New Delhi metallo-beta-lactamases (NDMs) are an uncommon but emerging cause of carbapenem resistance in the United States. Genomic factors promoting their domestic spread remain poorly characterized.
KEYWORDS: CRE, antibiotic resistance, carbapenem resistance, genomics, NDM
ABSTRACT
New Delhi metallo-beta-lactamases (NDMs) are an uncommon but emerging cause of carbapenem resistance in the United States. Genomic factors promoting their domestic spread remain poorly characterized. A prospective genomic surveillance program among Boston-area hospitals identified multiple new occurrences of NDM-carrying strains of Escherichia coli and Enterobacter cloacae complex in inpatient and outpatient settings, representing the first occurrences of NDM-mediated resistance since initiating genomic surveillance in 2011. Cases included domestic patients with no international exposures. PacBio sequencing of isolates identified strain characteristics, resistance genes, and the complement of mobile vectors mediating spread. Analyses revealed a common 3,114-bp region containing the blaNDM gene, with carriage of this conserved region among unique strains by diverse transposon and plasmid backbones. Functional studies revealed a broad capacity for blaNDM transmission by conjugation, transposition, and complex interplasmid recombination events. NDMs represent a rapidly spreading form of drug resistance that can occur in inpatient and outpatient settings and in patients without international exposures. In contrast to Tn4401-based spread of Klebsiella pneumoniae carbapenemases (KPCs), diverse transposable elements mobilize NDM enzymes, commonly with other resistance genes, enabling naive strains to acquire multi- and extensively drug-resistant profiles with single transposition or plasmid conjugation events. Genomic surveillance provides effective means to rapidly identify these gene-level drivers of resistance and mobilization in order to inform clinical decisions to prevent further spread.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) are an urgent public health threat, presenting both treatment and infection prevention challenges (1, 2). Within the United States, Klebsiella pneumoniae carbapenemases (KPCs), serine beta-lactamases of the Ambler class A group, are the most commonly identified carbapenemase. Several enzyme isoforms have been identified, largely carried by variants of the Tn4401 transposon embedded in multiple plasmid types within the Enterobacteriaceae (3). Outside the United States, the landscape of carbapenem resistance more commonly includes other serine beta-lactamases, such as the Ambler class D OXA family, and metallo-beta-lactamases, including the Ambler class B NDM and VIM families (4).
NDM beta-lactamases have been a source of particular concern due to their increasing incidence and frequent occurrence within a diverse assortment of mobile genetic elements harboring other resistance genes, leading to extensively drug resistant (XDR) and panresistant (PDR) strains (5–8). NDMs are thought to have been acquired by Acinetobacter baumannii, with carriage by the Tn125 transposon (9). Subsequent events leading to mobilization by transposable elements, including IS3000 and IS5, have facilitated spread among members of the Enterobacteriaceae, particularly by broad-host-range plasmids that have acquired these elements (5, 6, 10–12).
NDMs are still relatively rare in the United States, with 379 reported to the CDC as of December 2017 (13). A recent survey based on data from the National Health and Safety Network (NHSN) showed that of 4,247 phenotypic CRE isolates, 134 (3.2%) carried blaNDM (1). Recent studies have identified domestic carriage of NDM family enzymes in IncA/C and IncF plasmid backbones, although the resolution of genetic context remains poor (14, 15). While most U.S. isolates have been from patients with international exposures, concerns for domestic acquisition remain (14, 16–19).
The Centers for Disease Control and Prevention (CDC), as well as the President’s Council of Advisors on Science and Technology (23) have called for increased surveillance efforts to control spread of these enzymes, with an emphasis on genomic strategies to identify carriage and the vectors mediating spread. Genomic analyses can broadly identify the complement of resistance genes and mobilizing vectors within drug-resistant strains in order to provide further assessments of risks for transmission (1, 20–23).
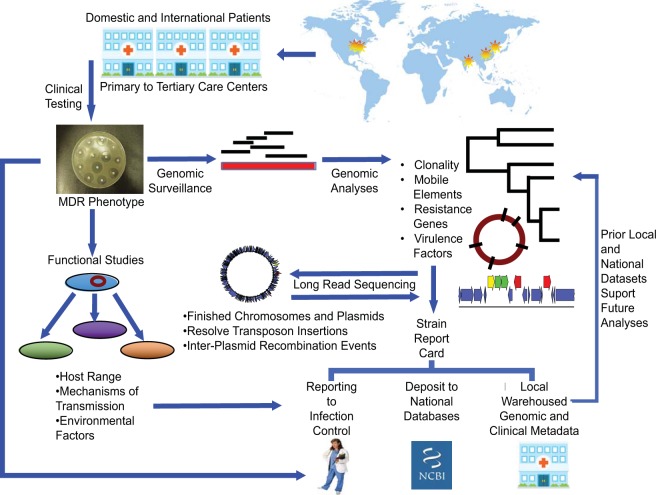
A prospective genomic surveillance program initiated in 2011 across multiple health care institutions within the Greater Boston area (Fig. 1) has provided real-time assessment and reporting to local infection prevention teams of the landscape of CRE resistance, with resolution to the level of genes promoting resistance, mobilizing vectors, and carrying strains (22).
