Bacterial membrane vesicle research has so far focused mainly on Gram-negative bacteria. Only recently have Gram-positive bacteria been demonstrated to produce and release extracellular membrane vesicles (MVs) that contribute to bacterial virulence.
KEYWORDS: Staphylococcus aureus, antibiotics, bacteriophages, membrane vesicles
ABSTRACT
Bacterial membrane vesicle research has so far focused mainly on Gram-negative bacteria. Only recently have Gram-positive bacteria been demonstrated to produce and release extracellular membrane vesicles (MVs) that contribute to bacterial virulence. Although treatment of bacteria with antibiotics is a well-established trigger of bacterial MV formation, the underlying mechanisms are poorly understood. In this study, we show that antibiotics can induce MVs through different routes in the important human pathogen Staphylococcus aureus. DNA-damaging agents and antibiotics inducing the SOS response triggered vesicle formation in lysogenic strains of S. aureus but not in their phage-devoid counterparts. The β-lactam antibiotics flucloxacillin and ceftaroline increased vesicle formation in a prophage-independent manner by weakening the peptidoglycan layer. We present evidence that the amount of DNA associated with MVs formed by phage lysis is greater than that for MVs formed by β-lactam antibiotic-induced blebbing. The purified MVs derived from S. aureus protected the bacteria from challenge with daptomycin, a membrane-targeting antibiotic, both in vitro and ex vivo in whole blood. In addition, the MVs protected S. aureus from killing in whole blood, indicating that antibiotic-induced MVs function as a decoy and thereby contribute to the survival of the bacterium.
INTRODUCTION
Bacterial membrane vesicles (MVs) affect diverse biological processes, including virulence, DNA transfer, export of cellular metabolites, phage decoy, and cell-to-cell communication, as shown in various bacterial species (1–3). Although MVs were originally shown to originate from blebbing of the outer membrane of Gram-negative bacteria, evidence has accumulated that various Gram-positive bacteria also release MVs composed of cytoplasmic membrane (2). A recent study has shown that MV formation in the Gram-positive bacterium Bacillus subtilis is stimulated by the expression of an endolysin encoded by a defective prophage (4). The hydrolytic activity of the endolysin creates small holes in the peptidoglycan layer through which cytoplasmic membrane (CM) material can protrude and is released as cytoplasmic MVs (CMVs). It has been observed that these cells lose CM integrity, as indicated by the formation of ghost cells containing many intracellular CMVs. Therefore, this vesicle biogenesis mechanism has been named “bubbling cell death” (5).
Prophages are integrated into the bacterial host genome and are passed on to daughter cells at each cell division. When the host cell experiences genotoxic stress, e.g., by exposure to DNA-damaging agents or UV radiation, the prophage induces the expression of lytic genes that promote DNA replication, phage particle assembly, DNA packaging, and, eventually, bacterial lysis to release the new phage particles, along with vesicles that are also formed in this process (4). Another route that stimulates CMV formation through the weakening of the cell wall is treatment with β-lactam antibiotics (4, 6, 7), which leads to protrusion of the cytoplasmic membrane into the extracellular space. While it is well known that MV formation is stimulated by antibiotic treatment, the underlying mechanisms are only poorly understood.
Staphylococcus aureus causes a wide spectrum of human infections, ranging from superficial cutaneous infections to systemic infections, including pneumonia, osteomyelitis, endocarditis, and bacteremia (8). The production of CMVs plays an important role in the delivery of S. aureus virulence factors to eukaryotic host cells (9, 10), stimulates biofilm formation (11), increases resistance to killing by whole blood, and activates purified human neutrophils ex vivo. Mice immunized with CMVs were protected against subcutaneous and systemic S. aureus infections (12). CMVs were also shown to increase antibiotic resistance, since they can contain β-lactamases (13). Here we used lysogenic S. aureus strains and their phage-devoid counterparts to investigate the effects of different antibiotics on CMV formation. Our results show that antibiotics can induce vesiculation by at least two routes, in a phage-dependent as well as a phage-independent fashion, depending on the mode of action of the antibiotic compound. We further examined how the different routes resulting in CMV production influence the content of the CMVs as well as their efficacy in protecting against antibiotic killing.
