Class D carbapenemases are enzymes of the utmost clinical importance due to their ability to confer resistance to the last-resort carbapenem antibiotics. We investigated the role of the conserved hydrophobic bridge in the carbapenemase activity of OXA-23, the major carbapenemase of the important pathogen Acinetobacter baumannii.
KEYWORDS: antibiotic resistance, β-lactamases, mechanisms of resistance
ABSTRACT
Class D carbapenemases are enzymes of the utmost clinical importance due to their ability to confer resistance to the last-resort carbapenem antibiotics. We investigated the role of the conserved hydrophobic bridge in the carbapenemase activity of OXA-23, the major carbapenemase of the important pathogen Acinetobacter baumannii. We show that substitution of the bridge residue Phe110 affects resistance to meropenem and doripenem and has little effect on MICs of imipenem. The opposite effect was observed upon substitution of the other bridge residue Met221. Complete disruption of the bridge by the F110A/M221A substitution resulted in a significant loss of affinity for doripenem and meropenem and to a lesser extent for imipenem, which is reflected in the reduced MICs of these antibiotics. In the wild-type OXA-23, the pyrrolidine ring of the meropenem tail forms a hydrophobic interaction with Phe110 of the bridge. Similar interactions would ensue with ring-containing doripenem but not with imipenem, which lacks this ring. Our structural studies showed that this interaction with the meropenem tail is missing in the F110A/M221A mutant. These data explain why disruption of the interaction between the enzyme and the carbapenem substrate impacts the affinity and MICs of meropenem and doripenem to a larger degree than those of imipenem. Our structures also show that the bridge directs the acylated carbapenem into a specific tautomeric conformation. However, it is not this conformation but rather the stabilizing interaction between the tail of the antibiotic and the hydrophobic bridge that contributes to the carbapenemase activity of class D β-lactamases.
INTRODUCTION
Antibiotic resistance poses a clinical threat of the utmost importance. One of the most common causes of resistance in bacteria is the presence of β-lactamases. These enzymes destroy β-lactam antibiotics, thus preventing their lethal effect. β-Lactamases are divided into four molecular classes based on the presence of conserved amino acid motifs and amino acid sequence similarity. Classes A, C, and D utilize an active-site serine for catalysis, while class B β-lactamases are metalloenzymes (1). Of the class D or OXA enzymes, more than 600 have been identified, with this number more than doubling in the last 5 years. Despite sharing several conserved motifs with the other active-site serine enzymes, the OXA enzymes are distinct in that they possess an N-carboxylated lysine that is essential for catalysis (2). Earlier discovered class D enzymes had narrow substrate specificity and hydrolyzed oxacillin more efficiently then benzylpenicillin (3). However, in recent years, many OXA enzyme variants have a broader spectrum of activity for β-lactams, with the carbapenem-hydrolyzing class D β-lactamases (CHDLs) being one of the most clinically challenging, due to their ability to destroy the last resort antibiotics the carbapenems (4, 5). Even more alarming, CHDLs are widely spread in Acinetobacter baumannii and Klebsiella pneumoniae, bacteria from the notorious Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (ESKAPE) list that includes pathogens most frequently associated with life-threatening nosocomial infections (3, 6).
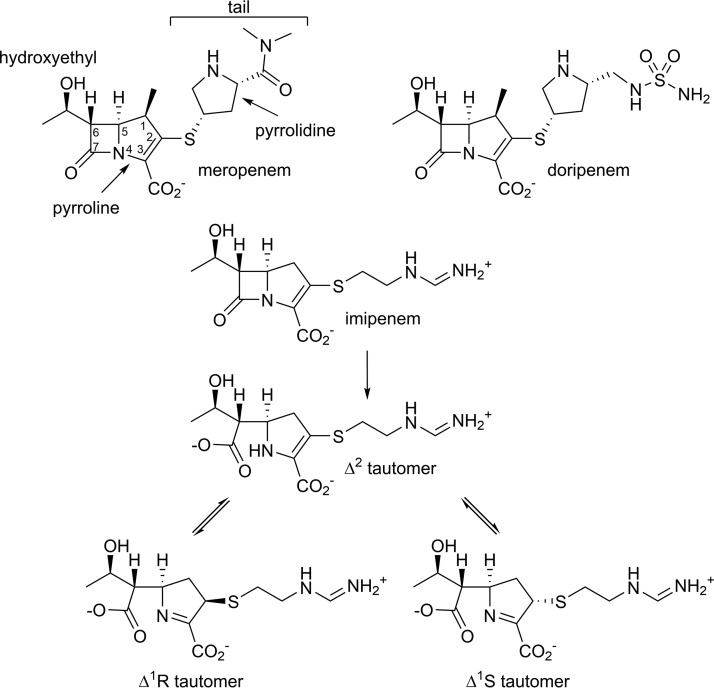
Although CHDLs are of high clinical importance, many aspects regarding the mechanism of their carbapenemase activity have yet to be elucidated. One of the unresolved issues is the function of the so-called hydrophobic bridge present in CHDLs of A. baumannii. This bridge was originally identified upon elucidation of the first CHDL structure, that of OXA-24/40. It is formed by residues Tyr112 and Met223, which create a tunnel-like entrance to the active site. Limited mutational analysis of these residues and molecular docking experiments suggested this bridge properly orients carbapenem antibiotics for hydrolysis (7). Further studies of OXA-24/40 deacylation-deficient mutants showed that upon formation of the acyl-enzyme species with doripenem, the pyrroline ring of the hydrolyzed carbapenem adopts the Δ2 tautomeric conformation (8). This was in contrast to the Δ1R tautomer observed in the structure of OXA-1, which lacks both this bridge and carbapenemase activity (Fig. 1 and Fig. S1) (9). Based on these results, it was suggested that, unlike the Δ2 tautomer, the Δ1 tautomer is not efficiently hydrolyzed, and the bridge in OXA-24/40 prevents its formation (8).
FIG 1.
Chemical structures of carbapenem antibiotics. Chemical structures of meropenem, doripenem, and imipenem, including its tautomeric conformations are shown.
