Commonly used antibiotics exert their effects predominantly on rapidly growing bacterial cells; yet, the growth dynamics taking place during infection in a complex host environment remain largely unknown. Hence, a means to measure in situ bacterial growth rate is essential to predict the outcome of antibacterial treatment.
KEYWORDS: bacterial growth, chromosome replication, Escherichia coli, experimental animal model
ABSTRACT
Commonly used antibiotics exert their effects predominantly on rapidly growing bacterial cells; yet, the growth dynamics taking place during infection in a complex host environment remain largely unknown. Hence, a means to measure in situ bacterial growth rate is essential to predict the outcome of antibacterial treatment. We have recently validated chromosome replication as a readout of in situ bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. By the use of two complementary methods (quantitative PCR and fluorescence microscopy) for differential genome origin and terminus copy number quantification, we demonstrated the ability to track bacterial growth rate, both on a population average level and on a single-cell level, from one single biological specimen. Here, we asked whether the in situ growth rate predicts antibiotic treatment effect during infection in the same model. Parallel in vitro growth experiments were conducted as a proof of concept. Our data demonstrate that the activities of the commonly used antibiotics ceftriaxone and gentamicin correlated with pretreatment bacterial growth rate; both drugs performed better during rapid growth than during slow growth. Conversely, ciprofloxacin was less sensitive to bacterial growth rate, both in a homogenous in vitro bacterial population and in a more heterogeneous in vivo bacterial population. The method serves as a platform to test any antibiotic’s dependency on active in situ bacterial growth. Improved insight into this relationship in vivo could ultimately prove helpful in evaluating future antibacterial strategies.
INTRODUCTION
Studies of antibacterial activity are largely based on laboratory models, where balanced bacterial populations propagate rapidly in well-defined batch cultures with a finite quantity of life-sustaining nutrients. These in vitro models fail to mirror the true growth dynamics of bacterial pathogens taking place during infection in vivo, where growth in a bacterial population appears to be both slower and less homogeneous (1–3). The dependency on active bacterial growth for most antibiotics to exert their effect is acknowledged and has been examined by other investigators, both in vitro and in vivo (4–11). However, these studies were largely based on bacterial count kinetics as a direct measure of bacterial growth rate. This measure could be misleading during infection in vivo, where the net change in the bacterial count is a function not only of growth but also of the elimination of bacterial cells by the host immune system, a factor not taken into account in the bacterial count kinetics method. Moreover, the method fails to report on any growth heterogeneity within the bacterial population. Hence, there is a need for refined means to measure bacterial growth that can extend into clinical use. To date, no gold standard method for measuring in vivo bacterial growth rate exists. In recent years, it has been demonstrated that it is possible to extract direct measures of in vivo bacterial growth rate by analyzing differential genome coverage from whole-genome sequencing data (12, 13). This method circumvents the limitation of a nonquantifiable bacterial elimination factor, as it reports directly on the bacteria’s physiological state. Nonetheless, it reports merely on the mean population growth rate.
Most bacterial chromosomes are circular with a single origin of replication (oriC), from where chromosome replication is initiated and carried out bidirectionally toward a single oppositely located terminus (terC) during bacterial growth (14, 15). In Escherichia coli, it is acknowledged, from decades of in vitro studies, that bacterial growth rate is a function of growth conditions and is precisely coordinated with genome replication (16–18). When growth conditions are favorable, overlapping rounds of synchronously initiated bidirectional chromosome replication occur, allowing for the presence of more than two oriCs (number of oriC copies = 2n [n = 2, 3, or 4]) in rapidly growing cells (17, 19). In contrast, when growth conditions are disadvantageous, no or merely one round of chromosome replication occurs, allowing for the presence of only one or a maximum of two oriCs (number of oriC copies = 2n [n = 0 or 1]) in nongrowing or slowly growing cells (19). Hence, the copy number ratio of oriC to terC (ori:ter) reflects the bacterial population growth rate: during rapid growth, larger fractions of cells undergo one or more rounds of chromosome replication (i.e., mean population ori:ter of ≥2) and during slow or no growth, only few cells will undergo chromosome replication (i.e., mean population ori:ter of ∼1) (20). By the use of two complementary methods for measuring ori:ter, quantitative PCR (qPCR) and fluorescence microscopy, we have been able to probe in situ growth rates of fluorescently labeled E. coli ATCC 25922, both on a population average level (by qPCR) and on a single-cell level (by fluorescence microscopy) during widespread infection in the mouse peritonitis model (3). We demonstrated a correlation between oriC and bacterial cell size at all growth rates and the ability of ori:ter to predict the development in net bacterial population size. Moreover, in this recent observation of growth dynamics during host infection, we found that growth rates were largely heterogeneous within the bacterial populations propagating both in the peritoneum and in the blood throughout the duration of infection (3). This finding is consistent with previous reports of Staphylococcus aureus growth rate heterogeneity in cystic fibrosis sputum, as measured by isotope tracing (1). These observations underscore the need for refined and easily accessible methods to measure in situ bacterial growth rate during various types of infection and its causal relationship with the outcome of antibacterial treatment.
Here, we extended the approach of using chromosome replication as a readout of in vivo bacterial growth rate in the mouse peritonitis model to explore its potential in predicting antibacterial treatment effect. For comparison, we chose a representative drug from each of three classes of commonly used bactericidal antibiotics with different cellular targets: ceftriaxone (CRO; a β-lactam), ciprofloxacin (CIP; a fluoroquinolone), and gentamicin (GEN; an aminoglycoside) (21). We hypothesized that the drugs tested would perform better when given during rapid than during slow bacterial growth.
RESULTS
We defined the minimal and maximal growth rates of E. coli during infection in the mouse peritonitis model and compared the activities of standardized antibacterial dosing regimens given during either rapid or slow bacterial growth, as defined by qPCR-derived ori:ter from infected body fluids. As a qualitative control, fluorescence microscopy was applied for the same materials to demonstrate any treatment-induced physiological change in cell morphology or chromosome replication at a single-cell level. As a proof of concept, corresponding in vitro treatment experiments using the same model infective organism, the EUCAST and CLSI reference strain E. coli ATCC 25922, were carried out.
Antibacterial activity as a function of growth rate in vitro.
