Candida glabrata can attach to various medical implants and forms thick biofilms despite its inability to switch from yeast to hyphae. The current in vivo C. glabrata biofilm models only provide limited information about colonization and infection and usually require animal sacrifice.
KEYWORDS: Candida glabrata, animal models, antifungal agents, biofilms, bioluminescence imaging
ABSTRACT
Candida glabrata can attach to various medical implants and forms thick biofilms despite its inability to switch from yeast to hyphae. The current in vivo C. glabrata biofilm models only provide limited information about colonization and infection and usually require animal sacrifice. To gain real-time information from individual BALB/c mice, we developed a noninvasive imaging technique to visualize C. glabrata biofilms in catheter fragments that were subcutaneously implanted on the back of mice. Bioluminescent C. glabrata reporter strains (lucOPT 7/2/4 and lucOPT 8/1/4), free of auxotrophic markers, expressing a codon-optimized firefly luciferase were generated. A murine subcutaneous model was used to follow real-time in vivo biofilm formation in the presence and absence of fluconazole and caspofungin. The fungal load in biofilms was quantified by CFU counts and by bioluminescence imaging (BLI). C. glabrata biofilms formed within the first 24 h, as documented by the increased number of device-associated cells and elevated bioluminescent signal compared with adhesion at the time of implant. The in vivo model allowed monitoring of the antibiofilm activity of caspofungin against C. glabrata biofilms through bioluminescent imaging from day four after the initiation of treatment. Contrarily, signals emitted from biofilms implanted in fluconazole-treated mice were similar to the light emitted from control-treated mice. This study gives insights into the real-time development of C. glabrata biofilms under in vivo conditions. BLI proved to be a dynamic, noninvasive, and sensitive tool to monitor continuous biofilm formation and activity of antifungal agents against C. glabrata biofilms formed on abiotic surfaces in vivo.
INTRODUCTION
Candida glabrata is an emerging human fungal pathogen causing mucosal and deep tissue infections, especially in elderly patients with a compromised immune system. Following Candida albicans, C. glabrata is the second most commonly isolated species from hospitalized patients in Europe (1). Although C. glabrata does not possess the ability to form true hyphae or secrete proteases, it still maintains many virulence factors, including a biofilm-forming capacity, which contribute to its potential to cause disease (2). The majority of life-threatening infections are associated with biofilm formation on biotic and abiotic substrates. Biofilms provide microorganisms with a shelter from external effects of therapeutics, giving them an opportunity to withstand very high concentrations of antifungal agents, for example azoles (3, 4). Because of biofilm resilience to antifungals, therapeutic approaches are very limited, often leading to the surgical removal of the implant material and its subsequent replacement as a sole solution. Recently, our group demonstrated that in vivo C. glabrata biofilms formed inside polyurethane catheter pieces implanted subcutaneously on the back of Sprague-Dawley rats (5). Biofilms were composed of multilayer structures of yeast cells embedded in extracellular matrix exhibiting increased tolerance to therapeutic concentration of fluconazole. Echinocandins proved to remain active against in vivo C. glabrata-biofilm-forming cells in this particular model (5). One disadvantage of almost all existing biofilm models is that the fungal load is traditionally analyzed postmortem, requiring host sacrifice. To avoid this, sensitive noninvasive techniques, such as bioluminescence imaging (BLI), provide new and often unexpected temporal and spatial information about infection development in individual animals in real time (6, 7). The monitoring of light emitted from luciferase-expressing reporter cells has been used to examine an extensive range of biomolecular functions (8). BLI has been successfully applied to monitor the development of oral candidiasis (6, 9) and C. albicans dissemination after tail vein injection in mice (10–12). This technique was also employed to follow superficial and subcutaneous infections caused by C. albicans (12). Moreover, it was employed to study the time-dependent development of C. albicans biofilm formation in a subcutaneous biofilm mouse model (13).
In this study, we present the construction of an auxotrophic marker-free bioluminescent C. glabrata reporter strain based on a C. glabrata-optimized firefly luciferase integrated into the HIS1 locus using HIS1 as selectable marker. This strain allowed us to study biofilm development in real-time with the use of BLI in a mouse subcutaneous model system. Furthermore, it allowed us to monitor the efficacy of fluconazole and caspofungin treatment in vivo and can thereby contribute to implementation of the replacement, reduction, and refinement of animal testing (3Rs) in therapeutic experiments.
RESULTS
In vitro characterization of bioluminescent strains.