However, later, another carbapenem, meropenem, was observed as the Δ1S tautomer in the crystal structure of the most prevalent A. baumannii CHDL OXA-23, which also possesses the hydrophobic bridge (Fig. 1 and Fig. S1). These results demonstrated that the presence of the bridge in this enzyme does not prevent formation of the Δ1 tautomer, and both the Δ2 and the Δ1S tautomers can be hydrolyzed by CHDLs (10). Moreover, the structure of the non-Acinetobacter enzyme OXA-48 showed the hydrophobic bridge is missing in this clinically important CHDL and, thus, is not a conserved feature required for the carbapenemase activity of class D β-lactamases (11). Evaluation of the carbapenemase activity of other class D enzymes further confirmed this assumption, as it was demonstrated that OXA-2 and OXA-10 enzymes, both lacking the hydrophobic bridge, possess potent carbapenemase activity that equals that of some known CHDLs (12).
To evaluate the role of the hydrophobic bridge in class D enzymes, mutagenesis of the amino acid residues forming the bridge in OXA-24/40 and OXA-58 has been performed. While these studies demonstrated that some of the substitutions led to decreased activity of the enzymes toward carbapenems, the mutagenesis was limited only to substitution with alanine in OXA-24/40 and alanine, threonine, or isoleucine in OXA-58 (7, 13). Moreover, no structural information of apo- or enzyme-substrate complexes of CHDLs with substituted bridge residues is currently available.
To further study the contribution of the bridge to the carbapenemase activity of CHDLs, we performed site-saturation mutagenesis of the bridge amino acids Phe110 and Met221 of the most prevalent CHDL of A. baumannii OXA-23. Here, we report the microbiological characterization of these mutants and steady-state kinetic parameters for the Ala/Ala double-bridge mutant of OXA-23. We also determined the X-ray crystal structures of this double mutant and its acyl-enzyme complexes with two carbapenems, imipenem and meropenem, and evaluated the impact of the bridge disruption on the substrate-binding mode and tautomeric state of the acylated antibiotics.
RESULTS AND DISCUSSION
Effect of substitutions of OXA-23 bridge residues on resistance to carbapenems.
To evaluate the impact of substitution of the bridge residues Phe110 and Met221 of OXA-23 on the levels of produced resistance to carbapenem antibiotics, we performed their site-saturation mutagenesis. We then determined the effects of these mutations by determining the MIC of these drugs against A. baumannii CIP 70.10 producing the mutant OXA-23 enzymes (Table 1). First, we evaluated whether mutations at these positions have a deleterious effect on the activity of the enzyme toward β-lactams in general. For this, we substituted both bridge residues with alanine, which has the smallest chiral sidechain and is commonly used for mutagenesis studies. The F110A/M221A double-mutant enzyme conferred only a 2-fold lower resistance to ampicillin than the parental OXA-23 β-lactamase (Table 1). These data demonstrate that disruption of the bridge in OXA-23 contributes very little to the conferred resistance to this antibiotic. Small changes in MICs of ampicillin were also observed with most of the 19 individual substitutions at positions 110 and 221. These results indicate that such mutations have an insignificant impact on the overall activity of the enzyme, its stability, or expression level. For the Arg, Asp, and Glu substitutions at these positions, where a larger, 4-fold change in the MICs of ampicillin was observed, the contribution of these factors cannot be ruled out.
TABLE 1.
MICs produced by OXA-23 mutant enzymes with substitutions of Phe110 or Met 221
| Substitution | MIC (μg/ml) fora: |
|||
|---|---|---|---|---|
| AMP | IPM | MEM | DOR | |
| Controlb | 64 | 0.25 | 0.5 | 0.25 |
| Wild typec | 16,384 | 32 | 64 | 32 |
| Ala-Alad | 8,192 | 16 | 8 | 8 |
| Ala | 8,192 (16,384)e | 32 (16) | 16 (32) | 16 (32) |
| Gly | 8,192 (16,384) | 16 (4) | 16 (64) | 8 (32) |
| Val | 8,192 (16,384) | 32 (32) | 16 (64) | 16 (32) |
| Leu | 16,384 (8,192) | 32 (16) | 32 (64) | 16 (32) |
| Ile | 16,384 (16,384) | 32 (16) | 16 (64) | 16 (32) |
| Met | 16,384 (NAf) | 32 (NA) | 16 (NA) | 16 (NA) |
| Pro | 8,192 (16,384) | 16 (4) | 32 (32) | 8 (8) |
| Ser | 8,192 (16,384) | 32 (8) | 32 (64) | 16 (32) |
| Thr | 16,384 (16,384) | 32 (16) | 16 (64) | 16 (32) |
| Cys | 8,192 (8,192) | 16 (16) | 16 (64) | 16 (16) |
| Gln | 8,192 (16,384) | 32 (16) | 16 (64) | 16 (32) |
| Asn | 16,384 (8,192) | 32 (8) | 8 (64) | 8 (16) |
| His | 16,384 (8,192) | 32 (16) | 32 (64) | 32 (16) |
| Lys | 8,192 (8,192) | 16 (32) | 8 (64) | 8 (32) |
| Arg | 4,096 (8,192) | 16 (32) | 16 (64) | 8 (32) |
| Asp | 4,096 (4,096) | 32 (2) | 16 (64) | 16 (16) |
| Glu | 4,096 (8,192) | 32 (4) | 16 (32) | 16 (16) |
| Phe | NA (16,384) | NA (16) | NA (64) | NA (32) |
| Trp | 8,192 (8,192) | 32 (16) | 32 (64) | 32 (32) |
| Tyr | 8,192 (8,192) | 32 (8) | 64 (64) | 32 (32) |
AMP, ampicillin; IPM, imipenem; MEM, meropenem; DOR, doripenem.
Parental A. baumannii CIP 70.10 with no β-lactamase.
Parental OXA-23 enzyme.
F110A/M221A double mutant.
The first value represents MICs of Phe110 mutants, while the second, in parentheses, those of Met221 mutants.
Not applicable (the residue represents the wild-type sequence).