E. coli growth experiments in batch cultures were performed to validate the methods applied in the experimental in vivo infection model. For these experiments, we used lysogeny broth (LB), a rich medium that supports rapid bacterial growth, allowing the cells to reach stationary phase due to exhaustion of utilizable carbon sources before any significant physiological alterations occurred (22). Here, stationary-phase bacterial cells were diluted in fresh medium and allowed to grow, with repeated sample collections at 4, 6, 8, and 10 h of incubation. All antibiotics were given as a single dose during rapid or slow bacterial growth. Antibiotic concentrations were standardized, defined from previous studies, and corresponded to serum concentrations observed in humans on standard dosing regimens: ceftriaxone (CRO), 30 mg/liter; ciprofloxacin (CIP), 1 mg/liter; gentamicin (GEN), 10 mg/liter (23–28). Sampling was performed after 2 h of antibiotic exposure. There was a difference in pretreatment bacterial count at the time points chosen for rapid and slow bacterial growth treatment induction, respectively. Hence, antibiotic activity was measured as the difference between pre- and posttreatment bacterial count relative to the pretreatment bacterial count (i.e., relative Δlog10 CFU/ml) for comparison.
We tested the wild-type bacterial strain (ATCC 25922) in parallel with the genetically modified derivative of the strain with chromosomally incorporated fluorescent oriC and terC labels (ALO 4783) to ensure the absence of growth retardation due to transgene insertion. As no growth differences were observed (Fig. 1a), in vitro data from both versions of the strain were pooled for analysis. In regard to the gentamicin treatment regimen, however, only wild-type ATCC 25922 was applied, as the gentamicin MIC was increased by the presence of the nonremovable kanamycin cassette encoding kanamycin phosphotransferase, used as a clonal selection marker, in ALO 4783 (3, 29) (Table 1).
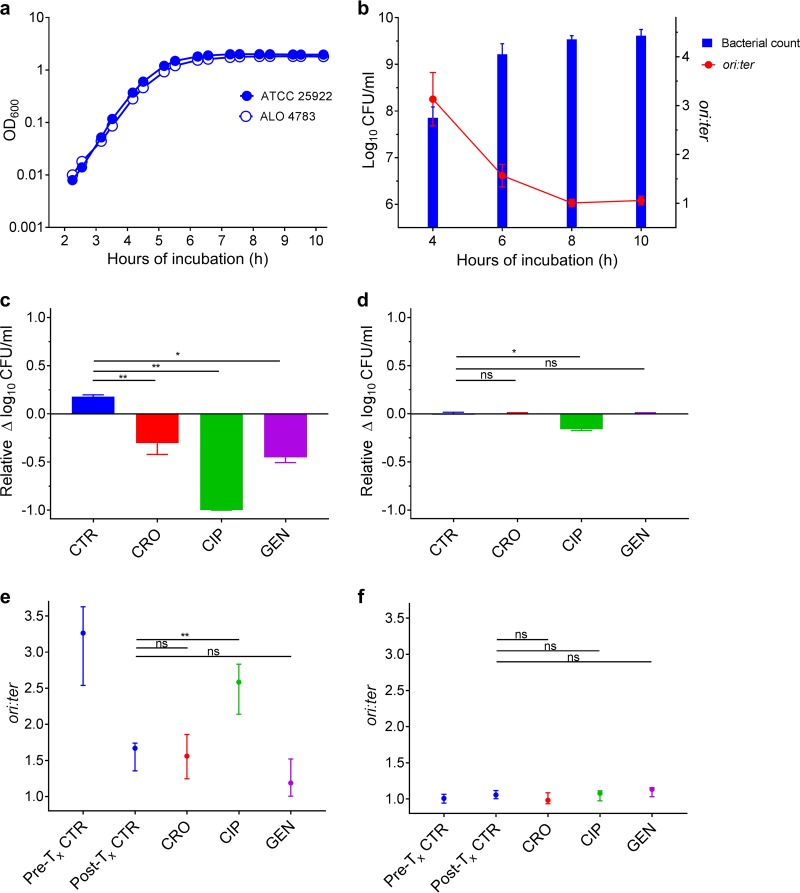
FIG 1.
Antibiotic activity as a function of bacterial growth rate in vitro. (a) Parallel growth of ALO 4783 relative to the ATCC 25922 wild type in LB revealed no growth retardation due to transgene insertions. Cell density measured as optical density at 600 nm (OD600). (b) Bacterial counts (CFU/ml) and growth rates (ori:ter) in untreated control batch cultures (ATCC 25922 and ALO 4783); n = 6. Data presented as means (SDs). (c) Bacterial count reductions after 2 h of antibiotic exposure in ceftriaxone (CRO), ciprofloxacin (CIP), and gentamicin (GEN) treatment batch cultures when therapy was induced during rapid bacterial growth (i.e., at 4 h of incubation). Controls (CTR) received no antibiotic therapy. (d) Bacterial count reductions after 2 h of antibiotic exposure in treatment batch cultures when therapy was induced during slow bacterial growth (i.e., at 8 h of incubation). CTR received no antibiotic therapy. For comparison of activities between treatment inductions during rapid and slow growth, data in panels c and d are presented as relative bacterial count reductions. (e) Bacterial growth rates (ori:ter) in pretreatment controls (Pre-Tx CTR) and posttreatment controls (Post-Tx CTR) (i.e., at 4 and 6 h of incubation, respectively) and in treatment batch cultures after 2 h of antibiotic exposure when therapy was induced during rapid bacterial growth. (f) Bacterial growth rates (ori:ter) in Pre-Tx and Post-Tx controls (i.e., at 8 and 10 h of incubation, respectively) and in treatment batch cultures after 2 h of antibiotic exposure when therapy was induced during slow bacterial growth. Data in panels c to f are presented as medians and interquartile ranges (IQRs). CTR, n = 6; CRO, n = 6; CIP, n = 6; GEN, n = 3. *, P < 0.05; **, P < 0.01; ns, P > 0.05 by Mann-Whitney U test.
TABLE 1.
Overview of MICs and susceptibility interpretation of the two isogenic strains utilized in in vitro and in vivo experiments
| Antibiotica | Phenotypeb | MIC (mg/liter) | Interpretive category (EUCAST/CLSI)c |
|---|---|---|---|
| CRO | WT | 0.125 | S/S |
| MUT | 0.125 | S/S | |
| CIP | WT | 0.016 | S/S |
| MUT | 0.016 | S/S | |
| GEN | WT | 0.125 | S/S |
| MUT | 1 | S/S |
CRO, ceftriaxone; CIP, ciprofloxacin; GEN, gentamicin.
WT, wild type (ATCC 25922); MUT, mutant (ALO 4783).
S, susceptible.
Antibiotics administered during rapid bacterial growth in vitro.
The bacterial populations propagating in batch cultures reached maximal growth rates at 4 h of incubation (mean and standard deviation [SD] ori:ter = 3.13 [0.55]) (Fig. 1b). At this stage, it has been demonstrated that E. coli population growth in LB is close to balanced and dominated by large cells growing with overlapping rounds of chromosome replication (3). This is exemplified in Fig. 2A, illustrating a large bacterial cell with multiple oriCs. As a consequence of rapid bacterial growth, there was a subsequent increase in population size (Fig. 1a and b). Between 4 and 6 h of incubation, the growth started to slow down, illustrated by a reduction of ori:ter at 6 h of incubation followed by a minimal increase in net population size (Fig. 1a and b).