C. glabrata reporter strains lucOPT 7/2/4 and lucOPT 8/1/4 were generated via integration of a C. glabrata-optimized firefly luciferase into the HIS1 locus using HIS1 as selectable marker. To ensure that C. glabrata lucOPT 7/2/4 and lucOPT 8/1/4 produced substantial bioluminescence and that luciferase expression did not affect growth rates, transformants were tested in series of in vitro experiments. We did not observe any changes in growth rates between reporter strains and wild type (WT) in RPMI 1640 (pH 7.0) and yeast extract-peptone-dextrose (YPD) medium at 37°C (see Fig. S1 in the supplemental material). Additionally, we prepared 10-fold dilution series of WT and bioluminescent strains to detect the signal. Signals from reporter strains correlated (Spearman’s r = 0.993, n = 24, P < 0.0001) well with the number of cells, with a detection limit of about 100 cells per well (Fig. S1).
In vitro Candida glabrata adhesion and biofilm formation monitored by BLI.
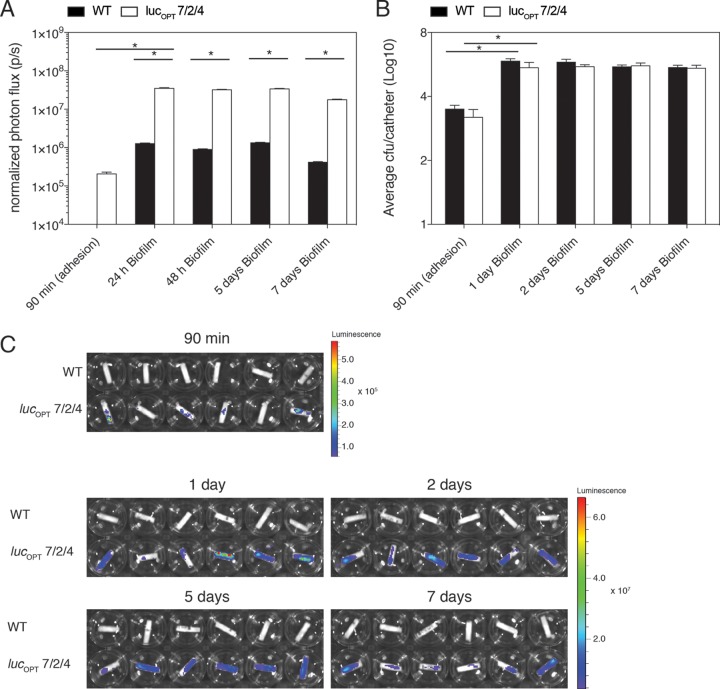
To evaluate the potential use of BLI in vitro to study fungal adhesion (90 min after inoculation) and biofilm formation (after 2 days), we inoculated C. glabrata strains on the serum-coated bottom of 96-well polystyrene plates, which represents a very simple biofilm system. The bioluminescent signal acquired from the reporter strains was significantly higher than the WT (Fig. 1A and C). A parallel 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt (XTT) assay showed a significant increase in metabolic activity for all biofilm-forming cells after 2 days (Fig. 1B). The BLI signal intensity from biofilms formed by luciferase-expressing strains on polyurethane catheters increased significantly compared with the signal determined from the WT strain (Fig. 1D and F). The number of CFU was significantly higher during biofilm formation than adhesion (Fig. 1E), which is in agreement with the BLI signal measurements. To determine whether BLI can accurately measure the kinetics of C. glabrata biofilm formation, we quantified bioluminescence and CFU after 90 min (adhesion period) and 1, 2, 5, and 7 days. At each imaging time point, the BLI signal retrieved from reporter strain C. glabrata lucOPT 7/2/4 cells was significantly higher than the signal emitted from the WT strain (Fig. 2A and C). The number of fungal cells significantly increased between adhesion and the first 24 h of biofilm formation (Fig. 2B). At later time points of biofilm formation, the BLI signal and the amount of CFU remained stable. In addition, we show that this reporter strain allows accurate in vitro biofilm quantification, as CFU and bioluminescence correlated well (Spearman’s r = 0.7027, n = 27, P < 0.0001). Taken together, these results indicate that BLI represents a sensitive tool to study in vitro C. glabrata adherence and biofilm formation on abiotic surfaces.
FIG 1.