RESULTS
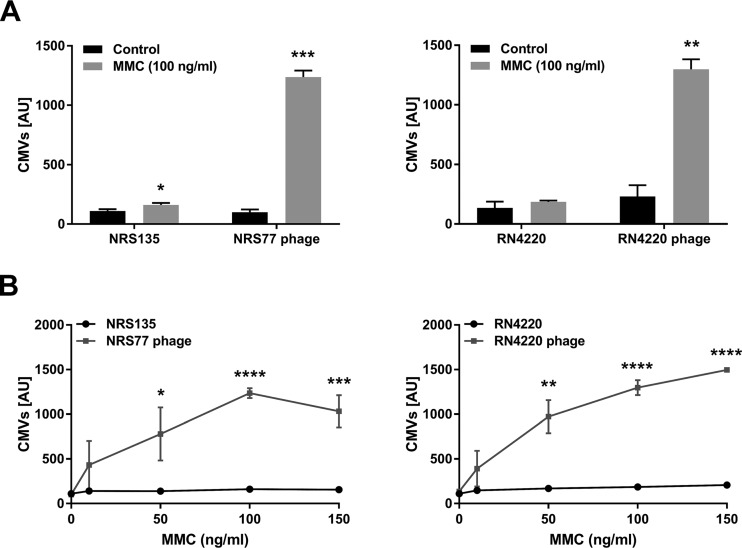
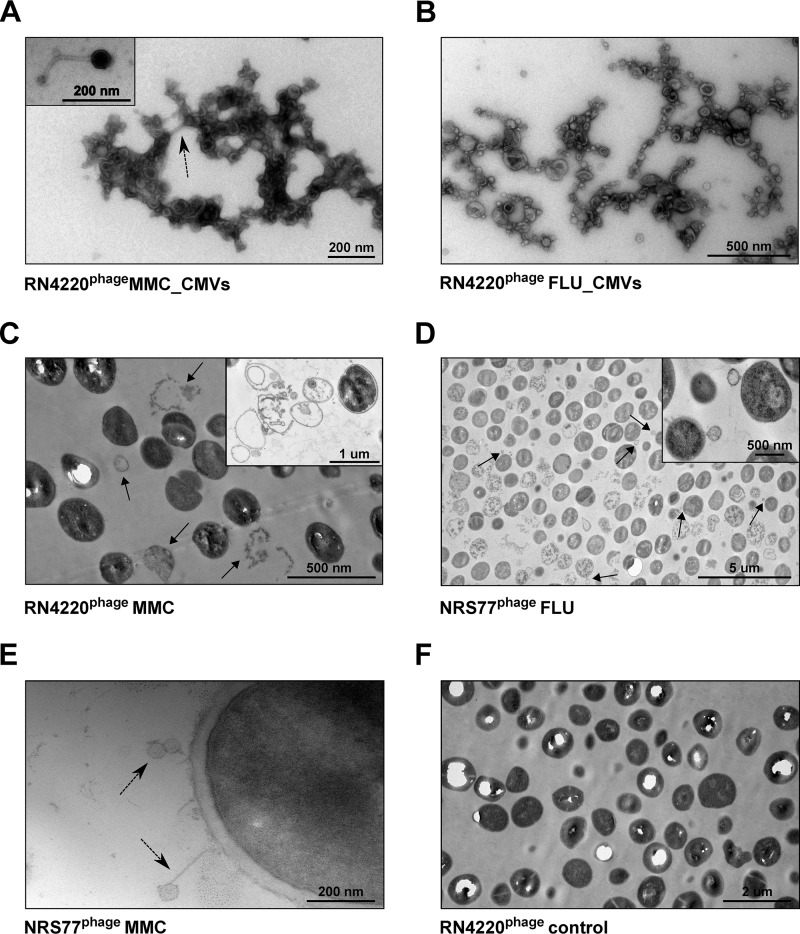
To test whether temperate phages affect vesicle formation in S. aureus, we compared the amounts of CMVs produced by the methicillin-susceptible S. aureus (MSSA) clinical strains NRS135 and RN4220 and their phage-bearing counterparts NRS77phage and RN4220phage, respectively (Table 1). Under standard growth conditions, no significant difference in CMV production was observed (Fig. 1). However, upon treatment with subinhibitory concentrations of the DNA-damaging agent mitomycin C (MMC), a well-characterized trigger of the SOS response (14), a strong increase in CMV formation was observed in the strains carrying a prophage but not in the cured strains devoid of phages (Fig. 1A). Moreover, CMV formation was stimulated in a concentration-dependent manner in the lysogenic strains (Fig. 1B). Using transmission electron microscopy (TEM), we observed CMVs, phages, and ghost cells, indicative of endolysin-triggered cell lysis, in samples of MMC-treated, phage-carrying strains (Fig. 2A, C, and E). Ghost cells were rarely observed in untreated controls (Fig. 2F) or cured strains. These data suggest that phage-triggered cell lysis is an important mechanism of CMV formation in S. aureus.
TABLE 1.
S. aureus strains and MICsa
| S. aureus strain | Source or references | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| MMC | DAP | CIP | FLU | CPT | ||
| RN4220phage | 28–30; P. François laboratory | 0.125 | 4 | 0.50 | 0.0625 | 4 |
| RN4220 | 28, 29 | 0.500 | 2 | 0.25 | 0.0625 | 4 |
| NRS77phage | 33, 34 | 0.250 | 4 | 0.25 | 0.0625 | 4 |
| NRS135 | 33, 34 | 0.500 | 4 | 0.50 | 0.1250 | 4 |
| CI1449 | A. Zinkernagel laboratory | NA | 2 | NA | NA | NA |
All strains were grown in LB (Lennox) broth (BD) at 37°C aerobically under constant shaking at 220 rpm or were plated on tryptic soy broth (TSB) agar (BD). MICs were determined in LB broth by microdilution test (35). MMC, mitomycin C; DAP, daptomycin; CIP, ciprofloxacin; FLU, flucloxacillin; CPT, ceftaroline. NA, not applicable.
FIG 1.
MMC treatment induces CMV formation in lysogenic S. aureus strains NRS and RN4220. Mitomycin C (MMC) was added at 100 ng/ml (A) or at the concentrations indicated (B) to log-phase cultures of bacterial strains NRS135 and RN4220, as well as their lysogenic variants NRS77phage and RN4220phage. After 4 h of incubation, the amounts of CMVs in the supernatants were measured using a fluorescent dye and were quantified in arbitrary units (AU). In our system, a value of 1,500 AU corresponds to a CMV preparation containing 250 μg protein/ml. The graphs show data from at least three independent experiments. Statistical analysis was carried out using the unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FIG 2.