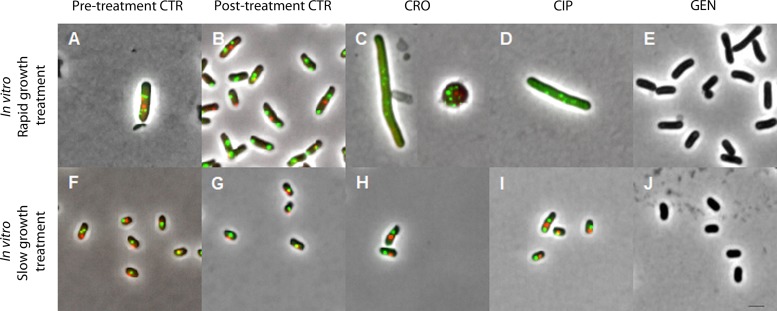
FIG 2.
Representative examples of pooled bacterial cells observed by fluorescence microscopy and isolated after antibiotic induction during rapid (top row) or slow (bottom row) bacterial growth in vitro. Images are shown at the same magnification (using a 100× objective) in phase contrast; intracellular oriC foci in green (green fluorescent protein [GFP]) and terC foci in red (mCherry) (ALO 4783). For GEN treatment experiments, ATCC 25922 without fluorescent foci was utilized. A total of 500 pooled cells were analyzed per time point from all cultures with the exception of rapid growth treatment cultures: CRO, n = 28; CIP, n = 112; GEN, n = 330. Due to the limited resolution of fluorescence microscopy for colocalizing oriCs, some bacterial cells with overlapping chromosome replication origins may appear with too few foci (33). Mean (SD) population medial axis cell lengths were as follows: (A) rapid bacterial growth, pretreatment CTR, 4.1 (0.98) μm; (B) rapid bacterial growth, posttreatment CTR, 3.51 (0.86) μm; (C) rapid bacterial growth, post-CRO treatment, not determined (due to overrepresentation of spherical cells); (D) rapid bacterial growth, post-CIP treatment, 7.39 (2.52) μm; (E) rapid bacterial growth, post-GEN treatment, 3.25 (0.84) μm; (F) slow bacterial growth, pretreatment CTR, 2.58 (0.66) μm; (G) slow bacterial growth, posttreatment CTR, 2.25 (0.52) μm; (H) slow bacterial growth, post-CRO treatment, 2.67 (0.67) μm; (I) slow bacterial growth, post-CIP treatment, 2.53 (0.69) μm; (J) slow bacterial growth, post-GEN treatment, 2.18 (0.45) μm. CTR, controls; CRO, ceftriaxone; CIP, ciprofloxacin; GEN, gentamicin. Scale bar, 2 μm.
When introduced into batch cultures of rapidly growing cells, all antibiotics resulted in significant bacterial count reduction compared to that of controls (CRO and CIP, P < 0.001; GEN, P < 0.01) (Fig. 1c). Correspondingly, single-cell microscopic analysis of pooled live bacterial cells from treatment batch cultures revealed that the majority of the cells isolated after exposure to either ceftriaxone or ciprofloxacin were affected by the treatment when it was introduced during rapid bacterial growth (Fig. 2C and D, respectively). These bacterial populations were represented predominantly by spherical and filamentous cells (ceftriaxone) or elongated cells (ciprofloxacin); all with multiple fluorescent foci, representing direct or indirect chromosome replication disturbance. Unfortunately, we were unable to accurately quantify the population distributions of oriC and terC due to multiple, overlapping fluorescent foci in these treatment groups. The morphological changes in the ceftriaxone-exposed bacterial populations suggest that ceftriaxone exerted its bactericidal effect through cell wall inhibition predominantly by binding to penicillin-binding proteins (PBPs) 2 and 3, in accordance with previous studies (30). We underscore that the microscopy data from bacterial cultures treated with antibiotics during rapid growth (Fig. 2C to E) are subject to uncertainty, given that only few live bacterial cells (n < 500) were isolated due to the efficient bacterial killing (Fig. 1c). Nevertheless, ori:ter values extracted from the qPCR data complemented the above-described microscopy findings regarding chromosome replication: high ori:ter ratios were observed where photomicrographs were dominated by large cells with multiple oriCs and low ori:ter ratios were observed where photomicrographs were dominated by small cells with few oriCs in both treatment and control groups (Fig. 1e and 2A to E). Only ciprofloxacin administered during rapid bacterial growth entailed significantly higher ori:ter ratios than those from posttreatment controls (P < 0.01) (Fig. 1e). As to ceftriaxone and gentamicin treatments administered during rapid bacterial growth, both induced decreases in ori:ter toward ∼1, similar to that of the control group (Fig. 1e). For gentamicin-treated bacterial cells, microscopic visualization of oriC and terC was not possible, as only the wild-type ATCC 25922 was utilized. However, the size and morphology of the cells did not appear to differ from those of the respective control population (Fig. 2E and B).
Antibiotics administered during slow bacterial growth in vitro.
At 8 h of incubation, minimal bacterial growth rates (mean [SD] ori:ter, 1.01 [0.07]) were reached due to nutrient starvation (Fig. 1b). At this stage, it has been demonstrated that the bacterial population is dominated by small bacterial cells without ongoing chromosome replication, i.e., a stage with complete or near complete cessation of growth (3). This is exemplified in Fig. 2F, illustrating small bacterial cells with predominantly one oriC/cell. Consequently, there was no significant subsequent net population size increase, and all parameters remained largely unchanged by 10 h of incubation (Fig. 1a and b and 2G). When antibiotic regimens identical to those applied during rapid bacterial growth were introduced into this population of slowly/nongrowing bacterial cells, only ciprofloxacin caused a significant bacterial count reduction (P < 0.01), albeit considerably less than when administered during rapid growth, where a near total clearance of cells was observed (Fig. 1c and d). Ceftriaxone and gentamicin treatment effects were absent (Fig. 1d). Correspondingly, photomicrographs were dominated by cells largely unaffected by all three antibiotics, when compared to posttreatment controls (Fig. 2G to J), and no significant change in ori:ter was observed in any treatment group compared to posttreatment controls (Fig. 1f). The ciprofloxacin-induced increase in ori:ter observed during rapid bacterial growth treatment (Fig. 1e and Fig. 2D) was absent when ciprofloxacin was added to a population of cells largely without ongoing chromosome replication (mean population ori:ter ∼1) (Fig. 1f and 2I).
