Chikungunya virus (CHIKV) causes a febrile disease associated with chronic arthralgia, which may progress to neurological impairment. Chikungunya fever (CF) is an ongoing public health problem in tropical and subtropical regions of the world, where control of the CHIKV vector, Aedes mosquitos, has failed.
KEYWORDS: chikungunya, chikungunya virus, arthralgia, antiviral, sofosbuvir, drug
ABSTRACT
Chikungunya virus (CHIKV) causes a febrile disease associated with chronic arthralgia, which may progress to neurological impairment. Chikungunya fever (CF) is an ongoing public health problem in tropical and subtropical regions of the world, where control of the CHIKV vector, Aedes mosquitos, has failed. As there is no vaccine or specific treatment for CHIKV, patients receive only palliative care to alleviate pain and arthralgia. Thus, drug repurposing is necessary to identify antivirals against CHIKV. CHIKV RNA polymerase is similar to the orthologue enzyme of other positive-sense RNA viruses, such as members of the Flaviviridae family. Among the Flaviviridae, not only is hepatitis C virus RNA polymerase susceptible to sofosbuvir, a clinically approved nucleotide analogue, but so is dengue, Zika, and yellow fever virus replication. Here, we found that sofosbuvir was three times more selective in inhibiting CHIKV production in human hepatoma cells than ribavirin, a pan-antiviral drug. Although CHIKV replication in human induced pluripotent stem cell-derived astrocytes was less susceptible to sofosbuvir than were hepatoma cells, sofosbuvir nevertheless impaired virus production and cell death in a multiplicity of infection-dependent manner. Sofosbuvir also exhibited antiviral activity in vivo by preventing CHIKV-induced paw edema in adult mice at a dose of 20 mg/kg of body weight/day and prevented mortality in a neonate mouse model at 40- and 80-mg/kg/day doses. Our data demonstrate that a prototypic alphavirus, CHIKV, is also susceptible to sofosbuvir. As sofosbuvir is a clinically approved drug, our findings could pave the way to it becoming a therapeutic option against CF.
INTRODUCTION
Chikungunya virus (CHIKV) is a member of the Togaviridae family, genus Alphavirus, which causes febrile debilitating illness associated with arthralgia and skin rash (1). Although prolonged and debilitating joint pain and edema differentiate CHIKV infection among contemporary arboviruses, such as dengue (DENV) and Zika (ZIKV) viruses, most often these agents display similar clinical signs and symptoms during the early phase of infection (1). Severe outcomes of CHIKV infection leading to acute and convalescent neurological impairment have also been described (2).
Chikungunya fever (CF) is an established public health problem with substantial impact in tropical and subtropical regions of the world where Aedes spp. mosquitoes are prevalent and vector control measures have failed (1). In the last 5 years, the Americas, Africa, and Eurasia have been severely affected by CHIKV (https://www.cdc.gov/chikungunya/geo/index.html). For example, the Asian and East/Central/South African (ECSA) genotypes of CHIKV have cocirculated since 2014 in Brazil (3–5), highlighting substantial viral activity in a country where DENV historically is hyperendemic. As there is no specific treatment or vaccine against CHIKV, repurposing clinically approved drugs, preferentially aiming at a viral target, is a necessary response against CF.
CHIKV has a positive-sense, single-stranded 11.8-kb RNA genome that encodes four non-structural (NsP1 to NsP4) and five structural proteins (C, E1, E2, E3, and 6K) (6). Among these proteins, NsP4 is coded for the viral RNA-dependent RNA polymerase (RdRp). Recent advances in studies on NsP4 activity and putative structure have been reported (7). As with other RNA polymerases from positive-sense RNA viruses, CHIKV NsP4 has well-conserved motifs, such as D-x(4,5)-D and GDD, which are spatially juxtaposed, wherein Asp binds Mg2+ and Asn selects ribonucleotide triphosphates over deoxynucleoside triphosphates (dNTPs), determining RNA synthesis (8). Moreover, as RdRp activity is absent from host cells, it constitutes a suitable target for antiviral intervention.
We and others have demonstrated that sofosbuvir (β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine), a clinically approved anti-hepatitis C virus (HCV) drug (9–11), also inhibits the replication of flaviviruses, such as ZIKV and DENV, and yellow fever virus (YFV) (12–17). Sofosbuvir is safe and well tolerated at 400 to 1,200 mg daily in a 24-week regimen. It is a UMP prodrug that requires the removal of phosphate protection to enter a pathway to yield sofosbuvir triphosphate (SFV), the pharmacologically active antiviral compound (9). Although hepatic cells have the most effective system for removing sofosbuvir phosphate protection, functional assays have revealed that other cells relevant to arbovirus infection also activate sofosbuvir (9, 14, 18). As expected for a nucleotide analogue, sofosbuvir inhibits the RNA polymerase from different Flaviviridae family members, i.e., HCV, ZIKV, DENV, and YFV (12–17). As the CHIKV NsP4 RdRp domain is likely conserved compared to that of other positive-sense virus RNA polymerases, we hypothesized that CHIKV could also be susceptible to sofosbuvir. Indeed, we are the first to demonstrate via cellular assays and animal models that sofosbuvir inhibits CHIKV replication.
RESULTS
CHIKV NsP4 as the predicted target of sofosbuvir.
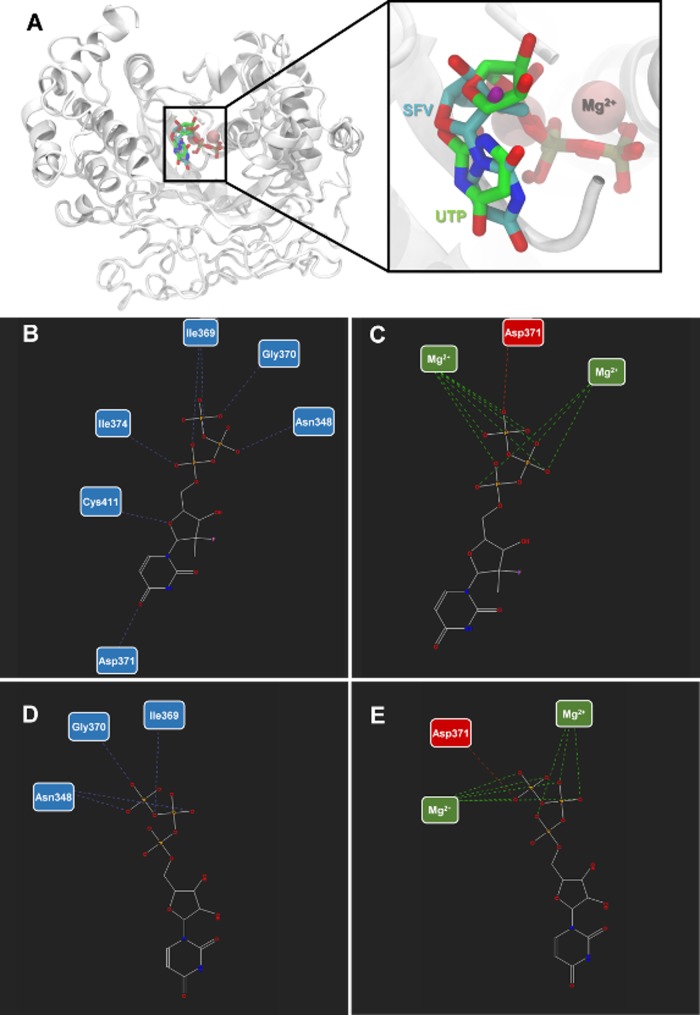
We considered the homology among viral RDRP to evaluate whether sofosbuvir docks on CHIKV NsP4. For comparison, the binding mode of SFV and the natural substrate uridine triphosphate (UTP) were analyzed on the NsP4 model. Three docking simulations per ligand (totaling 30 poses per ligand) were carried out. The poses with the lowest energy were selected for analysis (Table 1 and Fig. 1). SFV and UTP have similar modes of interaction but different energy values, −78.41 and −108.78 arbitrary units (a.u.) (with respect to MolDock scores), respectively (Table 1). Moreover, SFV interacted via H-bonds with Asn348, Ile369, Gly370, Asp371, and Cys411 (H-bond energy, −6.97 a.u.), whereas UTP formed H-bonds with Asn348, Ile369, and Gly370 (H-bond energy, −3.11 a.u.) (Table 1 and Fig. 1). Both SFV and UTP formed electrostatic attractive interactions with the two Mg2+ ions and repulsive interactions with Asp371. Consequently, SFV and UTP displayed electrostatic interaction energies of −117.12 a.u. and −112.84 a.u., respectively (Table 1 and Fig. 1). SFV and UTP use similar amino acid residues for steric interactions with Phe280, Asn344, Asn348, Ala367, Phe368, Ile369, Asp371, Asp372, Asn373, Ile374, and Cys411, resulting in energies equal to −24.50 a.u. and 48.76 a.u., respectively. Nevertheless, minor differences in steric interaction were observed: SFV docked onto Thr345 and Phe410, whereas UTP interacted with Leu250 and Phe251.
