Herein, we evaluated sustainability of humanized exposures of cefiderocol in vivo over 72 h against pathogens with cefiderocol MICs of 0.5 to 16 μg/ml in the neutropenic murine thigh model. In Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae displaying MICs of 0.5 to 8 μg/ml (n = 11), sustained kill was observed at 72 h among 9 isolates.
KEYWORDS: Gram-negative bacteria, pharmacodynamics, pharmacokinetics
ABSTRACT
Herein, we evaluated sustainability of humanized exposures of cefiderocol in vivo over 72 h against pathogens with cefiderocol MICs of 0.5 to 16 μg/ml in the neutropenic murine thigh model. In Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae displaying MICs of 0.5 to 8 μg/ml (n = 11), sustained kill was observed at 72 h among 9 isolates. Postexposure MICs revealed a single 2-dilution increase in one animal compared with controls (1/54 samples, 1.8%) at 72 h. Adaptive resistance during therapy was not observed.
INTRODUCTION
Standard pharmacodynamic assessment studies designed to quantify antimicrobial efficacy typically occur over 24 h. However, prolonged (i.e., 72-h) studies have demonstrated additional insight into the characterization of a drug’s rate, extent, and robustness of antimicrobial activity (1, 2). The effect of humanized doses of cefiderocol, a novel siderophore cephalosporin, was recently characterized across a wide array of clinical isolates in the neutropenic murine thigh infection model. Data from these 24-h pharmacodynamic studies revealed that cefiderocol humanized exposures produced predictable bacterial kill against multidrug resistant (MDR) Gram-negative isolates with MICs (MICs) of ≤4 μg/ml (3). Little is known about the sustained antimicrobial effect of cefiderocol beyond 24 h. Furthermore, previous studies with siderophore monobactams demonstrated adaptive resistance in the presence of antibiotic pressure (4, 5). The aim of the current study was to evaluate the sustainability of cefiderocol activity in vivo over 72 h against Gram-negative pathogens and to characterize the phenotypic and genotypic profiles of organisms recovered postexposure.
Shionogi & Co., Ltd., supplied cefiderocol vials (lot 12M01) for the purposes of in vivo testing. The cefiderocol dosing regimen utilized has been validated in several studies with the same mouse model to provide exposures equivalent to those in humans attained after administration of 2 g every 8 h (q8h) (3 h infusion) (3, 6, 7). Commercially available cefepime acquired from Cardinal Health, Inc. (1 g; lot 106014C; Sagent Pharmaceuticals) was also utilized. Cefepime vials were reconstituted and diluted with 0.9% normal saline and administered in volumes of 0.2 ml as subcutaneous injections at doses simulating those equivalent to 2 g q8h (3 h infusion) in humans (7).
A total of 12 Gram-negative isolates (2 Pseudomonas aeruginosa isolates, 4 Acinetobacter baumannii isolates, and 6 Enterobacteriaceae isolates) with cefiderocol MICs ranging from 0.5 to 16 μg/ml were provided by Shionogi & Co., Ltd., or IHMA, Inc. These isolates were selected from among those utilized in the previous 24-h pharmacodynamic study representing various species across a range of cefiderocol MICs, having shown modest efficacy despite lower MICs to cefiderocol (3). By use of iron-depleted cation-adjusted Mueller-Hinton broth (ID-CAMHB), as recommended by CLSI, isolates were screened for cefiderocol MICs in triplicate, and the modal value was reported (8). Cefepime MICs were determined in triplicate according to CLSI broth microdilution methods (9). For isolates shown to regrow at 72 h or to otherwise display an unexpected response (ranging from stasis to 4 log growth) despite cefiderocol MICs predictive of susceptibility (i.e., KP 543, EC 458, KP 539, AB 152, EC 462, KP 531), a bacterial sample from a single thigh of treated and control animals was collected and tested for its cefiderocol postexposure MIC at 24, 48, and 72 h. Any change in MIC of >2-fold compared with those in infected controls in the same period of exposure was considered meaningful. All retest MICs were performed in triplicate using ID-CAMHB.