In vitro BLI of Candida glabrata biofilm formation on 96-well plate and inside polyurethane catheters. Bioluminescence signal intensity (A) and XTT data measurements (B) were obtained after adherence (90 min, black columns, n = 18 wells or devices per strain) and biofilm formation (48 h, white columns, n = 18 wells or catheters per strain) by C. glabrata ATCC 2001 (WT), C. glabrata lucOPT 7/2/4, and lucOPT 8/1/4 in 96-well plate. (C) A characteristic bioluminescent image of 2-day-old biofilm formed on the surface of a 96-well plate. Measurements of BLI signal (D) and CFU counts (E) from C. glabrata adhered cells (90 min, black columns) and biofilms formed (48 h, white columns) inside catheters. (F) A typical picture of BLI acquired after 2 days of biofilm formation inside polyurethane fragments. Statistical analyses were performed using two-way analysis of variance (ANOVA) with post hoc Tukey’s honestly significant difference (HSD) test. Differences were considered significant at *P value of ≤0.05. Error bars indicate the standard error of the mean (SEM) of replicate samples.
FIG 2.
In vitro BLI of Candida glabrata ATCC 2001 (WT) and C. glabrata lucOPT 7/2/4 biofilm formation inside polyurethane catheters over time. (A) Bioluminescent signal intensity acquired at different time points (adhesion, 1, 2, 5, and 7 days) of C. glabrata ATCC 2001 (WT, black columns) and C. glabrata lucOPT 7/2/4 (white columns) biofilm development inside catheters. (B) BLI was accompanied with CFU counts after adherence and 1, 2, 5, and 7 days of in vitro biofilm formation inside polyurethane devices (n = 18 devices per strain and time point). (C) A BLI signal retrieved after adhesion (90 min) and later stages of biofilm formation. The two-way ANOVA with post hoc Tukey’s HSD test was used to perform statistical analyses. Differences were considered significant at *P values of ≤0.05, and error bars indicate the SEMs of replicate samples.
In vivo BLI of Candida glabrata biofilms developed in a subcutaneous mouse model.
Biofilm formation can impose serious medical problems in immunocompromised patients. First, we assessed the role of immunosuppression on C. glabrata biofilms and on dissemination into the surrounding tissue. One group of mice was treated with dexamethasone (0.4 mg/liter) in the drinking water, whereas the second group remained immunocompetent. Our preliminary experiment displayed no major differences in the number of CFU after devices were explanted from immunocompromised and immunocompetent mice (see Fig. S2A in the supplemental material). Similarly, the number of C. glabrata cells quantified from tissue surrounding catheters was almost identical (Fig. S2B). As no significant difference was observed among both groups, we continued our experiments in immunocompetent mice. Moreover, fungal cells did not disseminate into any vital organ of immunocompromised or immunocompetent mice, as no CFUs were retrieved from kidneys, liver, or spleen after homogenization (data not shown).
In this model, catheters infected with C. glabrata WT cells and the lucOPT 7/2/4-expressing strain were implanted subcutaneously on the left and right side, respectively, of the lower back of mice (Fig. 3A). In vivo imaging was performed after catheter implantation and continued on day 1, 5, and 7 (Fig. 3A). At every imaging time point, 3 mice were sacrificed and explanted catheters were used for ex vivo quantification. A significant BLI signal was detected only from C. glabrata lucOPT 7/2/4-biofilm-forming cells, where it increased significantly over time (Fig. 3A and B). The quantification of CFUs documented with the BLI signal in parallel was significantly higher at the first day of biofilm maturation than at adhesion (90 min) (Fig. 3C). Moreover, the number of biofilm-forming cells and bioluminescent signal retrieved from C. glabrata lucOPT 7/2/4 correlated well (Spearman’s r = 0.7576, n = 10, P < 0.0149) (Fig. 3A and B). These findings indicate that BLI can be used for the continuous detection and quantification of in vivo C. glabrata biofilm formation.
FIG 3.