In summary, during controlled bacterial growth in a closed rich medium batch culture, extreme situations of both rapid growth and complete or near complete cessation of growth were provoked. When identical antibiotic treatment regimens were introduced to cultures of bacterial populations growing at either a maximal or minimal growth rate, the dependency of active bacterial growth for all drugs to exert their effect became evident. The significant bacterial load reductions observed when ceftriaxone or gentamicin was added to rapidly growing bacterial populations were lost when identical treatment regimens were induced during slow bacterial growth. Ciprofloxacin, however, was less sensitive to active bacterial growth, as a significant reduction in bacterial load was observed upon administration during both rapid and slow growth.
Antibacterial activity as a function of growth rate in vivo.
In the experimental mouse peritonitis model, a total of 54 mice pooled from 4 independent experiments were challenged intraperitoneally with stationary-phase E. coli. All animals developed widespread infection within 2 h postchallenge. Maximal and minimal bacterial growth rates were successfully probed from infected body fluids (peritoneal lavage fluid [PLF] and blood) (3). During propagation in this infection model, bacterial growth was overall slower than that in vitro, yet never came to a complete cessation (i.e., ori:ter remained >1 at all times) during the experiment (Fig. 3a). All antibiotics were administered during rapid or slow bacterial growth as a single dose subcutaneously (s.c.) as follows (concentration and optimal pharmacokinetic/pharmacodynamic [PKPD] parameters mentioned in parentheses): ceftriaxone 5 mg (178 mg/kg, time that the drug concentration exceeds the MIC under steady-state pharmacokinetic conditions [T>MIC]); ciprofloxacin 0.4 mg (14 mg/kg, area under the concentration-time curve over 24 h in the steady state divided by the MIC [AUC/MIC]); and gentamicin 1 mg (36 mg/kg; AUC/MIC). Antibiotic concentrations were defined from previous studies (23–28) and all were greater than 10× MIC (Table 1). Doses were intended to simulate standard human doses of the same antibiotics. However, due to the rapid elimination in mice, the doses were chosen as a compromise between maximum concentration of drug in serum (Cmax) and either T>MIC (ceftriaxone) or AUC/MIC (ciprofloxacin and gentamicin). Infected biological specimens (PLF, blood, spleen, and kidneys) were harvested after 2 h of antibiotic exposure. For ceftriaxone and ciprofloxacin treatment experiments, ALO 4783 was used as the infective agent. For gentamicin treatment experiments, the ATCC 25922 wild type was applied, for reasons previously explained.
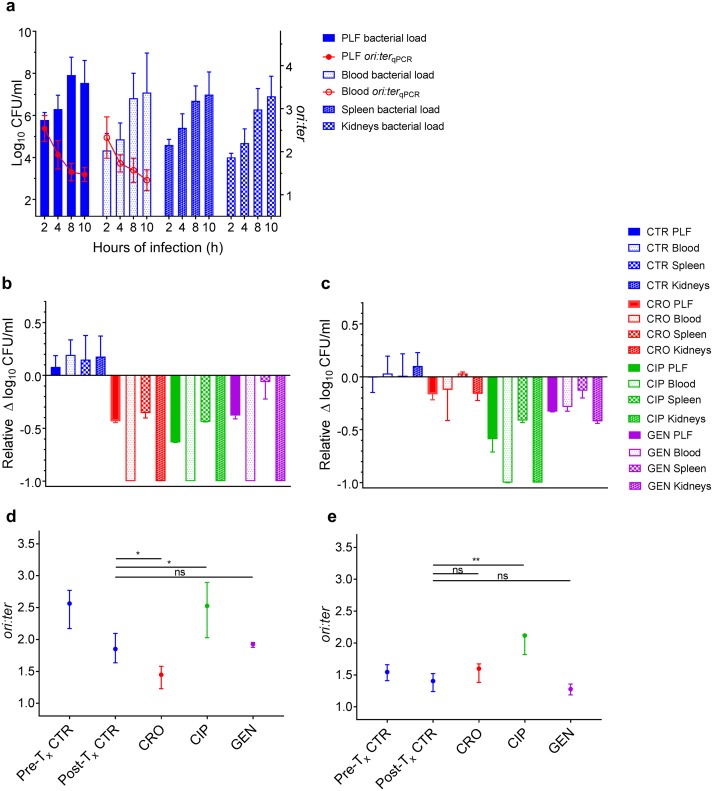
FIG 3.
Antibiotic activity as a function of bacterial growth rate during infection in vivo in the mouse peritonitis model. (a) Bacterial counts (CFU/ml; n = 9 per time point) and growth rates (ori:ter; 2, 8, and 10 h of infection, n = 9; 4 h of infection, n = 6) in untreated control groups (ATCC 25922 and ALO 4783) in the peritoneal lavage fluid (PLF), blood, spleen, and kidneys (bacterial counts only in the tissues). (b) Bacterial count reductions in PLF, blood, spleen, and kidneys after 2 h of antibiotic exposure in ceftriaxone (CRO), ciprofloxacin (CIP), and gentamicin (GEN) treatment groups when therapy was induced during rapid bacterial growth (i.e., at 2 h of infection). Controls (CTR) received no antibiotic therapy. (c) Bacterial count reductions in PLF, blood, spleen, and kidneys after 2 h of antibiotic exposure in treatment groups when therapy was induced during slow bacterial growth (i.e., at 8 h of infection). CTR received no antibiotic therapy. For comparison of activities between treatment induction during rapid and slow growth, data in panels b and c are presented as relative bacterial count reductions. CTR, n = 9; CRO, CIP, and GEN, n = 3. (d) Bacterial growth rates (ori:ter) in pretreatment controls (Pre-Tx CTR) and posttreatment controls (Post-Tx CTR) (i.e., at 2 and 4 h of infection, respectively) and in treatment groups after 2 h of antibiotic exposure when treatment was induced during rapid bacterial growth. As there was no significant difference in ori:ter values between control bacterial populations from PLF and blood, these were pooled for analysis. ori:ter values in treatment groups were only available from PLF, due to total bacterial elimination from the blood. Pretreatment CTR, n = 18; posttreatment CTR, n = 12; CRO, n = 3; CIP, n = 3; GEN, n = 3. (e) Bacterial growth rates (ori:ter) in Pre-Tx CTR and Post-Tx CTR (i.e., at 8 and 10 h of infection, respectively) and in treatment groups after 2 h of antibiotic exposure when treatment was induced during slow bacterial growth. PLF and blood ori:ter values were pooled for analysis. Pretreatment CTR, n = 18; posttreatment CTR, n = 18; CRO, n = 6; CIP, n = 3; GEN, n = 6. Data in panels b to e are presented as medians and interquartile ranges (IQRs). *, P < 0.05; **, P < 0.01; ns, P > 0.05 by Mann-Whitney U test.
Antibiotics administered during rapid bacterial growth in vivo.