Subsequent whole-genome sequencing was performed on bacterial isolates after cefiderocol exposure and on paired in vivo controls. All genotypic analyses were performed with the CLC genomics workbench (version 11; Qiagen). Reads from whole-genome shotgun sequencing on an Illumina HiSeq (150-bp paired-end reads) were sampled down to a predicted coverage depth of 75× for each genome, and de novo assembly was performed. β-Lactamase genes were identified by use of the “find resistance” module, which utilizes the same database as that utilized by ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/). All β-lactamase genes identified had 100% nucleotide sequence identity and length of the specified reference except for the endogenous class C β-lactamase gene of Escherichia coli, blaEC, which had 97% identity to the reference in each isolate. MLST was carried out on a guided assembly utilizing the most closely related closed genome in NCBI (Find Best Matches using K-mer Spectra tool) as the reference sequence. A minimum coverage depth of 30× for each of the 7 loci was obtained. MLST databases were acquired from PubMLST (https://pubmlst.org/), and the Achtman scheme was used for E. coli isolates. Genome assemblies from the two control isolates, 32a (E. coli) and 73a (Klebsiella pneumoniae), were annotated with the rapid annotation using subsystem technology (RAST) pipeline (10–12).
Reads from the treated-isolate genomes were aligned to the annotated contigs of their respective control isolates, and the Basic Variant Detection tool was used to identify single nucleotide polymorphisms (SNPs) and small in/del mutations in the treated isolates. Positions with <20× or >300× read depths were ignored. If the mutation had an impact on the sequence of an annotated gene, the resultant amino acid variation was indicated. Reads from samples 32a and 73a were aligned to their own annotated assemblies for mutational analysis as described above. Any mutations identified this way were considered spurious and were not reported when identified in the genomes of the treated strains.
The study protocol was reviewed and approved by the Institutional Animal Care and Use Committee at Hartford Hospital. Animals were prepared by using a previously described murine thigh infection model (13).
All isolates were stored frozen at −80°C in skim milk (BD BioSciences, Sparks, MD). Before mouse thigh inoculation, two bacterial transfers were performed onto Trypticase soy agar plates containing 5% sheep blood (TSA II; Becton, Dickinson & Co., Sparks, MD). Subsequently, a suspension containing ∼107 CFU/ml was made for inoculation. Thigh infection was established via intramuscular injection of the inoculum (0.1 ml) into each thigh 2 h before the initiation of antimicrobial therapy.
In total, 12 isolates (2 P. aeruginosa [MIC range, 2 to 4 μg/ml], 4 A. baumannii [MIC range, 0.5 to 16 μg/ml], and 6 Enterobacteriaceae [MIC range, 1 to 8 μg/ml]) were tested in the neutropenic thigh infection model. Cefiderocol preexposure and comparator MIC values are presented in Table 1. Control animals were dosed with vehicle. For isolates treated only with cefiderocol (i.e., no cefepime control), groups of 3 animals were assigned to 0-h control and to control, cefiderocol, or cefepime groups corresponding to 24-, 48-, and 72-h time points. Animals that did not survive to the time allotted were grouped with the interval corresponding to the time at which they were found deceased. After the animals were killed, the thighs were removed and individually homogenized in normal saline. Samples were serially diluted and plated on agar medium for enumeration of bacterial burden. Changes in CFU for each thigh relative to 0 h were determined at 24, 48, and 72 h, and average values (± standard deviations) were determined for each group. Efficacy was defined as a reduction in bacterial burden relative to 0 h control.
TABLE 1.
