We investigated the ability of several recent clinical viridans group streptococci (VGS) bloodstream isolates (Streptococcus mitis/S. oralis subgroup) from daptomycin (DAP)-naive patients to develop DAP resistance in vitro. All strains rapidly developed high-level and stable DAP resistance.
KEYWORDS: CdsA, PgsA, Streptococcus mitis, daptomycin resistance
ABSTRACT
We investigated the ability of several recent clinical viridans group streptococci (VGS) bloodstream isolates (Streptococcus mitis/S. oralis subgroup) from daptomycin (DAP)-naive patients to develop DAP resistance in vitro. All strains rapidly developed high-level and stable DAP resistance. Substitutions in two enzymes involved in the cardiolipin biosynthesis pathway were identified, i.e., CdsA (phosphatidate cytidylyltransferase) and PgsA (CDP-diacylglycerol-glycerol-3-phosphate-3-phosphatidyltransferase). These mutations were associated with complete disappearance of phosphatidylglycerol and cardiolipin from cell membranes. DAP interactions with the cell membrane differed in isolates with PgsA versus CdsA substitutions.
INTRODUCTION
A major problem associated with infections caused by viridans group streptococci (VGS) is their relative frequency of in vitro antimicrobial resistance to β-lactams and tolerance to vancomycin (1–4). Daptomycin (DAP) has been proposed as a potential alternative for severe infections caused by β-lactam-resistant Streptococcus mitis/S. oralis strains. We previously detailed the ability of a well-known strain of S. oralis (351) to rapidly develop stable, high-level DAP resistance (DAP-R) on exposure to the drug both in vitro and in an in vivo model of experimental infective endocarditis (5–7). (We use the term “daptomycin resistance” [DAP-R] rather than “daptomycin nonsusceptibility” in this paper for ease of presentation.)
The mechanism of resistance was a substitution in CdsA, a phosphatidate cytidylyltransferase that is a key enzyme for the first committed biosynthetic step within the cardiolipin (CL) biosynthesis pathway. The link between CdsA and DAP-R was shortly thereafter reported by a different group, thereby confirming our findings (8).
In the current study, we evaluated whether changes in CdsA would also be readily induced during in vitro DAP exposures of DAP-susceptible (DAP-S) clinical isolates of S. mitis/S. oralis recovered from the bloodstream of patients who had never received DAP (DAP naive). We included three recent clinical strains (S. mitis strains VGS007 and VGS008 and S. oralis strain 32364) obtained from bloodstream infections in DAP-naive patients (Table 1). All isolates were subjected to in vitro passage using brain heart infusion (BHI) broth with DAP 20 μg/ml and CaCl2 50 mg/liter to select for DAP-R derivatives. Spontaneous frequency of DAP-R evolution was also evaluated by using BHI agar (supplemented with CaCl2) with increasing concentrations of DAP (0 to 32 μg/ml). Selected DAP-R derivatives and their DAP-S parental strains were then compared in regard to MICs, cell surface charge, membrane fluidity, cell membrane phospholipid compositional profiling, and DAP binding assays, as described previously (5). Whole-genome sequencing (WGS) was performed in DAP-S and DAP-R pairs to evaluate the genetic basis for DAP-R (see details in the supplemental material).
TABLE 1.
Predicted amino acid changes in PgsA and CdsA and phospholipid analyses of S. mitis/oralis strains
| Species and strain | DAP MIC (μg/ml) | Predicted amino acid changes and enzyme | % total phospholipid content (mean ± SD) of: |
||
|---|---|---|---|---|---|
| Phosphatidylglycerol | Cardiolipin | Phosphatidic acid | |||
| S. mitis | |||||
| VGS007 | 0.38 | Parental | 38 ± 5 | 47 ± 5 | 15 ± 7 |
| VGS007-D8 | >256 | D249E in CdsA | 0a | 0a | 100a |
| VGS008 | 0.38 | Parental | 29 ± 2 | 69 ± 4 | 2 ± 1 |
| VGS008-D12 | >256 | R230H in CdsA | 0a | 0a | 100a |
| S. oralis | |||||
| 32364 | 1 | Parental | 19 ± 5 | 70 ± 3 | 11 ± 4 |
| 32364-D1 | >256 | G221V in CdsA, G52S in PgsA | 0a | 0a | 100a |
| 32364-D5 | >256 | G52S in PgsA | 0a | 0a | 100a |
P value was <0.001 DAP-R for S. mitis/S. oralis strains versus DAP-S S. mitis/S. oralis parental strains.