The bacterial populations reached maximal in situ growth rates (mean [SD]: ori:ter PLF, 2.54 [0.30]; ori:ter blood, 2.37 [0.46]) at 2 h of infection (Fig. 3a). At this stage of infection, it has been demonstrated that bacterial population growth is heterogeneous (i.e., the population is made up of bacterial cells of various sizes and DNA contents) (3). Figure 4A illustrates a representative large cell with multiple oriCs, isolated from the PLF. As a consequence of average high growth rates, there were subsequent increases in net bacterial population sizes in all biological specimens (Fig. 3a). After 2 h of infection, there was a gradual decrease in ori:ter, resulting in overall net population stagnation between 8 and 10 h of infection (Fig. 3a).
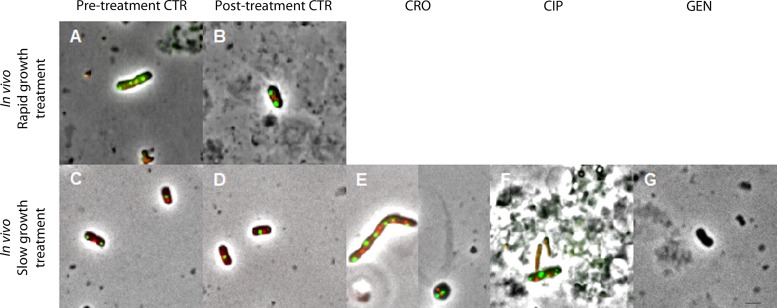
FIG 4.
Representative examples of bacterial cells observed by fluorescence microscopy and isolated after antibiotic induction during rapid (top row) or slow (bottom row) bacterial growth in vivo (blood and peritoneal lavage fluid [PLF] bacterial cells pooled). Images are shown at the same magnification (using a 100× objective) in phase contrast; intracellular oriC foci in green (GFP) and terC foci in red (mCherry) (ALO 4783). For GEN treatment experiments, ATCC 25922 without fluorescent foci was utilized. Examples shown are bacteria isolated from the PLF. Due to total or near total elimination of bacterial cells during rapid growth treatment, no bacterial cells were available from these treatment groups. A total of 500 cells were pooled and analyzed per time point from pre- and posttreatment controls during slow growth. For pre- and posttreatment controls during rapid growth, fewer cells were isolated due to low bacterial counts during early hours of infection: n = 142 and n = 66, respectively. For slow growth treatment induction: CRO, n = 170; CIP, n = 35; GEN, n = 228. Due to the limited resolution of fluorescence microscopy for colocalizing oriCs, some bacterial cells with overlapping chromosome replication origins may appear with too few foci (33). Mean (SD) population medial axis cell lengths were as follows: (A) rapid bacterial growth, pretreatment CTR, 3.96 (1.15) μm; (B) rapid bacterial growth, posttreatment CTR, 3.73 (1.13) μm; (C) slow bacterial growth, pretreatment CTR, 3.19 (0.73) μm; (D) slow bacterial growth, posttreatment CTR, 2.77 (0.86) μm; (E) slow bacterial growth, post-CRO treatment, not determined (due to overrepresentation of spherical cells); (F) slow bacterial growth, post-CIP treatment, 3.25 (0.68) μm; (G) slow bacterial growth, post-GEN treatment, 2.56 (0.76) μm. CTR, controls; CRO, ceftriaxone; CIP, ciprofloxacin; GEN, gentamicin. Scale bar, 2 μm.
When administered during rapid bacterial growth, all antibiotics (CRO, CIP, and GEN) caused significant bacterial count reductions at all anatomical sites examined (PLF, blood, spleen, and kidneys; P < 0.01), including a total elimination in blood and kidneys, compared to those in the respective controls (Fig. 3b). The anatomical site-specific differences in antibiotic activities observed (e.g., between the blood and PLF) (Fig. 3b) cannot be explained by differences in site-specific in situ pretreatment ori:ter, as these were not significantly different (P > 0.5) (Fig. 3a). Rather, these differences are conceivably attributable to other pharmacodynamic and/or host immune parameters.
We were able to isolate only a few live bacterial cells for fluorescence microscopy from infected body fluids in pre- and posttreatment control groups during rapid bacterial growth (Fig. 4A and B) due to low bacterial counts during the early hours of infection (Fig. 3a). Given the substantial or total clearance of bacteria from the PLF and blood following antibiotic exposure during rapid bacterial growth, we were unable to isolate live bacterial cells from these treatment groups (Fig. 4). However, the overall ori:ter developments in these treatment groups were similar to those observed during rapid growth in vitro: ciprofloxacin treatment induced significantly higher ori:ter ratios compared to posttreatment controls (P < 0.05), and ori:ter developments in ceftriaxone and gentamicin treatment groups were similar to those of the posttreatment controls (i.e., reduction toward ∼1), with ceftriaxone being most efficient (P < 0.05) (Fig. 3d).
Antibiotics administered during slow bacterial growth in vivo.
Minimal bacterial growth rates (mean [SD]: ori:ter PLF,1.53 [0.2]; ori:ter blood, 1.57 [0.29]) were observed starting from 8 h of infection (Fig. 3a). Marginally lower ori:ter levels were observed in bacteria isolated from both PLF and blood at 10 h of infection (mean [SD]: PLF, 1.48 [0.17]; ori:ter blood, 1.34 [0.24]) (Fig. 3a). However, these differences were not statistically significant (P > 0.5), and at this time, the criteria for euthanasia were met for control animals. Hence, 8 h of infection was chosen as the time point for slow bacterial growth treatment induction.
When administered during slow bacterial growth, only ciprofloxacin treatment caused significant bacterial count reductions at all anatomical sites (PLF, blood, spleen, and kidneys; P < 0.01) (Fig. 3c). The activity of gentamicin was overall reduced, and ceftriaxone activity was substantially reduced, at all anatomical sites when antibiotics were administered during slow bacterial growth compared to that during rapid bacterial growth (Fig. 3b and c).
Photomicrographs of live bacterial cells exposed to antibiotics during slow bacterial growth indicated that the drugs exerted their effects similar to that in vitro: ceftriaxone inhibited cell wall synthesis predominantly via PBP 2 (demonstrated by the presence of spherical cells) and PBP 3 (demonstrated by the presence of filamentous cells) (30) (Fig. 4E), and ciprofloxacin interrupted natural bacterial growth by inducing cell enlargement, with multiple fluorescent foci, due to interference with ongoing chromosome replication (Fig. 4F) (31). Due to multiple, overlapping fluorescent foci in these treatment groups, we were unable to accurately quantify the population distribution of oriC and terC. For gentamicin-treated bacterial cells, microscopic visualization of oriC and terC was not possible, as the wild-type ATCC 25922 was utilized. However, the size and morphology of the bacterial cells did not appear to differ from those of the respective control population, as observed after antibiotic administration during slow bacterial growth (Fig. 4D and G). We emphasize the uncertainty in these microscopy data (Fig. 4E to G), as only few live bacteria (n < 500) were isolated due to antibiotic-induced bacterial killing (Fig. 3c).