TABLE 1.
Summary of the interactions of SFV and UTP with NsP4 model of CIKV
| Virus or substrate | Value(s) for NsP4 model of CHIKV |
||||||
|---|---|---|---|---|---|---|---|
| H-bond energya | Residues (H-bond) | Electrostatic interaction energy | Residue and cofactor (electrostatic) | Steric interaction energy by PLPb | Residues (steric) | MolDock score | |
| SFV | −6.97 | Asn348, Ile369, Gly370, Asp371, Cys411 | −117.24 | Asp371, Mg2+ | −24.50 | Phe280, Asn344, Thr345, Asn348, Ala367, Phe368, Ile369, Gly370, Asp371, Asp372, Asn373, Ile374, Phe410, Cys411 | −78.41 |
| UTP | −3.11 | Asn348, Ile369, Gly370 | −112.84 | Asp371, Mg2+ | −48.76 | Leu250, Phe251, Phe280, Asn344, Thr345, Asn348, Ala367, Ile369, Gly370, Asp371, Asp372, Asn373, Ile374, Cys411 | −108.78 |
In arbitrary units.
Piecewise linear potential (57).
FIG 1.
Sofosbuvir triphosphate and NsP4 interactions. (A) Structural representation of the nsP4 model of CHIKV and its interaction with SFV and UTP. Hydrogen bonds and electrostatic interactions between SVF (B and C) and UTP (D and E) and nsP4 model of CHIKV. The interactions are represented by blue (H-bonds), green (attractive electrostatic interactions), and red (repulsive electrostatic interactions) interrupted lines. The nitrogen atoms are shown in blue, oxygen in red, fluor in pink, and the carbon chain in gray.
CHIKV was susceptible to sofosbuvir in vitro.
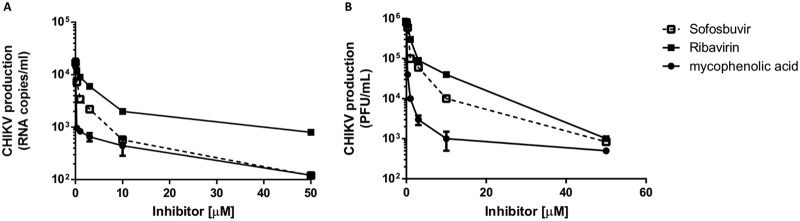
To evaluate if sofosbuvir has anti-CHIKV activity, phenotypic antiviral assays were performed in human cells previously associated with peripheral virus replication and nervous system invasion (19, 20), hepatoma Huh-7 cells and astrocytes derived from induced pluripotent stem cells (iPSCs), respectively. Supernatants from infected cultures were harvested, and virus RNA levels and infectivity were determined. We observed dose-dependent inhibition of CHIKV production in the hepatoma cells (Fig. 2 and Table 2), which have the machinery for converting the sofosbuvir prodrug to the pharmacologically active metabolite (10). Sofosbuvir had intermediate pharmacological activity compared to the two positive controls, i.e., ribavirin and mycophenolic acid (Fig. 2). Sofosbuvir and the controls consistently inhibited viral replication, as measured by RNA levels (Fig. 2A) and virus titers (Fig. 2B). The antiviral potencies varied 2-fold when the two methodologies were compared (Table 2). Sofosbuvir and mycophenolic acid were 25% less cytotoxic than ribavirin (Table 2). Consequently, the selectivity indexes (SI) for sofosbuvir and mycophenolic acid were >3 times better than that for ribavirin. The ribavirin and mycophenolic acid potencies were consistent with some previous reports (21–25). Although mycophenolic acid was marginally more potent than sofosbuvir (Fig. 2 and Table 2), the uridine analog is a clinically approved and direct-acting antiviral with a safe history for patients, motivating further investigation.
FIG 2.
Inhibition of CHIKV replication in hepatoma cells. Huh-7 cells (104 cells/well in 96-well plate [A] and 2.105 cells/well in 24-well plate [B]) were infected with CHIKV at an MOI of 0.1 and exposed to various concentrations of sofosbuvir, ribavirin, or mycophenolic acid for 24 h. Supernatant was harvested, and virus content was determined by measuring RNA levels (A) or titers (B) in Vero cells. The data represent means ± standard errors of the means (SEM) from five independent experiments.
TABLE 2.
Pharmacological parameters associated with drug inhibition of CHIKV replication in hepatoma cells
| Drug | EC50 (μM) |
CC50 (μM) | SIa |
||
|---|---|---|---|---|---|
| RNA level | Virus titer | RNA level | Virus titer | ||
| Sofosbuvir | 1.0 ± 0.1 | 2.7 ± 0.5 | 402 ± 32 | 402 | 149 |
| Ribavirin | 2.5 ± 0.3 | 5.5 ± 1.5 | 298 ± 22 | 120 | 54 |
| Mycophenolic acid | 0.8 ± 0.05 | 1.1 ± 0.2 | 370 ± 55 | 463 | 336 |
SI, selectivity index (CC50/EC50).
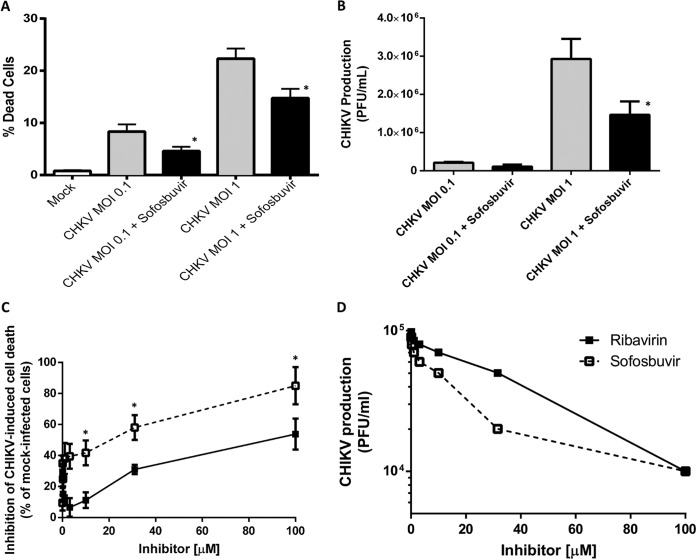
We next tested if sofosbuvir, which inhibits ZIKV replication in neural stem cells (NSCs) and brain organoids (14), also inhibits CHIKV in iPSC-derived human astrocytes. The astrocytes succumbed to CHIKV infection in a multiplicity of infection (MOI)-dependent manner, and sofosbuvir prevented cell mortality significantly (Fig. 3A). Accordingly, 10 μM sofosbuvir decreased CHIKV replication on astrocytes by half at an MOI of 1.0 (Fig. 3B). Subsequently, we compared the pharmacology of sofosbuvir and ribavirin on CHIKV-infected astrocytes at an MOI of 1.0 in terms of mortality and replication. Sofosbuvir was three times more efficient than ribavirin in preventing CHIKV-induced astrocyte death (Fig. 3C). Sofosbuvir’s and ribavirin’s 50% effective concentrations (EC50) were 17 ± 5 μM and 58 ± 12 μM, respectively (Fig. 3D). Altogether, these data demonstrate that sofosbuvir, as well as other pan-antivirals, inhibits CHIKV replication in a cell- and MOI-dependent manner.