In vivo Candida glabrata ATCC 2001 (WT) and Candida glabrata lucOPT 7/2/4 biofilm formation studied by longitudinal BLI. (A) In vivo BLI of C. glabrata biofilm formation after implant (90 min, period of adhesion) and 1, 5, and 7 days of biofilm formation. Three polyurethane devices infected with C. glabrata ATCC 2001 (WT) strain were implanted on left side of the back, whereas 3 devices challenged with lucOPT 7/2/4 were inserted on the ride side of the back of the animal. (B) Quantification of bioluminescence signal intensity acquired at different time points (adhesion, 1, 5, and 7 days, n = 6 animals per time point, except for a group with noninfected catheters with n = 1) of noninfected devices (black columns), C. glabrata ATCC 2001 (gray columns), and C. glabrata lucOPT 7/2/4 (white columns) biofilm formation. (C) CFU counts retrieved from catheters infected with C. glabrata ATCC 2001 (black columns) and with C. glabrata lucOPT 7/2/4 (white columns) after adhesion and 1, 5, and 7 days of in vitro biofilm formation inside polyurethane devices. (C) A BLI signal intensity retrieved after adhesion (90 min) and later stages of biofilm formation. Statistical analyses were performed using the two-way ANOVA with post hoc Tukey’s HSD test. Differences were considered significant at *P values of ≤0.05, and error bars indicate SEM of replicate samples.
In vivo BLI of fluconazole and caspofungin activity against C. glabrata biofilms.
Prior to in vivo experiments, we performed a few tests to determine the efficacy of an azole fluconazole and an echinocandin caspofungin on free-living cells and in vitro biofilms. C. glabrata planktonic cells showed decreased susceptibility to fluconazole (MIC50 ranging from 8 to 16 μg/ml), whereas all strains were susceptible to caspofungin (MIC50, 0.0625 mg/liter). In vitro C. glabrata biofilms did not respond to therapy with fluconazole (MIC50, >64 mg/liter), while caspofungin (MIC50, 0.25 to 0.5 mg/liter) significantly decreased the number of C. glabrata-biofilm-forming cells of all strains tested (see Fig. S3 in the supplemental material). Furthermore, we evaluated the activity of fluconazole (125 mg/kg of body weight/day) and caspofungin (10 mg/kg of body weight/day) against in vivo C. glabrata biofilms. Daily in vivo imaging and the number of CFUs retrieved from catheters ex vivo on day 9 revealed that fluconazole was not active against C. glabrata-biofilm-forming cells (Fig. 4A to D). In contrast, the signal detected in mice treated with caspofungin significantly decreased from day 4 of treatment until day 7 (Fig. 4A and B). Furthermore, treatment with caspofungin resulted in significantly decreased numbers of CFUs recovered from polyurethane devices ex vivo (Fig. 4C and D). In conclusion, these data support the use of BLI to demonstrate activities of antifungal agents against biofilms under in vivo conditions.
FIG 4.
BLI of fluconazole (FLC) and caspofungin (CAS) efficacy against in vivo Candida glabrata ATCC 2001 (WT) and Candida glabrata lucOPT 7/2/4 biofilm-associated infections. (A) Representative images of a signal acquired from Candida glabrata ATCC 2001 (WT, 3 devices implanted on the left side of the back of mice) and C. glabrata lucOPT 7/2/4 (3 catheters inserted on the right side of the back of a host) biofilms. Biofilms were formed for 24 h, and afterward, treatment with fluconazole (125 mg/kg of body weight/day, n = 4) and caspofungin (10 mg/kg of body weight/day, n = 4) was initiated. Drugs were administered once daily for 7 days. The control-treated group of animals (n = 4) was injected with sterile saline. (B) Quantification of BLI signal intensity, which started to decrease 4 days after animal treatment with caspofungin. Data show an average ± SEM of a BLI signal acquired from catheters implanted in animals treated with fluconazole or caspofungin on different days. C. glabrata ATCC 2001 (C) and C. glabrata lucOPT 7/2/4 (D) CFU counts retrieved from individual catheters after biofilm formation. Each symbol represents the log10 CFU ± SEM of saline-treated (control) and treated fungal cells retrieved from catheter pieces from individual mice. The bar represents the average log10 CFU ± SEM of the number of fungal cells retrieved from a drug-free group and from a treated group of animals. The two-way ANOVA with post hoc Tukey’s HSD test was used to calculate statistical significance at a *P value of ≤0.05.