The bacterial populations (both in PLF and blood) exposed to antibiotics during late stage of infection (i.e., at 8 h of infection) (Fig. 3a) differed from those observed after prolonged propagation in vitro (i.e., at 8 h of incubation) (Fig. 1b) in that there was no complete cessation of growth in the former. Here, fractions of the population were still undergoing chromosome replication (as expressed by a mean ori:ter of >1). Hence, the effect of ciprofloxacin on ori:ter was apparent also upon treatment during slow bacterial growth (Fig. 3e).
In summary, in vivo bacterial growth rates at the time points representing rapid and slow bacterial growth did not differ to the same extent as those during propagation in a rich medium in vitro. Consequently, the difference in antibacterial activity as a function of bacterial growth rate became less explicit. Nevertheless, the overall trends were similar to those observed in vitro: only ciprofloxacin treatment entailed significant bacterial load reduction in all examined body fluids and tissues, both during rapid and slow bacterial growth. Contrary to the in vitro results, however, ceftriaxone and gentamicin both caused a certain bacterial load reduction when administered during slow bacterial growth, albeit less overall than that during rapid bacterial growth. This difference is likely due to the fact that bacterial growth at 8 h of infection was not at a (near) complete arrest, as the ori:ter remained >1.
DISCUSSION
In this study, we determined the activities of three commonly used bactericidal antibiotics with different antibacterial targets as a function of in situ bacterial growth rate, expressed by differential genome origin and terminus copy number quantification (ori:ter) by qPCR. We demonstrated that the overall activities of both ceftriaxone and gentamicin were substantially lower when administered during slow bacterial growth than when administered during rapid bacterial growth in vivo. Contrarily, ciprofloxacin was less sensitive to bacterial growth rate, as the overall activity remained largely unchanged when going from rapid to slow bacterial growth rate treatment induction in vivo. The findings of ciprofloxacin being less sensitive to bacterial growth rate than β-lactams and aminoglycosides has been demonstrated by others, however, with the limitation of bacterial growth rate being estimated from net bacterial population kinetics (9, 10). In the parallel in vitro experiments, the difference between rapid and slow bacterial growth rate was more explicit, including complete or near complete cessation of growth as the lower extreme growth rate. When administered during near cessation of bacterial growth, only ciprofloxacin exerted a significant bacterial load reduction, while ceftriaxone and gentamicin lost their effect, in agreement with previous observations where bacterial growth rates were extracted from population kinetics (4, 8). The increase in ori:ter observed after the administration of ciprofloxacin to populations of rapidly growing cells, both in vivo and in vitro, confirms the drug’s mode of action. Ciprofloxacin exerts its effect predominantly through DNA gyrase inhibition, which results in the formation of double-strand DNA breaks during chromosome replication, prohibiting the replication forks from reaching the terminus (i.e., the copy number of oriC relative to terC will be high) (31). As anticipated, this effect was lost when ciprofloxacin was introduced into a population without ongoing chromosome replication during slow growth treatment in vitro, as opposed to the slowly growing bacterial populations in vivo, where fractions of cells were still undergoing chromosome replication. As to ceftriaxone and gentamicin, both drugs induced decreases in ori:ter toward ∼1 in rapidly growing bacterial populations, both in vitro and in vivo. Contrary to the respective posttreatment control bacterial populations, however, where similar reductions in ori:ter were observed, these decreases cannot be explained by natural reduction of bacterial growth rate due to population entrance into stationary phase (i.e., starvation of life-sustaining nutrients due to high population density), as bacterial counts were reduced during antibiotic treatment. Rather, the ceftriaxone- or gentamicin-induced ori:ter reduction toward ∼1 is likely the result of preferential elimination of fractions of rapidly growing bacterial cells (i.e., those with ori:ter of >1). Hence, besides allowing for measurement of pretreatment in situ bacterial growth rate, ori:ter may, to some extent, demonstrate the antimicrobial mode of action by analysis of posttreatment ori:ter. Fluorescence microscopy can complement these findings by direct single-cell visualization, as demonstrated, yet is limited by the absence of live bacterial cells after efficient bacterial elimination.
The growth rate scenarios observed during bacterial propagation in vitro were not representative of the bacterial growth dynamics taking place during infection in a complex host environment. In the latter, a complete or near complete cessation of bacterial growth was not observed as long as the host was alive and bacterial life-sustaining nutrients presumably not exhausted. Hence, for more meaningful prediction of antibiotic activity in vivo, it is important to be able to test this in relation to the in situ bacterial growths rate taking place during host infection rather than extrapolating from in vitro studies. ori:ter provides predictive value in informing on the likelihood of antibiotic activity, both during E. coli propagation in vitro and during host infection in vivo. There are, however, limitations to be considered in this study. Maximal bacterial growth rates were observed after a few hours of propagation, while minimal growth rates were only observed after prolonged propagation, both in vitro and in vivo. Consequently, the net bacterial population sizes were larger during slow than during rapid bacterial growth. For a meaningful comparison of the effect between identical antibacterial treatment regimens applied, we calculated the relative killing effect in both scenarios. We cannot exclude the possibility of an inoculum effect as a factor contributing to the lower antibiotic activity observed during slow bacterial growth. This is, however, a phenomenon mainly observed in β-lactams and rather unlikely to have occurred at the high antibiotic concentrations that were applied, both in vitro and in vivo (32). Moreover, we were unable to adequately purify bacterial DNA from spleen and kidney tissues. Hence, the difference in spleen and kidney antibiotic treatment effect between the two scenarios in vivo is only assumed to result from different in situ bacterial growth rates in these tissues. Yet, as the temporal development of the net bacterial population size in these tissues followed those in the PLF and blood, we find it likely that bacterial growth rates in the tissues would also be higher in the early hours of infection than after prolonged propagation.