TEM images of membrane vesicles and S. aureus after stimulation with MMC or FLU. (A through E) TEMs of CMVs and cells of S. aureus RN4220phage and NRS77phage treated with either 100 ng/ml MMC (A, C, E) or 10 times the MIC of FLU (B and D). Phages are indicated by dashed arrows. In samples of MMC-treated cultures, the presence of ghost cells (C) (indicated by arrows and shown in the inset) and of phages (E) was observed. In cultures treated with FLU, high numbers of ghost cells and cells with blebs (indicated by arrows and shown in the inset) were observed (D). (F) Untreated RN4220phage cells.
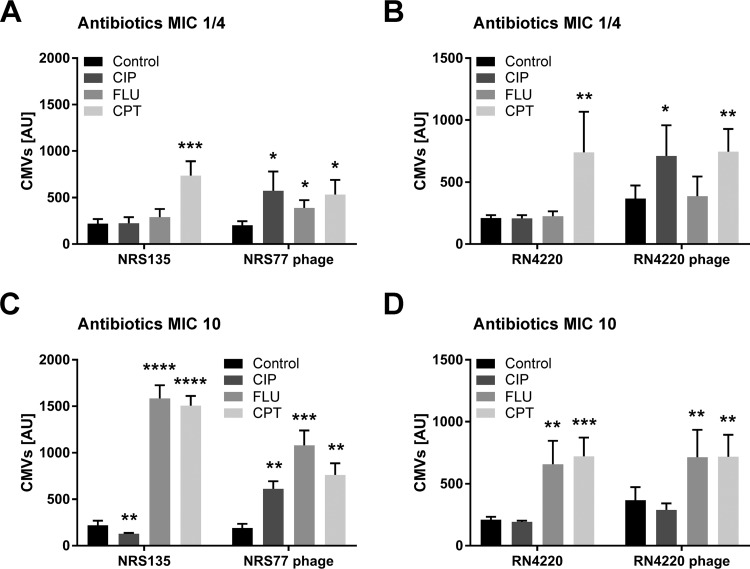
Since some antibiotics, particularly quinolones, are known to induce the cellular SOS response (15, 16), we tested next whether treatment with ciprofloxacin (CIP) concentrations equivalent to ¼ of the MIC or 10× MIC would affect CMV production. In the lysogenic strain NRS77phage, but not in the phage-free background strain NRS135, we observed a clear increase in vesicle production (Fig. 3A and C). With strain RN4220, we observed a similar trend at ¼ of the MIC, while the differences were not statistically significant at 10× MIC (Fig. 3B and D).
FIG 3.
Antibiotics triggered phage-dependent and -independent CMV induction in S. aureus strains NRS135 and RN4220. Log-phase cultures of strains NRS135 and RN4220, as well as their lysogenic variants NRS77phage and RN4220phage, were treated with ciprofloxacin (CIP), flucloxacillin (FLU), or ceftaroline (CPT) at concentrations equivalent to ¼ of the MIC (A and B) or 10× MIC (C and D). The amounts of CMVs in the supernatants were measured after 6 h of induction using a fluorescent dye and were quantified in arbitrary units (AU). In our system, a value of 1,500 AU corresponds to a CMV preparation containing 250 μg protein/ml. The graphs show data from at least three independent experiments. Statistical analysis was carried out using the unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Another possibility for stimulating vesicle formation in Gram-positive bacteria is to inhibit cell wall biosynthesis with β-lactam antibiotics (4, 6, 7). Treatment with β-lactam antibiotics is thought to weaken the peptidoglycan such that cytoplasmic membrane material can protrude into the extracellular space and is released as CMVs. Exposing the S. aureus strains to 10 times the MIC of the β-lactam antibiotic flucloxacillin (FLU) or ceftaroline (CPT) strongly increased vesicle formation independently of the presence or absence of the prophage (Fig. 3C and D). Upon exposure to ¼ of the MIC, a clear, phage-independent effect was observed for CPT but not for FLU, which, at this low concentration, caused a modest increase of vesiculation only in the lysogenic strain NRS77phage (Fig. 3A and B). While TEMs showed large amounts of CMVs, phages could not be observed when bacteria were treated with 10 times the MIC of FLU (Fig. 2B), indicating that these vesicles do not originate from SOS-induced bubbling cell death but likely through an alternative blebbing mechanism (Fig. 2D).
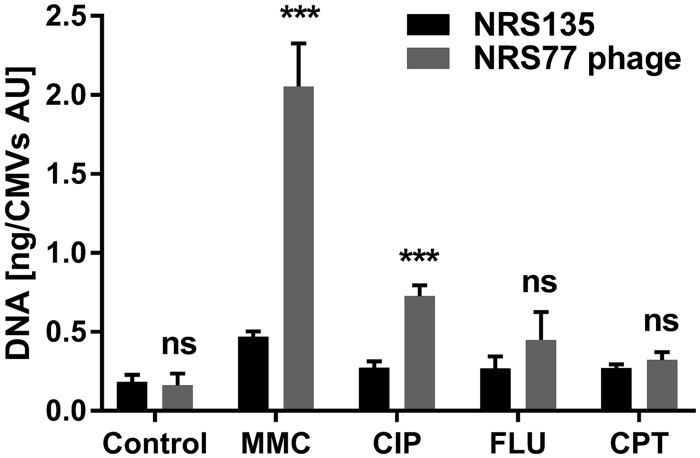
To investigate whether the cargoes carried by CMVs originating from blebbing or SOS-induced cell lysis are different, we determined the amounts of DNA associated with the CMVs (Fig. 4). These data provided clear evidence that CMVs originating from phage lysis have a generally higher DNA content than CMVs originating from a blebbing mechanism.