DISCUSSION
The usage of indwelling medical devices has become indispensable to medical care. Unfortunately, the increased risk of biofilm-related infections in hospitalized patients is associated with the use of these devices. One of the key challenges in biofilm research is the development of in vivo models that allow assessment of the efficacy of existing or novel therapeutics. So far, only a few in vitro C. glabrata biofilm model systems have been introduced to elucidate the different stages of biofilm and its architecture and response to antifungal treatment (2). However, these models do not take the host immune system and changes at the host site of infection into consideration, which are crucial during infections in humans. To date, in vivo C. glabrata biofilms have been studied inside central venous catheters, as described by Nett et al. (14). Recently, in vivo C. glabrata biofilms have been established subcutaneously in immunocompromised Sprague-Dawley rats (5). In this study, we established an in vivo C. glabrata infection model that mimics nonbloodstream biofilm-associated infections in immunocompetent mice. This model is of interest given the frequent development of such biofilms in endotracheal tubes, intracardiac prosthetics, and prosthetic joints (15). Our flow cytometry experiments (data not shown) displayed a trend toward more neutrophil recruitment in the subcutaneous area surrounding catheter pieces 72-h postsurgery in animals carrying C. glabrata-infected catheters than in those carrying noninfected devices. These findings suggest that the host immune system is triggered upon C. glabrata infection; however, the predominant innate immune response is likely not driven solely by neutrophils. Further research elucidating the specific innate immune response toward these biofilms has to be conducted. The translation of the originally developed rat model to mice improved the cost-efficiency of in vivo experiments and presents the potential to study biofilm formation in different transgenic mice, allowing us to discover different host factors that may affect biofilm development.
In the majority of animal experiments, the number of CFUs is determined postmortem which requires a large number of animals in order to perform time course experiments. This is also the case if one wants to study the effect of antifungals on catheter-associated biofilm infections in animals. Because of this issue, we searched for an alternative tool which allowed us to study the development of infection repeatedly in the same animal. Bioluminescence imaging (BLI) has been previously described as a sensitive, reliable, and noninvasive technique to follow systemic candidiasis caused by C. albicans and its dissemination into kidneys (10–12). Additionally, BLI has been applied to study subcutaneous C. albicans infections (10) and vulvovaginal candidiasis (11). It was used to monitor C. albicans biofilm formation in a subcutaneous catheter mouse model (13). In that particular scenario, Gaussia princeps luciferase was expressed on the cell wall (12) and emitted light upon contact with the substrate coelenterazine, whereby light emission correlated with CFUs (13). Gabrielli et al. (16) used C. albicans strains with extracellularly expressed G. princeps luciferase to study the development of oropharyngeal candidiasis. They found that in vivo BLI was more reliable than CFU counts detecting early infection in the oral cavity of the host. Very recently, red-shifted firefly luciferase has been introduced to test the fate of C. albicans in different animal models (17). In our study, the use of firefly luciferase is known for its glow-type bioluminescence over a period of time; therefore, it is advantageous compared with other reporter systems (18). Renilla luciferase and beetle reporter systems have been used to study eukaryotes. In the case of bacteria, it is advantageous to use luminescent reporters based on the lux operon with the intracellular expression of luciferase (6). Furthermore, the strains generated here are otherwise not auxotrophic and show no altered phenotype compared with the parental WT. We have clearly demonstrated that BLI is a sensitive and powerful tool to study long-term infection development on abiotic surfaces within living animals.
It is known that the extracellular matrix serves as a protective barrier, allowing microorganisms to withstand high concentrations of antimicrobial agents. The treatment of C. glabrata can be especially challenging because of its acquired resistance to the “gold-standard” treatment antimycotic fluconazole (4). In our study, the daily administration of fluconazole (125 mg/kg of body weight/day) for 7 days did not cause any significant decline in the number of C. glabrata catheter-associated cells as shown by comparable BLI and CFU results. However, the daily administration of caspofungin (10 mg/kg of body weight/day) significantly decreased the number of CFUs retrieved from implanted catheters compared with that of saline-treated animals. Remarkably, the daily monitoring of animals resulted in decreased signal from day 4 in caspofungin-treated animals. These studies are in agreement with our previous fluconazole and caspofungin efficacy testing in a rat model (5, 19).
In conclusion, bioluminescence imaging is a reliable and noninvasive method not only to visualize but also to quantify fungal biofilms. Even more importantly, this technique is cost-effective, offering the capability to study the activity of novel or existing antimicrobial agents over the course of time and avoiding animal sacrifice at each time point.
MATERIALS AND METHODS
Ethics.
All animal experiments were performed in accordance with the KU Leuven animal care guidelines and were approved by the ethical committee of KU Leuven (project number P090/2013). All animals were given a standard ad libitum diet and housed at random with 4 animals in filter-top cages in a dedicated animal room where temperature, light, and humidity were regulated.
Generation of C. glabrata histidine auxotrophic mutants.