FIG 1.
In the Pathogen Genomic Surveillance Program, patients from local, regional, and international sites receive care at participating institutions. Clinically ordered testing identifies carbapenem-resistant Enterobacteriaceae, which are flagged for genome analyses with rapid-turnaround short-read sequencing to define clonal relationships, resistance genes, and mobile elements carrying resistance. Novel strains undergo long-read sequencing to refine analyses. Functional studies define the host range of mobile vectors and mechanisms of transmission. Strain report cards to local infection control teams summarize clinical, microbiologic, genomic, and functional data to evaluate outbreak potential and the landscape of strain, vector, and gene-level causes of resistance. Local warehousing of the combined information supports ongoing and future surveillance efforts. Depositing of deidentified strain antibiogram and genomic data sets into national databases supports national and international surveillance efforts.
Starting in 2015, multiple Escherichia coli and one Enterobacter cloacae NDM-producing strains occurred both in patients with international exposure and, alarmingly, domestic patients with no prior travel history or epidemiologic links to affected patients. Genomic analyses of the NDM-producing strains assessed clonality among isolates and conducted a detailed assessment of the transposons and plasmids responsible transmitting the blaNDM and other linked resistance genes. Functional studies further refined the mechanisms by which the blaNDM gene could be spread within and across species. Clinically, this information rapidly ruled out clonal associations among the sporadically occurring isolates but identified common vectors and gene-level cassettes mediating the drug-resistant phenotype and its capacity for mobilization.
RESULTS
Patient demographics and phenotypic resistance.
Table 1 shows the patient demographics of those diagnosed with infections caused by extensively drug-resistant organisms (XDR). Isolates were all cultured within 24 h of admission or at an initial outpatient visit. Three E. coli isolates, ECO-222, ECO-165, and ECO-219, were isolated from international patients. E. cloacae complex (ECC)-174 and E. coli (ECO)-190 occurred in patients living in the northeastern United States with no reported travel history in the past 10 years but with prior hospitalizations across multiple health care centers. Inpatients were put on contact precautions upon identification of CREs. CRE surveillance cultures of other hospitalized patients in the same units detected no transmission events to other patients.
TABLE 1.
NDM strain and patient demographic data
| NDM strain | Yr of isolation | Setting | Species | MLST | Source | Patient origin |
|---|---|---|---|---|---|---|
| ECO-222 | 2015 | Outpatient | E. coli | 405 | Urine | India |
| ECO-165 | 2015 | Inpatient | E. coli | 101 | Abscess | China |
| ECC-174 | 2015 | Inpatient | E. cloacae complex | 231 | Blood | United States |
| ECO-190 | 2016 | Inpatient | E. coli | 90 | Urine | United States |
| ECO-219 | 2016 | Inpatient | E. coli | 167 | Blood | India |
The patient isolates demonstrated resistance to multiple classes of antibiotics (Table 2). Strains ECC-174 and ECO-190 remained susceptible to tetracyclines, and all strains were susceptible to glycylcyclines, and polymyxins.
TABLE 2.
Phenotypic and genomic antibiogram of NDM isolatesa
| Drug class | Drug | AST method and call | ECO-222 | ECO-165 | ECC-174 | ECO-190 | ECO-219 |
|---|---|---|---|---|---|---|---|
| Beta-lactams | Aztreonam | KB resistance | R | S | R | R | R |
| Cefoxitin | MIC (μg/ml), resistance | 64, R | 64, R | 64, R | 64, R | 64, R | |
| Ceftriaxone | KB resistance | R | R | R | R | R | |
| Cefepime | KB resistance | R | R | R | R | R | |
| Ertapenem | KB resistance | R | R | R | R | R | |
| Imipenem | KB resistance | R | R | R | I | R | |
| Meropenem | KB resistance | R | R | R | R | R | |