FIG 3.
Sofosbuvir inhibits CHIKV-induced cell death and replication in iPSC-derived human astrocytes. (A and B) Astrocytes were infected at the indicated MOIs and treated with sofosbuvir at 10 μM. After 3 days, cells were labeled for activated caspase-3/7 and propidium iodide (A) and titers of virus in the supernatant were determined in Vero cells (B). (C and D) Astrocytes were infected at an MOI of 1.0 and treated with sofosbuvir or ribavirin at the indicated concentrations. After 3 days, cells death (C) and virus titers in Vero cells (D) were determined. The data represent means ± SEM from five independent experiments performed with five technical replicates. *, P < 0.05 for comparisons between infected, untreated (gray bars) and treated (black bars) groups (A and B) and for comparisons between groups infected/treated with sofosbuvir (open squares) and ribavirin (black squares) (C and D).
Sofosbuvir had a protective effect in CHIKV-infected mouse models of arthralgia and severe acute infection.
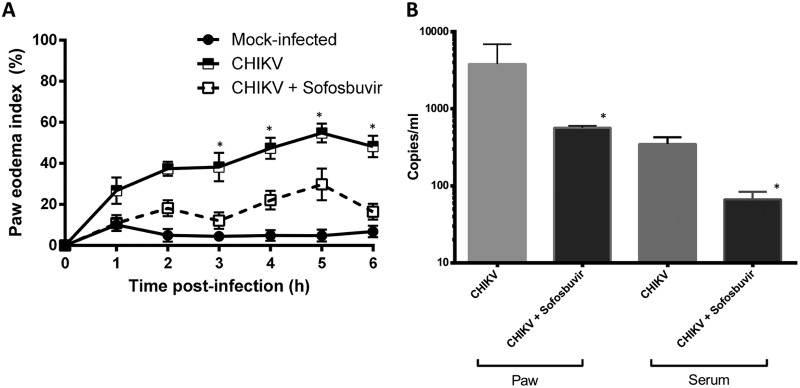
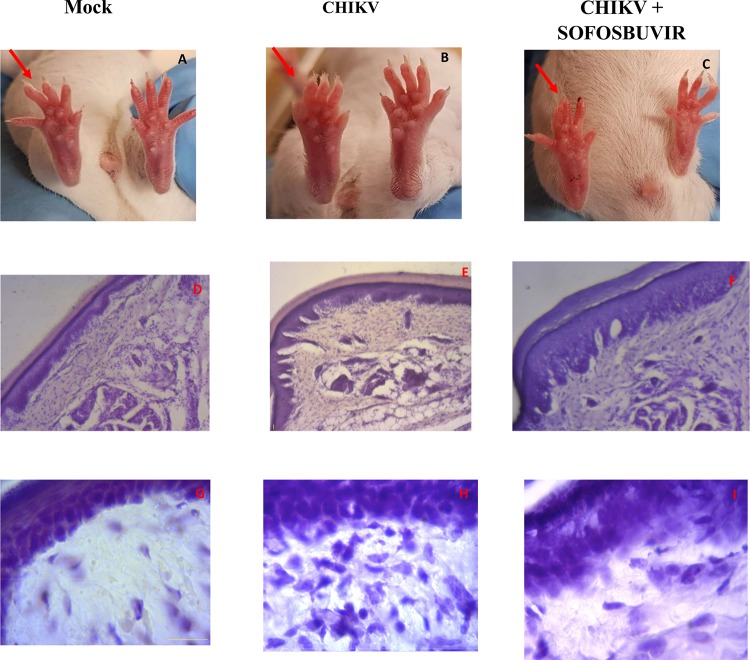
To analyze whether the in vitro results would translate into systemic protection, we treated CHIKV-infected mice with sofosbuvir. Considering that the major chronic problem associated with CHIKV infection is arthralgia, we used sofosbuvir (20 mg/kg of body weight/day orally at 1 h prior to infection) to treat adult Swiss mice whose right hind paws were infected with 2 × 105 PFU. CHIKV-induced paw edema was ameliorated by sofosbuvir at day 3 postinfection (Fig. 4A) and continued thereafter. Improvement in the paw condition was consistent with inhibition of virus replication at the infection site and peripherally (Fig. 4B). As replication was impaired in the sofosbuvir-treated CHIKV-infected mice, the paw inflammation in these animals was also reduced (Fig. 5). Tissue histology showed that CHIKV infection in the paw was characterized by disorganization of the epithelial and conjunctive tissues and by a substantial number of infiltrating inflammatory cells, mainly neutrophils. Sofosbuvir prevented the increase in inflammatory cell numbers and the disorganization of epithelial/conjunctive thickness (Fig. 5).
FIG 4.
Sofosbuvir ameliorated CHIKV-induced paw edema. Male Swiss Webster mice (20 to 25 g) received RPMI medium (mock infected) or 2 × 105 PFU CHIKV in 50 μl per paw in the ventral side of the right hind foot. Oral treatment with sofosbuvir (20 mg/kg/day) started 1 h prior to infection and continued thereafter daily. (A) Paw volume was measured in a hydropletismometer and normalized to the paw volume of each animal before injection. *, P < 0.05 by Tukey's multiple-comparison test (n = 8/group). (B) On the sixth day after infection, animals were euthanized and quantitative RT-PCR was performed with RNA from the serum or macerated paw. *, P < 0.05 by Student's t test (n = 3/group).
FIG 5.
Representative CHIKV-associated paw edema and sofosbuvir protection. The hind paws of mock-infected, CHIKV-infected (untreated), and CHIKV-infected/treated animals at the 6th day after infection are displayed. Red arrows indicate the infected paw (A to C). Histopathology data representing H&E-stained sections of paws at 40× (D to F) and 100× (G to I) are presented. Panels A, D, and G represent the mock-infected group. Panels B, E, and H represent the CHIKV-infected and untreated group. Panels C, F, and I represent the CHIKV-infected group treated with sofosbuvir. Images are representative of at least 5 animals per group.
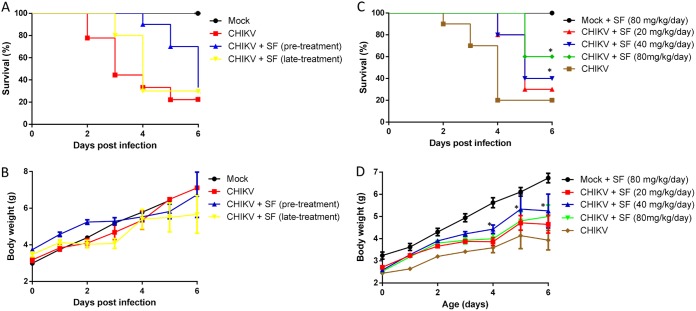
Subsequently, we studied the ability of sofosbuvir to enhance the survival of CHIKV-infected neonatal mice. Three-day-old Swiss mice were infected with CHIKV (2 × 102 PFU intraperitoneally). Sofosbuvir was administered daily, initially with an intraperitoneal injection of 20 mg/kg sofosbuvir, beginning 1 day prior to infection (pretreatment) or on day 2 after infection (late treatment). Although pretreatment doubled the mean survival time (T50) compared to that of mock-infected animals, all infected mice died 6 days after infection (Fig. 6A). Late treatment enhanced the T50 marginally (Fig. 6A). Postnatal development of the infected mice varied only marginally among groups (Fig. 6B). Under the same experimental conditions of infection, we next performed pretreatment with different sofosbuvir doses. Sofosbuvir at 40 and 80 mg/kg/day doubled and tripled, respectively, the percentage of animal survival (Fig. 6C). Mice that received 80 mg/kg/day sofosbuvir had significantly superior postnatal development compared to that of the infected controls (Fig. 6D).