MICs of preexposure cefiderocol and comparators against a collection of 12 P. aeruginosa, A. baumannii, and Enterobacteriaceae isolates
| Isolate | MIC (μg/ml) fora: |
||||
|---|---|---|---|---|---|
| Cefiderocol | FEP | MEM | CAZ | TZP | |
| AB 135 | 0.5 | ND | 128 | ND | ND |
| AB 152 | 1 | 64 | 128 | ND | ND |
| EC 462 | 1 | >64 | 0.06 | >128 | >128 |
| EC 458 | 1 | 512 | 0.06 | >128 | >64 |
| PSA 1574 | 2 | 8 | 16 | ND | ND |
| PSA 1595 | 4 | 2 | 1 | ND | ND |
| KP 531 | 4 | >64 | 256 | >64 | >32 |
| KP 519 | 4 | >64 | 512 | >32 | >32 |
| KP 539 | 4 | >512 | 0.12 | 128 | 128 |
| AB 87 | 4 | >16 | 256 | >16 | >64 |
| KP 543 | 8 | >512 | ND | ND | ND |
| AB 84 | 16 | >64 | 64 | >16 | >64 |
MEM, meropenem; FEP, cefepime; CAZ, ceftazidime; TZP, piperacillin-tazobactam; ND, no data.
Average initial bacterial burden at 0 h among the isolates was 6.08 ± 0.25 log10 CFU/thigh for A. baumannii, 5.49 ± 0.23 log10 CFU/thigh for P. aeruginosa, 5.91 ± 0.44 log10 CFU/thigh for E. coli, and 5.92 ± 0.24 log10 CFU/thigh for K. pneumoniae. In isolates displaying initial kill at 24 h after human-simulated exposures of cefiderocol, a sustained antimicrobial effect was observed over the subsequent 48-h dosing period (Fig. 1 to 4). Cefepime-resistant isolates (EC 458, KP 539, KP 543; MIC, ≥512 μg/ml) showed kill on cefiderocol therapy, while validation of the model was confirmed as comparator animals receiving human-simulated exposures of cefepime grew >3 log10 CFU at 72 h (Fig. 4).
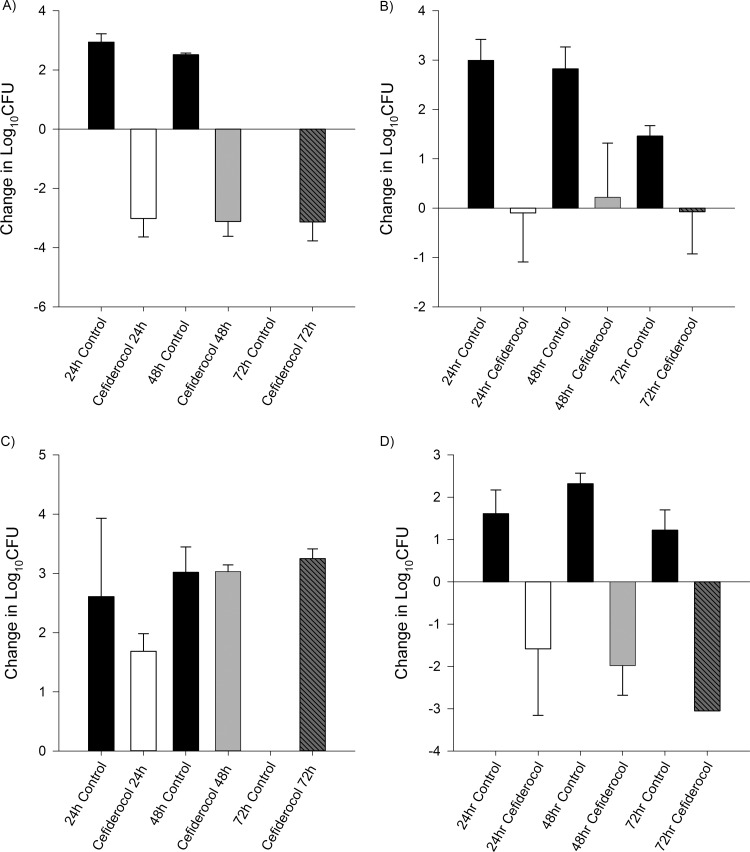
FIG 1.