We next examined the effects of substitutions of the bridge residues in the OXA-23 CHDL on the MICs of carbapenems. We observed significant differences in resulting MICs for imipenem versus those for meropenem and doripenem. The carbapenems meropenem and doripenem differ from imipenem by having a methyl group at C1 and bulkier tails harboring a pyrrolidine ring (Fig. 1). With imipenem, we observed at most 2-fold decreases in MICs with five of the substitutions at position 110. In contrast, we observed larger changes (up to 16-fold) when the other bridge residue Met221 was mutated (Table 1). Substitution of Met221 with the small, flexible residue glycine or proline, which forms a rigid ring structure with the protein backbone, resulted in an 8-fold decrease of the imipenem MIC compared with wild-type OXA-23. We also observed similar large drops in the MIC of imipenem when the charged residues aspartate and glutamate were introduced at position 221. While we observed no change in the MIC of imipenem for the F110A mutant of OXA-23, substitution of the analogous residue Y112A in OXA-24/40 resulted in the MIC decreasing 8-fold (7). However, for both enzymes, substitution of methionine forming the other side of the bridge produced the same 2-fold decrease in the MIC of imipenem.
Next, we evaluated the MICs of carbapenems with bulky tails, meropenem and doripenem, for the OXA-23 mutants and, in contrast to imipenem, observed that MICs of these antibiotics were more affected by substitutions at position 110. The MICs of meropenem dropped up to 8-fold (from 64 down to 8 μg/ml) with the polar and charged residues asparagine and lysine. Similarly, for doripenem, the MICs decreased from 32 to 8 to 16 μg/ml for 16 of the 19 substitutions, with the largest changes seen with glycine, proline, asparagine, lysine, and arginine (Table 1). In contrast, most mutations of the bridge position Met221 resulted in no changes in the MICs of both meropenem and doripenem (Table 1). A similar trend was observed when the bridge residues were substituted with alanine in the OXA-24/40 CHDL; however, the magnitude of the change was quite different. For F110A OXA-23 we observed a 4-fold decrease in the MIC of meropenem, while a 64-fold drop was reported for the OXA-24/40 Y112A mutant (7). Structural differences between these two enzymes might be responsible for the observed differences in the levels of resistance to meropenem. As the OXA-23 and OXA-24/40 enzymes and their mutants were expressed in different A. baumannii hosts, the amount of the produced enzyme could differ, which also can have an effect on the levels of resistance. When we compared the results for the other bridge mutant M221A OXA-23 to those of M223A OXA-24/40, the changes were more similar, with the MIC of meropenem decreasing 2-fold and 4-fold, respectively (Table 1) (7).
Next, we examined the effect of the complete disruption of the bridge in the F110A/M221A double mutant on resistance to carbapenems. We found that the MIC of imipenem for this mutant consistently decreased only 2-fold compared with wild-type OXA-23, the same as we observed for the single M221A mutant (Table 1). These results suggest this small change in the resistance level of imipenem is due to substitution in this enzyme of residue Met221 and not Phe110. Different results were observed with the Y112A/M223A mutant of OXA-24/40, where the MIC of imipenem decreased 8-fold, from 32 to 4 μg/ml (7, 14). The same decrease in the MIC was observed for the single Y112A mutant of this enzyme, indicating that, contrary to OXA-23, residue Tyr112 plays a bigger role in resistance to imipenem in OXA-24/40.
Unlike our results with imipenem, we observed that the MICs of meropenem and doripenem for the double OXA-23 mutant decreased more significantly, from 64 to 8 μg/ml and 32 to 8 μg/ml, respectively (Table 1). These changes in MICs are greater than those for the single alanine mutants, suggesting that both bridge residues of OXA-23 contribute to the observed resistance levels for meropenem and doripenem. For Y112A/M223A OXA-24/40, there was a much larger, 128-fold, decrease in the MIC of meropenem, from 64 to 0. μg/ml. In contrast to OXA-23, residue Tyr112 of OXA-24/40 provides the major contribution to the observed resistance, as its mutation lowers the MIC of meropenem from 64 to 1 μg/ml (7). Later, another publication reported a much smaller, 16-fold change in the MIC of meropenem for the Y112A/M223A mutant of OXA-24/40 (from 32 to 2 μg/ml), which is in line with our results for OXA-23, where we observed an 8-fold decrease in the MIC of meropenem. However, the effect of the single Y112A substitution was not evaluated in this study (14).
The analysis of our MIC data shows that substitution of the bridge residues Phe110 and Met221 in OXA-23 by most of the 19 residues does not dramatically decrease the resistance to carbapenem antibiotics. More importantly, elimination of the bridge in the double Ala/Ala mutant does not decrease the MICs of carbapenems below the clinically significant levels (15). These data demonstrate that while the bridge formed by Phe110 and Met221 in OXA-23 plays a role in the resistance to carbapenems in A. baumannii, it is not the only or even the major contributor to the carbapenemase activity of this enzyme.
Steady-state kinetics of F110A/M221A OXA-23.
To further evaluate the contribution of the bridge to the carbapenemase activity of OXA-23, we purified the F110A/M221A mutant of OXA-23 and measured the steady-state kinetic parameters (Table 2). To verify the integrity of the catalytic machinery of this enzyme, we measured its activity against ampicillin, which is a good substrate of wild-type OXA-23. We observed that there was an almost 2-fold increase in kcat compared with OXA-23, which shows the mutant is at least as active as the parental enzyme. We also observed a 3-fold increase in the Km value and an overall 2-fold decrease in the enzyme’s catalytic efficiency, kcat/Km, compared with OXA-23 (Table 2). This 2-fold change in the catalytic activity correlates perfectly with the observed 2-fold decrease in the MIC of ampicillin. Our kinetic results correlate well with data for the Y112A/M223A bridge mutant of OXA-24/40, where a 2-fold increase in the kcat value and a 5-fold increase in the Km value for ampicillin were observed (7).
TABLE 2.