FIG 4.
Amounts of DNA associated with CMVs. The DNA contents of CMVs isolated from S. aureus NRS135 and NRS77phage cultures treated either with 100 ng/ml MMC or with the indicated antibiotics at 10× MIC were determined. The graphs show data from at least three independent experiments. Statistical analysis was carried out using the unpaired t test. ***, P < 0.001; ns, not significant.
We next tested whether S. aureus CMVs can protect cells from the last-resort antibiotic daptomycin (DAP), which inhibits cell envelope synthesis by interfering with fluid membrane microdomains (17). Due to its intrinsic resistance to infection with the phages produced by the RN4220phage and NRS77phage strains, the clinical S. aureus strain CI1449, a bloodstream isolate recovered from a patient with S. aureus endocarditis, was chosen. Incubation of the CI1449 strain with a phage lysate obtained from the NRS77phage strain did not result in phage binding to the bacterial surface (see Fig. S1C and D in the supplemental material), while binding of phages was observed for strain NRS135 (Fig. S1A and B), indicating that strain Cl1449 lacks a functional phage receptor. This allowed us to test the protective effect of MMC-induced CMVs as they copurified with phages (Fig. 2A), which would lyse sensitive strains. We assessed DAP killing in the presence or absence of purified CMVs both in vitro and ex vivo using freshly drawn human whole blood.
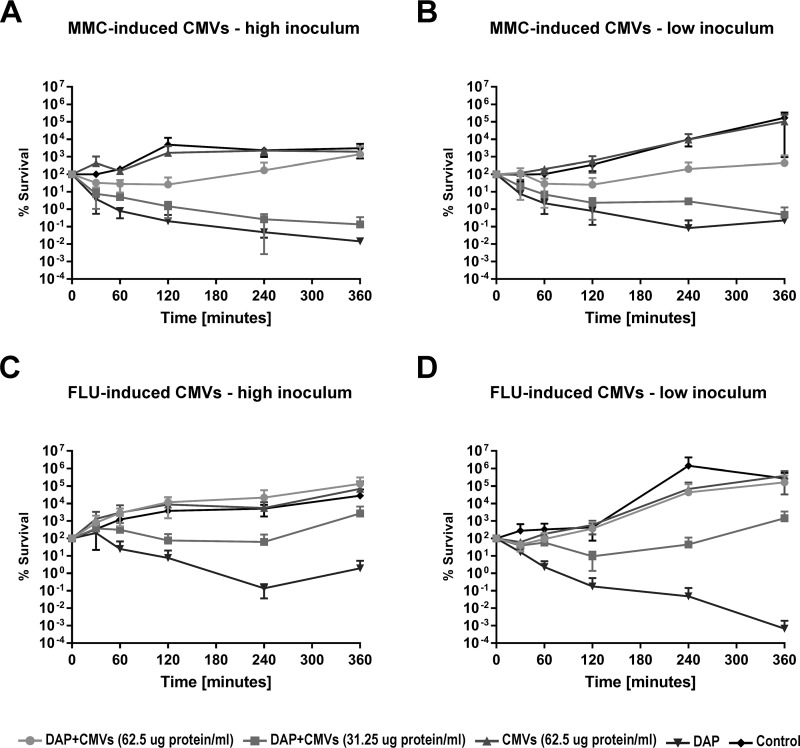
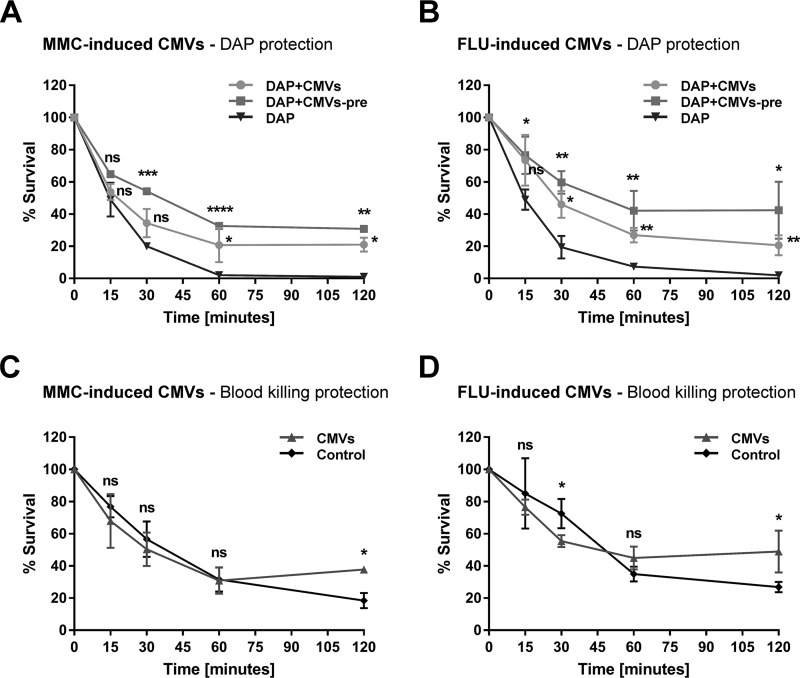
For the in vitro experiments, exponentially growing phage-resistant S. aureus strain CI1449 (at a high inoculum [5 × 107 CFU] or a low inoculum [105 CFU]) were challenged with two different concentrations of CMVs. FLU-induced CMVs exhibited a clear concentration-dependent protective effect against DAP as early as 30 min after the start of the assay for both inoculum sizes tested (Fig. 5C and D). Although slightly reduced, a protective effect was also observed for MMC-induced CMVs (Fig. 5A and B). For the ex vivo experiments, 2.3 × 104 S. aureus CI1449 cells were suspended in whole blood, and DAP was added with or without CMVs (with or without preincubation for 30 min in whole blood). Preincubation resulted in an enhanced protective effect against DAP challenge, which was observed as early as 15 min after the start of the assay for both MMC- and FLU-induced CMVs (Fig. 6A and B). Without preincubation, the protective effect was slightly reduced.