Of note, we were unable to recover DAP-R colonies of streptococci on any of the DAP-containing BHI agar plates as part of the spontaneous selection experiment (frequency <1 × 10−8 for all 3 strains). For the in vitro passage experiment, all S. mitis/S. oralis strains developed DAP-R (MIC, >256 μg/ml) rapidly on DAP exposure (Table 1). DAP-R derivatives arose after 1 to 8 days of exposure to DAP. The DAP-R phenotype was stable after 5 days of passage in antibiotic-free medium. We selected DAP-R derivatives at the end of the in vitro passage experiments for S. mitis strains VGS007 and VGS008. Two DAP-R derivatives of S. oralis strain 32364, D1 and D5, were included in this study to capture any genetic and phenotypic changes associated with early emergence of resistance. Growth curves indicated that the DAP-R derivatives had a marked decrease in fitness (see Fig. S2 in the supplemental material). We did not find any significant differences in cell surface charge or membrane fluidity between parental and DAP-R derivatives (data not shown). Comparative analysis of the relative content of the major cell membrane phospholipids indicated complete disappearance of phosphatidylglycerol (PG) and CL in all DAP-R derivatives compared with the parental controls (Table 1). The loss of the PG and CL content in DAP-R strains was accompanied by a significant increase in the relative content of phosphatidic acid (PA) (Table 1), as previously described (5).
WGS of DAP-S parental versus respective DAP-R derivatives disclosed several relevant mutations (Table 1; see also Table S1 in the supplemental material). Importantly, we identified substitutions in two major phospholipid enzymes, namely, CdsA and PgsA (Table 1). These enzymes are necessary for the synthesis of the major cell membrane phospholipids PG and CL, using PA as the initial substrate (9). We identified three substitutions in CdsA among the DAP-R derivatives of S. mitis/S. oralis, namely, G221V, R230H, and D249E. Based on the crystal structure of Thermotoga maritima (10), D249E is located within the HGGxxDRxD motif (amino acids [aa] 244 to 252), which is important for the binding of CTP and phosphatidate for the synthesis of CDP-diacylglycerol (a precursor of PG and CL), along with the SPxKxxEG motif (aa 169 to 176). Similarly, the predicted R230H substitution is located in a highly conserved residue located between these two important binding motifs. The G221V substitution is located adjacent to D222, which is predicted to form part of a cation-binding Asp-Asp dyad, previously shown to be essential for enzymatic activity (10). Taken together, these data led us to predict that these substitutions resulted in a major decrease in (or abolishment of) CdsA catalytic activity.