Efficacy of human-simulated exposures of cefiderocol compared with same-period untreated controls at 24, 48, and 72 h among A. baumannii isolates. Isolate cefiderocol preexposure MICs (μg/ml): A. baumannii 135, 0.5 (A); A. baumannii 152, 1 (B); A. baumannii 84, 16 (C); A. baumannii 87, 4 (D). Absence of control data at any given interval indicates zero survival for that isolate.
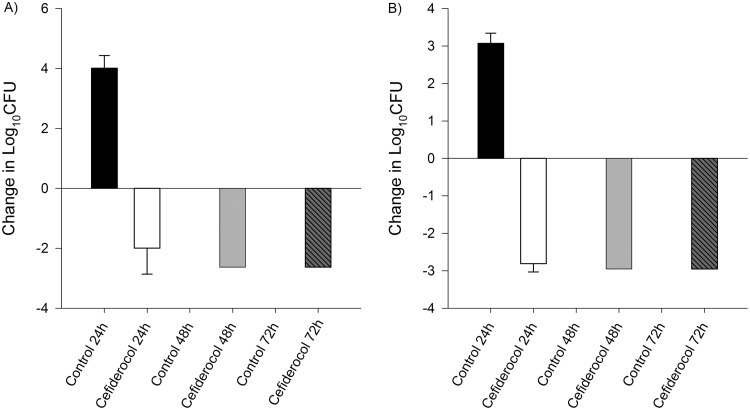
FIG 2.
Efficacy of human-simulated exposures of cefiderocol compared with same-period untreated controls at 24, 48, and 72 h among P. aeruginosa isolates. Isolate cefiderocol preexposure MICs (μg/ml): P. aeruginosa 1574, 2 (A); P. aeruginosa 1595, 4 (B). Absence of control data at any given interval indicates zero survival for that isolate.
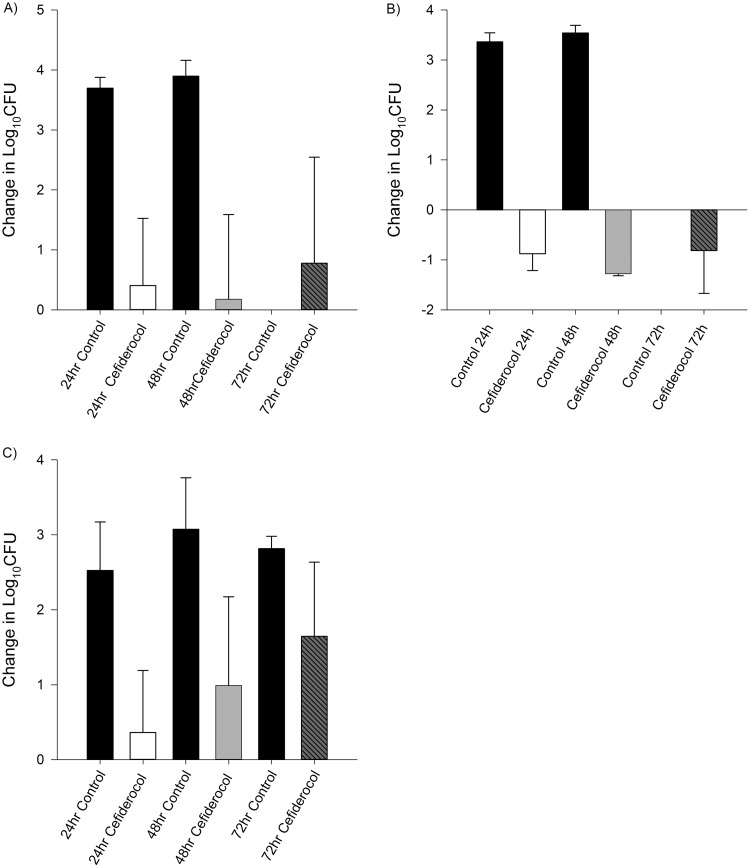
FIG 3.