| All beta-lactams | Genotype | blaTEM-1 blaCTX-M-15 blaNDM-5 blaOXA-1 | blaTEM-1 (2 copies) blaNDM-5 | blaACT25 blaTEM-1 blaSHV-7 blaSHV-12 (2 copies) blaNDM-1 | blaCTX-M-15 blaCMY-6 blaTEM-1b blaOXA-1 blaNDM-1 | blaCTX-M-15 blaTEM-1 blaOXA-1 blaNDM-5 | |
| Glycylcyclines | Tigecyclineb | Etest (μg/ml) | 0.25 | 0.75 | 0.75 | 0.125 | 0.38 |
| Tetracyclines | Tetracycline | KB resistance | R | Not done | Not done | Not done | S |
| Tetracycline | MIC (μg/ml), resistance | Not done | 16, R | 4, S | 2, S | Not done | |
| All tetracyclines | Genotype | tetB tetD | tetA | None detected | None detected | None detected | |
| Aminoglycosides | Gentamicin | MIC (μg/ml), resistance | 16, R | 4, R | 16, R | 16, R | 16, R |
| All aminoglycosides | Genotype | aac(3)-Id aac(6′)-Ib-cr ant(3″)-Ia (2 copies) | rmtB aac(6′)Ib-cr ant(3″)-Ia family aph(3″) family aph(6)-I family aac(3)-Id | aph(3′)-Ia ant(3″) family ant(3″)-Ia aph(6)-Id aph(3″) family ant(2″)-Ia aac(6)-Ib | rmtC aph(6)-Id aph(3″)-Ib aac(6′)Ib-cr aph(3′)-Ia aph(3′)-VI | rmtB ant(3)-Ia aac(6′)-Ib-cr | |
| Folate antagonists | Trimethoprim-sulfamethoxazole | MIC (μg/ml), resistance | 16/304, R | 16/304, R | 16/304, R | 16/304, R | 16/304, R |
| All folate antagonists | Genotype | sul1 (2 copies) dfrA17 dfrA12 | sul1 sul2 dfrA14 (2 copies) | sul1 (3 copies) dfrA8 dfrA19 | sul1 sul2 dfrA14 | sul1 dfrA12 | |
| Fluoroquinolones | Ciprofloxacin | MIC (μg/ml), resistance | 4, R | 64, R | 4, R | 4, R | 4, R |
| All fluoroquinolones | Genotype | aac(6′)Ib-cr qnrS1 | aac(6′)Ib-cr | qnrS1 qnrB2 | aac(6′)Ib-cr qnrB1 | aac(6′)-Ib-cr | |
| Phenicol | Chloramphenicol | KB resistance | I | R | R | R | I |
| All phenicols | Genotype | None detected | catA1 | catA1 | cm/family | None detected | |
| Polymyxins | Colistinc | MIC (μg/ml) resistance | 0.06 wild type | 0.06 wild type | 0.06 wild type | 0.12 wild type | 0.12 wild type |
| All polymyxins | Genotype | None detected | None detected | None detected | None detected | None detected |
AST, antimicrobial susceptibility testing method; KB, Kirby-Bauer disk diffusion testing. MICs are by concentration cutoff for growth. Genotype indicates resistance genes identified by next-generation sequencing and gene annotation. R, resistant; I, intermediate; S, susceptible.
Tigecycline Etest does not have CLSI guidelines for reporting resistance or susceptibility.
Colistin MICs were assessed by broth microdilution as described in the CLSI guidelines (M100-ED28 [37]). Results are reported as epidemiological cutoff values (ECVs).
Genomic surveillance and resistance markers.
As part of the CRE surveillance program, all isolates with phenotypic resistance to carbapenem antibiotics undergo whole-genome sequencing to assess resistance genes, mobile vectors, and relatedness of carrying strains (22, 24). The incorporation of phenotypic and epidemiologic data, with comparison against previously sequenced strains, identifies genomic causes of drug resistance and relatedness among isolates for communication with local infection control teams (Fig. 1).
Analyses identified blaNDM-5 in isolates ECO-222, ECO-165, and ECO-219 and blaNDM-1 in ECC-174 and ECO-190 (Fig. 1 and Table 2). In contrast, all prior CRE isolates analyzed by the surveillance program had identified blaKPC genes or a combination of extended-spectrum beta-lactamases (ESBLs) or upregulated ampC beta-lactamases in the presence of mutated porins, as underlying causes of carbapenem resistance (22).
In addition to the NDM beta-lactamase, several additional resistance genes were detected, including those encoding extended-spectrum and AmpC beta-lactamases, as well as resistance genes for aminoglycosides, fluoroquinolones, phenicols, sulfonamides, and trimethoprim (Table 2).
Single nucleotide polymorphism (SNP) and multilocus sequence typing (MLST) analyses evaluated clonal relationships among the NDM-carrying isolates. The E. coli isolates were all different MLSTs (ST405, ST101, ST90, and ST167) and were further separated by a large number of pairwise SNPs. No clonal clusters were identified, suggesting sporadic entry of these strains into the health care system.
Genomic contexts of NDMs.