FIG 5.
CMVs derived from S. aureus NRS77phage stimulated with MMC or FLU protect a clinical S. aureus strain from DAP treatment. Log-phase cultures of the clinical S. aureus isolate CI1449 were treated with 10 times the MIC of DAP in the presence or absence of two different concentrations of CMVs (62.5 or 31.25 μg protein/ml) derived from S. aureus NRS77phage stimulated with either MMC (100 ng/ml) (A and B) or FLU (10× MIC) (C and D). The viability of the S. aureus clinical isolate CI1449 was followed over 24 h. Strain CI1449 is resistant to infection by phages produced by strain NRS77phage upon treatment with MMC. High inoculum, 5 × 107 CFU/ml; low inoculum, 105 CFU/ml. Missing data points indicate no detectable live bacteria; the lower limit of detection for this assay is 103 CFU/ml. The graphs show data from at least three independent experiments.
FIG 6.
CMVs derived from S. aureus NRS77phage stimulated with MMC or FLU protect a clinical S. aureus strain from killing by daptomycin in whole blood and from whole-blood killing. The clinical S. aureus isolate CI1449 (2.3 × 104 CFU/ml) was incubated in 400 μl blood in the presence or absence of DAP (10× MIC) and/or CMVs (14.4 μg protein/ml) induced by either MMC (A and C) or FLU (B and D). pre, 30-min preincubation in whole blood. Data from at least three independent experiments are shown. Statistical analysis was carried out using the unpaired t test. The results of statistical analysis are indicated on the graphs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We also observed significant protective effects of both MMC- and FLU-induced CMVs against whole-blood killing after 120 min of incubation, confirming that CMVs can also protect against the host’s innate immune system (Fig. 6C and D). In agreement with the in vitro data, the level of protection conferred by MMC-induced CMVs ex vivo was lower than that conferred by FLU-induced CMVs.
DISCUSSION
This study demonstrates that treatment of S. aureus with the DNA-damaging agent MMC or specific antibiotics routinely used in clinics increases CMV production both in a phage-dependent and in a phage-independent fashion, depending on the mode of action of the compound. We provide evidence that at least two different mechanisms account for this stimulatory effect. (i) MMC and CIP induce the SOS response and consequently increase vesicle formation through endolysin-triggered cell death, provided the strain harbors a temperate phage. (ii) The β-lactams FLU and CPT, at 10 times the MIC, weaken the cell wall and thereby stimulate CMV production through a phage-independent blebbing mechanism. Significant induction of vesiculation was observed already at ¼ the MIC of CPT, while this low concentration of FLU only weakly increased vesicle formation in the lysogenic strain NRS77phage. No difference in CMV production was observed for untreated strains, independently of the presence of prophages. This is in agreement with recent studies showing that under noninducing growth conditions, phage-encoded endolysins are not essential for CMV formation, which depends on the host’s autolysin Sle1 (14, 18).
We also observed a difference in the DNA cargoes harbored by CMVs produced by strains NRS135 and NRS77phage after stimulation with the DNA-damaging agents MMC and CIP. This difference possibly occurs because the phage lytic cycle involves the degradation of the host chromosome, resulting in smaller DNA fragments that may be more efficiently packed into vesicles. This idea is supported by a recent report showing that treatment of Stenotrophomonas maltophilia with CIP stimulates not only the production of MVs with high DNA contents but also the production of large numbers of phages, both of which are a consequence of SOS response induction in this bacterium (19). In contrast, no significant difference in DNA load was observed between lysogenic and cured strains for CMVs produced by induction with the cell-membrane-damaging agents FLU and CPT.
Previous work has shown that CMVs can provide protection against host defense factors such as antimicrobial peptides from mammalian tissue and complement system factors of the blood (20–22). In addition, it was recently shown that vesicles can serve as decoys for phages and membrane-targeting antibiotics (23, 24). In agreement with these findings, we show that both FLU- and MMC-induced CMVs protect cells from the membrane-active antibiotic DAP not only in vitro but also ex vivo in whole blood. This result suggests that the protective effect is relevant under a more physiological condition, which may more closely reflect the situation of an infected patient. The lower degree of protection we observed for MMC-induced CMVs than for FLU-induced CMVs in the bacterial protection assay could be due to the fact that CMVs induced by SOS response activation carry phage endolysins that can affect cell viability (4, 25, 26). Askarian et al. recently showed that CMVs naturally produced by S. aureus protect cells from whole-blood killing (12). We observed a similar effect for CMVs produced by S. aureus in response to DNA damage or antibiotic stress, suggesting that the mechanism of CMV genesis does not affect the protective function of CMVs against the host innate immune defense (12). Since phage binding to the surface of strain CI1449 is impaired, the presence of phages in MMC-induced CMV preparations is not responsible for the protective effect observed against killing by DAP and whole blood.
