 9.39 (4.93–17.91) | 0.000 |
| Acute renal failureg (n [%]) | 4 (2.7) | 30 (43.5) | 27.7 (9.2–83.3) | 0.000 |
| Maximum no. of daily bowel movements (median [IQR]) | 5.0 (3.0–7.0) | 6.0 (4.0–8.25) | 0.01 | |
| Concomitant antibiotic during CDI-specific treatment (n [%]) | 97 (61.8) | 61 (87.1) | 4.2 (1.94–9.05) | 0.000 |
| Recurrence in the following 8 weeks (n [%]) | 20 (12.7) | 16 (22.9) | 2.03 (0.98–4.21) | 0.05 |
| Positive for binary toxin (n [%]) | 24 (15.3) | 17 (24.3) | 1.78 (0.88–3.57) | 0.1 |
| Positive EIA result for A/B toxin (n [%]) | 63 (40.1) | 41 (58.6) | 2.11 (1.19–3.74) | 0.01 |
| Toxin B CT value (mean ± SD) | 26.05 ± 4.47 | 24.9 ± 4.24 | 0.63 (−0.09–2.4) | 0.07 |
| Delay from symptom onset to start of treatment (days) (median [IQR]) | 5.0 (2.0–9.75) | 3.0 (1.0–7.0) | 0.03 | |
| Type of therapy and delay from sample submission to lab | ||||
| No treatment (n [%]) | 33 (21.0) | 3 (4.3) | 5.94 (1.76–20.1) | 0.001 |
| Empirical treatment (n [%]) | 18 (11.5) | 16 (22.9) | 2.29 (1.1–4.81) | 0.026 |
| Advance treatment (days) (median ± SD) | 2.33 ± 1.84 | 2.25 ± 1.61 | 0.59 (−1.3–1.13) | 0.89 |
| Targeted treatment (n [%]) | 103 (65.6) | 51 (72.9) | 1.41 (0.75–2.61) | 0.28 |
| Delay (days) (median ± SD) | 2.23 ± 3.13 | 1.49 ± 2.24 | 0.49 (−0.22–1.71) | 0.13 |
| Unknown (n [%]) | 3 (1.9) | 0 (0.0) | 0.68 (0.63–0.75) | 0.55 |
| Treatment (n [%]) | ||||
| Oral metronidazole | 98 (62.8) | 26 (37.1) | 0.35 (0.19–0.62) | 0.000 |
| i.v. metronidazole | 21 (13.3) | 32 (45.7) | 5.4 (2.80–10.45) | 0.000 |
| Oral vancomycin | 34 (21.8) | 43 (61.4) | 5.7 (3.1–10.55) | 0.000 |
| Enemas of vancomycin | 0 (0.0) | 1 (1.4) | 0.31 (0.25–0.37) | 0.31 |
| Fidaxomicin | 0 (0.0) | 0 (0.0) | ||
| Rifaximin | 6 (3.8) | 1 (1.4) | 0.36 (0.04–3.1) | 0.44 |
| Probiotics | 12 (7.6) | 5 (7.1) | 0.93 (0.31–2.75) | 0.89 |
| Polyclonal gamma globulin | 0 (0.0) | 1 (1.4) | 0.30 (0.25–0.37) | 0.31 |
| i.v. tigecycline | 2 (1.3) | 7 (10.0) | 8.61 (1.74–42.6) | 0.004 |
| Surgical treatment | 0 (0.0) | 0 (0.0) | ||
Within the 6 months prior to the diagnosis of CDI.
Daily dose of >20 mg of prednisone or equivalent for more than 3 weeks at diagnosis.
Within the 4 weeks prior to the diagnosis of CDI.
Within the 12 weeks prior to the admission in which the diagnosis of CDI was made.
To the primary care physician, emergency department, or outpatient facility.
White blood cell count of ≥15,000 cells/ml.
Increase of serum creatinine of ≥1.5-fold compared to the premorbid level.
CCI, age-adjusted Charlson comorbidity index; CDI, Clostridium difficile infection; CI, confidence interval; CT, threshold cycle; EIA, enzyme immunoassay; IQR, interquartile range; OR, odds ratio; PPI, proton pump inhibitors; SD, standard deviation. Statistically significant results are shown in bold.
TABLE 4.