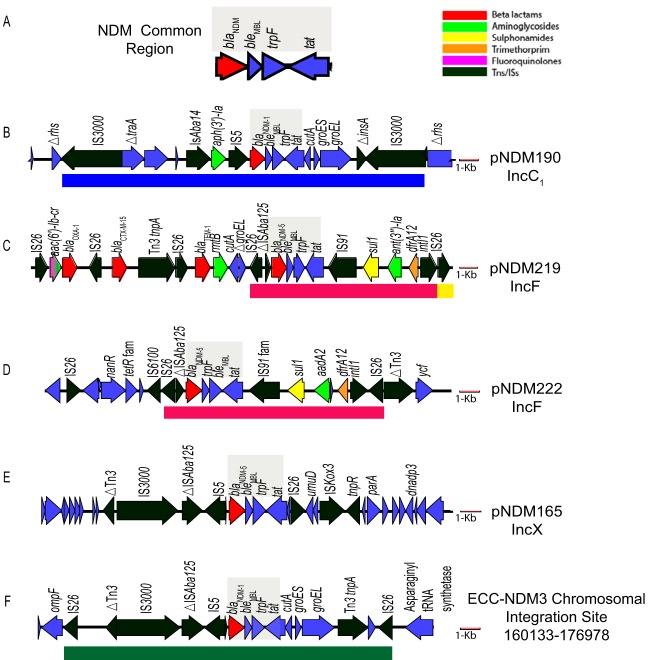
To define the genomic context of blaNDM carriage, closed genomes and plasmids were generated using PacBio sequencing. A 3,114-bp region that was common to all of the strains carried blaNDM (Fig. 2A) (25). This element was bordered on the 5′ end by an incomplete insertion sequence ISAba125 and on the 3′ end by the bleomycin resistance genes bleMBL, trpF (involved in tryptophan biosynthesis), and tat, a gene encoding a protein with the twin-arginine transport signal sequence. This conserved segment has been reported within Tn125, the transposon of origin for blaNDM in Acinetobacter spp. (26, 27).
FIG 2.
NDM-carrying vectors. (A) Shaded area is common blaNDM region found among strains. Subsequent panels show 20 kb of surrounding sequence with resistance gene and mobile element sequences for each clinical isolate. (B) blaNDM-1 in pNDM190_177 from strain ECO-190. (C) blaNDM-5 in pNDM219_158 from strain ECO-219. (D) blaNDM-5 region (shaded) in pNDM222_110 from E. coli strain ECO-222. (E) blaNDM-5 in pNDM165_46 from strain ECO-165. (F) IS26-mediated chromosomal insertion of blaNDM-1 in strain ECC-174. A 1-kb scale bar shown in red. Tns, transposons.
Plasmids mobilizing the blaNDM cassette, however, were quite diverse and included multiple carrying transposons, many of which were further embedded in broad- and narrow-host-range plasmids (Fig. 2). Genomic analyses of transposon boundaries and duplications associated with unique insertion sites of transposons identified mechanisms of acquisition in some strains but not in others.
The blaNDM-1 gene in ECO-190 resided in an IncC1 plasmid among a series of insertion sequences within the conserved ARI-A resistance island, flanked by IS3000 elements (Fig. 2B). Genes encoding NDMs have been found inserted into this hot spot region from international isolates (6, 28). The flanking IS3000 elements defined a 19.8-kb region that included 5-bp direct repeats (ACATT), indicating an intact transposition event into the IncC1 plasmid (Fig. 2B, blue box).
Strains ECO-219 (Fig. 2C) and ECO-222 (Fig. 2D) carried blaNDM-5 within an identical 10,494-bp region (red boxes) but in the distinct and novel IncF plasmids pNDM219_158 and pNDM222_109. In both strains, an intact IS26 element flanked the 5′ end of this 10.4-kb region, while a complex class I integron, interrupted by a second IS26 element, flanked the 3′ end. However, the 3′ IS26 insertions occurred in opposite orientations between the strains (Fig. 2C, yellow box) and also lacked matching 8-bp direct repeats, characteristic of intact IS26 insertions, suggesting that the paired IS26 elements may have integrated via separate transposition events, had prior partner elements that were lost in subsequent genetic recombination events, or integrated as part of a translocatable unit (TU) element (29).
In strain ECO-165 (Fig. 2E), unpaired IS3000 IS5, IS26, and ISKox3 flanked the blaNDM-5 cassette in an IncX plasmid, suggesting multiple prior transposition or recombination events into this region, leading to loss of the original transposon boundaries. NDM carriage in IncX plasmids has been identified in other isolates from China, from where this patient had received prior health care (30).
In contrast to the other isolates, strain ECC-174 harbored an IS26-bounded chromosomal insertion of the blaNDM-1 gene, a rarely described location for blaNDM genes (31). Eight-base-pair direct repeats (TTATTTAT), characteristic of a duplication at the IS26 insertion site, flanked the mobilizing IS26 elements (green box, Fig. 2F).
In addition to the diverse plasmid backbones carrying the blaNDM genes (IncX, IncC1, IncFII, and IncFI) (Fig. 2 and Table S3), isolates also carried additional plasmids, many of which harbored other resistance genes that contributed to their highly resistant phenotypes (see Fig. S1 in the supplemental material and Table S3). While a few of these had close (>99% identical) relatives in GenBank, several had large regions without close homology to published plasmids.
Dynamics of inter- and intraspecies transfer of NDM resistance.