Steady-state kinetic parameters for hydrolysis of β-lactam antibiotics by OXA-23 and its F110A/M221A mutant
| Antibiotic | Parameters (mean ± SD) for: |
|||||
|---|---|---|---|---|---|---|
| Wild-type OXA-23 |
F110A/M221A OXA-23 |
|||||
| kcat (s−1) | Km (or Ks) (μM) | kcat/Km (M−1s−1) | kcat (s−1) | Km (or Ks) (μM) | kcat/Km (M−1s−1) | |
| Ampicillin | 580 ± 20 | 100 ± 10 | (5.8 ± 0.6) × 106 | 930 ± 60 | 320 ± 50 | (2.9 ± 0.5) × 106 |
| Imipenem | 0.40 ± 0.01 | 0.19 ± 0.02a | (2.1 ± 0.2) × 106 | 0.29 ± 0.01 | 0.40 ± 0.07b | (7.3 ± 1.3) × 105 |
| Meropenem | 0.074 ± 0.001 | 0.060 ± 0.010a | (1.2 ± 0.2) × 106 | 0.067 ± 0.001 | 1.7 ± 0.1 | (3.9 ± 0.2) × 104 |
| Doripenem | 0.040 ± 0.001 | 0.025 ± 0.003a | (1.6 ± 0.2) × 106 | 0.060 ± 0.001 | 1.5 ± 0.1 | (4.0 ± 0.3) × 104 |
These data were previously reported (10).
Km values less than 1 μM could not be reliably measured; therefore, Ks values were determined using competition experiments.
Next, we evaluated the steady-state kinetic parameters of the F110A/M221A OXA-23 mutant enzyme for carbapenem antibiotics. In general, there were only small (less than 2-fold) increases or decreases in the kcat values compared with the parental enzyme (Table 2). The Km values of the F110A/M221A OXA-23 mutant for meropenem and doripenem were significantly increased (30- and 60-fold, respectively), indicating a weaker apparent binding affinity for these antibiotics relative to the wild-type enzyme (Table 2). For imipenem, the Km value was below the detection limit; therefore, the dissociation constant Ks was measured. The Ks value of the mutant enzyme increased only 2-fold for imipenem, revealing that it has a slightly weaker binding affinity for this substrate than the parental OXA-23 (Table 2). Changes in affinities of the mutant enzyme for carbapenems are reflected in the decreases in the conferred MIC values. Due to close similarities between the kcat values of the wild-type and F110A/M221A mutant OXA-23 enzymes for carbapenem antibiotics, the changes in their calculated catalytic efficiencies (kcat/Km) are mainly the result of changes in their affinity for these substrates (Table 2).
Kinetic data for the Y112A/M223A OXA-24/40 mutant with imipenem were reported in two studies (7, 14), while those for meropenem in one study (7). In these studies, small changes in kcat values were reported for the mutant OXA-24/40 with meropenem and imipenem, which are similar to what we observed with the double mutant of OXA-23. For the Y112A/M223A OXA-24/40 mutant, the Km values were increased 300-fold for meropenem and 10- to 16-fold for imipenem, which are even more significant changes than we observed for the OXA-23 double mutant.
Structural studies of apo and acyl-enzyme carbapenem complexes of F110A/M221A OXA-23.
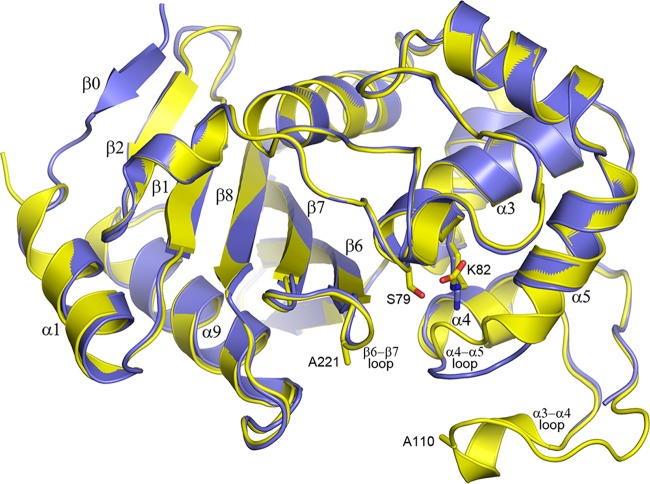
Currently, no structures of bridge-deficient CHDLs have been reported. To analyze the structural implications of the substitution of the bridge residues of OXA-23, the crystal structures of F110A/M221A OXA-23 were determined at neutral pH (7.0) and low pH (4.1) (Fig. 2). The overall fold of the enzyme is the same at both pHs, and the root mean square deviation (RMSD) after superposition is 0.43 Å (for 216 matching Cα positions). The main structural deviations occur in the loops between helices α4 and α5 and between helices α3 and α4 (Fig. 2). In addition, a difference is observed at the N terminus where refolding of this region in the low pH structure forms an additional β strand (β0). At low pH, there is no electron density for residues 103 to 115 of the α3-α4 loop, which carries the F110A substitution, indicating that this part of the structure has become highly mobile and is sampling multiple conformations. Inspection of the electron density maps in the active site of both structures shows the presence of a large piece of residual electron density at the end of the Lys82 side chain in the neutral pH form and lack of this density in the low pH form, showing that the lysine residue is fully carboxylated only in the former (Fig. S2).
FIG 2.
Superposition of the F110A/M221A OXA-23 mutant structures at pH 4.1 and 7.0. The structure at pH 4.1 (blue, PDB code 6N6T) is superimposed on the pH 7.0 structure (yellow, PDB code 6N6W), showing the location of the two mutations (Ala110 and Ala221), the active site serine (Ser79), and the lysine (Lys82) which is carboxylated at neutral (yellow sticks) but not at low pH (blue sticks).
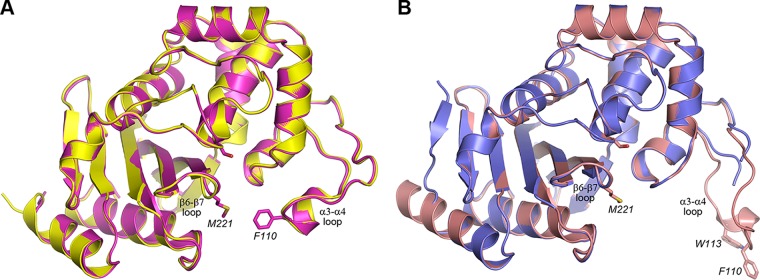
Superposition of the neutral pH form of the F110A/M221A OXA-23 mutant onto the wild-type structure of OXA-23 obtained at the same pH (Protein Data Bank [PDB] code 4JF6) gives an RMSD of 0.35 Å for 238 matching Cα atoms. Inspection of the superimposed structures shows that the loss of the two bridge residues does not affect the positions of the loops carrying these two residues (the α3-α4 loop, residues 103 to 117; the β6-β7 loop, residues 220 to 227; Fig. 3A). The distance between the Cα atoms of residues 110 and 221 is essentially the same in both structures (13. Å in the wild-type structure and 14.2 Å in the mutant). However, the distance of closest approach between the side chains of these residues changes drastically, from 4.3 Å in the wild-type enzyme to over 12.5 Å in the mutant. As the result, the substrate-binding pocket is significantly more open and accessible in the mutant enzyme (Fig. S3).
