The objective of this study was to assess the antimicrobial resistance of enteroaggregative Escherichia coli (EAEC) and enterotoxigenic E. coli (ETEC) strains causing traveler’s diarrhea (TD) and to investigate the molecular characterization of antimicrobial resistance genes to third-generation cephalosporins, cephamycins, and quinolones. Overall, 39 EAEC and 43 ETEC clinical isolates were studied.
KEYWORDS: cephalosporin, enteroaggregative E. coli, enterotoxigenic E. coli, quinolones, resistance, traveler’s diarrhea
ABSTRACT
The objective of this study was to assess the antimicrobial resistance of enteroaggregative Escherichia coli (EAEC) and enterotoxigenic E. coli (ETEC) strains causing traveler’s diarrhea (TD) and to investigate the molecular characterization of antimicrobial resistance genes to third-generation cephalosporins, cephamycins, and quinolones. Overall, 39 EAEC and 43 ETEC clinical isolates were studied. The susceptibilities of EAEC and ETEC against ampicillin, amoxicillin-clavulanic acid, cefotaxime, imipenem, chloramphenicol, tetracycline, co-trimoxazole, nalidixic acid, ciprofloxacin, azithromycin, and rifaximin were determined. All genes encoding resistance determinants were detected by PCR or PCR plus DNA sequencing. The epidemiology of selected EAEC and ETEC strains was studied using multilocus sequence typing (MLST). The resistance to quinolones of EAEC and ETEC strains causing TD has significantly increased over the last decades, and high percentages have been found especially in patients traveling to India and sub-Saharan Africa. Sequence type 38 (ST38) and ST131, carrying the blaCTX-M-15 and blaCTX-M-27 genes, respectively, are highly prevalent among extended-spectrum β-lactamase (ESBL)-producing EAEC and ETEC strains. The cephamycinase ACT-20 is described in the present study for the first time in EAEC and ETEC strains causing TD in patients who had traveled to Central America. The percentages of resistance to azithromycin in EAEC and ETEC isolates from patients to Southeast Asia/India and Africa are above 25%. Meanwhile, rifaximin is still active against EAEC and ETEC, with the prevalence of resistant strains not being high. In conclusion, fluoroquinolones should no longer be considered the drugs of choice for the prevention or treatment in TD for travelers traveling to India and Africa. Azithromycin and rifaximin are still a good alternative to treat TD caused by EAEC or ETEC.
INTRODUCTION
Traveler’s diarrhea is the most frequent infection presented by travelers attending a travel medicine unit following a trip to low- or middle-income countries (1, 2). Enteroaggregative Escherichia coli (EAEC) and enterotoxigenic E. coli (ETEC) are two of the most frequent bacteria causing traveler’s diarrhea (TD), together with Shigella spp. and Campylobacter spp. (3–5). Other nonbacterial enteric pathogens identified as etiological agents of TD in minor proportions (between 28% and 35%) are Norovirus, Giardia, and Cryptosporidium spp. (5).
In the cases of TD where antimicrobial treatment is suggested, fluoroquinolones, azithromycin, and rifaximin are the recommended antibiotics (6, 7). In addition, fluoroquinolones have been considered an option in the prevention of TD in travelers at high risk, such as immunocompromised patients in whom chemoprophylaxis is considered essential. However, other views concerning the antibiotic use for TD (especially for mild and moderate diarrhea) have emerged recently, as antimicrobials have been shown to be an independent risk factor that predisposes travelers to contracting resistant strains, such as extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (8).
Information in the scientific literature regarding the antimicrobial susceptibilities of EAEC and ETEC strains is scarce; therefore, we do not know whether the guidelines recommending empirical treatment remain valid. In addition, the prevalence of ESBLs as a mechanism of resistance to third-generation cephalosporins has mainly been investigated in extraintestinal E. coli (ExPEC) strains, but there is little research into diarrheagenic E. coli. The main purpose of this study was to assess the antimicrobial resistance of EAEC and ETEC strains causing traveler’s diarrhea during the period of 2011 to 2017 and investigate the mechanisms of resistance to third-generation cephalosporins, cephamycins, and quinolones.