Multivariate models for predicting poor outcomec
| Factor | Value for clinical model |
Statistical analysis result |
||||||
|---|---|---|---|---|---|---|---|---|
| Clinical model |
Clinical model + positive toxin EIA |
Clinical model + toxin B PCR CT |
||||||
| Not poor outcome (n = 157) | Poor outcome (n = 70) | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Male gender (n [%]) | 81 (51.6) | 26 (37.1) | 1.52 (0.78–2.96) | 0.21 | 1.46 (0.74–2.9) | 0.27 | 1.51 (0.77–2.97) | 0.23 |
| CCI (median [IQR]) | 4.0 (1.0–6.0) | 5.5 (4.0–7.0) | 1.09 (0.98–1.21) | 0.09 | 1.01 (0.98–1.22) | 0.09 | 1.09 (0.98–1.21) | 0.10 |
| Chemotherapya (n [%]) | 12 (7.6) | 15 (21.4) | 1.96 (0.76–5.02) | 0.16 | 2.02 (0.76–5.35) | 0.15 | 2.47 (0.9–6.79) | 0.08 |
| Inflammatory bowel disease (n [%]) | 15 (9.6) | 1 (1.4) | 0.08 (0.01–0.7) | 0.02 | 0.07 (0.009–0.67) | 0.02 | 0.07 (0.009–0.65) | 0.018 |
| Solid organ transplantation (n [%]) | 19 (12.1) | 2 (2.9) | 0.18 (0.04–0.92) | 0.04 | 0.17 (0.03–0.85) | 0.03 | 0.21 (0.04–1.12) | 0.06 |
| CDI symptoms as main reason for consultationb (n [%]) | 68 (43.3) | 39 (55.7) | 3.16 (1.55–6.46) | 0.002 | 2.99 (1.44–6.22) | 0.003 | 3.02 (1.45–6.29) | 0.003 |
| Maximum no. of daily bowel movements (median [IQR]) | 5.0 (3.0–7.0) | 6.0 (4.0–8.25) | 1.07 (0.97–1.18) | 0.18 | 1.07 (0.96–1.18) | 0.21 | 1.06 (0.96–1.18) | 0.20 |
| Concomitant antibiotic during CDI-specific treatment (n [%]) | 97 (61.8) | 61 (87.1) | 4.54 (1.88–10.9) | 0.001 | 5.25 (2.12–12.98) | 0.000 | 5.11 (2.01–12.54) | 0.000 |
| Positive EIA result for A/B toxin (n [%]) | 63 (40.1) | 41 (58.6) | 2.51 (1.29–4.91) | 0.007 | ||||
| Toxin B CT of <27.55 (n [%]) | 101 (64.3) | 55 (78.6) | 2.55 (1.18–5.49) | 0.017 | ||||
Within the 6 months prior to the diagnosis of CDI.
To the primary care physician, emergency department, or outpatient facility.
CCI, age-adjusted Charlson comorbidity index; CDI, Clostridium difficile infection; CI, confidence interval; CT, threshold cycle; EIA, enzyme immunoassay; IQR, interquartile range; OR, odds ratio; PPI, proton pump inhibitors; SD, standard deviation. Statistically significant results are shown in bold.
Additional predictive models for both outcomes.
The “PCR-including models” using a prespecified high-sensitivity criterion of >95% and the CT cutoff of 26.35 reported by Senchyna et al. (5) did not improve the predictive values of the “EIA-including models” for any of the outcomes (see the supplemental material).
DISCUSSION
In the present single-center cohort of patients with a first episode of CDI, we compared the performances of several models for predictions of recurrence and poor outcome. Clinical variables were consecutively combined with the information obtained from two of the recommended diagnostic methods in the current guidelines (toxin A/B EIA result and PCR CT). In accordance with previous studies (5–8, 10), patients who suffered from recurrence or a poor outcome were more frequently toxin EIA positive and had a significantly lower mean tcdB gene CT value. However, our results suggest that the inclusion of the latter variable in the prediction process provides only low incremental value compared to that of models based on clinical features and detection of C. difficile toxin by nonmolecular methods.
The optimal CT cutoff value for recurrence was set at 25.65 for our cohort. Unlike those in other studies (7), the PPV for this cutoff point is very poor (21.54% [95% CI, 18.1 to 25.5%]), and although the NPV is acceptable (91.7% [95% CI, 85.5 to 95.4%]), 22.2% of patients (8/36 patients) still had a recurrence despite having a PCR CT value of >25.65. The auROC for the “PCR-including model” was similar to that for the “EIA-including model” (0.785 versus 0.775, respectively).