FIG 6.
Sofosbuvir, at concentrations of 40 and 80 mg/kg/day, increased survival and inhibited weight loss of CHIKV-infected mice. Three-day-old Swiss mice were infected with CHIKV (2 × 102 PFU) and treated with sofosbuvir (SF) either 1 day before (pretreatment) or 2 days after infection (late treatment). Survival (A and C) and weight variation (B and D) were assessed during the course of treatment. (A and B) Experiments of both pretreatment and late treatment with sofosbuvir at 20 mg/kg/day. (C and D) Pretreatment with the indicated concentrations of sofosbuvir. Survival was statistically assessed by log-rank (Mentel-Cox) test. Differences in weight are displayed as the means ± SEM, and two-way ANOVA for each day was used to assess significance. Independent experiments were performed with at least 10 mice/group (n = 30). *, P < 0.01.
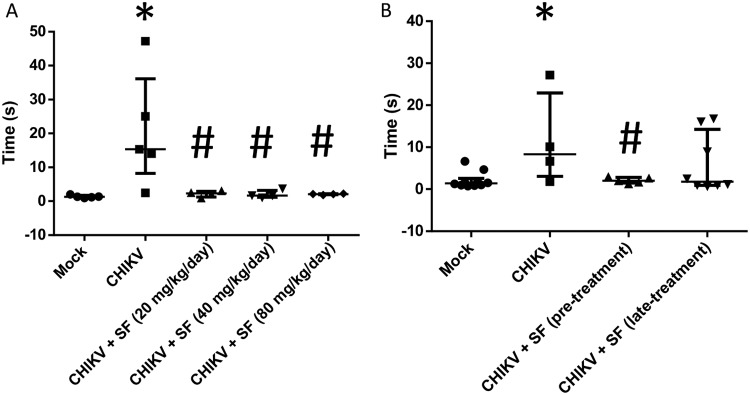
As some infected and untreated animals survived the experiments described in Fig. 6, we examined them for neuromotor sequelae and compared them to treated survivors. The animals were placed supine with all four paws facing up and then released. The time taken to flip over onto the stomach with all four paws touching the surface was measured as a proxy for neuromotor function. CHIKV-infected mice took a median of 10 to 20 s to get upright, whereas mock-infected animals did so immediately (Fig. 7). Importantly, CHIKV-infected and sofosbuvir-pretreated animals did not present neuromotor sequelae, indicating that they were healthier than the infected controls (Fig. 7). Of note, although late treatment diminished the median time associated with neuromotor sequelae, some animals displayed behavior similar to that of CHIKV-infected animals, making these groups statistically indistinguishable. Altogether, our data suggest that sofosbuvir also inhibits CHIKV replication in vivo, ameliorating arthralgia in the animals, enhancing survival, and preserving neuromotor function.
FIG 7.
Sofosbuvir prevented neuromotor impairment of CHIKV-infected mice. Three-day-old Swiss mice were infected with CHIKV (2 × 102 PFU) and treated with sofosbuvir (SF) beginning at 1 day before infection (pretreatment) or 2 days after infection (late treatment). (A) Treatment was performed with a dose of 20 mg/kg/day. (B) Pretreatment was performed with the indicated concentrations. At the sixth day after infection, animals were turned on their backs and allowed up to 60 s to return upright. The results are presented as means ± SEM. This was a routine measure, and at least 10 animals per group were analyzed. Student's t test was used to compare untreated CHIKV-infected mice with other groups individually. *, P < 0.01 mock- versus CHIKV-infected animals. #, P < 0.01 untreated versus treated animals.
DISCUSSION
CHIKV is among the reemergent arboviruses of the early 21st century. Although first characterized in the 1950s in Tanzania (26), CHIKV activity has increased worldwide since the 2000s and has reached the New World (27). CF is estimated to cause around 45.26 disability-adjusted life years (DALYs) lost per million people (28). In Brazil, CHIKV was introduced in 2014 (4, 5), when the Asian genotype was confirmed in the northern region (Oiapoque, Amapá state), and the ECSA genotype was identified in the northeastern region (Feira de Santana, Bahia state). The ECSA genotype was subsequently detected throughout Brazil. To the best of our knowledge, Brazil is a rare case, where two genotypes of CHIKV cocirculate. Since early 2016 (summer in the Southern Hemisphere), DENV, ZIKV, and CHIKV have cocirculated in the Americas, and CHIKV has become the most prevalent arbovirus in overcrowded Brazilian cities, such as Rio de Janeiro (29). Due to the absence of a vaccine and specific antiviral treatment, CHIKV prevention essentially depends on vector control, whereas patients with CF receive palliative care with nonsteroidal anti-inflammatory drugs (NSAIDs) or corticoids, depending on the phase of the disease (30).
CHIKV pathogenesis is complex and not fully understood; the virus replicates in the peripheral organs and may invade the nervous system and synovial fluid (19, 31). Recent efforts to identify substances against CHIKV have led to the discovery of chloroquine, berberine, mycophenolic acids, abamectin, and ivermectin (25, 32, 33). Among these substances, ivermectin inhibits flavivirus NS3 helicase activity (34). By analogy, it may also target a CHIKV protein, such as NsP2, which appears to have helicase activity (35). Berberine and mycophenolic acid target cellular rather than viral pathways (24, 36). We and others have shown that sofosbuvir has antiviral activity against flaviviruses (13–15). The recent advances in the CHIKV NsP4 RDRP core domain function and putative structure highlight the presence of conserved motifs among RNA polymerases from positive-sense RNA viruses (7). Indeed, sofosbuvir docked onto the putative CHIKV NsP4 using conserved amino acid residues also required for binding UTP, the natural substrate. Sofosbuvir inhibited CHIKV replication in human Huh-7 hepatoma cells. These cells were used because they represent the most efficient in vitro model for converting sofosbuvir (prodrug) to sofosbuvir-triphosphate (active) metabolite (37). Moreover, the liver is a relevant organ in CHIKV pathogenesis (19, 31). Growing evidence indicates that CHIKV impairs neurological function by directly invading the nervous system (20, 31, 38). We used a sophisticated iPSC-derived human astrocyte culture to show that sofosbuvir inhibits CHIKV replication and virus-associated cell mortality, although it does so less effectively than Huh-7 cells. Notably, sofosbuvir potency varied according to the MOI, cell system, and assay readout (RNA levels or PFU titers). Similarly, other RNA polymerase inhibitors, such as favipiravir and ribavirin, inhibit CHIKV replication at different potencies based on the factors analyzed here plus the viral strain/genotype used (21, 22, 24, 25, 39–42). For example, the potency of ribavirin has been reported to be 341 μM (42), 142 μM (with a substantial error of ± 126 μM) (40), 10.95 μM (22), 3.06 μM (21), and 2.05 μM (23). We understand that the EC50 values of ribavirin, which range from 2 to 15 μM, are consistent, because different types of assay readouts, both objective and empirical, produce these values. In our review of the literature, high EC50 values for ribavirin tend to be described with empirical assay readouts, that is, dependent on a human reader, such as by quantifying cytopathic effects under a light microscope. Under our experimental conditions, the potencies of sofosbuvir and the control varied within an acceptable range for each cell type. The EC50 values described here are consistent, and the orders of magnitude are different from the 50% cytotoxic concentration (CC50) values.