Efficacy of human-simulated exposures of cefiderocol compared with same-period untreated controls at 24, 48, and 72 h among Enterobacteriaceae. Isolate cefiderocol preexposure MICs (μg/ml): K. pneumoniae 531, 4 (A); K. pneumoniae 519, 4 (B); E. coli 462, 1 (C). Absence of control data at any given interval indicates zero survival for that isolate.
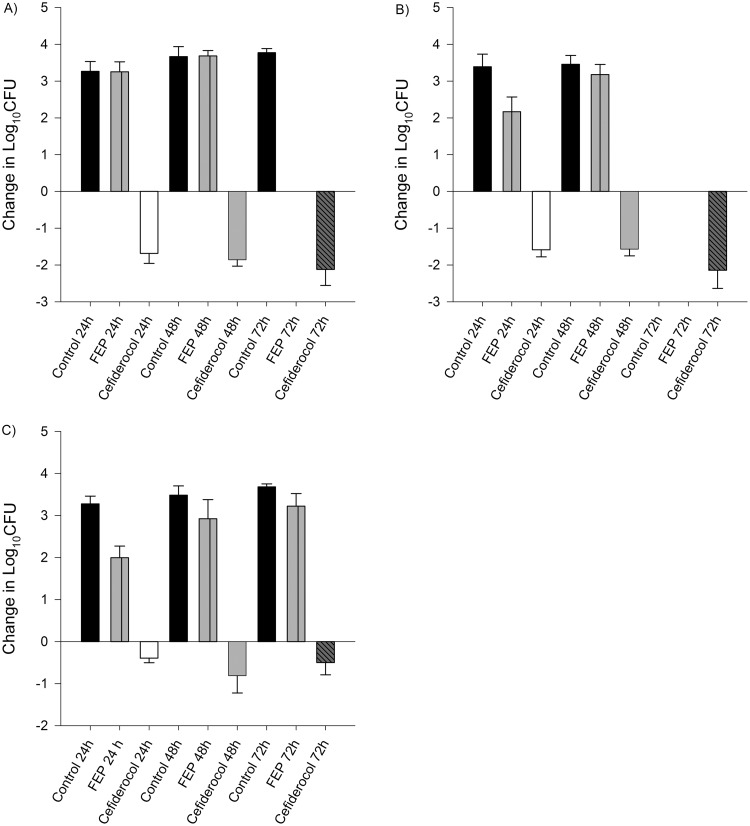
FIG 4.
Efficacy of human-simulated exposures of cefiderocol and cefepime compared with same-period untreated controls at 24, 48, and 72 h among Enterobacteriaceae. Isolate cefiderocol preexposure MICs (μg/ml): E. coli 458, 1/512 (A); K. pneumoniae 539, 4/>512 (B); K. pneumoniae 543, 8/>512 (C). Absence of control data at any given interval indicates zero survival for that isolate.
Results from the retest MICs after cefiderocol exposure showed a 2-dilution change in only one treated animal (1/54 samples [1.8%]). The MIC in this single sample increased from 1 to 4 μg/ml for isolate E. coli 462 at 72 h, while two additional samples from similarly infected animals with E. coli 462 showed no change after 72 h of cefiderocol exposure.
As expected, isolates 40a, 41a, and 42b shared the same sequence type and horizontally transferred β-lactamases as isolate 32a, namely, ST410 (ST23 complex) and blaCMY-42, respectively (Table 2). Similarly, isolate 82a had the same ST and β-lactamase genes as 73a, i.e., ST258 and blaSHV-12 and blaKPC-2, respectively.
TABLE 2.