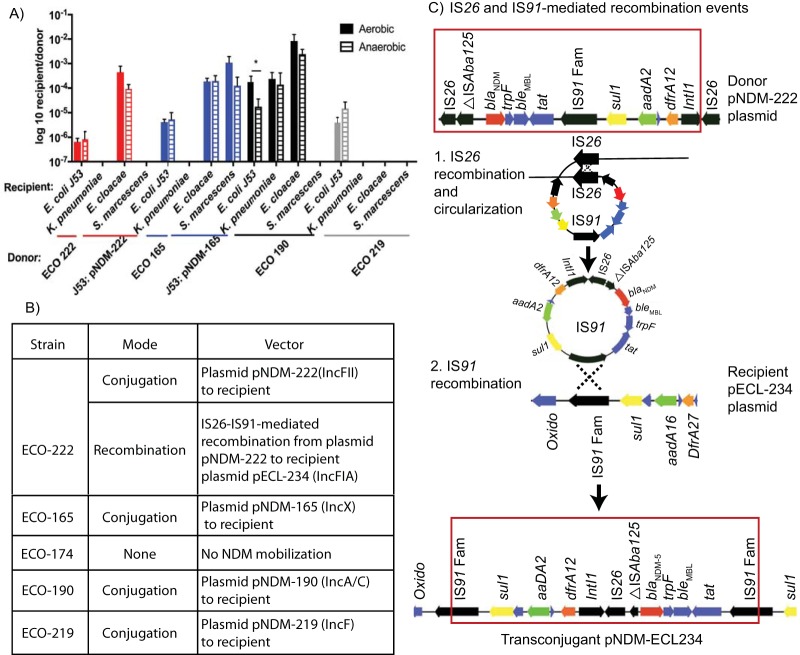
Functional studies evaluated the host range and capacity for transposons and plasmids to mediate further transmission (Fig. 3), including under aerobic and anaerobic conditions to replicate environments occurring in the environment and the gut, respectively.
FIG 3.
Host range and transfer of NDM resistance. (A) In vitro mating of NDM donors to clinical isolates of K. pneumoniae, E. cloacae, and S. marcescens or the azide-resistant E. coli strain J53, under aerobic (solid bar) or anaerobic (striped bar) conditions. For highly drug-resistant ECO-222 and ECO-165, E. coli J53 NDM transconjugants were used as donors. Asterisk indicates transfer of the pNRM-190 plasmid to the J53 recipient that showed a log difference of more effective mating under aerobic conditions. All other matings showed no significant differences between aerobic and anaerobic conditions. (B) Table illustrating modes of NDM transmission to recipients. (C) Proposed mechanism for the movement of an IS26-bounded composite transposon from the pNDM-222 donor plasmid in strain ECO-222 to an IncF1A plasmid in the recipient E. cloacae. Boxed region shows the IS26-bounded sequences. 1 and 2, IS26 appears to have mobilized out of the donor plasmid (1) and integrated into the recipient plasmid via homologous recombination of the identical IS91 family insertion sequences (2).
Among carrying plasmids, the broad host-range IncC1 plasmid in ECO-190 and IncX3 plasmid in ECO-165 demonstrated the broadest promiscuity to spread resistance to other members of the Enterobacteriaceae. Transfers from strain ECO-190 occurred to E. coli, K. pneumoniae, and E. cloacae at high efficiencies (Fig. 3A), while the IncX3 plasmid in ECO-165 transferred broadly and at high efficiency to recipient E. coli, E. cloacae, and Serratia marcescens. In contrast, the IncF family plasmids in ECO-222 and ECO-219 demonstrated a narrower host range. The IncFII plasmid in ECO-222 transferred to recipient E. coli and to E. cloacae, while the IncFI plasmid in strain ECO-219 transferred only to recipient E. coli.
Among recipients, E. cloacae demonstrated the greatest capacity to acquire broad-range NDM plasmids, with efficiencies ranging from 10−4 to 10−2 conjugation events per recipient. Conjugations occurred comparably under aerobic and anaerobic conditions. The IS26 sequences flanking the chromosomal transposon insertion in ECC-174 failed to mobilize under any conditions (data not shown).
Transfer of NDM-mediated carbapenem resistance occurred most commonly via plasmid-based conjugation (Fig. 3B). However, interplasmid recombination events mediated cross-species transfer of the IS26-flanked blaNDM-5 gene cassette in ECO-222 to plasmid pECL-234 in the recipient E. cloacae. High-resolution plasmid maps derived from PacBio sequencing identified a complex NDM-containing region in the new recipient plasmid likely resulting from two recombination events at different insertion sequences (Fig. 3C). In this model, a circularization and excision event between the IS26 insertion sequences releases the ndm gene cassette from the donor plasmid. The circularized molecule includes the blaNDM-5 gene as well as a single IS91 region. A subsequent recombination event between the circular molecule’s IS91 and homologous region in the recipient plasmid enabled integration into the new plasmid vector. This phenomenon has been described for IS26-flanked regions and is referred to as a translocatable unit (TU) (29). While TUs are known to integrate into recipient constructs containing additional IS26 elements, this example identified that other prevalent elements can support homologous recombination of TUs (Fig. 3C). This interplasmid recombination event resulted in a recipient plasmid containing only one copy of IS26 and lacking characteristic duplications of adjacent sequences seen with replicative movement of IS26. The original pNDM-222 donor plasmid was not detected in the recipient.