Accordingly, in the neonatal mouse model of CHIKV infection, sofosbuvir enhanced survival at doses at least 2-fold higher than required to produce the same effect on ZIKV-infected pups (14). Nevertheless, 1,200 mg/day sofosbuvir, which represents a 3-fold higher dose than the reference for routine treatment of HCV-infected patients, i.e., 400 mg/day, is considered safe for humans (10). We observed a narrow window of opportunity for treating CHIKV-infected mice, similar to what we have seen for ZIKV (43). In other acute virus infections, such as influenza, mortality is dramatically reduced when neuraminidase inhibitors are administered early in the time course of infection, e.g., within 2.5 days of infection (44). Identifying higher-risk individuals for receiving sofosbuvir prophylactically or very early after infection represents one of the challenges to translating our data into public health intervention.
Importantly, we observed that, under experimental infection conditions milder than those required for animal mortality, a reference dose for preclinical studies of 20 mg/kg/day sofosbuvir (11) protected against arthralgia-related paw edema, reducing viral replication, inflammation, and tissue damage. Thus, further study of whether sofosbuvir can act synergistically with anti-inflammatory drugs to improve the quality of life of patients with CHIKV-associated chronic arthralgia is important. Patients with CF very often present with arthralgia and impaired neuromuscular function, which cause a debilitating condition that contributes to the burden of disease (31). Using survivor animals from lethal challenge, we observed that sofosbuvir, also at reference dose, protected CHIKV-infected mice from neuromotor sequalae compared to untreated animals. Based on this behavior test, it is likely that both direct neuromotor sequelae and/or problems in articulation have contributed to the longer time required for the CHIKV-infected mice to right themselves from an upside down position.
Altogether, our data reveal that CHIKV is susceptible to sofosbuvir, highlighting the possibility that other genetically distinct and clinically important viruses phylogenetically distributed among members of the Togaviridae and Flaviviridae families could also be susceptible to this drug. We advocate generic sofosbuvir beyond treatment of HCV-infected individuals, as it represents a safer and more effective antiviral option than ribavirin. Finally, in the context of this study, our findings encourage phase II clinical investigations on the novel use of sofosbuvir as treatment against CHIKV.
MATERIALS AND METHODS
Reagents.
The antiviral sofosbuvir (β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine) was donated by the BMK Consortium: Blanver Farmoquímica Ltd., Microbiológica Química e Farmacêutica Ltd., and Karin Bruning & Cia Ltd. (Taboão da Serra, São Paulo, Brazil). Ribavirin was donated by the Instituto de Tecnologia de Farmacos (Farmanguinhos, Fiocruz, Rio de Janeiro, Brazil). All small-molecule inhibitors were dissolved in 100% dimethyl sulfoxide (DMSO) and subsequently diluted at least 104-fold in culture before each assay. The final DMSO concentrations showed no cytotoxicity. The materials for cell culture were purchased from Thermo Scientific Life Sciences (Grand Island, NY, USA) unless otherwise mentioned.
Cells.
African green monkey kidney (Vero) and human hepatoma (Huh-7) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM). The culture medium for each cell type was supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (45, 46) at 37°C in 5% CO2.
Virus.
CHIKV (Asian strain) was donated by Amilcar Tanuri and propagated in the Vero cells at an MOI of 0.1. Infection was carried out for 1 h at 37°C. The residual virus particles next were removed by washing with phosphate-buffered saline (PBS), and the cells were cultured for 2 to 5 days. After each period, the cells were lysed by freeze-thawing and centrifuged at 1,500 × g at 4°C for 20 min to remove cellular debris. Virus titers were determined by PFU.
To titer virus PFU, 24-well plates containing 105 cells per well were exposed to virus inoculum at 10-fold serial dilutions. After 1 h, unbound viruses were washed out with PBS and the cells were covered with overlay medium (DMEM with 1% FBS and 1.5% carboxymethylcellulose). After 2 to 5 days of incubation at 37°C, the monolayers were fixed with 10% formaldehyde in PBS and stained with 0.1% crystal violet solution in 70% methanol when titers were scored.
Cytotoxicity assay.
Cell monolayers (2 × 104 to 5 × 104 cells/well) in 96-well plates were treated for 5 days with sofosbuvir or ribavirin (control). Five mg/ml 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) in DMEM was added to the cells in the presence of 0.01% N-methyl dibenzopyrazine methyl sulfate (PMS). After a 4-h incubation at 37°C, the plates were read in a spectrophotometer at 492 nm and 620 nm (47). The CC50 was calculated by nonlinear regression analysis of the dose-response curves.
Yield reduction assay.
Huh-7 cell monolayers (5 × 104 cells/well) in 96-well plates were infected with CHIKV at an MOI of 0.1 for 1 h at 37°C. The cells were washed with PBS to remove residual viruses and then incubated with sofosbuvir or positive controls in DMEM containing 1% FBS. After 24 h, the cells were lysed, the cellular debris cleared by centrifugation, and the virus titers in the supernatant were determined in Vero cells as PFU/ml. The EC50 was determined using nonlinear regression analysis of the dose-response curves.
Generation of human iPSC-derived astrocyte lines.
Astrocytes were differentiated from NSCs (20 × 103 cells/well in a 96-well plate) derived from human iPSCs of three control cell lines obtained from healthy subjects (48). These cell lines were previously used in other studies by our research group (14). The three cell lines were from one female subject (GM23279A; available from the Coriell Institute, https://www.coriell.org) and two male subjects (CF1 and CF2; cells reprogrammed at the D’Or Institute for Research and Education, Rio de Janeiro, Brazil). The NSCs were differentiated into astrocytes as described by Yan et al. (49). Briefly, the NSCs were cultured in differentiation medium (1% N2 supplement and 1% FBS in DMEM-F12 medium) for 21 days with medium change every other day and weekly passage. Subsequently, glial cells were grown for 5 weeks in 10% FBS in DMEM-F12 with medium change twice a week prior to use. Cells were infected at an MOI of 1.0 or 10 for 2 h at 37°C.The cells next were washed, and fresh medium containing sofosbuvir was added. The cells were treated daily with sofosbuvir. Virus titers were determined from the culture supernatant. Cell death was measured by adding 2 μM CellEvent caspase-3/7 reagent and the fluorescent dye ethidium homodimer (ThermoFischer Scientific) (50) when the culture supernatants were collected. Images were acquired with an Operetta high-content imaging system with a 20× objective and high numerical apertures (NA) (PerkinElmer, Waltham, MA, USA). The data were analyzed using Harmony 5.1 high-content image analysis software (PerkinElmer). Seven independent fields were evaluated from triplicate wells per experimental condition.
RNA extraction and reverse transcription-PCR (RT-PCR).
Viral RNA was extracted from the culture supernatant, serum, or tissue homogenate using RNA purification kits (QIAamp RNA viral minikit; Qiagen, Hilden, Germany) and eluted to a final volume of 60 μl. To evaluate cross-contamination, negative controls were included at all stages.
CHIKV RNA was quantified by real-time RT-PCR using QuantiTect/QuantiNova probe RT-PCR kits (Qiagen) with previously described primers and probes (51). Samples with cycle threshold (CT) values of <37 and with sigmoid curves were considered positive. Copies per milliliter were determined using a synthetic RNA fragment to amplify the target region (GenScript, Nanjing, China). CHIKV positive and negative controls, including a nontemplate control, were used in the RT-PCR.
3D modeling of CHIKV NsP4.