All oligonucleotides used in this study are described in Table S1 in the supplemental material. For generation of a histidine auxotrophic mutant of C. glabrata ATCC 2001 (WT), the HIS1 locus was selected for gene deletion. A 750-bp promoter region (P1 + P2) and a 693-bp downstream region (P3 + P4) of the HIS1 gene were amplified from C. glabrata genomic DNA. Oligonucleotides P1 and P4 contained overlapping sequences to the BamHI-digested pUC19 vector. Oligonucleotides P2 and P3 contained flanking sequences to the nourseothricin resistance cassette that was amplified with oligonucleotides P5 and P6 from plasmid pJK863 (20). Primer P5 contained an overlap to the upstream and P6 to the downstream flanking region of the HIS1 gene. PCRs were performed with Phusion Hot Start II polymerase (Thermo), and PCR fragments were gel purified using the GeneJet gel purification kit (Thermo). All fragments were mixed with the BamHI-digested pUC19 plasmid and assembled by in vitro recombination using the InFusion HD cloning kit (TaKaRa/Clonetech). After transformation of Escherichia coli DH5α cells, oligonucleotides P1 + P7 and P4 + P8 were used to screen colonies for correct plasmid assembly. Plasmid DNA was isolated, digested with KpnI, and used for electroporation of C. glabrata WT cells (21). Transformants were regenerated on YPD medium with 200 μg/ml nourseothricin. Individual colonies were tested for histidine auxotrophy. Integration of the deletion construct into the HIS1 locus was confirmed by diagnostic PCR with oligonucleotides P7 + P9. The deletion mutants Δ2/4 and Δ2/12 were used in downstream experiments for the generation of bioluminescent strains.
Generation and characterization of luciferase-expressing C. glabrata strains.
A codon-optimized synthetic firefly luciferase gene adapted to the codon usage of C. glabrata was used for the generation of bioluminescent C. glabrata strains. To ensure constitutive gene expression, the promoter of the enolase gene (pENO1) was selected. The enolase promoter was amplified with oligonucleotides P10 + P11 from genomic DNA of WT and ligated into the NdeI site of the luciferase gene containing plasmid pUC57. The ENO1 promoter in combination with the luciferase gene was amplified with oligonucleotides P12 + P13, whereby P12 contained an overhang to a short terminator region of the HIS1 gene and P13 an overhang to a downstream region of the HIS1 gene. To integrate the luciferase construct into the original HIS1 locus of the ΔHIS1 strains, the HIS1 gene, with a 720-bp promoter region and a short 60-bp terminator sequence, was amplified from genomic DNA of the WT with oligonucleotides P14 + P15. Oligonucleotide P14 contained an overhang to the BamHI site of the pUC19 plasmid, and P15 contained an overhang to the ENO1 promoter. Finally, to enable a homologous integration into the HIS1 locus, a 668-bp downstream region of the HIS1 gene was amplified from C. glabrata genomic DNA with oligonucleotides P16 and P17, whereby P16 overlapped with the 3′ end of the luciferase and P17 overlapped with the BamHI-digested puC19 vector. All PCR fragments were gel purified and mixed with the BamHI-digested puC19 vector for in vitro recombination using the InFusion HD cloning kit. E. coli DH5α cells were transformed with the reaction mixture, and the correct plasmids were identified by colony PCR using oligonucleotides P18 + P19 for the fusion of the luciferase cassette with the HIS1 gene and P20 + P21 for the combination of the luciferase gene with the HIS1 downstream region. Correctly assembled plasmids were purified, and the transformation cassette was excised from the pUC19 backbone by BamHI restriction and used for transformation of C. glabrata HIS1 deletion mutants using histidine prototrophy as selection marker. Transformants were regenerated on yeast nitrogen base (YNB) with ammonium but without amino acids and niacin as supplements. Transformants were prescreened for the loss of nourseothricin resistance, which was expected by homologous integration of the reporter cassette into the original HIS1 locus of HIS1 deletion mutants. Integration of the cassette into the correct locus was additionally confirmed by diagnostic PCR using oligonucleotides P9 + P22. Finally, transformants were streaked onto YNB agar plates containing 0.4 mM d-luciferin and analyzed for bioluminescence using an IVIS 100 system (Perkin Elmer). Two independent transformants, namely C. glabrata lucOPT 7/2/4 and lucOPT 8/1/4, producing similar signal intensities were selected for further work. An overview of the strains used in this study is provided in Table S2 of the supplemental material.