RESULTS
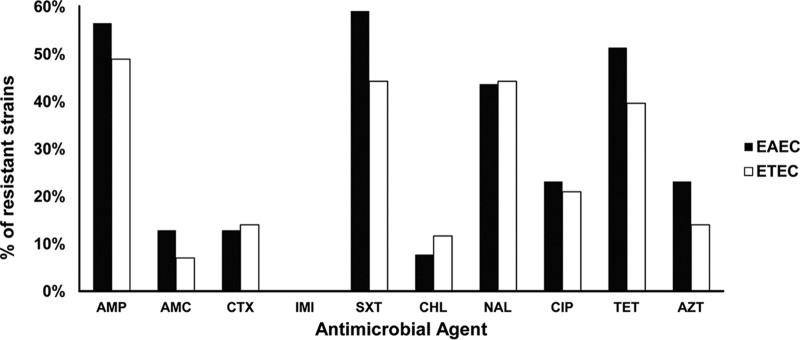
The susceptibilities of 39 EAEC and 43 ETEC clinical isolates were determined by disk diffusion, Etest, or microdilution methods and are shown in Fig. 1 and in Table S1 in the supplemental material. Overall, EAEC showed greater resistance than ETEC, without significant differences. EAEC presented the following percentages of resistance: ampicillin (AMP), 56.4%; amoxicillin-clavulanic acid (AMC), 12.8%; cefotaxime (CTX), 12.8%; co-trimoxazole (SXT), 59%; chloramphenicol (CHL), 7.7%; tetracycline (TET), 51.3%; nalidixic acid (NAL), 43.6%; ciprofloxacin (CIP), 23%; and azithromycin (AZT), 23%. Meanwhile, the percentages of resistance of the ETEC clinical isolates were AMP, 48.9%; AMC, 7%; CTX, 14%; SXT, 44.2%; CHL, 11.6%; TET, 39.5%; NAL, 44.2%; CIP, 21%; and AZT, 14%. All EAEC and ETEC clinical isolates were susceptible to imipenem. Since no breakpoints are defined for rifaximin, we determined the MIC50 and MIC90 for EAEC and ETEC, being 8 and 16 μg/ml, respectively, for the two E. coli pathotypes.
FIG 1.
Percentages of resistance to different antibacterial agents in EAEC and ETEC strains. AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; IMI, imipenem; SXT, co-trimoxazole; CHL, chloramphenicol; NAL, nalidixic acid; CIP, ciprofloxacin; TET, tetracycline; AZT, azithromycin.
The distribution of the percentages of resistance according to the E. coli pathotype and the geographical area visited is shown in Table 1. The resistance percentages were similar between EAEC and ETEC, with levels of resistance to cefotaxime greater than 38% in strains isolated from patients traveling to Southeast Asia/India than patients traveling to other areas. The prevalence of strains resistant to co-trimoxazole was higher in Africa than in Southeast Asia/India and Latin America. The prevalence of nalidixic acid-resistant strains was greater than 27% in all areas, being above 64% in Southeast Asia/India. The high level (>40%) of strains resistant to ciprofloxacin in Southeast Asia/India is worthy of mention, while in Africa, the percentage was intermediate (between 11 and 19%), and in Latin America, it was below 10%. Azithromycin-resistant strains were also more frequent in Southeast Asia/India than in Africa and Latin America, with resistance rates of 33.3%, 25%, and 9.1%, respectively, for EAEC and 28.6%, 11.1%, and 0%, respectively, for ETEC. It is important to highlight that 58% of the patients with TD from Southeast Asia/India visited India, and among these, the percentages of resistance to nalidixic acid were 75% and 71.4% for EAEC and ETEC, respectively; ciprofloxacin resistance rates were 62.5% and 43% for EAEC and ETEC, respectively; and rates of resistance to azithromycin were 37.5% and 28.6% for EAEC and ETEC, respectively (data not shown). However, statistical analysis was not performed due to the low population size obtained when stratifying the strains according to pathotype and geographical origin.
TABLE 1.
Percentages of resistance of EAEC and ETEC strains to different antimicrobial agents according to three geographical areas
| Antimicrobial agent | % resistant isolates (no.) for: |
|||||
|---|---|---|---|---|---|---|
| EAEC |
ETEC |
|||||
| Southeast Asia/India (n = 12) | Africa (n = 16) | Latin America (n = 11) | Southeast Asia/India (n = 14) | Africa (n = 18) | Latin America (n = 11) | |
| Cefotaxime | 33.3 (4)a | 6.3 (1)a | 0 (0) | 42.9 (6)a | 0 (0) | 0 (0) |
| Co-trimoxazole | 50(6) | 81.3 (13) | 36.4 (4) | 35.7 (5) | 61.1 (11) | 27.3 (3) |
| Nalidixic acid | 66.7 (8) | 37.5 (6) | 27.3 (3) | 64.3 (9) | 38.9 (7) | 27.3 (3) |
| Ciprofloxacin | 41.7 (5) | 18.8 (3) | 9.1 (1) | 42.9 (6) | 11.1 (2) | 9.1 (1) |
| Azithromycin | 33.3 (4) | 25 (4) | 9.1 (1) | 28.6 (4) | 11.1 (2) | 0 (0) |
ESBL-producing strains.