The setup we used in this study aimed to reflect the clinical situation. S. aureus infections due to methicillin-susceptible S. aureus are typically treated with FLU alone or in combination with a fluoroquinolone such as CIP, which leads to CMV release, as shown in our study. DAP is used for treating infections with multiresistant S. aureus strains, including methicillin-resistant S. aureus (MRSA) strains. In addition, DAP may be added to FLU in the case of persisting bacteremia. Our data show that antibiotic-induced CMVs could hinder the action of DAP against S. aureus and thus might counteract the clearance of the infection. Interestingly, a recent study showed that DAP can trigger the release of membrane vesicles in S. aureus, which may protect the bacterial cells (27). Further work will be required to unravel the differences between DAP- and FLU-triggered CMVs that account for their different DAP binding affinities.
MATERIALS AND METHODS
Strains and media.
S. aureus strains (Table 1) were grown in LB (Lennox) medium (BD) with shaking (220 rpm) at 37°C in the presence of MMC (10, 50, 100, or 150 ng/ml) or DAP (Cubicin), CIP (Ciproxin), FLU (Floxapen), or CPT (Zinforo) at a concentration equivalent to ¼ of the MIC, 2.3× MIC, or 10× MIC, depending on the assay (MIC values are given in Table 1).
The RN4220 strain was derived from strain NCTC8325-4 by UV and chemical mutagenesis (28, 29). Its prophage-positive counterpart, strain RN4220phage, was created by phage transduction using the MRSA bacteremia strain S. aureus 300-169 (30) as the prophage donor. To obtain phage preparations, cultures of strain 300-169 were treated with mitomycin C at a concentration of 0.5 μg/ml, as described previously (31). The filtered supernatants corresponding to putative lysates were stored at −80°C before utilization. Phages were propagated using the receiver isolate RN4220. Selection was performed by PCR, since prophages are devoid of any resistance markers (Table 2). Titrated phage preparations were kept at −80°C. The induced phage preparations were centrifuged on a sucrose density gradient. Phage particles were negatively stained with 2% uranyl acetate, examined in a JEOL 1230 transmission electron microscope at an accelerating voltage of 120 kV, and photographed (32). Morphological types were defined on the basis of phage tail length. The NRS77 strain (shown as NRS77phage in this work) was cured from phages to give rise to strain NRS135 (33, 34). CI1449 is an invasive S. aureus strain isolated at the University Hospital Zurich. It was chosen for bacterial protection and whole-blood killing assays because it is a bloodstream isolate and because of its resistance to infection by the phages harbored in strains NRS77phage and RN4220phage, as confirmed by a plaque assay. MIC values were determined using the broth microdilution test in LB medium, as described previously (35).
TABLE 2.
Primers used for the selection of the RN4220phage straina
| Primer name | Sequence |
|---|---|
| F1_phageKati | TCCATTGCATGTTGTCACCT |
| R1_phageKati | ATTTCAGCGGCTTGTTTTGT |
| F2_phageKati | TAAATTGGTGCGTCAGCTTG |
| R2_phageKati | ATCAGCATTTGATGGCGTTT |
| SAM_F35 | ATGACCCATGGGAAGCATAT |
| SAM_R1124 | GTTTGTGCATATGACGCTCA |
The primers were used to screen for the presence of prophages in the RN4220 strain genome.
CMV induction and quantification.
S. aureus was grown overnight (O/N) in 10 ml of LB (Lennox) medium in a 50-ml tube with shaking at 220 rpm, diluted in 10 ml of fresh medium in a 50-ml tube to an optical density at 600 nm (OD600) of 0.1, and then cultured in the presence or absence of MMC for 4 h or in the presence or absence of antibiotics for 6 h. CMVs were isolated and quantified as described previously (36). Briefly, cell cultures were centrifuged for 10 min at 4,600 × g, and the supernatant was filtered through a 0.22-μm-pore-size filter and was ultracentrifuged for 1 h at 150,000 × g and 4°C. The resulting pellets were resuspended in double-distilled water for CMV quantification by staining with the fluorescent dye FM1-43 (Life Technologies, USA). Fluorescence was evaluated by using a Varioskan Flash fluorimeter (Thermo Scientific) or a Synergy HT plate reader (MWG Biotech). The protein contents of CMV preparations were assessed using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). In our system, a value of 1,500 arbitrary units (AU) corresponds to a CMV preparation of 250 μg protein/ml.
Transmission electron microscopy.
CMVs were isolated from bacterial cultures stimulated with either MMC at 100 ng/ml or FLU at 10× MIC, as described above. The isolated CMVs were absorbed on glow-discharged, Formvar-coated 300-mesh copper grids and were negatively stained with 1% uranyl acetate for visualization. Thin sections of bacteria were prepared as described previously (37) from bacterial cultures stimulated with either MMC at 100 ng/ml or FLU at 10× MIC.
DNA content measurement.