The amino acid sequence of CHIKV NsP4 (UniProtKB entry F2YI10) was obtained from the ExPASy server (52). The region between Met1 and Lys516, part of the NsP4 sequence that includes the whole catalytic core, was considered to construct the model using the I-TASSER server (53). The I-TASSER methodology is very accurate for constructing protein models when the sequence identity between the target sequence and the template protein is <30%, where the lack of a high-quality structure match may yield substantial alignment errors and, consequently, low-quality models (53, 54). Thus, the final model was validated using the PROCHECK (55) and VERIFY3D (56) programs. PROCHECK analyzes the stereochemical quality and VERIFY3D performs compatibility analysis between the three-dimensional (3D) model and its own amino acid sequence by assigning a structural class based on its location and environment and by comparing the results with that of crystal structures with good resolution (55, 56).
Molecular docking.
The SFV and UTP structures were built in Spartan’14 software (Wavefunction, Inc., Irvine, CA, USA). The docking of the two ligands to the NsP4 model was performed using the Molegro Virtual Docker 6.0 (MVD) program (CLC Bio, Aarhus, Denmark) (57), which uses a heuristic search algorithm that combines differential evolution with a cavity prediction algorithm (57). The MolDock scoring function used is based on a modified piecewise linear potential (PLP) with new hydrogen bonding and electrostatic terms included. The full description of the algorithm and its reliability compared to that of other common docking algorithms have been described (57). The two Mg2+ ions were set as the center of the search for space with a radius of 10 Å. In addition, the search algorithm MolDock optimizer was used with a minimum of 100 runs, and the parameter settings were the following: population size, 500; maximum iteration, 2,000; scaling factor, 0.50; offspring scheme, scheme 1; termination scheme, variance based; crossover rate, 0.90. Due to the stochastic nature of algorithm search, three independent simulations per ligand were performed to predict the binding mode. Consequently, the complexes with the lowest interaction energy were evaluated. The interactions between the NsP4 model and each ligand were analyzed using the ligand map algorithm, a standard algorithm in the MVD program (57). The usual threshold values for H-bonds and steric interactions were used.
All figures of NsP4 modeling and molecular docking results were edited using the Visual Molecular Dynamics 1.9.3 (VMD) program (http://www.ks.uiuc.edu/Research/vmd/vmd-1.9.3/) (58).
Animals.
Swiss albino mice (Mus musculus, pathogen free) from the Oswaldo Cruz Foundation breeding unit (Instituto de Ciência e Tecnologia em Biomodelos [ICTB]/Fiocruz, Rio de Janeiro, Brazil) were used for the studies. The animals were kept at a constant temperature (25°C) with free access to chow and water in a 12-h light/dark cycle. The experimental laboratory received pregnant mice (at approximate gestational day 14) from the breeding unit. The pregnant mice were observed daily until delivery to accurately determine the postnatal day. We established a litter size of 10 animals for all experimental replicates.
The Animal Welfare Committee of the Oswaldo Cruz Foundation (CEUA/Fiocruz) approved and covered (license numbers L-016/2016 and CEUA L-002/2018) the experiments in this study. The procedures described here were in accordance with the local guidelines and the guidelines published in the National Institutes of Health Guide for the Care and Use of Laboratory Animals (59). The study is reported in accordance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines (60). If necessary, euthanasia was performed to alleviate animal suffering. The criteria for euthanasia were (i) difference in weight gain between infected and control groups of >50%, (ii) ataxia, (iii) loss of gait reflex, (iv) absence of righting reflex within 60 s, and (v) separation, with no feeding, of moribund offspring by the female adult mouse.
Experimental infection and treatment.
(i) Neonate model. Three-day-old mice were infected intraperitoneally with 2 × 102 PFU (61, 62) unless otherwise mentioned. Sofosbuvir treatments were administered with 20 mg/kg/day sofosbuvir intraperitoneally. Treatment was started 1 day prior to infection (pretreatment) or 2 days after infection (late treatment). In both cases, treatment was administered for 6 days. For comparison, mock-infected and mock-treated groups were used as controls. The animals were monitored daily for survival, weight gain, and virus-induced short-term sequelae (righting in up to 60 s).
(ii) Arthralgia model. The arthralgia model was adapted from previous publications (63, 64). Male Swiss Webster mice (8 weeks old; 20 to 25 g) were infected with 2 × 105 PFU in the right hind paw toward the ankle. Sofosbuvir was administered orally (20 mg/kg) beginning 1 h before the first virus injection. Treatment was conducted for 6 days. The control group was injected with 50 μl RPMI. Paw edema was evaluated from day 1 to 6 after infection by hydropletismometer for small volumes (Ugo Basile, Milan, Italy), and data were acquired as the paw volume (in milliliters).
On day 6 after infection, the animals were euthanized and samples processed for RT-PCR detection of virus RNA and for histopathology study of the paw. For RT-PCR, serum was collected from cardiac puncture, and the paw was dissected, the fingers and ankle were removed, and tissues were homogenized in PBS. For histopathology, the paws were fixed in 4% buffered paraformaldehyde for 2 days and decalcified using 10% EDTA for 15 days, with gentle rocking and daily replacement of the solution. The decalcified paws were cryoprotected in 20% sucrose at 4°C, embedded in OCT (Tissue-Tek Sakura Finetek, Taipei, Taiwan) for frozen sectioning on a cryotome (5 μm; Leica Microsystems, Wetzlar, Germany), stained with hematoxylin and eosin (H&E), and photographed using an Axio Imager microscope (Zeiss, Jena, Germany).
(iii) Behavioral tests. To test the righting reflex, the mice were tested daily during the course of acute infection. The animals were placed in a supine position with all four paws facing up for 5 s. They were then released, and the time taken to flip over onto the stomach with all four paws touching the surface was recorded. Each trial spanned a maximum of 60 s, and the animals were tested twice a day with a 5-min minimum interval between trials. For each animal, the lowest time was plotted in a graph. Animals that failed the test were included in the graph with a time of 60 s.
Statistical analysis.
All assays were performed and codified by one professional. Subsequently, a different professional analyzed the results before the experimental groups were identified. This approach was used to retain blinding of the pharmacological assays. All experiments, including technical replicates in each assay, were performed at least three times independently. The dose-response curves used to calculate the EC50 and CC50 values were generated by Excel for Windows using GraphPad Prism 5.0 software. The significance of the survival curves was evaluated using the log-rank (Mantel-Cox) test. The equations to fit the best curve were generated based on an R2 of ≥0.9. Analysis of variance (ANOVA), followed by Tukey’s post hoc test, was also used, with P values of <0.05 considered statistically significant. The statistical analyses specific to each software program used in the bioinformatics analysis are described above.
ACKNOWLEDGMENTS
Funding was provided by the National Council for Scientific and Technological Development (CNPq), Ministry of Science, Technology, Information and Communications (grants including, but not limited to, 465313/2014-0), Ministry of Education/CAPES (465313/2014-0), Research Foundation of the State of Rio de Janeiro (FAPERJ) (grants including, but not limited to, 465313/2014-0), and the Oswaldo Cruz Foundation (Fiocruz). A competing financial interest exists for K.B., who is partner in a consortium able to produce generic sofosbuvir. The consortium and her participation were limited to providing sofosbuvir for this study. The BMK Consortium is comprised of Blanver Farmoquímica Ltd., Microbiológica Química e Farmacêutica Ltd., and Karin Bruning & Cia Ltd. A.C.F., P.A.R., C.S.D.F., C.Q.S., L.V.B.H., M.M.B., M.M., N.R., I.G.A.Q., C.S.G.P., L.R.Q.S., E.C.L., P.T., Y.R.V., and G.B.-L. performed the experiments. H.C.C.F.N., N.B., S.K.R., K.B., F.A.B., P.T.B., and T.M.L.S. conceptualized the experiments and raised funds. K.B. provided critical material. T.M.L.S. organized the study. All authors revised and approved the manuscript. Funders had no role in the experiment design or interpretation.