FIG 3.
Superposition of the wild-type and mutant OXA-23 structures. (A) Superposition of the wild-type OXA-23 (magenta, PDB code 4JF6) and its F110A/M221A mutant (yellow, PDB code 6N6W) at pH 7.0. The side chains of the catalytic serine (Ser79) and the hydrophobic bridge residues (Phe110 and Met221) are shown as magenta sticks for wild-type OXA-23. (B) Superposition of wild-type OXA-23 (pink, PDB code 4JF5) and the F110A/M221A OXA-23 mutant (blue, PDB code 6N6T) at pH 4.1. The side chains of the catalytic serine (Ser79), the hydrophobic bridge residues (Phe110 and Met221), and Trp113 on the α3-α4 loop are shown as pink sticks for wild-type OXA-23.
A similar superposition of the low pH mutant structure on the low pH wild-type structure (PDB code 4JF5) gives an RMSD of 0.40 Å for 224 matching Cα atoms. The β6-β7 bridge loop, which harbors the M221A substitution, occupies the same position in both enzymes (Fig. 3B). As noted above, the loop carrying the F110A mutation is disordered in the low pH mutant structure, which is in contrast to the low pH wild-type structure where this loop is fully ordered and swings outward from the active site by over 16 Å to adopt an open conformation (Fig. 3) (10). It is likely that in wild-type OXA-23, the Phe110 side chain provides structural stability to the loop by forming an aromatic interaction with the side chain of Trp113, and the loss of the phenylalanine in the mutant enzyme destabilizes the end of the loop.
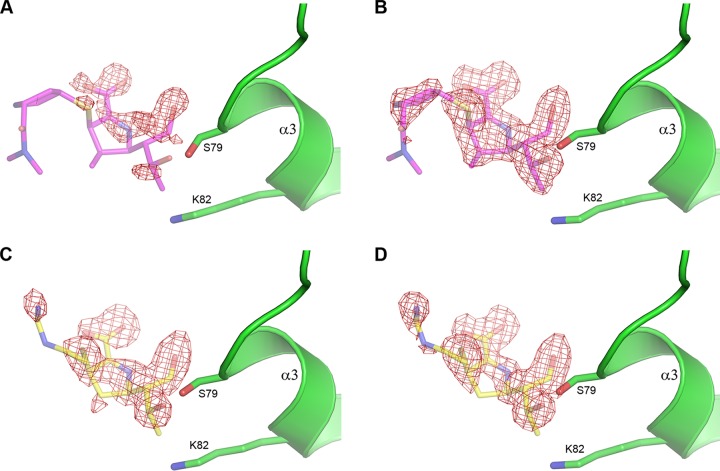
Acyl-enzyme complexes of meropenem and imipenem with the bridge-deficient mutant at both neutral and low pH were prepared by soaking preformed apo crystals in solutions containing the carbapenems. In earlier substrate soaking experiments with the low pH form of wild-type OXA-23, it was found that a tightly bound citrate anion present in the active site precluded the formation of acyl-enzyme species (10). Therefore, in order to facilitate acylation in the low pH crystals of the mutant, the citrate was removed by transferring crystals of the apo enzyme into a synthetic mother liquor containing acetate buffer at pH 4.6 prior to soaking experiments. In the neutral pH case, crystals were soaked directly in mother liquor supplemented with carbapenem. For the low pH structures, with a soak time of 5 min or less, the electron density for meropenem was weak and discontinuous, indicative of partial occupancy for the acyl-enzyme intermediate (Fig. 4A) and, perhaps, the presence of some nonhydrolyzed carbapenem, although this was not modeled into the density. A highly occupied meropenem complex was obtained with a 10-min soak time (Fig. 4B) and was still present after 30 min of soaking (data not shown). With imipenem, a 5-min soak produced a highly occupied acyl-enzyme intermediate (Fig. 4C), which was still present after 10 min of soaking (Fig. 4D) and persisted even after 6 hours of soaking (data not shown). These results show that the imipenem acyl-enzyme intermediate forms more rapidly than the meropenem intermediate, likely due to the higher affinity of imipenem for the mutant OXA-23 enzyme (Table 2). With the neutral pH form, shorter soak times of 1 to 3 min were sufficient to obtain highly occupied acyl-enzyme complexes (Fig. S4). Longer soak times resulted in significantly weaker electron density of the acylated substrate, and this can be attributed to substrate hydrolysis, since at pH 7.0 the catalytic lysine is carboxylated and the enzyme is fully active.
FIG 4.
Residual Fo-Fc electron density for carbapenem-soaked structures of the F110A/M221A OXA-23 mutant at pH 4.6 contoured at 2.75 σ. (A) Five-minute soak with meropenem. (B) Ten-minute soak with meropenem. (C) Five-minute soak with imipenem. (D) Ten-minute soak with imipenem. In all panels the corresponding carbapenem is shown as semitransparent sticks based on the final refined positions from the 10-min soak structures (meropenem, PDB code 6N6V, magenta; imipenem, PDB code 6N6U, yellow). The side chains of Ser79 and Lys82 are also shown.
Hydrolysis of the β-lactam ring of a carbapenem by a β-lactamase first yields a Δ2 configuration of the pyrroline, where the C2 carbon is sp2 hybridized, the S1 sulfur atom is coplanar with the pyrroline, and the double bond is between the C2 and C3 carbon atoms (Fig. 1). This tautomerizes to form the more stable Δ1 isomer, where the double bond is now between the C3 carbon and the N4 nitrogen, and the proton has moved from the N4 to the C2 atom, which becomes sp3 hybridized. The S1 sulfur can be on either side of the plane of the pyrroline ring (Fig. 1), giving two possible diastereomers (Δ1R and Δ1S) which are interchangeable via the Δ2 tautomer, and it has been suggested that in solution both Δ1 tautomers are equally stable (16). An analysis of the crystal structures of carbapenem complexes of β-lactamases available in the PDB shows that the Δ2 isomer predominates in the class A enzymes, the Δ2 and Δ1 isomers are present equally in the class D enzymes, and the Δ1S isomer is more prevalent in the class B metallo-β-lactamases (data not shown).