The DNA loads of CMVs derived from strains NRS135 and NRS77phage stimulated with either MMC at 100 ng/ml or antibiotics at 10× MIC were quantified as described previously (38), with some minor modifications. Briefly, CMVs were resuspended in phosphate-buffered saline (PBS) and were incubated for 1 h in the presence of DNase I (NEB) to degrade extravesicular DNA. The DNase was then inactivated at 75C° for 15 min. DNA was released from CMVs by lysis, using 0.125% Triton X-100. DNA was quantified using the PicoGreen dsDNA kit (Invitrogen) and was normalized against CMV concentrations.
Bacterial protection assay.
CMVs derived from either MMC (100 ng/ml)- or FLU (10× MIC)-stimulated NRS77 bacteria were incubated with DAP (10× MIC) for 2 h in LB (Lennox) medium at a final concentration of 62.5 or 31.25 μg protein/ml. An O/N CI1449 culture was diluted 1:10 into fresh LB medium, grown for 2 h, diluted to an OD600 of 0.2 in LB medium plus CaCl2 (final concentration, 50 μg/ml), and added at 5 × 107 (high inoculum) or 105 (low inoculum) CFU/ml to the CMV-DAP mixture. CFU counts were determined at 30 min, 1 h, 2 h, 3 h, 4 h, 6 h, and 24 h.
Whole-blood killing.
To assess the role of CMVs in protecting bacteria from whole-blood killing and DAP challenge in whole blood, we used two experimental setups. CMVs and DAP were either preincubated in 400 μl of freshly drawn blood for 30 min at 37°C prior to the addition of bacteria (Fig. 6, DAP+CMVs-pre) or added to 400 μl of freshly drawn blood at the same time as the bacteria (Fig. 6, DAP+CMVs). The CMVs used for this assay were derived from NRS77 cultures, which were stimulated with either MMC (100 ng/ml) or FLU (10× MIC). CMVs were used at a final concentration of 14.4 μg protein/ml, and DAP was used at 2.3× MIC.
Strain CI1449 was prepared as described above for the bacterial protection assay and was added to whole blood to a concentration of 2.3 × 104 CFU/ml. Bacterial survival was assessed at 15, 30, 60, and 120 min by spotting bacterial suspensions of serial dilutions onto tryptic soy broth (TSB) agar plates. Bacteria were enumerated after O/N growth at 37°C. Blood was drawn into heparin tubes (BD).
Phage lysates.
Lysates of phages carried by strain NRS77 were prepared as follows. A 100-μl volume of a CMV sample obtained by stimulation of strain NRS77 with 100 ng/ml MMC was added to 300 μl of an O/N culture of the recipient strain NRS135 after the addition of 5 mM CaCl2 to the growth medium. Following incubation for 15 min at room temperature, 3 ml of warm LB soft agar (LB broth plus 0.6% agar) was added, and samples were spread onto 5% sheep blood plates (bioMérieux). The plates were incubated O/N at 37°C, and the phage lysate was harvested in 2 ml LB medium plus 5 mM CaCl2 and was stored at 4°C after filtration through a 0.45-μm filter.
Phage binding assay.
A 100-μl volume of phage lysate was incubated with 300 μl of an O/N culture of the recipient strain NRS135 or CI1449 after the addition of 5 mM CaCl2 to the growth medium. Bacteria were incubated for 15 min at room temperature, washed once in PBS, and resuspended in 2.5% glutaraldehyde in 0.1 mM cacodylate buffer. Samples were processed for electron microscopy as described above.
Study approval.
The collection of healthy volunteers’ blood complied with the current version of the Declaration of Helsinki. The national legal and regulatory requirements and sample collection were approved by the Canton Ethics Committee (Kantonale Ethikkommission Zurich, Switzerland, KEK—2010—0126).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to L.E. (Swiss National Foundation [SNF] grant 31003A_169307), A.S.Z. (SNF grant 31003A_176252), and M.T. (JSPS KAKENHI grant 16H06189 and JST ERATO grant PMJER1502). Imaging was performed with the support of the Center for Microscopy and Image Analysis, University of Zurich.
We thank Kati Seidl for helpful advice.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01439-18.