REFERENCES
- 1.Pialoux G, Gaüzère B-A, Jauréguiberry S, Strobel M. 2007. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 7:319–327. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 2.Pinheiro TJ, Guimarães LF, Silva MTT, Soares CN. 2016. Neurological manifestations of Chikungunya and Zika infections. Arq Neuropsiquiatr 74:937–943. doi: 10.1590/0004-282X20160138. [DOI] [PubMed] [Google Scholar]
- 3.Souza TMA, Azeredo EL, Badolato-Corrêa J, Damasco PV, Santos C, Petitinga-Paiva F, Nunes PCG, Barbosa LS, Cipitelli MC, Chouin-Carneiro T, Faria NRC, Nogueira RMR, de Bruycker-Nogueira F, dos Santos FB. 2017. First report of the East-Central South African genotype of chikungunya virus in Rio de Janeiro, Brazil. PLoS Curr 9:ecurrents.outbreaks.4200119978d62ccaa454599cd2735727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlys da Costa A, Thézé J, Komninakis SCV, Sanz-Duro RL, Felinto MRL, Moura LCC, Barroso IM, Santos LEC, Nunes MA, Moura AA, Lourenço J, Deng X, Delwart EL, Guimarães MR, Pybus OG, Sabino EC, Faria NR. 2017. Spread of chikungunya virus East/Central/South African genotype in northeast Brazil. Emerg Infect Dis 23:1742–1744. doi: 10.3201/eid2310.170307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes MRT, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, Azevedo RDSDS, Dea DS, Evp DS, da Silva SP, Carvalho VL, Coelho GE, Cruz ACR, Rodrigues SG, da Silva Gonçalves Vianez JL, Nunes BTD, Cardoso JF, Tesh RB, Hay SI, Pybus OG, da Costa Vasconcelos PF. 2015. Emergence and potential for spread of chikungunya virus in Brazil. BMC Med 13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen MW, Tan YB, Zheng J, Zhao Y, Lim BT, Cornvik T, Lescar J, Ng LFP, Luo D. 2017. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antiviral Res 143:38–47. doi: 10.1016/j.antiviral.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem 275:25523–25532. doi: 10.1074/jbc.M002671200. [DOI] [PubMed] [Google Scholar]
- 9.Singh H, Bhatia H, Grewal N, Natt N. 2014. Sofosbuvir: a novel treatment option for chronic hepatitis C infection. J Pharmacol Pharmacother 5:278. doi: 10.4103/0976-500X.142464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilead Sciences. 2015. Product monograph, Pr sovaldi (sofosbuvir) tablets 400 mg sofosbuvir. Gilead Sciences, Foster City, CA. [Google Scholar]
- 11.European Medicines Agency. 2013. Committee for Medicinal Products for Human Use (CHMP) assessment report. European Medicines Agency, London, United Kingdom. [Google Scholar]
- 12.Ferreira AC, Zaverucha-do-Valle C, Reis PA, Barbosa-Lima G, Vieira YR, Mattos M, Silva PDP, Sacramento C, de Castro Faria Neto HC, Campanati L, Tanuri A, Brüning K, Bozza FA, Bozza PT, Souza TML. 2017. Sofosbuvir protects Zika virus-infected mice from mortality, preventing short- and long-term sequelae. Sci Rep 7:9409. doi: 10.1038/s41598-017-09797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullard-Feibelman KM, Govero J, Zhu Z, Salazar V, Veselinovic M, Diamond MS, Geiss BJ. 2017. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res 137:134–140. doi: 10.1016/j.antiviral.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sacramento CQ, de Melo GR, de Freitas CS, Rocha N, Hoelz LVB, Miranda M, Fintelman-Rodrigues N, Marttorelli A, Ferreira AC, Barbosa-Lima G, Abrantes JL, Vieira YR, Bastos MM, de Mello Volotão E, Nunes EP, Tschoeke DA, Leomil L, Loiola EC, Trindade P, Rehen SK, Bozza FA, Bozza PT, Boechat N, Thompson FL, de Filippis AMB, Brüning K, Souza TML. 2017. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci Rep 7:40920. doi: 10.1038/srep40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, Han W, Pletikos M, Gao T, Zhu Y, Bichsel C, Varela L, Szigeti-Buck K, Lisgo S, Zhang Y, Testen A, Gao X-B, Mlakar J, Popovic M, Flamand M, Strittmatter SM, Kaczmarek LK, Anton ES, Horvath TL, Lindenbach BD, Sestan N. 2016. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep 16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H-T, Colby-Germinario SP, Hassounah SA, Fogarty C, Osman N, Palanisamy N, Han Y, Oliveira M, Quan Y, Wainberg MA. 2017. Evaluation of sofosbuvir (β-D-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine) as an inhibitor of dengue virus replication. Sci Rep 7:6345. doi: 10.1038/s41598-017-06612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitas CD, Higa L, Sacramento C, Ferreira A, Reis P, Delvecchio R, Monteiro F, Barbosa-Lima G, Vieira Y, Mattos M, Hoelz L, Leme R, Bastos M, Bozza F, Bozza P, Boechat N, Tanuri A, Souza T. 2018. Yellow fever virus is susceptible to sofosbuvir both in vitro and in vivo. bioRxiv 10.1101/266361. [DOI] [PMC free article] [PubMed]
- 18.Vicenti I, Boccuto A, Giannini A, Dragoni F, Saladini F, Zazzi M. 2018. Comparative analysis of different cell systems for Zika virus (ZIKV) propagation and evaluation of anti-ZIKV compounds in vitro. Virus Res 244:64–70. doi: 10.1016/j.virusres.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Ganesan VK, Duan B, Reid SP. 2017. Chikungunya virus: pathophysiology, mechanism, and modeling. Viruses 9:368. doi: 10.3390/v9120368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inglis FM, Lee KM, Chiu KB, Purcell OM, Didier PJ, Russell-Lodrigue K, Weaver SC, Roy CJ, MacLean AG. 2016. Neuropathogenesis of Chikungunya infection: astrogliosis and innate immune activation. J Neurovirol 22:140–148. doi: 10.1007/s13365-015-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangeetha K, Purushothaman I, Rajarajan S. 2017. Spectral characterisation, antiviral activities, in silico ADMET and molecular docking of the compounds isolated from Tectona grandis to chikungunya virus. Biomed Pharmacother 87:302–310. doi: 10.1016/j.biopha.2016.12.069. [DOI] [PubMed] [Google Scholar]
- 22.Rothan HA, Bahrani H, Mohamed Z, Teoh TC, Shankar EM, Rahman NA, Yusof R. 2015. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS One 10:e0126360. doi: 10.1371/journal.pone.0126360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur P, Thiruchelvan M, Lee RCH, Chen H, Chen KC, Ng ML, Chu JJH. 2013. Inhibition of chikungunya virus replication by harringtonine, a novel antiviral that suppresses viral protein expression. Antimicrob Agents Chemother 57:155–167. doi: 10.1128/AAC.01467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz DJM, Bonotto RM, Gomes RGB, da Silva CT, Taniguchi JB, No JH, Lombardot B, Schwartz O, Hansen MAE, Freitas-Junior LH. 2013. Identification of novel compounds inhibiting chikungunya virus-induced cell death by high throughput screening of a kinase inhibitor library. PLoS Negl Trop Dis 7:e2471. doi: 10.1371/journal.pntd.0002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan M, Dhanwani R, Patro IK, Rao PVL, Parida MM. 2011. Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antiviral Res 89:1–8. doi: 10.1016/j.antiviral.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Mason PJ, Haddow AJ. 1957. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53; an additional note on Chikungunya virus isolations and serum antibodies. Trans R Soc Trop Med Hyg 51:238–240. doi: 10.1016/0035-9203(57)90022-6. [DOI] [PubMed] [Google Scholar]
- 27.Weaver SC. 2014. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 8:e2921. doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnamoorthy K, Harichandrakumar KT, Krishna Kumari A, Das LK. 2009. Burden of chikungunya in India: estimates of disability adjusted life years (DALY) lost in 2006 epidemic. J Vector Borne Dis 46:26–35. [PubMed] [Google Scholar]
- 29.Svs MS. 2018. Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 8 de 2018. http://portalarquivos2.saude.gov.br/images/pdf/2018/outubro/08/BE-N---40-Monitoramento-dos-casos-de-dengue--febre-de-chikungunya-e-febre-pelo-v--rus-Zika-at---a-Semana-Epidemiol--gica-SE-36-de-2018.pdf.