In contrast to the multiple mutations identified in CdsA, we found that the two DAP-R derivatives of S. oralis strain 32364 harbored the same G52S substitution in PgsA (Table 1). This enzyme is responsible for the synthesis of PG, using phosphatidylglycerol 3-phosphate as the substrate (9). Because PG is the precursor of CL, alterations in PgsA are also expected to modulate CL synthesis. Interestingly, a crystal structure of a representative CDP-alcohol phosphotransferase (CDP-AP) from Archaeoglobus fulgidus identified a conserved CDP-AP signature motif (D1xxD2G1xxAR…G2xxxD3xxxD4) within the active site of such enzymes (11). We performed an alignment of CDP-AP from A. fulgidus and PgsA sequences from Staphylococcus aureus (12), Bacillus subtilis (13), and Escherichia coli isolates (14) (see Fig. S1 in the supplemental material). The alignment revealed that the G52S substitution is located in this conserved hydrophilic subdomain, likely exposed to the cytoplasm, and present in several phospholipid synthesis enzymes, such as diacylglycerol cholinephosphotransferase from E. coli and ethanolaminephosphotransferase in Saccharomyces cerevisiae, among others (14). Structural characterization of a representative member of CDP-AP from A. fulgidus showed that Gly218, which is represented by G1 of the motif, provides the necessary flexibility to the enzyme active site (11). Because the absence of PG in membrane preparations of S. oralis 32364-D5 was evident (Table 1), we postulated that the G52S substitution (G1) abolishes the enzymatic synthesis of PG. Indeed, Peleg et al. (12) and Hachmann et al. (13) found that an adjacent substitution (A55V, using S. oralis 32364 numbering) was associated with DAP-R in S. aureus and B. subtilis isolates (Fig. S1); of note, decreased PG content has been strongly associated with the DAP-R phenotype in these latter organisms (13). Given the potential importance of the G52S mutation, we aimed to create a ΔpgsA or pgsAG52S variant of S. oralis strain 32364 by allelic replacement. After multiple attempts, this strain was found to be nontransformable, so mutagenesis was not feasible. We next applied the same strategies to S. oralis strain 351, which was used previously for the study of DAP-R (5). Again, introduction of pgsAG52S and deletion of pgsA were unsuccessful in this strain. These results suggest that pgsA plays an essential role in the cellular function of streptococci, as has been described for Streptococcus sanguinis and several other species (15). In our effort to establish a link between pgsA and DAP-R in S. oralis, we searched our collection of streptococci for mutated alleles in pgsA. Among isolates of streptococci, a DAP-R derivative (MIC, >256 μg/ml) of S. oralis SF100 was found to harbor a G65E mutation in pgsA (unpublished data). Thus, we attempted to introduce pgsAG65E into S. mitis 351, using the methods described above. This approach was eventually successful, but generation of the pgsAG65E mutation required DAP selection (8 μg/ml). Note that Gly65 (represented by G2 in the conserved signature motif of CDP-AP) and Gly52 are conserved residues that form the surface of the ligand-binding pocket of CDP-APs. Mutations in these residues were found to render the enzyme inactive in S. cerevisiae isolates (16). The difficulty we encountered in mutating pgsA is consistent with previous studies in S. sanguinis, Streptococcus mutans, E. coli, and B. subtilis isolates (13, 15, 17, 18). Indeed, work in E. coli isolates has shown that a pgsA-null mutant was only viable when a functional copy of the pgsA gene was carried on a plasmid (19). Likewise, an allelic replacement of pgsAA55V was not possible in a wild-type strain of B. subtilis until a wild-type pgsA was introduced (reversion of pgsAA55V to wild type) to the DAP-R derivative, which harbors other changes associated with the resistance phenotype (13). Nevertheless, our effort to introduce a mutated pgsA into an S. oralis isolate resulted in a DAP-R phenotype. Although we cannot rule out changes in other genes that contribute to the phenotype, our results and others published previously confirm the link between pgsA and DAP-R (12, 13, 20). Our data also suggest the occurrence of multiple pathways of phospholipid adaptation leading to marked changes in cell membrane content of streptococci on antibiotic challenge.