REFERENCES
- 1.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orench-Rivera N, Kuehn MJ. 2016. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyofuku M, Carcamo-Oyarce G, Yamamoto T, Eisenstein F, Hsiao CC, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L. 2017. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun 8:481. doi: 10.1038/s41467-017-00492-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyofuku M, Nomura N, Eberl L. 2018. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 6.Biagini M, Garibaldi M, Aprea S, Pezzicoli A, Doro F, Becherelli M, Taddei AR, Tani C, Tavarini S, Mora M, Teti G, D'Oro U, Nuti S, Soriani M, Margarit I, Rappuoli R, Grandi G, Norais N. 2015. The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol Cell Proteomics 14:2138–2149. doi: 10.1074/mcp.M114.045880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wichgers Schreur PJ, Rebel JM, Smits MA, van Putten JP, Smith HE. 2011. Lgt processing is an essential step in Streptococcus suis lipoprotein mediated innate immune activation. PLoS One 6:e22299. doi: 10.1371/journal.pone.0022299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 9.Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. 2011. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 6:e27958. doi: 10.1371/journal.pone.0027958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thay B, Wai SN, Oscarsson J. 2013. Staphylococcus aureus alpha-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One 8:e54661. doi: 10.1371/journal.pone.0054661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Yuan F, Lu F, Yin Y, Cao J. 2017. Vancomycin-induced biofilm formation by methicillin-resistant Staphylococcus aureus is associated with the secretion of membrane vesicles. Microb Pathog 110:225–231. doi: 10.1016/j.micpath.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Askarian F, Lapek JD Jr, Dongre M, Tsai CM, Kumaraswamy M, Kousha A, Valderrama JA, Ludviksen JA, Cavanagh JP, Uchiyama S, Mollnes TE, Gonzalez DJ, Wai SN, Nizet V, Johannessen M. 2018. Staphylococcus aureus membrane-derived vesicles promote bacterial virulence and confer protective immunity in murine infection models. Front Microbiol 9:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, Kim YK, Roh TY, Gho YS. 2013. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrob Agents Chemother 57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohn MT, Kjelgaard P, Frees D, Penades JR, Ingmer H. 2011. Clp-dependent proteolysis of the LexA N-terminal domain in Staphylococcus aureus. Microbiology 157:677–684. doi: 10.1099/mic.0.043794-0. [DOI] [PubMed] [Google Scholar]
- 15.Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol 189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Power EG, Phillips I. 1992. Induction of the SOS gene (umuC) by 4-quinolone antibacterial drugs. J Med Microbiol 36:78–82. doi: 10.1099/00222615-36-2-78. [DOI] [PubMed] [Google Scholar]
- 17.Muller A, Wenzel M, Strahl H, Grein F, Saaki TN, Kohl B, Siersma T, Bandow JE, Sahl HG, Schneider T, Hamoen LW. 2016. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci U S A 113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Thompson CD, Weidenmaier C, Lee JC. 2018. Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat Commun 9:1379. doi: 10.1038/s41467-018-03847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devos S, Van Putte W, Vitse J, Van Driessche G, Stremersch S, Van Den Broek W, Raemdonck K, Braeckmans K, Stahlberg H, Kudryashev M, Savvides SN, Devreese B. 2017. Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ Microbiol 19:3930–3937. doi: 10.1111/1462-2920.13793. [DOI] [PubMed] [Google Scholar]
- 20.Aung KM, Sjostrom AE, von Pawel-Rammingen U, Riesbeck K, Uhlin BE, Wai SN. 2016. Naturally occurring IgG antibodies provide innate protection against Vibrio cholerae bacteremia by recognition of the outer membrane protein U. J Innate Immun 8:269–283. doi: 10.1159/000443646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Codemo M, Muschiol S, Iovino F, Nannapaneni P, Plant L, Wai SN, Henriques-Normark B. 2018. Immunomodulatory effects of pneumococcal extracellular vesicles on cellular and humoral host defenses. mBio 9:e00559-18. doi: 10.1128/mBio.00559-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duperthuy M, Sjostrom AE, Sabharwal D, Damghani F, Uhlin BE, Wai SN. 2013. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog 9:e1003620. doi: 10.1371/journal.ppat.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharina A, Podolich O, Faidiuk I, Zaika S, Haidak A, Kukharenko O, Zaets I, Tovkach F, Reva O, Kremenskoy M, Kozyrovska N. 2015. Temperate bacteriophages collected by outer membrane vesicles in Komagataeibacter intermedius. J Basic Microbiol 55:509–513. doi: 10.1002/jobm.201400711. [DOI] [PubMed] [Google Scholar]
- 25.Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178:2767–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Clarke AJ, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 180:5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, Edwards AM. 2016. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat Microbiol 2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 28.Nair D, Memmi G, Hernandez D, Bard J, Beaume M, Gill S, Francois P, Cheung AL. 2011. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol 193:2332–2335. doi: 10.1128/JB.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez D, Seidl K, Corvaglia AR, Bayer AS, Xiong YQ, Francois P. 2014. Genome sequences of sequence type 45 (ST45) persistent methicillin-resistant Staphylococcus aureus (MRSA) bacteremia strain 300-169 and ST45 resolving MRSA bacteremia strain 301-188. Genome Announc 2:e00174-14. doi: 10.1128/genomeA.00174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Gialluly C, Loulergue J, Bruant G, Mereghetti L, Massuard S, van der Mee N, Audurier A, Quentin R. 2003. Identification of new phages to type Staphylococcus aureus strains and comparison with a genotypic method. J Hosp Infect 55:61–67. [DOI] [PubMed] [Google Scholar]
- 32.van der Mee-Marquet N, Corvaglia A-R, Valentin A-S, Hernandez D, Bertrand X, Girard M, Kluytmans J, Donnio P-Y, Quentin R, François P. 2013. Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect Genet Evol 18:299–308. doi: 10.1016/j.meegid.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Bæk KT, Frees D, Renzoni A, Barras C, Rodriguez N, Manzano C, Kelley WL. 2013. Genetic variation in the Staphylococcus aureus 8325 strain lineage revealed by whole-genome sequencing. PLoS One 8:e77122. doi: 10.1371/journal.pone.0077122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 36.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Carcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schilcher K, Andreoni F, Dengler Haunreiter V, Seidl K, Hasse B, Zinkernagel AS. 2016. Modulation of Staphylococcus aureus biofilm matrix by subinhibitory concentrations of clindamycin. Antimicrob Agents Chemother 60:5957–5967. doi: 10.1128/AAC.00463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Cruz C, Carrión O, Delgado L, Martinez G, López-Iglesias C, Mercade E. 2013. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microbiol 79:1874–1881. doi: 10.1128/AEM.03657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.