The mechanisms of resistance to third-generation cephalosporins are usually associated with the production of ESBLs. This analysis was determined in all 11 isolates presenting cefotaxime resistance by the disk diffusion test (diameters obtained ranged from 8 to 18 mm, considering the criteria resistance [R], ≤ 22 mm; intermediate [I], 23 to 25 mm; and susceptibility [S], ≥ 26 mm) and a positive result by double-disk synergy test. The MIC of cefotaxime was determined, showing a range from 6 to >256 μg/ml in EAEC isolates and from 6 to 96 μg/ml in ETEC isolates. All the isolates were positive for CTX-M ESBL; eight of them belonged to CTX-M-15 and three to the CTX-M-27. The multilocus sequence type (MLST) analysis generated a high heterogeneity of types. Indeed, it was of note that three strains from India belonging to sequence type 38 (ST38) (two EAEC and one ETEC) carried the blaCTX-M-15 gene; however, the plasmid typing was K for one EAEC strain, FIB (allele 33) and FII (allele 1) for the other EAEC strain, and Y for the ETEC strain. In addition, two ETEC strains carrying the blaCTX-M-27 gene belonging to the high-risk clone ST131 had the same plasmid type profile (FIB, allele 20; FIA, allele 2). One of the patients with TD caused by the ST131 clone had visited India, while another had traveled to Vietnam and Cambodia (Table 2). Moreover, the blaCTX-M-27 gene was also found in a singleton (ST1193) strain of EAEC carrying the same plasmid replicon types as the ST131-belonging strains but with different alleles. Five EAEC and three ETEC isolates were resistant to amoxicillin-clavulanic acid, with MICs from 12 to 64 μg/ml (data not shown). The presence of genes encoding OXA, TEM, SHV, and plasmid mediated AmpC-type β-lactamases was determined. The blaACT-20 gene was detected in two strains, one EAEC and one ETEC, showing the highest MICs for amoxicillin-clavulanic acid of 24 and 64 μg/ml, respectively. The MIC for cefoxitin for both strains was >256 μg/ml. These patients had visited Guatemala and the Dominican Republic (Table S1). The remaining six strains (four EAEC and two ETEC), with an MIC of 12 μg/ml to amoxicillin-clavulanic acid and MICs between 6 and 16 μg/ml to cefoxitin, presented the blaOXA-1-like gene, which explains the moderate level of resistance to amoxicillin-clavulanic acid. Two EAEC strains isolated from patients who had traveled to India also harbored the blaSHV-like gene, and none of the overall strains were found to carry the blaTEM gene.
TABLE 2.
Main features of the EAEC and ETEC clinical isolates carrying ESBLs
| Isolate no. by type | Geographical origin | MLST |
Plasmid typing (allele no.) | CTX-M type | CTX MIC (μg/ml) | |
|---|---|---|---|---|---|---|
| ST | CC | |||||
| EAEC | ||||||
| 5 | India | ST1193 | Singleton | FIB (10), FIA (6) | CTX-M-27 | 8 |
| 11 | India | ST38 | ST38 | FIB (33), FII (1) | CTX-M-15 | >256 |
| 20 | India | ST7615 | Singleton | I1, K, B/O | CTX-M-15 | 6 |
| 70 | India | ST38 | ST38 | K | CTX-M-15 | 128 |
| 84 | Togo | ST44 | ST10 | I1, FIA (6) | CTX-M-15 | >256 |
| ETEC | ||||||
| 36 | India/Nepal | ST23 | ST23 | FIB (not described) | CTX-M-15 | 64 |
| 38 | India | ST1284 | Singleton | FIA (20) | CTX-M-15 | 96 |
| 39 | India | ST131 | ST131 | FIA (2), FIB (20) | CTX-M-27 | 6 |
| 43 | Vietnam/Cambodia | ST131 | ST131 | FIA (2), FIB (20) | CTX-M-27 | 12 |
| 102 | China | ST5584 | Singleton | Y | CTX-M-15 | 12 |
| 107 | India | ST38 | ST38 | Y | CTX-M-15 | 64 |
Two phenotypes could be defined among quinolone-resistant EAEC (17 strains) and ETEC (20 strains) strains. One was nalidixic acid resistant (NALr) but ciprofloxacin intermediate or susceptible (CIPi-s) (considering the criteria resistant [R], <24 mm; intermediate [I], 24 to 25 mm; and susceptible [S], ≥ 26 mm). The second phenotype corresponded to the strains which were resistant to both nalidixic acid and ciprofloxacin (NALr CIPr). Both chromosome- and plasmid-mediated quinolone resistances were found (Table 3). All eight EAEC strains with the NALr CIPi-s phenotype showed a mutation in the gyrA gene, whereas only one EAEC strain with the NALr CIPr phenotype presented a mutation in amino acid codon Ser-83 of the gyrA gene. The remaining eight strains with the NALr CIPr phenotype showed the following mechanisms of resistance: four strains had a mutation in the same position in the gyrA gene and in the amino acid codon Ser-80 of the parC gene, two strains had the same double mutation plus the qnrS gene, and two strains also had this double mutation and the aac(6′)-Ib-cr gene. In the 11 ETEC strains with the Nalr CIPi-s phenotype, the mechanisms of resistance to quinolones found were eight strains with a mutation in the amino acid codon Ser-83 of the gyrA gene, one with a mutation in the amino acid codon Asp-87, and only the qnrS gene was detected in the last strain (the only one NALi CIPi). The mechanisms of resistance to quinolones in the nine ETEC strains with the NALr CIPr phenotype were one strain with a mutation in the amino acid codon Ser-83 of the gyrA gene; three strains with a double mutation in the gyrA and parC genes, as mentioned above; two strains with a gyrA gene mutation and the presence of the qnrS gene; and finally, three strains with the a double mutation and the aac(6′)-Ib-cr gene.