All strains were grown on YPD (1% yeast extract, 2% peptone, and 2% glucose) agar plates at 30°C. C. glabrata planktonic cultures were incubated in liquid YPD medium for 72 h at 30°C (stationary phase). The growth of planktonic cells was followed by growth measurement on a Bioscreen C (Labsystems) system at 30°C and 37°C under continuous shaking for 24 h, and measurements were taken every 30 min.
In vivo C. glabrata biofilm formation in a murine subcutaneous biofilm model.
We translated a rat subcutaneous biofilm model (5) to specific-pathogen-free immunocompetent BALB/c mice (6 to 8 weeks old, 20 g). C. glabrata (1 × 106 cells/ml) attached onto the surface of serum-coated polyurethane catheter pieces during in vitro adhesion (90 min, 37°C) followed by a washing step with PBS. Mice were anesthetized as previously described (5). In order to compare bioluminescence from the WT control strain and the bioluminescent strain in a single animal, two small incisions were made, one on the left and one on the right side of the back of a mouse, as previously demonstrated (22). Three catheter pieces seeded with the WT strain were implanted on the left, and 3 catheters infected with the bioluminescent strain were inserted on the right side of the back. For catheter explant, animals were sacrificed by cervical dislocation. Catheter fragments were removed, washed twice with PBS, and placed in microcentrifuge tubes containing PBS. Devices were sonicated, vigorously vortexed, and plated to enumerate the number of CFUs. The dissemination of C. glabrata cells from biofilms into the kidneys, liver, spleen, and the tissue surrounding the catheter pieces was examined after 48 and 144 h of biofilm formation.
In vivo BLI.
Isoflurane anesthesia and the BLI procedure were performed as described before (13, 22). Prior to BLI, fresh d-luciferin solution (33.3 mg/ml) was dissolved in sterile saline (0.9% NaCl). A photograph was taken to ensure that the animals were in the desired position for imaging. Subsequently, 100 μl of the d-luciferin substrate was injected subcutaneously at both sides of the back, at the regions surrounding the implanted catheter pieces. During substrate injection, animals were kept anaesthetized through a nose cone providing gas anesthesia. Consecutive series of scans were acquired at 60-s exposure time (medium binning) until the maximum signal was reached (around 15 min after injection). Rectangular region of interests (ROIs) were drawn around each trio of catheters and reported as photon flux per second (p s−1). The background BLI signal was measured by using the same ROI after injecting d-luciferin subcutaneously on the upper back of the mouse where no catheters were implanted.
In vivo antifungal treatment.
A stock solution of fluconazole (Sigma-Aldrich, USA) was dissolved in sterile water, whereas caspofungin was prepared in pure dimethyl sulfoxide (DMSO). Intraperitoneal administration of fluconazole (125 mg/kg of body weight/day) and caspofungin (10 mg/kg of body weight/day) were initiated 24 h postimplant. Solutions for injection were prepared in sterile saline and administered intraperitoneally once daily for 7 days. In the case of caspofungin, the concentration of DMSO used for the injection was 0.5%. The control group of mice was injected with sterile saline.
Additional materials and methods are described in supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Celia Lobo Romero for her excellent technical assistance during in vitro and in vivo experimental procedures. We express our gratitude to Tinne Buelens and Ann Van Santvoort for their technical assistance during animal experiments and bioluminescence imaging, performed at the Molecular Small Animal Imaging Facility (MoSAIC) of KU Leuven. We acknowledge Nico Vangoethem for his help with figures and tables.
This work was funded by the Merck Investigator Studies Program number 40312 awarded to P.V.D.; by a research grant (2013) from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) awarded to S.K.; and by the Fund for Scientific Research Flanders (FWO research community on biofilms, WO.009.16N). S.K. is a postdoctoral fellow of the Fund for Scientific Research Flanders (FWO). S.K. was also supported by a postdoctoral grant from KU Leuven (PDMK 11/089). O.R. is supported by Strategic Basic Research (SB), Research Foundation-Flanders (FWO), and grant number 1S55817N.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01555-18.