We conclude that chromosome replication as a means to measure bacterial growth rate can predict antibacterial treatment outcome. To some extent, it can also elucidate the antibiotic mode of action, as exemplified by the increase in ori:ter caused by ciprofloxacin-induced double-strand DNA breaks and the decrease in ori:ter caused by preferential elimination of rapidly growing bacterial cells by both ceftriaxone and gentamicin. While our findings are in agreement with a previously demonstrated causal relationship between in vivo bacterial growth rate and antibiotic activity, previous studies were limited by the methodology; e.g., the bacterial population kinetics method fails to take into account the host elimination factor, and the tracking of bacterial growth by isotope trace incorporation is largely inconvenient when it comes to pursuing the method in clinical practice. Tracking bacterial growth rate by differential genome origin and terminus quantification by qPCR has the advantage of being accessible and inexpensive and reports directly on the bacterial physiology, circumventing the limitation of the bacterial count kinetics method. Also, growth rates can be probed from a single biological sample, which is convenient in a clinical setting where repeated sample measurement often is difficult. The method could serve as a platform for testing any antimicrobial’s activity as a function of pretreatment bacterial growth rate in experimental infection models and could be pursued in a clinical setting to examine bacterial growth rates in infected biological materials. This could in turn prove helpful in evaluating future antibacterial strategies.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli ATCC 25922, a clinical isolate from the American Type Culture Collection (Manassas, VA, USA) and CLSI and EUCAST control strain for antibiotic susceptibility testing, was used throughout the study. This strain was utilized both as a wild type and as a genetically modified version expressing fluorescent fusion proteins at chromosomal sites corresponding to oriC and terC (ALO 4783) (3).
Antimicrobial agents and susceptibility testing.
The antimicrobial agents used in this study were procured as the commercial products registered for parenteral use in Denmark: ciprofloxacin (CIP) as ciprofloxacin 2 mg/ml (Fresenius Kabi, Germany), ceftriaxone (CRO) as ceftriaxone Stragen 1 g (Stragen Nordic, Denmark), and gentamicin (GEN) as hexamycin 40 mg/ml (Sandoz, Denmark). Ceftriaxone was dissolved in sterile physiological saline immediately before use. The MICs were detected by antimicrobial gradient strips (Etest; bioMérieux, France) according to the manufacturer’s instructions using a standard inoculum size; i.e., McFarland standard of 0.5, corresponding to >108 CFU/ml.
In vitro batch culture experiments.
For in vitro experiments, bacteria were grown in lysogeny broth (LB) as previously described (3). Antibiotics were added to each batch culture at either maximal bacterial growth rate (i.e., at 4 h of incubation) or at minimal bacterial growth rate (i.e., at 8 h of incubation). Samples for quantification of bacterial count, qPCR analysis, and fluorescence microscopy were withdrawn pretreatment (i.e., at 4 or 8 h of incubation at the maximal or minimal growth rate, respectively) and after 2 h of antibiotic exposure (i.e., at 6 or 10 h of incubation at the maximal or minimal growth rate, respectively). All samples were immediately set on ice after withdrawal. Control cultures without antibiotic treatment exposure were included in every experiment.
Both treatment studies were performed in duplicates, including both the ATCC 25922 wild type and the genetically modified ALO 4783, in three independent experiments. Both versions of the strain were tested in parallel to ensure the absence of any alterations in growth or antibiotic treatment effect attributable to the transgene insertions. As both growth curves and ori:ter values were similar for both versions of the strain, these results were pooled for statistical analyses.
Regarding gentamicin treatment, only the wild-type ATCC 25922 was utilized, as the gentamicin MIC was affected by the presence of a nonremovable kanamycin (KAN) cassette (encoding kanamycin phosphotransferase) used as clonal selection marker in ALO 4783.
Mouse peritonitis model in vivo experiments.
The mouse peritonitis model was carried out as previously described, using outbred female NMRI mice (weight 28 ± 2 g; Taconic, Denmark) (3). Animals were kept in cages in groups of three; each cage constituting one experimental unit that would be randomly assigned to treatment (CRO, CIP, or GEN) or no treatment (control [CTR]). Antibiotics were administered as a single bolus injection subcutaneously (s.c.) at either the maximal bacterial growth rate (i.e., at 2 h of infection) or the minimal bacterial growth rate (i.e., at 8 h of infection). Sample collections (peritoneal lavage fluid [PLF], blood, spleen, and kidneys) were performed pretreatment (i.e., at 2 or 8 h of infection at the maximal or minimal growth rate, respectively) in control groups and after 2 h of antibiotic exposure (i.e., at 4 or 10 h of infection at the maximal or minimal growth rate, respectively) in both treatment and control groups. Euthanasia and harvesting of biological specimens were carried out as previously described (3). All biological specimens were immediately placed in an insulated 4°C cooling box for transportation and kept on ice at 4°C until application in subsequent tests. Temporary storage of E. coli cultures on ice has previously been demonstrated not to induce any alteration in in situ bacterial growth parameters (cell size, oriC/cell, or ori:ter) postharvesting (3).
The mouse peritonitis model was repeated in 4 independent experiments, including a total of 54 animals. Data from repeated experiments were pooled for statistical analyses.
ALO 4783 was utilized in all experiments, except for the gentamicin treatment experiment where the ATCC 25922 wild type was used as the infecting agent due to the altered gentamicin MIC in ALO 4783, as mentioned above.
Ethics statement.
All animal experiments were approved by the Danish Animal Experimentation Inspectorate (license no. 2014-15-0201-00171) and performed according to institutional guidelines. The mice were regularly observed and scored for signs of distress. Humane endpoints constituted signs of irreversible sickness; the mice would be euthanized upon presentation of any of these signs.
Quantification of antibacterial activity.
Bacterial count measurements from in vitro and in vivo experiments were performed as previously described (3). Antibacterial activity was measured as the difference between bacterial counts pre- and posttherapy (Δlog10 CFU/ml). For meaningful comparison between identical treatment regimens administered at different growth rates (i.e., different pretreatment bacterial loads), antibacterial activity was reported as Δlog10 CFU/ml relative to the pretreatment bacterial count.
Quantitative real-time PCR.
ori:ter was calculated as the population mean level of qPCR amplified oriC relative to that of terC from purified bacterial DNA, as previously described (3).
Fluorescence microscopy.
Fluorescence microscopy was used to verify the qPCR data, whenever possible, by direct observation of live single cells of ALO 4783 carrying fluorescent markers corresponding to the oriC and terC sites. Fluorescence microscopy analysis was carried out as previously described (3). For gentamicin treatment experiments, only cell size and morphology were analyzed, as the ATCC 25922 wild-type strain was utilized. Live bacterial cells were isolated at each sampling time point (i.e., 4, 6, 8, and 10 h of incubation in the in vitro experiments, and 2, 4, 8, and 10 h of infection in the in vivo experiments). We have previously shown that PLF and blood bacterial population growth rates in this in vivo model do not differ (3). Hence, isolated bacteria from PLF and blood were pooled for analysis. We aimed at isolating 500 bacterial cells at each time point, both with and without antibiotic exposure, but this was not always possible due to substantial or total bacterial clearance in many of the treatment groups, as annotated in Fig. 2 and 4.
Statistical analyses.
Bacterial count data were log10 transformed prior to analysis. D’Agostino and Pearson omnibus normality tests were applied to all data sets. In general, the control group bacterial counts and qPCR data sets represented normal distributions; those of the treatment groups did not. Statistical significance was evaluated by unpaired t tests for parametric data and by Mann-Whitney U tests for nonparametric data. A two-tailed P value of <0.05 was considered significant. GraphPad Prism version 7 (GraphPad Software, CA, USA) was applied for statistical analyses and illustrations.