When we evaluated the combined variable poor outcome (severe CDI, severe-complicated CDI, or death within 8 weeks of the completion of therapy), the optimal CT cutoff value was set at 27.55, and the auROC for the “PCR-including model” was again similar to that for the “EIA-including model” (0.804 versus 0.801), respectively.
Thus, the clinical usefulness of NAAT-based algorithms in terms of predicting unfavorable events and tailoring CDI-specific therapeutic approaches is questionable (11).
Several clinical scores have been developed over recent years (10, 12–16) for early identification of which CDI patients are at higher risk of recurrence or a complicated course and may benefit from expensive or laborious therapies; however, the use of complicated scores in the rush of daily clinical practice is sometimes challenging, and therefore the search for an unbiased, quantifiable, and specific parameter (such as the PCR CT value) is tempting.
Previous studies showed a significant inverse correlation between PCR CT values and bacterial loads measured by quantitative culture (17) or toxin EIA detection (5, 6), as well as a significant inverse correlation between CT values and severity of CDI (6, 7, 18). Therefore, it has been suggested that assessment of CT values might obviate a two-step algorithm for the diagnosis of CDI. Despite the undeniable appeal of this idea, differences in mean PCR CT values among patients with and without recurrence or a poor outcome were too subtle to be the basis of clinical decisions. The optimal PCR CT value varies across different studies and is different depending on the predicted endpoint, e.g., ≤23.5 to correlate with a high risk of poor outcome according to Reigadas et al. (7), 26.35 for prediction of the presence of free toxin by use of a rapid membrane EIA according to Senchyna et al. (5), and 28.0 for use of a toxin A/B plate EIA as the reference method for patients with cancer according to Kamboj et al. (6).
In our study, the cutoff points were different for predictions of recurrence (25.65) and severity (27.55), and neither of them added much to a positive toxin EIA result. Both had poor specificity and PPV and insufficient sensitivity and NPV.
According to our results, a generalizable optimal cutoff point for the PCR CT value cannot be established and, in any case, does not add much to the demonstration of free toxin by use of EIA. Therefore, in our opinion, the systematic implementation and reporting of the PCR CT to the clinician should not be encouraged.
Strengths of our study include the careful review of medical and nursing records by experienced investigators blinded to diagnostic methods, inclusion of a relatively large number of cases, and real-life evaluation of a heterogeneous group of patients with ages and comorbidities typically associated with CDI.
On the other hand, the study has several limitations, some of them inherent to its retrospective nature. The results concerning some variables should be taken with caution, as they rely on retrospective assessment of medical records. Criteria used by physicians from our institution to order a CDI test might have been different from those used at other sites. Although systematic ribotyping of C. difficile isolates was not performed, there were no CDI episodes due to ribotype 027 during the study period, and therefore our findings might not be applicable to areas with different predominant clones.
In conclusion, although there is an inverse correlation between the toxin B PCR CT value and the CDI severity and recurrence risk, this determination does not represent a relevant contribution to a model based on clinical variables plus a positive toxin EIA, and we do not suggest basing medical and therapeutic decisions on this value alone.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest.
This study was supported by the MSD Investigator Initiated Studies Program (MISP) (grant 55012). J.O. held a Rio Hortega research training contract (CM13/00180) from the Spanish Ministry of Economy and Competitiveness, Instituto de Salud Carlos III. M.F.-R. held a Juan Rodés clinical research contract (JR14/00036) from the Spanish Ministry of Economy and Competitiveness, Instituto de Salud Carlos III.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
J.O. and L.C. retrospectively reviewed the clinical data.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01125-18.