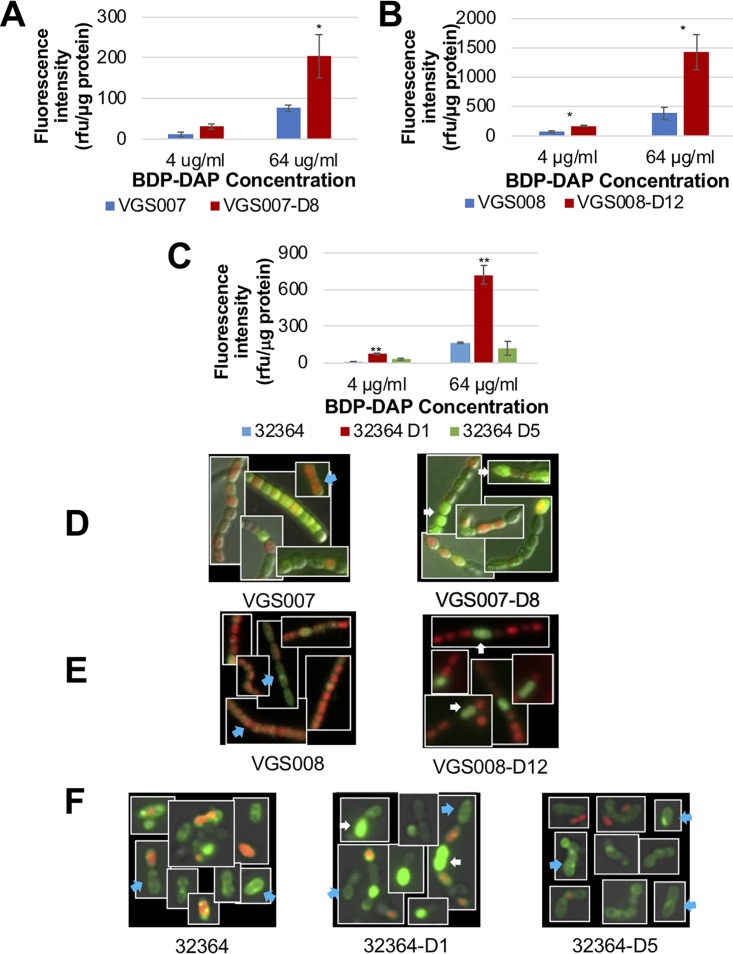
We previously showed that mutations in CdsA were associated with increased DAP binding but only to select cells within the overall population. This unique phenotype had not been described in relation to cationic antimicrobial peptide resistance (5). Thus, using Bodipy FL-labeled DAP (BDP-DAP), we investigated the ability of this antibiotic to bind to cell membranes of parental clinical isolates and their DAP-R derivatives, as measured by fluorescence intensities. Figure 1A to C shows that all DAP-R derivatives of S. mitis strains (which harbor mutations in cdsA) exhibited a statistically significant increase in binding of BDP-DAP, especially at higher DAP concentrations (64 μg/ml). In contrast, the DAP-R derivatives of S. oralis strain 32364 (D1 and D5, Table 1), exhibited a distinct phenotype of BDP-DAP binding. DAP-R isolate obtained from day 1 of in vitro serial passage (D1) had a pattern of increased BDP-DAP similar to that seen in our other DAP-R S. mitis strains. However, the binding pattern of the DAP-R derivative obtained after 5 days of in vitro passage (D5), which lacks mutations in CdsA but harbors a G52S substitution in PgsA, was quite different, with a trend toward lower DAP binding, albeit not statistically significant (Fig. 1C). Our results suggest that, as seen with CdsA, mutations in PgsA result in loss of PG and CL synthesis. Unlike CdsA mutations, however, the PgsA mutation additionally appears to minimally impact the binding of DAP within the target cell membrane.
FIG 1.
BDP-DAP binding of S. mitis and S. oralis. Fluorescence intensity was normalized to protein ratio, using BDP-DAP binding to S. mitis VGS007 and VGS007-D8 (A), S. mitis VGS008 and VGS008-D12 (B), and S. oralis 32364, 32364-D1, and 32364-D5 (C). The increased binding of BDP-DAP is seen in DAP-R derivatives with mutations in cdsA compared to its DAP-S parental isolate. BDP-DAP staining (64 μg/ml) of Streptococcus cells is shown in green, propidium iodide (red), and overlay of DAP-S parental and DAP-R derivatives of VGS007 (D), VGS008 (E), and 32364 (F). Representative of uniform/septal binding is indicated by blue arrows, and hyperaccumulative binding is indicated by white arrows. *, P < 0.05; **, P < 0.01.