ACKNOWLEDGMENTS
We thank Jytte Mark Andersen and colleagues at Statens Serum Institut for technical assistance with the animal experiments.
The study was partly funded by an EU-IMI Joint Undertaking, grant agreement no. 115583 (ENABLE), by the Scandinavian Society for Antimicrobial Chemotherapy Foundation and by the Danish National Research Foundation (DNRF120) through the Centre for Bacterial Stress Response and Persistence (BASP).
We declare no competing interests.
REFERENCES
- 1.Kopf SH, Sessions AL, Cowley ES, Reyes C, Van Sambeek L, Hu Y, Orphan VJ, Kato R, Newman DK. 2016. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olm MR, Brown CT, Brooks B, Firek B, Baker R, Burstein D, Soenjoyo K, Thomas BC, Morowitz M, Banfield JF. 2017. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res 27:601–612. doi: 10.1101/gr.213256.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haugan MS, Charbon G, Frimodt-Møller N, Løbner-Olesen A. 2018. Chromosome replication as a measure of bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. Sci Rep 8:14961. doi: 10.1038/s41598-018-33264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AJ, Wang S, Meredith HR, Zhuang B, Dai Z, You L. 2018. Robust, linear correlations between growth rates and β-lactam-mediated lysis rates. Proc Natl Acad Sci U S A 115:4069–4074. doi: 10.1073/pnas.1719504115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by β-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 6.Brown MR, Collier PJ, Gilbert P. 1990. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob Agents Chemother 34:1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozens RM, Tuomanen E, Tosch W, Zak O, Suter J, Tomasz A. 1986. Evaluation of the bactericidal activity of beta-lactam antibiotics on slowly growing bacteria cultured in the chemostat. Antimicrob Agents Chemother 29:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35:1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantin B, Leggett J, Ebert S, Craig WA. 1991. Correlation between in vitro and in vivo activity of antimicrobial agents against Gram-negative bacilli in a murine infection model. Antimicrob Agents Chemother 35:1413–1422. doi: 10.1128/AAC.35.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeiler HJ, Voigt WH. 1987. Efficacy of ciprofloxacin in stationary-phase bacteria in vivo. Am J Med 82:87–90. [PubMed] [Google Scholar]
- 11.Zeiler HJ. 1985. Evaluation of the in vitro bactericidal action of ciprofloxacin on cells of Escherichia coli in the logarithmic and stationary phases of growth. Antimicrob Agents Chemother 28:524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, Sirota-Madi A, Thaiss CA, Pevsner-Fischer M, Sorek R, Xavier RJ, Elinav E, Segal E. 2015. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34:1256–1263. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper S, Helmstetter CE. 1968. Chromosome replication and the division cycle of Escherichia coli Br. J Mol Biol 31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang JD, Levin PA. 2009. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol 7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donachie WD. 1968. Relationship between cell size and time of initiation of DNA replication. Nature 219:1077–1079. [DOI] [PubMed] [Google Scholar]
- 17.Helmstetter CE, Cooper S. 1968. DNA synthesis during the division cycle of rapidly growing Escherichia coli Br. J Mol Biol 31:507–518. [DOI] [PubMed] [Google Scholar]
- 18.Hill NS, Kadoya R, Chattoraj DK, Levin PA. 2012. Cell size and the initiation of DNA replication in bacteria. PLoS Genet 8:e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skarstad K, Boye E, Steen HB. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 5:1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bremer H, Churchward G. 1977. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J Theor Biol 69:645–654. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen S, Ladefoged K, Frimodt-Møller N. 1997. Experience with once daily dosing of gentamicin: considerations regarding dosing and monitoring. Chemotherapy 43:442–450. doi: 10.1159/000239604. [DOI] [PubMed] [Google Scholar]
- 24.Jakobsen L, Cattoir V, Jensen KS, Hammerum AM, Nordmann P, Frimodt-Møller N. 2012. Impact of low-level fluoroquinolone resistance genes qnrA1, qnrB19 and qnrS1 on ciprofloxacin treatment of isogenic Escherichia coli strains in a murine urinary tract infection model. J Antimicrob Chemother 67:2438–2444. doi: 10.1093/jac/dks224. [DOI] [PubMed] [Google Scholar]
- 25.Frimodt-Møller N, Bentzon MW, Thomsen VF. 1986. Experimental infection with Streptococcus pneumoniae in mice: correlation of in vitro activity and pharmacokinetic parameters with in vivo effect for 14 cephalosporins. J Infect Dis 154:511–517. [DOI] [PubMed] [Google Scholar]
- 26.Knudsen JD, Frimodt-Møller N, Espersen F. 1995. Experimental Streptococcus pneumoniae infection in mice for studying correlation of in vitro and in vivo activities of penicillin against pneumococci with various susceptibilities to penicillin. Antimicrob Agents Chemother 39:1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espersen F, Frimodt-Møller N, Corneliussen L, Riber U, Rosdahl VT, Skinhøj P. 1994. Effect of treatment with methicillin and gentamicin in a new experimental mouse model of foreign body infection. Antimicrob Agents Chemother 38:2047–2053. doi: 10.1128/AAC.38.9.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frimodt-Møller N, Frølund Thomsen V. 1987. The pneumococcus and the mouse-protection test: correlation of in vitro and in vivo activity for beta-lactam antibiotics, vancomycin, erythromycin and gentamicin. Acta Path Microbiol Immunol Scand B 95:159–165. doi: 10.1111/j.1699-0463.1987.tb03106.x. [DOI] [PubMed] [Google Scholar]
- 29.Stouf M, Meile J-C, Cornet F. 2013. FtsK actively segregates sister chromosomes in Escherichia coli. Proc Natl Acad Sci U S A 110:11157–11162. doi: 10.1073/pnas.1304080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontana R, Aldegheri M, Ligozzi M, Lo Cascio G, Cornaglia G. 1998. Interaction of ceftriaxone with penicillin-binding proteins of Escherichia coli in the presence of human serum albumin. J Antimicrob Chemother 42:95–98. [DOI] [PubMed] [Google Scholar]
- 31.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob Agents Chemother 52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano F, Santamaría M, Ponte C, Castilla C, Fernández-Roblas R. 1988. In vivo significance of the inoculum effect of antibiotics on Escherichia coli. Eur J Clin Microbiol Infect Dis 7:410–412. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen HJ, Hansen FG. 2010. An automated and highly efficient method for counting and measuring fluorescent foci in rod-shaped bacteria. J Microsc 239:194–199. doi: 10.1111/j.1365-2818.2010.03374.x. [DOI] [PubMed] [Google Scholar]