REFERENCES
- 1.Kullberg BJ, Arendrup MC. 2015. Invasive Candidiasis. N Engl J Med 373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues CF, Rodrigues ME, Silva S, Henriques M. 2017. Candida glabrata biofilms: how far have we come? J Fungi (Basel) 3:11. doi: 10.3390/jof3010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.d' Enfert C, Janbon G. 2016. Biofilm formation in Candida glabrata: what have we learnt from functional genomics approaches? FEMS Yeast Res 16:fov111. doi: 10.1093/femsyr/fov111. [DOI] [PubMed] [Google Scholar]
- 4.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. 2017. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17:e383–e392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 5.Kucharíková S, Neirinck B, Sharma N, Vleugels J, Lagrou K, Van Dijck P. 2015. In vivo Candida glabrata biofilm development on foreign bodies in a rat subcutaneous model. J Antimicrob Chemother 70:846–856. doi: 10.1093/jac/dku447. [DOI] [PubMed] [Google Scholar]
- 6.Brock M. 2012. Application of bioluminescence imaging for in vivo monitoring of fungal infections. Int J Microbiol 2012:956794. doi: 10.1155/2012/956794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vecchiarelli A, d' Enfert C. 2012. Shedding natural light on fungal infections. Virulence 3:15–17. doi: 10.4161/viru.3.1.19247. [DOI] [PubMed] [Google Scholar]
- 8.Close DM, Xu T, Sayler GS, Ripp S. 2010. In vivo bioluminescence imaging (BLI): noninvasive visualization and interrogation of biological processes in living animals. Sensors (Basel) 11:180–206. doi: 10.3390/s110100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosci P, Pericolini E, Gabrielli E, Kenno S, Perito S, Bistoni F, d’Enfert C, Vecchiarelli A. 2013. A novel bioluminescence mouse model for monitoring oropharyngeal candidiasis in mice. Virulence 4:250–254. doi: 10.4161/viru.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen ID, Lüttich A, Kurzai O, Hube B, Brock M. 2014. In vivo imaging of disseminated murine Candida albicans infection reveals unexpected host sites of fungal persistence during antifungal therapy. J Antimicrob Chemother 69:2785–2796. doi: 10.1093/jac/dku198. [DOI] [PubMed] [Google Scholar]
- 11.Doyle TC, Nawotka KA, Kawahara CB, Francis KP, Contag PR. 2006. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb Pathog 40:82–90. doi: 10.1016/j.micpath.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Enjalbert B, Rachini A, Vediyappan G, Pietrella D, Spaccapelo R, Vecchiarelli A, Brown AJ, d' Enfert C. 2009. A multifunctional synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect Immun 77:4847–4858. doi: 10.1128/IAI.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vande Velde G, Kucharíková S, Schrevens S, Himmelreich U, Van Dijck P. 2014. Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cell Microbiol 16:115–130. doi: 10.1111/cmi.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nett J, Lincoln L, Marchillo K, Andes D. 2007. Beta-1,3 glucan as a test for central venous catheter biofilm infection. J Infect Dis 195:1705–1712. doi: 10.1086/517522. [DOI] [PubMed] [Google Scholar]
- 15.Ramage G, Martinez JP, Lopez-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 16.Gabrielli E, Roselletti E, Luciano E, Sabbatini S, Mosci P, Pericolini E. 2015. Comparison between bioluminescence imaging technique and CFU count for the study of oropharyngeal candidiasis in mice. Cytometry 87:428–436. doi: 10.1002/cyto.a.22666. [DOI] [PubMed] [Google Scholar]
- 17.Dorsaz S, Coste AT, Sanglard D. 2017. Red-shifted firefly luciferase optimized for Candida albicans in vivo bioluminescence imaging. Front Microbiol 8:1478. doi: 10.3389/fmicb.2017.01478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badr CE. 2014. Bioluminescence imaging: basics and practical limitations. Methods Mol Biol 1098:1–18. doi: 10.1007/978-1-62703-718-1_1. [DOI] [PubMed] [Google Scholar]
- 19.Vande Velde G, Kuchariková S, Van Dijck P, Himmelreich U. 2018. Bioluminescence imaging increases in vivo screening efficiency for antifungal activity against device-associated Candida albicans biofilms. Int J Antimicrob Agents 52:42–51. doi: 10.1016/j.ijantimicag.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Guo W, Köhler JR. 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun 73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner IE, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldón T, Hube B, d' Enfert C, Rupp S, Cormack BP, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e1004211. doi: 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kucharíková S, Vande Velde G, Himmelreich U, Van Dijck P. 2015. Candida albicans biofilm development on medically-relevant foreign bodies in a mouse subcutaneous model followed by bioluminescence imaging. J Vis Exp 95:52239. doi: 10.3791/52239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.