In the parallel fluorescence microscopy studies of DAP-R derivatives of strains VGS007 and VGS008 (harboring CdsA substitutions alone), DAP localization was similar to that seen in our previously published studies (5), namely, selected hyperaccumulation of BDP-DAP (64 μg/ml) in individual cells throughout the chain length compared to the uniform or septal binding of BDP-DAP in DAP-S parentals (Fig. 1D and E). We also used propidium iodide (PI) to assess cell viability after exposure to DAP. The PI images revealed that in contrast to its rather uniform uptake in DAP-S cells exposed to the antibiotic, DAP-R strains exhibited more focal uptake of PI, with a higher proportion of cells failing to take the dye, suggesting that these cells remained viable after DAP exposure (Fig. 1D and E). In contrast, for the S. oralis 32364-D5 derivative, which has only a substitution in PgsA, uniform binding of BDP-DAP with occasional cells stained with PI was identified (Fig. 1F). This pattern, along with data from the BDP-DAP binding assay (Fig. 1C), suggested a reduction in DAP binding to the cell membrane, in a manner similar to that of DAP-R B. subtilis associated with PgsA mutations (13). The DAP-R 32364-D1 derivative, which harbors substitutions in both CdsA and PgsA, exhibited a mixed pattern of hyperaccumulation and uniformed binding of BDP-DAP (Fig. 1F). Taken together, our observations suggest that distinct mechanisms of DAP-R associated with alteration in CdsA (hyperaccumulation) versus PgsA in different S. mitis/S. oralis strains. The exact mechanism of DAP-R (diversion, repulsion, or hyperaccumulation) associated with pgsA is unclear and is the object of future investigations.
In summary, our findings highlight the malleability of the VGS cell membrane in response to cationic peptide-induced stress and the multiple complex pathways involved in the emergence of DAP-R in these organisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by University El Bosque and the National Institutes of Health (NIAID K24 AI121296 and R01 AI134637 to C.A.A., K08 AI113317 to T.T.T., 1R01130056-01 to A.S.B., and R01 AI41513 to P.M.S.; M.J.R. is supported in part by R01 AI121400 and R21 AI109266). C.A.A. is supported by the UTHealth Presidential Award and University of Texas STARS Award. J.M.M. is supported by grants REIPI RD06/0008 (the Spanish Network for Research in Infectious Diseases) and PI11/01131 (Instituto Carlos III, Ministerio de Economía y Competitividad). J.M.M. received a research grant from Institut d’Investigacions Biomèdiques August Pi I Sunyer (IDIBAPS) in 2017 to 2019 and the European Regional Development Fund (ERDF). N.N.M. was supported by LABiomed-Harbor UCLA intramural research grant 31604-01.
C.A.A. has received grant support from Merck Pharmaceuticals and MeMed Diagnostics. A.S.B. has received grant support from ContraFect Corporation and Intron Pharmaceuticals. N.N.M. has received grant support from Merck Pharmaceuticals. M.J.R. received research support, consulted, or was part of a speaker bureau for Allergan, Bayer, Cempra, Merck, The Medicines Company, Sunovian, and Theravance Biopharmaceutics. J.M.M. has received consulting honoraria and/or research grants from AbbVie, Angelini, Bristol-Myers Squibb, Jansen, Genentech, Medtronic, Merck, Novartis, Gilead Sciences, and ViiV HealthCare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01531-18.