Whole-genome sequencing for selected isolates from infected animals after 72 h of no treatment or cefiderocol exposure
| Isolate | Sample | Regimen at 72 h | MLST | β-Lactamase | Lesion relative to control |
|---|---|---|---|---|---|
| E. coli 462 | 32a | Control | 410 | blaEC; blaCMY-42 | Not applicable |
| E. coli 462 | 40a | Cefiderocol | 410 | blaEC; blaCMY-42 | None identified |
| E. coli 462 | 41a | Cefiderocol | 410 | blaEC; blaCMY-42 | ΔpcnB, S100A in EMU65688.1 |
| E. coli 462 | 42b | Cefiderocol | 410 | blaEC; blaCMY-42 | None identified |
| K. pneumoniae 531 | 73a | Control | 258 | blaSHV-12; blaKPC-2 | Not applicable |
| K. pneumoniae 531 | 82a | Cefiderocol | 258 | blaSHV-12; blaKPC-2 | None identified |
The single sample, E. coli 462 (sample 41a [Table 2]), which displayed elevated MICs to cefiderocol compared to the unexposed control isolate (sample 32a), is the only isolate in which mutations were identified coding for changes in annotated protein sequences. Notably, a 4-base deletion of “CCAG” was identified that resulted in a frameshift at amino acid position 54 in the RNA poly(A) polymerase gene pcnB. The other mutation found in 41a was an SNP in a transposase 38 nucleotides from the edge of contig 59. While the mutation did code for an amino acid change (S100A), it is notable that transposases and other multiple copy genes will be prone to erroneous mapping in de novo assembly. No deduced amino acid sequence changes were identified in the other cefiderocol-exposed samples of E. coli 462 (40a or 42b relative to 32a), nor were any changes found in K. pneumoniae 531 isolates (sample 82a relative to 73a [Table 2]), leaving the predicted deletion of pcnB in EC 462 sample 41 as the sole lesion identified in any of the postexposure bacteria.
Examples of adaptive resistance following siderophore-conjugated monobactam exposure are well documented (4, 5). In such cases, despite supratherapeutic exposure of the siderophore antibiotic, bacterial growth similar to that in control animals was observed at 24 h. Moreover, resistant mutants rapidly evolved (4). In the current study, following human-simulated exposure of cefiderocol at 24 h, regrowth to the level of control was not observed. Furthermore, continued treatment at 48 and 72 h showed sustained kill in most isolates. For bacteria demonstrating cumulative regrowth (i.e., E. coli 462 and K. pneumoniae 531), the pattern of regrowth over the initial 24-h treatment period was inconsistent with the emergence of resistance observed with other siderophores (4, 5). Moreover, the phenomenon of adaptive resistance was not observed over the extended 72-h treatment period. The underlying mechanism behind this discordance between in vitro susceptibility and in vivo regrowth is not well understood. Documented cases of persister cells in the context of Gram-negative organisms against conventional antibiotic agents have been described in the literature (14).
Previous investigations into the mutations noted in our single isolate revealed that deletion of this gene led to a large regulatory switch, leading to changes in expression in multiple systems, including downregulation of the ferrichrome receptor operon fhuABCD, although these mutational changes have failed to demonstrate a link to cefiderocol resistance (15, 16).
Among a diverse group of Gram-negative isolates with a range of cefiderocol MICs, humanized exposures of the compound displayed sustained in vivo antibacterial effects over a 72-h period. Moreover, postexposure MIC studies revealed that the phenotypic profile for cefiderocol remained largely unchanged. Taken together, our current findings further illuminate the pharmacodynamic profile of this novel siderophore against resistant Gram-negative organisms.