REFERENCES
- 1.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang FC, Polage CR, Wilcox MH. 2017. Point-counterpoint: what is the optimal approach for detection of Clostridium difficile infection? J Clin Microbiol 55:670–680. doi: 10.1128/JCM.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O’Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senchyna F, Gaur RL, Gombar S, Truong CY, Schroeder LF, Banaei N. 2017. Clostridium difficile PCR cycle threshold predicts free toxin. J Clin Microbiol 55:2651–2660. doi: 10.1128/JCM.00563-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamboj M, Brite J, McMillen T, Robilotti E, Herrera A, Sepkowitz K, Babady NE. 2018. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer. J Infect 76:369–375. doi: 10.1016/j.jinf.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reigadas E, Alcala L, Valerio M, Marin M, Martin A, Bouza E. 2016. Toxin B PCR cycle threshold as a predictor of poor outcome of Clostridium difficile infection: a derivation and validation cohort study. J Antimicrob Chemother 71:1380–1385. doi: 10.1093/jac/dkv497. [DOI] [PubMed] [Google Scholar]
- 8.Origuen J, Corbella L, Orellana MA, Fernandez-Ruiz M, Lopez-Medrano F, San Juan R, Lizasoain M, Ruiz-Merlo T, Morales-Cartagena A, Maestro G, Parra P, Villa J, Delgado R, Aguado JM. 2018. Comparison of the clinical course of Clostridium difficile infection in glutamate dehydrogenase-positive toxin-negative patients diagnosed by PCR to those with a positive toxin test. Clin Microbiol Infect 24:414–421. doi: 10.1016/j.cmi.2017.07.033. [DOI] [PubMed] [Google Scholar]
- 9.Bagdasarian N, Rao K, Malani PN. 2015. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 313:398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobo J, Merino E, Martínez C, Cózar-Llistó A, Shaw E, Marrodán T, Calbo E, Bereciartúa E, Sánchez-Muñoz LA, Salavert M, Pérez-Rodríguez MT, García-Rosado D, Bravo-Ferrer JM, Gálvez-Acebal J, Henríquez-Camacho C, Cuquet J, Pino-Calm B, Torres L, Sánchez-Porto A, Fernández-Félix BM, Romero J, Muriel A, Giner L, Boix V, Ramos-Martínez A, Martínez R, Martos P, Arch O, Sardiña C, Aguirre E, Badía C, Boix L, Perales I, De Santos-Castro PA, Bratos-Pérez MA, Cuellar S, González E, Soto A, Sousa A, Llinares P, Castelo L, Morales I, Sojo J, Delgado-Iribarren A, Martí C, Vázquez R, Mairal P, Nosocomial Infection Study Group. 2018. Prediction of recurrent Clostridium difficile infection at the bedside: the GEIH-CDI score. Int J Antimicrob Agents 51:393–398. doi: 10.1016/j.ijantimicag.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Wilmore S, Goldenberg SD. 2018. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer: a reply. J Infect 76:424–426. doi: 10.1016/j.jinf.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 12.D’Agostino RB Sr, Collins SH, Pencina KM, Kean Y, Gorbach S. 2014. Risk estimation for recurrent Clostridium difficile infection based on clinical factors. Clin Infect Dis 58:1386–1393. doi: 10.1093/cid/ciu107. [DOI] [PubMed] [Google Scholar]
- 13.Hu MY, Katchar K, Kyne L, Maroo S, Tummala S, Dreisbach V, Xu H, Leffler DA, Kelly CP. 2009. Prospective derivation and validation of a clinical prediction rule for recurrent Clostridium difficile infection. Gastroenterology 136:1206–1214. doi: 10.1053/j.gastro.2008.12.038. [DOI] [PubMed] [Google Scholar]
- 14.Miller MA, Louie T, Mullane K, Weiss K, Lentnek A, Golan Y, Kean Y, Sears P. 2013. Derivation and validation of a simple clinical bedside score (ATLAS) for Clostridium difficile infection which predicts response to therapy. BMC Infect Dis 13:148. doi: 10.1186/1471-2334-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson SM, Slain D. 2015. Evaluation of a bedside scoring system for predicting clinical cure and recurrence of Clostridium difficile infections. Am J Health Syst Pharm 72:1871–1875. doi: 10.2146/ajhp150076. [DOI] [PubMed] [Google Scholar]
- 16.Zilberberg MD, Reske K, Olsen M, Yan Y, Dubberke ER. 2014. Development and validation of a recurrent Clostridium difficile risk-prediction model. J Hosp Med 9:418–423. doi: 10.1002/jhm.2189. [DOI] [PubMed] [Google Scholar]
- 17.Dionne LL, Raymond F, Corbeil J, Longtin J, Gervais P, Longtin Y. 2013. Correlation between Clostridium difficile bacterial load, commercial real-time PCR cycle thresholds, and results of diagnostic tests based on enzyme immunoassay and cell culture cytotoxicity assay. J Clin Microbiol 51:3624–3630. doi: 10.1128/JCM.01444-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jazmati N, Hellmich M, Licanin B, Plum G, Kaasch AJ. 2016. PCR cycle threshold value predicts the course of Clostridium difficile infection. Clin Microbiol Infect 22:e7–e8. doi: 10.1016/j.cmi.2015.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.