DISCUSSION
NDM carbapenemases represent an emerging form of carbapenem resistance in the United States. To date, little is known about the blaNDM-carrying transposon and plasmids that may be present in U.S. institutions.
The E. coli NDM-expressing strains occurred sporadically and represented diverse MLSTs (ST405, ST101, ST90, and ST167) that have been associated with NDM carriage in other countries and, in some instances, the United States (32, 33). The sole isolate of E. cloacae complex was MLST 231, in which infrequent blaNDM-1 and blaKPC-3 carriage have been reported (34). None of these MLSTs had previously been detected among CREs by the Partners prospective pathogen surveillance program (4).
Each extensively drug-resistant isolate (XDR) carried plasmid-borne resistance genes to multiple classes of antibiotics. Furthermore, diverse transposable elements, including class I integrons as well as IS3000, IS26, and IS5 elements, participated in the mobilization the blaNDM common region.
Genomic and functional studies demonstrated the capacity of these elements to transfer resistance via plasmid conjugation and transposition into new plasmid and chromosomal locations. Plasmid incompatibility group (Inc) typing refines strain clusters in genomic surveillance and further informs risks for spread. The IncC1 and IncX3 plasmids readily transferred NDM resistance to other species, while the E. coli IncFI plasmid pNDM219-158 showed more limited host range. All plasmids conjugated effectively under aerobic and anaerobic conditions, highlighting the potential for spread both within the gastrointestinal tract and in environmental reservoirs.
Isolates of Enterobacter cloacae demonstrated the broadest capacity to acquire blaNDM genes via plasmid conjugation, de novo transposition, and interplasmid recombination. The interplasmid recombination mechanism appeared to leverage homologous IS26 and IS91 insertion sequences accrued in multiple plasmid vectors seen in clinical isolates to facilitate introduction of the blaNDM gene cassette into a new carrying plasmid. This mechanism highlights the risk for spread of drug resistance genes and multigene cassettes by homologous recombination without the need for transposition events, or in cases of plasmid incompatibility between a donor plasmid and the recipient strain.
These patterns occur in contrast to KPC carbapenemases, carried primarily in plasmid-borne insertions of transposon Tn4401. Though relatively few NDM-producing strains have been reported in the United States, our results highlight the diverse machinery capable of mobilizing resistance, which has also been illustrated by NDM-producing isolates characterized outside the United States (5, 11).
Incorporating genomic analyses in clinical surveillance programs for MDR organisms enables efforts to track strain-, mobile vector-, and gene-level dynamics of resistance within and across institutions. Analyses using data from short-read sequencing platforms provide timely information to clinical infection control teams, while subsequent analysis with long-read sequencing platforms, such as PacBio, provides high-resolution chromosomal and plasmid maps to clearly define resistance gene-carrying vectors and events mediating spread. Warehousing of this combined information further supports more rapid and improved analyses of future patient isolates as they occur.
All NDM-producing isolates occurred sporadically in patients seen in institutions participating in the surveillance program. The epidemiology of NDM occurrence followed that observed for KPC carriage among participating institutions (4) with a dominance of sporadically occurring strains and carrying vectors, largely from patients with prior admissions to different health care facilities. In this setting, genomic surveillance enables continued vigilance for drug-resistant isolates with capacity to identify both intra- and extrainstitutional reservoirs. Findings assist infection control actions while also informing efforts at regional, national, and international levels to further prevent spread.
MATERIALS AND METHODS
Strain collection.
The Partners Prospective Pathogen Surveillance Program (institutional review board [IRB] protocol 2011-P-002287) pulls multidrug-resistant isolates from patients seen at Brigham & Women’s Hospital, the Dana-Farber Cancer Institute, Faulkner Hospital, Newton Wellesley Hospital, and outpatient centers associated with these institutions (4). Isolates (Table 1) with resistance to any carbapenem were collected for genomic analyses. Phenotypic resistance was determined by MIC, disk diffusion testing, or Etest according to clinical lab protocols. MICs were assessed by Vitek 2 using criteria set out by the CLSI in documents M100-S25 (35) and M100-S26 (36). Colistin MICs were assessed using manual broth microdilution and reported as epidemiological cutoff values (ECVs) of wild-type or non-wild-type according to CLSI document M100-S28 (37). Total DNA was extracted with the Qiagen EZ1 DNA tissue kit (Germantown. MD). Organisms were handled under biohazard level 2 containment.
Genome sequencing and data preprocessing.
Illumina MiSeq libraries were prepared, sequenced, and analyzed as described previously (22). For fine resolution of transposons and plasmids, isolates were sequenced on the Pacific Biosciences (PacBio) RSII sequencer, as described previously (38, 39). Size selection was performed with BluePippin (Sage Science, Beverly, MA). Analysis was done using SMRT Analysis 2.3.0. De novo assembly was established with the PacBio Hierarchical Genome Assembly Process (HGAP3.0) program using the continuous-long-reads from four single-molecule real-time (SMRT) cells. The assembly outputs from HGAP contain overlapping regions at the end which can be identified using dot plots in Gepard (40). Genomes were checked manually for even coverage. The improved consensus sequence was uploaded in SMRT Analysis 2.3.0 to determine the final consensus and accuracy scores using the Quiver consensus algorithm (41).