REFERENCES
- 1.Prabhu RM, Piper KE, Baddour LM, Steckelberg JM, Wilson WR, Patel R. 2004. Antimicrobial susceptibility patterns among viridans group streptococcal isolates from infective endocarditis patients from 1971 to 1986 and 1994 to 2002. Antimicrob Agents Chemother 48:4463–4465. doi: 10.1128/AAC.48.11.4463-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doern GV, Ferraro MJ, Brueggemann AB, Ruoff KL. 1996. Emergence of high rates of antimicrobial resistance among viridans group streptococci in the United States. Antimicrob Agents Chemother 40:891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabella C, Murphy D, Drummond-Webb J. 2001. Endocarditis due to Streptococcus mitis with high-level resistance to penicillin and ceftriaxone. JAMA 285:2195. [DOI] [PubMed] [Google Scholar]
- 4.Safdar A, Rolston KVI. 2006. Vancomycin tolerance, a potential mechanism for refractory Gram-positive bacteremia observational study in patients with cancer. Cancer 106:1815–1820. doi: 10.1002/cncr.21801. [DOI] [PubMed] [Google Scholar]
- 5.Mishra NN, Tran TT, Seepersaud R, Garcia-de-la-Maria C, Faull K, Yoon A, Proctor R, Miro JM, Rybak MJ, Bayer AS, Arias CA, Sullam PM. 2017. Perturbations of phosphatidate cytidylyltransferase (CdsA) mediate daptomycin resistance in Streptococcus mitis/oralis by a novel mechanism. Antimicrob Agents Chemother 61:e02435-16. doi: 10.1128/AAC.02435-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-de-la-Maria C, Xiong YQ, Pericas JM, Armero Y, Moreno A, Mishra NN, Rybak MJ, Tran TT, Arias CA, Sullam PM, Bayer AS, Miro JM. 2017. Impact of high-level daptomycin resistance in the Streptococcus mitis group on virulence and survivability during daptomycin treatment in experimental infective endocarditis. Antimicrob Agents Chemother 61:e02418-16. doi: 10.1128/AAC.02418-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-de-la-Maria C, Pericas JM, Del Rio A, Castaneda X, Vila-Farres X, Armero Y, Espinal PA, Cervera C, Soy D, Falces C, Ninot S, Almela M, Mestres CA, Gatell JM, Vila J, Moreno A, Marco F, Miro JM. Hospital Clinic Experimental Endocarditis Study Group. 2013. Early in vitro and in vivo development of high-level daptomycin resistance is common in mitis group streptococci after exposure to daptomycin. Antimicrob Agents Chemother 57:2319–2325. doi: 10.1128/AAC.01921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams HM, Joyce LR, Guan Z, Akins RL, Palmer KL. 2017. Streptococcus mitis and S. oralis lack a requirement for CdsA, the enzyme required for synthesis of major membrane phospholipids in bacteria. Antimicrob Agents Chemother 61:e02552-16. doi: 10.1128/AAC.02552-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohlenkamp C, Geiger O. 2016. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 40:133–159. doi: 10.1093/femsre/fuv008. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Yin Y, Wu J, Liu Z. 2014. Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis. Nat Commun 5:4244. doi: 10.1038/ncomms5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sciara G, Clarke OB, Tomasek D, Kloss B, Tabuso S, Byfield R, Cohn R, Banerjee S, Rajashankar KR, Slavkovic V, Graziano JH, Shapiro L, Mancia F. 2014. Structural basis for catalysis in a CDP-alcohol phosphotransferase. Nat Commun 5:4068. doi: 10.1038/ncomms5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RCJ, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachmann A-B, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother 55:4326–4337. doi: 10.1128/AAC.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usui M, Sembongi H, Matsuzaki H, Matsumoto K, Shibuya I. 1994. Primary structures of the wild-type and mutant alleles encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Bacteriol 176:3389–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P, Ge X, Chen L, Wang X, Dou Y, Xu JZ, Patel JR, Stone V, Trinh M, Evans K, Kitten T, Bonchev D, Buck GA. 2011. Genome-wide essential gene identification in Streptococcus sanguinis. Sci Rep 1:125. doi: 10.1038/srep00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JG, McMaster CR. 1998. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J Biol Chem 273:13482–13487. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Tan BK, Zhao J, Guan Z. 2016. In vivo and in vitro synthesis of phosphatidylglycerol by an Escherichia coli cardiolipin synthase. J Biol Chem 291:25144–25153. doi: 10.1074/jbc.M116.762070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacGilvray ME, Lapek JDJ, Friedman AE, Quivey RGJ. 2012. Cardiolipin biosynthesis in Streptococcus mutans is regulated in response to external pH. Microbiology 158:2133–2143. doi: 10.1099/mic.0.057273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heacock PN, Dowhan W. 1987. Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J Biol Chem 262:13044–13049. [PubMed] [Google Scholar]
- 20.Goldner NK, Bulow C, Cho K, Wallace M, Hsu F-F, Patti GJ, Burnham CA, Schlesinger P, Dantas G. 2018. Mechanism of high-level daptomycin resistance in Corynebacterium striatum. mSphere 3:e00371-18. doi: 10.1128/mSphereDirect.00371-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.