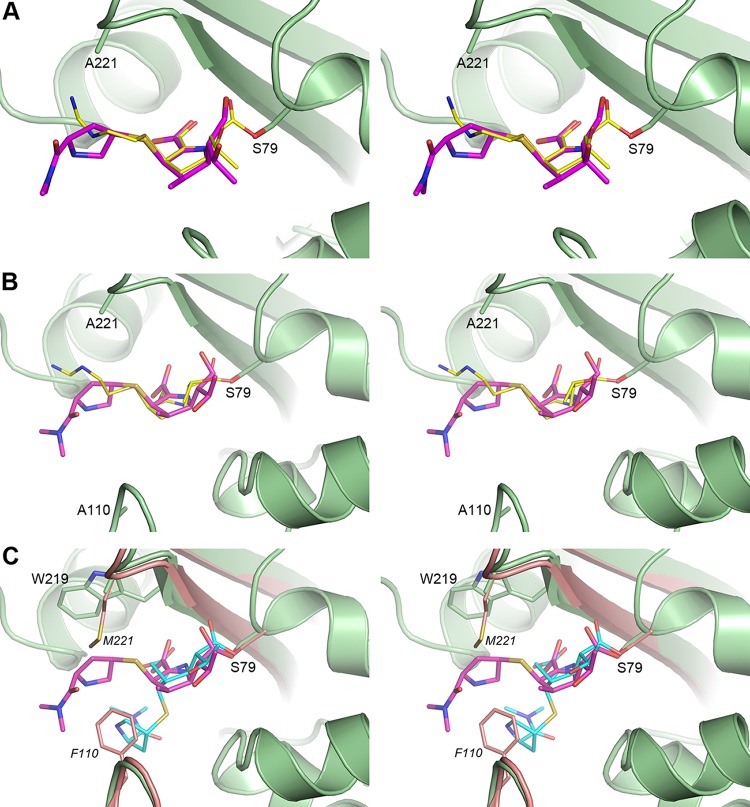
Our structures show that in the F110A/M221A OXA-23 mutant, the acylated species of both meropenem and imipenem are present as the Δ1R tautomers at both acidic and neutral pH (Fig. S4; Fig. 5A and B). This is in contrast to the wild-type OXA-23 enzyme, where the acylated meropenem is present as the Δ1S tautomer (10). Superposition of the meropenem complexes of the bridge-deficient mutant and the wild-type OXA-23 enzymes shows that the core part of the meropenem substrate (which includes the pyrroline ring, the C7 carbonyl, and the hydroxyethyl group; Fig. 1) is anchored in the same way and in the same orientation in both complexes (Fig. 5C). Due to differences in the tautomer conformation at C2, significant variation is observed in the orientation of the tail of the meropenem. In the wild-type complex, the pyrrolidine ring of the tail forms a hydrophobic interaction with Phe110 of the bridge (10). In the bridge-deficient mutant complex, the stabilizing interaction with the phenylalanine residue is lost, and the meropenem side group undergoes tautomerization to the Δ1R isomer. Subsequently, the tail projects toward the opposite side of the active site cleft, forming a weak hydrophobic interaction with the side chain of Trp219 (Fig. 5C). Such interactions are also expected for doripenem, which also has a pyrrolidine ring attached to the S1 sulfur (Fig. 1). Our kinetic studies with the F110A/M221A mutant show that there is a very minor change in the turnover rate of meropenem and doripenem compared with the parental OXA-23 enzyme. These results are consistent with our structural data, which show that the changes to the conformation of meropenem, upon transition from Δ1S to Δ1R, occur far from the region where deacylation takes place. On the other hand, the affinity of the mutant OXA-23 enzyme for meropenem and doripenem has decreased dramatically (Table 2), which likely results from the loss of the strong hydrophobic interaction between Phe110 of the enzyme and the pyrrolidine ring of the carbapenem tail.
FIG 5.
Stereoviews of the active site of the carbapenem complexes of the F110A/M221A OXA-23 mutant. (A) Superposition of the imipenem (PDB code 6N6U, yellow sticks) and meropenem (PDB code 6N6V, magenta sticks) structures at pH 4.6. The F110A/M221A OXA-23 mutant is shown as green ribbons and sticks for the imipenem complex only for clarity. (B) Superposition of the imipenem (PDB code 6N6X, yellow sticks) and meropenem (PDB code 6N6Y, magenta sticks) structures at pH 7.0. Only the F110A/M221A OXA-23 structure (green) for the imipenem complex is shown. (C) Superposition of the meropenem complexes of the F110A/M221A OXA-23 mutant (PDB code 6N6Y, pH 7.0, green ribbons and sticks) and wild-type OXA-23 (PDB code 4JF4, pH 4.1, pink ribbons and sticks). The meropenem Δ1R tautomer in the mutant complex is shown as magenta sticks, and the meropenem Δ1S tautomer in the wild-type complex is shown as cyan sticks.
While in our crystal structure of the OXA-23 mutant we observed the Δ1R tautomer of meropenem, in silico modeling shows that the active site of this enzyme could accommodate this substrate in any of the three tautomeric conformations (data not shown). However, in the wild-type enzyme, the presence of the hydrophobic bridge prohibits formation of the Δ1R tautomer, as in this conformation the pyrrolidine ring of meropenem would severely clash with the side chain of Met221 (Fig. 5C). This suggests that in the presence of the hydrophobic bridge, other carbapenems with bulky tails (such as doripenem; Fig. 1) would also be limited to either the Δ1S or the Δ2 tautomers upon acylation. The analysis of the carbapenem complexes of the class D β-lactamases whose structures are available (Table S1) supports this assumption. In the doripenem complexes of OXA-239 (an OXA-23 variant) and OXA-51 and the meropenem complex of OXA-13, which all have a hydrophobic bridge, the carbapenem adopts the Δ2 tautomer. Available structural data indicate that although the hydrophobic bridge in CHDLs prevents formation of the Δ1R tautomer of carbapenems with bulky side groups due to steric hindrance, all three tautomeric forms could be accommodated when the bridge is absent based on in silico modeling (data not shown). However, in all such known cases, the acylated carbapenems exclusively form the Δ1R tautomer, as observed in the wild-type OXA-1, the Gram-positive class D enzyme BPU-1, and the F110A/M221A OXA-23 mutant (Table S1). These data indicate that the Δ1R conformation provides the most favorable stabilizing interaction with the protein, while differences in the active site architecture of these enzymes determine the nature of such interactions. In the OXA-23 mutant, the pyrrolidine ring makes a hydrophobic interaction with the side chain of Trp219 (Fig. 5C), in OXA-1 the pyrrolidine ring forms a hydrogen bond with Leu255, while in BPU-1 the substrate is anchored by water-mediated hydrogen bonds.