Sequence analysis.
Sequences were annotated with RAST (42) and antibiotic resistance elements with CARD (43). Full names were reported for perfect matches. Genes were annotated as “family” members if they were at least 99% identical to proteins in the CARD or NCBI RefSeq databases. Transposons were annotated using ISFinder (44). Plasmid incompatibility type was determined with PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) (45). IncA and IncC plasmids were typed according to Ambrose et al. (46) Sequence maps were generated with MacVector (Apex, NC). To determine transposon insertion sites after matings, recipient contigs were ordered in MAUVE against donor sequences and the recipient prior to mating, followed by raw-read alignment to the ordered recipient contigs and donor plasmid to determine coverage and transposon boundaries. Gap regions were confirmed by Sanger sequencing. High-resolution plasmid maps from Pacific Biosciences long-read sequencing identified the context of blaNDM.
Infection prevention and control response and monitoring.
Patients with Enterobacteriaceae isolates resistant to any carbapenem were flagged as CRE positive in the electronic health record and placed onto contact precautions (patient consigned to a private room and health care personnel required to don a gown and gloves and perform hand hygiene) (47). After identification of the CRE isolate, rectal swabs were obtained from all patients on the same hospital unit to screen for CRE. All CRE isolates were reported to the Massachusetts Department of Public Health.
NDM transmission and host range.
Tetracycline-susceptible NDM-producing isolates were mated to tetracycline-resistant clinical isolates of Enterobacter cloacae, Klebsiella pneumoniae, Serratia marcescens, or to azide-resistant Escherichia coli strain J53 (48) (Fig. 3). For panresistant strains ECO-222 and ECO-165, the J53 NDM-producing transconjugant was used as the donor in matings to the other recipients. Donors and recipients were grown to exponential-growth phase in Luria-Bertani (LB) broth under aerobic or anaerobic conditions and spotted 1:1 on LB agar for incubation at 37°C for 18 h in a 5% CO2 incubator or anaerobic chamber. For each incubation condition and donor-recipient pair, a minimum of 4 experimental replicates were performed. Transconjugants were selected on KPC plates (CHROMagar, Springfield, NJ) containing 100 μg/ml sodium azide for the E. coli J53 selection or 20 μg/ml tetracycline for the clinical isolate recipients (Sigma, St. Louis, MO). Transfer of blaNDM was confirmed in recipients by PCR using ndm, plasmid, and strain-specific primers (Table 2). For matings with the E. cloacae complex 174 strain, selections were also conducted with and without meropenem selection (0.125 to 0.50 μg/ml) during the mating period. Statistical testing was performed in Prism 6.0 (GraphPad, La Jolla, CA) using the Mann-Whitney test. A P value of <0.05 was considered significant.
Data availability.
The Illumina sequencing data have been deposited to the NCBI Sequence Read Archive under BioProject number PRJNA278886. Complete genome and plasmid sequences are available in GenBank (Table 1).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants P30 DK034854 and T32 HL007627), the Food and Drug Administration’s GenomeTrakr Program, the Massachusetts Life Sciences Center, BWH Clinical Laboratories, and an NLM ORISE fellowship.
We thank David Hooper at Massachusetts General Hospital for providing the J53 azide-resistant E. coli strain, Georg Gerber (BWH), Alex McAdam (Boston Children’s Hospital), and Maura Pavao (Worcester State) for critical reading of the manuscript, and Dwight Hardy (URMC) for assistance with colistin MIC testing.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02040-18.