ACKNOWLEDGMENTS
We thank Safa Abuhussain, Kamilia Abdelraouf, Elizabeth Cyr, Sara Giovagnoli, Kimelyn Greenwood, Mordechai Grupper, Alissa Padgett, Jennifer Tabor-Rennie, Debora Santini, and Christina Sutherland of the Center for Anti-Infective Research and Development, Hartford, CT, for assistance with the conduct of the study. Additionally, we thank International Health Management Associates (IHMA), Mark Estabrook, and Mark Wise for assistance with the analysis of genotypic data.
Funding for this study was provided by Shionogi & Co. Ltd. Osaka, Japan.
REFERENCES
- 1.Tessier PR, Nicolau DP. 2013. Tigecycline displays in vivo bactericidal activity against extended-spectrum-β-lactamase-producing Enterobacteriaceae after 72-hour exposure period. Antimicrob Agents Chemother 57:640–642. doi: 10.1128/AAC.01824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keel RA, Tessier PR, Crandon JL, Nicolau DP. 2012. Comparative efficacies of human simulated exposures of tedizolid and linezolid against Staphylococcus aureus in the murine thigh infection model. Antimicrob Agents Chemother 56:4403–4407. doi: 10.1128/AAC.00122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monogue ML, Tsuji M, Yamano Y, Echols R, Nicolau DP. 2017. Efficacy of humanized exposures of cefiderocol (S-649266) against a diverse population of Gram-negative bacteria in a murine thigh infection model. Antimicrob Agents Chemother 61:e01022-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim A, Kutschke A, Ehmann DE, Patey SA, Crandon JL, Gorseth E, Miller AA, McLaughlin RE, Blinn CM, Chen A, Nayar AS, Dangel B, Tsai AS, Rooney MT, Murphy-Benenato KE, Eakin AE, Nicolau DP. 2015. Pharmacodynamic profiling of a siderophore-conjugated monocarbam in Pseudomonas aeruginosa: assessing the risk for resistance and attenuated efficacy. Antimicrob Agents Chemother 59:7743–7752. doi: 10.1128/AAC.00831-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomaras AP, Crandon JL, McPherson CJ, Banevicius MA, Finegan SM, Irvine RL, Brown MF, O'Donnell JP, Nicolau DP. 2013. Adaptation-based resistance to siderophore-conjugated antibacterial agents by Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4197–4207. doi: 10.1128/AAC.00629-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghazi IM, Monogue ML, Tsuji M, Nicolau DP. 2018. Humanized exposures of cefiderocol, a siderophore cephalosporin, display sustained in vivo activity against siderophore-resistant Pseudomonas aeruginosa. Pharmacology 101:278–284. doi: 10.1159/000487441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghazi IM, Monogue ML, Tsuji M, Nicolau DP. 2018. Pharmacodynamics of cefiderocol, a novel siderophore cephalosporin, in a Pseudomonas aeruginosa neutropenic murine thigh model. Int J Antimicrob Agents 51:206–212. doi: 10.1016/j.ijantimicag.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2017. In vitro activity of the siderophore cephalosporin, cefiderocol, against a recent collection of clinically relevant Gram-negative bacilli from North America and Europe, including carbapenem nonsusceptible isolates (SIDERO-WT-2014 study). Antimicrob Agents Chemother 61:e00093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute 2017. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 26:1–10. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 14.Grassi L, Di Luca M, Maisetta G, Rinaldi AC, Esin S, Trampuz A, Batoni G. 2017. Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front Microbiol 8:1917. doi: 10.3389/fmicb.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes A, Gracia C, Innocenti N, Zhang K, Aurell E, Hajnsdorf E. 2017. Landscape of RNA polyadenylation in E. coli. Nucleic Acids Res 45:2746–2756. doi: 10.1093/nar/gkw894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito A, Sato T, Ota M, Takemura M, Nishikawa T, Toba S, Kohira N, Miyagawa S, Ishibashi N, Matsumoto S, Nakamura R, Tsuji M, Yamano Y. 2017. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria antimicrobial agents and chemotherapy. Antimicrob Agents Chemother 62:e01454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]