Unlike the carbapenems with bulky side groups, it seems that the hydrophobic bridge restricts the movement of the smaller tail of imipenem to a lesser degree. Indeed, although imipenem binds to OXA-13 as the Δ2 tautomer, it adopts the Δ1R configuration in OXA-239 (Table S1). It is plausible that in this enzyme, the less bulky tail of imipenem is capable of overcoming restrictions imposed by the side chain of the Met221 residue of the bridge. Also, in the wild-type OXA-23 enzyme, the iminoamine tail of imipenem would not be expected to make as strong an interaction with Phe110, and this is reflected in the weaker affinity of this substrate for the wild-type enzyme than for meropenem (Table 2). As a result, the loss of the bridge residues in the mutant enzyme is expected to have a smaller effect on the affinity for imipenem, which indeed is reflected in only a 2-fold decrease in the Ks value of the mutant for this substrate (Table 2).
CONCLUSIONS
Our microbiological, kinetic, and structural studies, complemented with analyses of the available class D β-lactamase structures, provide novel insights into the role of the hydrophobic bridge in CHDLs. We demonstrate that substitutions of individual bridge residues in OXA-23 have contrasting effects on resistance to carbapenems with different structures. For carbapenems with bulky tails (meropenem and doripenem), the greatest changes in MICs were observed upon Phe110 substitution, while for imipenem, upon Met221 substitution. This is in contrast to OXA24/40, where regardless of the carbapenem, MICs decreased significantly only upon substitution of Tyr112 (7). Despite these differences, the observed MIC decreases for both enzymes were due to their reduced affinity for carbapenem substrates.
We also provide the first structures of bridge-deficient mutants of class D CHDLs (that of OXA-23), both in apo form and in complex with carbapenem antibiotics. These structures, combined with available data for other wild-type CHDLs, demonstrate that the bridge directs carbapenem substrates (at least those with large bulky side groups such as doripenem or meropenem) into one of two specific tautomeric conformations, Δ2 or Δ1S, and prohibits the formation of the Δ1R tautomeric configuration due to steric hindrance. We show that disruption of the bridge in OXA-23 results in the Δ1R tautomer of both imipenem and meropenem, and this is also the only conformation observed in class D enzymes lacking the bridge.
Kinetic studies of the OXA-23 and OXA-24/40 enzymes demonstrated that their catalytic efficiencies for carbapenems with bulky tails (meropenem and doripenem) are almost identical (12). However, the carbapenems are found in the structures of these enzymes in two different tautomeric configurations (Δ1S for OXA-23 and Δ2 for OXA-24/40), an indication that the degree of the contribution (or lack thereof) of these tautomers to the carbapenemase activity of these enzymes is very similar. Data presented here show that disruption of the bridge in the OXA-23 CHDL results not only in the loss of its interaction with the carbapenem antibiotic but also in the change of the tautomeric configuration of the substrate from Δ1S to Δ1R. While this transition results in a significant decrease of the catalytic activity of F110A/M221A OXA-23, the turnover rate of the mutant is very similar to those of parental OXA-23 and OXA-24/40 enzymes where carbapenems are bound as the Δ1S and Δ2 tautomers. However, there is a significant decrease in the affinity of the mutant enzyme for meropenem and doripenem. These data strongly indicate that the hydrophobic bridge contributes to the activity of CHDLs against carbapenems with bulky tails not by defining the tautomeric state of the acylated substrate but rather by providing additional stabilizing interactions between its hydrophobic side chains and the tails of carbapenem substrates.
MATERIALS AND METHODS
Mutagenesis and cloning.
The majority of amino acid substitutions at residues Phe110 and Met221 were generated using site-directed mutagenesis PCR with degenerative primers. The missing substitutions were obtained in the same manner using specific primers. All mutations were confirmed by sequencing, subcloned into the previously described shuttle vector (10) under the ISAba3 insertion sequence promoter, and electroporated into A. baumannii CIP 70.10 for MIC testing. For protein expression, the gene for the mature F110A/M221A OXA-23 enzyme was optimized for expression in Escherichia coli, custom synthesized, and subcloned into pET24a(+) (GenScript). The protein was expressed and purified as previously described for wild-type OXA-23 (10).
Antibiotic susceptibility testing.
MICs were determined in triplicate by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (15). Briefly, microtiter plates were inoculated with 5 × 105 CFU/ml bacteria in Mueller-Hinton II broth (Difco) and incubated at 37°C for 20 to 24 h before interpreting the results.
Steady-state kinetics.
A Cary 60 (Agilent) spectrophotometer was used to collect all kinetic data. Reaction mixtures containing various concentrations of β-lactams, 50 mM NaHCO3, and 0.2 mg/ml bovine serum albumin (BSA) in 100 mM NaPi (pH 7.0) were initiated at room temperature by the addition of F110A/M221A OXA-23. The change in absorbance was observed, and the following extinction coefficients were used to analyze the data: ampicillin Δε235, −670 M−1cm−1; imipenem Δε297, −10,930 M−1cm−1; meropenem Δε298, −7,200 M−1cm−1; and doripenem Δε299, −11,540 M−1cm−1. Steady-state velocities (v) were calculated from the linear phase of each reaction. Subsequently, the observed rate constant was determined using the relationship kobs = v/[E]. The parameters kcat and Km were determined by plotting kobs as a function of the substrate concentration and fitting the data using nonlinear regression to the Michaelis-Menten equation in Prism 7 (GraphPad). All measurements were made in triplicate.