REFERENCES
- 1.Woodworth KR, W MS, Weiner LM, Edwards J, Brown AC, Huang JY, Malik S, Slayton RB, Paul P, Capers C, Kainer MA, Wilde N, Shugart A, Mahon G, Kallen AJ, Patel J, McDonald LC, Srinivasan A, Craig M, Cardo DM. 2018. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms—United States, 2006–2017. MMWR Morb Mortal Wkly Rep 67:396–401. doi: 10.15585/mmwr.mm6713e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 6.Wailan AM, Paterson DL. 2014. The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev Anti Infect Ther 12:91–115. doi: 10.1586/14787210.2014.856756. [DOI] [PubMed] [Google Scholar]
- 7.Wailan AM, Sidjabat HE, Yam WK, Alikhan NF, Petty NK, Sartor AL, Williamson DA, Forde BM, Schembri MA, Beatson SA, Paterson DL, Walsh TR, Partridge SR. 2016. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY Plasmids. Antimicrob Agents Chemother 60:4082–4088. doi: 10.1128/AAC.00368-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnin RA, Poirel L, Nordmann P. 2014. New Delhi metallo-beta-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol 9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
- 10.Campos JC, da Silva MJF, dos Santos PRN, Barros EM, Pereira M. d O, Seco BMS, Magagnin CM, Leiroz LK, de Oliveira TGM, de Faria-Júnior C, Cerdeira LT, Barth AL, Sampaio SCF, Zavascki AP, Poirel L, Sampaio JLM. 2015. Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob Agents Chemother 59:7387–7395. doi: 10.1128/AAC.01458-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Pesesky MW, Hussain T, Wallace M, Wang B, Andleeb S, Burnham CA, Dantas G. 2015. KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg Infect Dis 21:1034–1037. doi: 10.3201/eid2106.141504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2017. Tracking CRE. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/hai/organisms/cre/trackingcre.html. [Google Scholar]
- 14.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J-J, Munoz-Price LS, Spychala CN, DePascale D, Doi Y. 2016. New Delhi Metallo-beta-lactamase-1-producing Klebsiella pneumoniae, Florida, USA. Emerg Infect Dis 22:744–746. doi: 10.3201/eid2204.151176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janelle SJ, Kallen A, de Man T, Limbago B, Walters M, Halpin A, Xavier K, Knutsen J, Badolato E, Bamberg WM. 2016. Notes from the field: New Delhi Metallo-beta-lactamase-producing carbapenem-resistant Enterobacteriaceae identified in patients without known health care risk factors—Colorado, 2014–2016. MMWR Morb Mortal Wkly Rep 65:1414–1415. doi: 10.15585/mmwr.mm6549a6. [DOI] [PubMed] [Google Scholar]
- 17.Aitken SL, Tarrand JJ, Deshpande LM, Tverdek FP, Jones AL, Shelburne SA, Prince RA, Bhatti MM, Rolston KVI, Jones RN, Castanheira M, Chemaly RF. 2016. High rates of nonsusceptibility to ceftazidime-avibactam and identification of New Delhi Metallo-beta-lactamase production in Enterobacteriaceae bloodstream infections at a major cancer center. Clin Infect Dis 63:954–958. doi: 10.1093/cid/ciw398. [DOI] [PubMed] [Google Scholar]
- 18.Epstein L, Hunter JC, Arwady MA, Tsai V, Stein L, Gribogiannis M, Frias M, Guh AY, Laufer AS, Black S, Pacilli M, Moulton-Meissner H, Rasheed JK, Avillan JJ, Kitchel B, Limbago BM, MacCannell D, Lonsway D, Noble-Wang J, Conway J, Conover C, Vernon M, Kallen AJ. 2014. New Delhi metallo-beta-lactamase-producing carbapenem-resistant Escherichia coli associated with exposure to duodenoscopes. JAMA 312:1447–1455. doi: 10.1001/jama.2014.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, Perez F, Endimiani A, Bonomo RA. 2016. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev 29:1–27. doi: 10.1128/CMR.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutgring JD, Limbago BM. 2016. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. 2015. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio 6:e01030-15. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.President's Council of Advisors on Science and Technology (PCAST). 2014. Report to the president on combating antibiotic resistance. PCAST, Washington, DC: Obamawhitehouse.archives.gov. [Google Scholar]
- 24.Nudel K, Zhao X, Basu S, Dong X, Hoffmann M, Feldgarden M, Allard M, Klompas M, Bry L. 2018. Genomics of Corynebacterium striatum, an emerging multidrug-resistant pathogen of immunocompromised patients. Clin Microbiol Infect 24:1016.e7–1016.e13. doi: 10.1016/j.cmi.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 56:1693–1697. doi: 10.1128/AAC.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. 2015. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. doi: 10.1371/journal.pone.0129454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pannaraj PS, Bard JD, Cerini C, Weissman SJ. 2015. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr Infect Dis J 34:11–16. doi: 10.1097/INF.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Chan EW, Zhang R, Chen S. 2016. Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother 60:4364–4368. doi: 10.1128/AAC.00859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang YW, Kreiswirth BN. 2016. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother 60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI document M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.CLSI. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Hoffmann M, Muruvanda T, Allard MW, Korlach J, Roberts RJ, Timme R, Payne J, McDermott PF, Evans P, Meng J, Brown EW, Zhao S. 2013. Complete genome sequence of a multidrug-resistant Salmonella enterica serovar Typhimurium var. 5- strain isolated from chicken breast. Genome Announc 1:e01068-13. doi: 10.1128/genomeA.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao K, Muruvanda T, Roberts RJ, Payne J, Allard MW, Hoffmann M. 2016. Complete genome and methylome sequences of two Salmonella enterica spp. Genome Announc 4:e01599-15. doi: 10.1128/genomeA.01599-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 41.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 42.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrose SJ, Harmer CJ, Hall RM. 2018. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention. 2017. Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE), November 2015 update CRE toolkit. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/hai/pdfs/cre/CRE-guidance-508.pdf. [Google Scholar]
- 48.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother 47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina sequencing data have been deposited to the NCBI Sequence Read Archive under BioProject number PRJNA278886. Complete genome and plasmid sequences are available in GenBank (Table 1).