- 30.World Health Organization. 2008. Guidelines on clinical management of chikungunya fever. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 31.Gasque P, Bandjee MCJ, Reyes MM, Viasus D. 2016. Chikungunya pathogenesis: from the clinics to the bench. J Infect Dis 214:S446–S448. doi: 10.1093/infdis/jiw362. [DOI] [PubMed] [Google Scholar]
- 32.Abdelnabi R, Neyts J, Delang L. 2015. Towards antivirals against chikungunya virus. Antiviral Res 121:59–68. doi: 10.1016/j.antiviral.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghese FS, Kaukinen P, Gläsker S, Bespalov M, Hanski L, Wennerberg K, Kümmerer BM, Ahola T. 2016. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res 126:117–124. doi: 10.1016/j.antiviral.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Mastrangelo E, Pezzullo M, De Burghgraeve T, Kaptein S, Pastorino B, Dallmeier K, de Lamballerie X, Neyts J, Hanson AM, Frick DN, Bolognesi M, Milani M. 2012. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J Antimicrob Chemother 67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varghese FS, Thaa B, Amrun SN, Simarmata D, Rausalu K, Nyman TA, Merits A, McInerney GM, Ng LFP, Ahola T. 2016. The antiviral alkaloid berberine reduces chikungunya virus-induced mitogen-activated protein kinase signaling. J Virol 90:9743–9757. doi: 10.1128/JVI.01382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang H-R, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HMM, Niu C, Otto MJ, Furman PA. 2010. Discovery of a β-d-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem 53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 38.Brizzi K. 2017. Neurologic manifestation of Chikungunya virus. Curr Infect Dis Rep 19:6. doi: 10.1007/s11908-017-0561-1. [DOI] [PubMed] [Google Scholar]
- 39.da Silva-Júnior EF, Leoncini GO, Rodrigues ÉES, Aquino TM, Araújo-Júnior JX. 2017. The medicinal chemistry of chikungunya virus. Bioorg Med Chem 25:4219–4244. doi: 10.1016/j.bmc.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallegos KM, Drusano GL, D Argenio DZ, Brown AN. 2016. Chikungunya virus: in vitro response to combination therapy with ribavirin and interferon alfa 2a. J Infect Dis 214:1192–1197. doi: 10.1093/infdis/jiw358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delang L, Segura Guerrero N, Tas A, Quérat G, Pastorino B, Froeyen M, Dallmeier K, Jochmans D, Herdewijn P, Bello F, Snijder EJ, de Lamballerie X, Martina B, Neyts J, van Hemert MJ, Leyssen P. 2014. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J Antimicrob Chemother 69:2770–2784. doi: 10.1093/jac/dku209. [DOI] [PubMed] [Google Scholar]
- 42.Briolant S, Garin D, Scaramozzino N, Jouan A, Crance JM. 2004. In vitro inhibition of Chikungunya and Semliki Forest viruses replication by antiviral compounds: synergistic effect of interferon-alpha and ribavirin combination. Antiviral Res 61:111–117. doi: 10.1016/j.antiviral.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira A, Valle C, Reis P, Barbosa-Lima G, Vieira Y, Mattos M, Silva P, Sacramento C, Neto H, Campanati L, Tanuri A, Bruning K, Bozza F, Bozza P, Souza TML. 2017. Sofosbuvir protects Zika virus-infected mice from mortality, preventing short- and long-term sequela. Sci Rep 6:9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muthuri SG, Venkatesan S, Myles PR, Leonardi-Bee J, Al Khuwaitir TSA, Al Mamun A, Anovadiya AP, Azziz-Baumgartner E, Báez C, Bassetti M, Beovic B, Bertisch B, Bonmarin I, Booy R, Borja-Aburto VH, Burgmann H, Cao B, Carratala J, Denholm JT, Dominguez SR, Duarte PAD, Dubnov-Raz G, Echavarria M, Fanella S, Gao Z, Gérardin P, Giannella M, Gubbels S, Herberg J, Iglesias ALH, Hoger PH, Hu X, Islam QT, Jiménez MF, Kandeel A, Keijzers G, Khalili H, Knight M, Kudo K, Kusznierz G, Kuzman I, Kwan AMC, Amine IL, Langenegger E, Lankarani KB, Leo Y-S, Linko R, Liu P, Madanat F, Mayo-Montero E, McGeer A, Memish Z, Metan G, Mickiene A, Mikić D, Mohn KGI, Moradi A, Nymadawa P, Oliva ME, Ozkan M, Parekh D, Paul M, Polack FP, Rath BA, Rodríguez AH, Sarrouf EB, Seale AC, Sertogullarindan B, Siqueira MM, Skręt-Magierło J, Stephan F, Talarek E, Tang JW, To KKW, Torres A, Törün SH, Tran D, Uyeki TM, Van Zwol A, Vaudry W, Vidmar T, Yokota RTC, Zarogoulidis P, Nguyen-Van-Tam JS, PRIDE Consortium Investigators, Nguyen-Van-Tam JS. 2014. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med 2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G-Y, Li B-G, Yang T, Liu G-Y, Zhang G-L. 2006. Chaetoindicins A-C, three isoquinoline alkaloids from the fungus Chaetomium indicum. Org Lett 8:3613–3615. doi: 10.1021/ol061525k. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Wang Y-F, Shen C-H, Agniswamy J, Rao KV, Xu C-X, Ghosh AK, Harrison RW, Weber IT. 2013. Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease. J Med Chem 56:1074–1083. doi: 10.1021/jm301519z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, Boyd MR. 1988. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res 48:4827–4833. [PubMed] [Google Scholar]
- 48.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y, Shin S, Jha BS, Liu Q, Sheng J, Li F, Zhan M, Davis J, Bharti K, Zeng X, Rao M, Malik N, Vemuri MC. 2013. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med 2:862–870. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauter NK, Hanson JE, Glick GD, Brown JH, Crowther RL, Park SJ, Skehel JJ, Wiley DC. 1992. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry 31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 51.Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, Campbell GL. 2007. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gazos-Lopes F, Oliveira MM, Hoelz LVB, Vieira DP, Marques AF, Nakayasu ES, Gomes MT, Salloum NG, Pascutti PG, Souto-Padrón T, Monteiro RQ, Lopes AH, Almeida IC. 2014. Structural and functional analysis of a platelet-activating lysophosphatidylcholine of Trypanosoma cruzi. PLoS Negl Trop Dis 8:e3077. doi: 10.1371/journal.pntd.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 56.Eisenberg D, Lüthy R, Bowie JU. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404. doi: 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 57.Thomsen R, Christensen MH. 2006. MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 59.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 60.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. 2012. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. Osteoarthritis Cartilage 20:256–260. doi: 10.1016/j.joca.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 61.van den Pol AN, Mao G, Yang Y, Ornaghi S, Davis JN. 2017. Zika virus targeting in the developing brain. J Neurosci 37:2161–2175. doi: 10.1523/JNEUROSCI.3124-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W-C, Abraham R, Shim B-S, Choe H, Page DT. 2016. Zika virus infection during the period of maximal brain growth causes microcephaly and corticospinal neuron apoptosis in wild type mice. Sci Rep 6:34793. doi: 10.1038/srep34793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, Morrison TE. 2013. Chronic joint disease caused by persistent chikungunya virus infection is controlled by the adaptive immune response. J Virol 87:13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya Virus arthritis in adult wild-type mice. J Virol 84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]