Determination of dissociation constants.
The dissociation constant Ks was determined using the method of Dixon (17). In this experiment, the chromogenic compound nitrocefin (Δε500, +15,900 M−1cm−1) was used as a substrate, while the carbapenems were treated as inhibitors. Briefly, reaction mixtures containing various concentrations of carbapenems, 50 mM NaHCO3, 0.2 mg/ml bovine serum albumin (BSA) in 100 mM NaPi pH 7.0, and 100, 200, or 400 μM nitrocefin were mixed with F110A/M221A OXA-23. The absorbance was monitored at room temperature, and the steady-state velocities were determined from the linear portion of each progress curve. Data were collected in triplicate. As k2 and k−1 values of the mutant enzyme are not known, the data could represent Ks′ instead of Ks if the value of k2 is equal to or larger than k−1.
Crystallographic analysis of the F110A/M221A OXA-23 mutant.
Crystals were grown using the same conditions which gave diffraction-quality crystals of the wild-type OXA-23, either from 0.06 M citric acid, 0.04 M bis-Tris propane (BTP; pH 4.1), and 16% polyethylene glycol (PEG) 3350 to give a low pH crystal form, or from 0.2 M succinic acid (pH 7.0) and 20% PEG 3350 to give a neutral pH form (10). Crystals grew after 1 to 2 days of incubation at room temperature. Diffraction data sets were collected for both crystal forms at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 (BL9-2), using X-rays at 12,658 eV (0.9795 Å). Data collection statistics are given in Tables S2 and S3. Both crystal forms contained a single molecule in the asymmetric unit, with solvent contents of 52% and 78% for the low pH and neutral pH forms, respectively. The low pH and neutral pH apo structures were solved by molecular replacement (MR) using the refined low pH and neutral pH structures from the PDB (PDB codes 4JF5 and 4JF6). Models were manually rebuilt using Coot (18) and refined with phenix.refine (19). Refinement statistics are given in Tables S4 and S5.
Carbapenem complexes of the F110A/M221A OXA-23 were prepared by soaking preformed crystals in modified crystallization buffer supplemented with 50 mM meropenem or 50 mM imipenem. For the low pH conditions, the citrate-BTP buffer was replaced with 0.1 M acetate (pH 4.6). Crystals were soaked for various times ranging from 5 to 30 min at low pH and 2 to 5 min at neutral pH. Data were collected from the soaked crystals at BL9-2, and data collection statistics are given in Tables S2 and S3. All structures were solved by MR using the refined apo structure at the appropriate pH as the starting model. Acylated carbapenem substrates were built into residual electron density in the active site using Coot (18), and the structures refined with phenix.refine (19). Refinement statistics are given in Tables S4 and S5.
Data availability.
The following coordinates and structure factors for the F110A/M221A OXA-23 mutant structures used in this study have been deposited in the Protein Data Bank (accession numbers): apo pH 4.1, resolution 1.25 Å (6N6T); imipenem complex pH 4.6, resolution 1.55 Å (6N6U); meropenem complex pH 4.6, resolution 1.55 Å (6N6V); apo pH 7.0, resolution 3.25 Å (6N6W); imipenem complex pH 7.0, resolution 3.1 Å (6N6X); and meropenem complex pH 7.0, resolution 3.5 Å (6N6Y).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant R01AI114668 from the NIH/NIAID (to S.B.V.) and the Pilot Project Grant Program from the Eck Institute for Global Health (to S.B.V.). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02191-18.
REFERENCES
- 1.Leonard DA, Bonomo RA, Powers RA. 2013. Class D β-lactamases: a reappraisal after five decades. Acc Chem Res 46:2407–2415. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher JF, Meroueh SO, Mobashery S. 2005. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev 105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 3.Antunes NT, Fisher JF. 2014. Acquired class D β-lactamases. Antibiotics (Basel) 3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans BA, Amyes SG. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Docquier JD, Mangani S. 2016. Structure-function relationships of class D carbapenemases. Curr Drug Targets 17:1061–1071. doi: 10.2174/1389450116666150825115824. [DOI] [PubMed] [Google Scholar]
- 6.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 7.Santillana E, Beceiro A, Bou G, Romero A. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci U S A 104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. 2011. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J Mol Biol 406:583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider KD, Karpen ME, Bonomo RA, Leonard DA, Powers RA. 2009. The 1.4 Å crystal structure of the class D β-lactamase OXA-1 complexed with doripenem. Biochemistry 48:11840–11847. doi: 10.1021/bi901690r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. 2013. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem Biol 20:1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. 2009. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol 16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratap S, Katiki M, Gill P, Kumar P, Golemi-Kotra D. 2016. Active-site plasticity is essential to carbapenem hydrolysis by OXA-58 class D β-lactamase of Acinetobacter baumannii. Antimicrob Agents Chemother 60:75–86. doi: 10.1128/AAC.01393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bou G, Santillana E, Sheri A, Beceiro A, Sampson JM, Kalp M, Bethel CR, Distler AM, Drawz SM, Pagadala SR, van den Akker F, Bonomo RA, Romero A, Buynak JD. 2010. Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase. J Am Chem Soc 132:13320–13331. doi: 10.1021/ja104092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Ratcliffe RW, Wildonger KJ, Di Michele L, Douglas AW, Hajdu R, Goegelman RT, Springer JP, Hirshfield J. 1989. Studies on the structures of imipenem, dehydropeptidase I–hydrolyzed imipenem, and related analogs. J Org Chem 54:653–660. doi: 10.1021/jo00264a028. [DOI] [Google Scholar]
- 17.Dixon M. 1953. The determination of enzyme inhibitor constants. Biochem J 55:170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following coordinates and structure factors for the F110A/M221A OXA-23 mutant structures used in this study have been deposited in the Protein Data Bank (accession numbers): apo pH 4.1, resolution 1.25 Å (6N6T); imipenem complex pH 4.6, resolution 1.55 Å (6N6U); meropenem complex pH 4.6, resolution 1.55 Å (6N6V); apo pH 7.0, resolution 3.25 Å (6N6W); imipenem complex pH 7.0, resolution 3.1 Å (6N6X); and meropenem complex pH 7.0, resolution 3.5 Å (6N6Y).