TABLE 3.
Different mechanisms of resistance to quinolones in EAEC and ETEC isolates
| Quinolone resistance mechanism(s) | No. of quinolone-resistant ETEC isolates by phenotype |
|||
|---|---|---|---|---|
| Nalr Cipi-s (n = 8) | Nalr Cipr (n = 9) | Nalr Cipi-s (n = 11) | Nalr Cipr (n = 9) | |
| gyrA mutation | 8 | 1 | 10 | 1 |
| gyrA + parC mutations | 0 | 4 | 0 | 3 |
| gyrA mutations + qnrS | 0 | 0 | 0 | 2 |
| gyrA + parC mutations + qnrS | 0 | 2 | 0 | 0 |
| gyrA + parC mutations + aac(6′)-Ib-cr | 0 | 2 | 0 | 3 |
| qnrS | 0 | 0 | 1 (Nali Cipi) | 0 |
DISCUSSION
ETEC and EAEC cause not only TD but also high morbidity in children in developing countries, mainly in those under 5 years of age (9). More than 50% of the patients attending the tropical medicine unit of our hospital presented TD, and antimicrobial therapy is needed due to the severity or persistence of the symptoms in around 35% of those in whom diarrhea is caused by ETEC or EAEC (10, 11). Nowadays, with the incorporation of rapid diagnostic tests based mainly on multiplex PCR, the etiology of the TD can be determined on the same working day, and therefore, a more adequate treatment can be implemented (12). Knowledge of the antimicrobial susceptibility of the most frequent etiological agents causing TD, such as EAEC or ETEC, will help in administering the most adequate treatment before the antimicrobial susceptibility of the bacteria isolated is generated.
Overall, in this study, the antimicrobial resistance rate of EAEC was slightly higher than that of ETEC, without significant differences. However, for both, the resistance to the classical and less expensive antibiotics used in developing countries, such as ampicillin, co-trimoxazole, and tetracycline, was greater than 39%. On stratifying the ETEC and EAEC strains according to the geographical area visited by the patient with TD, it was of note that the strains from Latin America were less resistant than those from Southeast Asia/India or Africa, but significance could not be calculated since the population was not large enough. This reflects the situation of antimicrobial resistance in different countries in Latin America versus Southeast Asia/India. Southeast Asia and India present high rates of resistance to the most available and inexpensive antibiotics, including quinolones, whereas in Latin America, ETEC and EAEC strains remain susceptible to these antimicrobial agents (13–19). The high prevalence of quinolone-resistant EAEC and ETEC isolates from Southeast Asia and India is worthy of mention, with rates of resistance for ciprofloxacin at higher than 40% for both EAEC and ETEC. In a previous study performed by our group during the period from 2001 to 2007, the overall percentages of resistance were 15 and 22% for nalidixic acid and were 4 and 8% for ciprofloxacin for EAEC and ETEC, respectively (20). Therefore, a significant increment (P < 0.0001 for both nalidixic acid and ciprofloxacin among EAEC strains and P = 0.0013 for nalidixic acid and P = 0.0062 for ciprofloxacin among ETEC strains) has been observed, being more dramatic in strains isolated from patients who had traveled to Southeast Asia/India, especially India, where 53.3% of the total strains were resistant to ciprofloxacin.
Four (44%) out of nine EAEC strains and two (33%) out of six ETEC strains with an MIC of azithromycin greater than 16 μg/ml showed an MIC of ≥256 μg/ml. Azithromycin reaches rectal concentrations of a mean of 133 μg/g with a single 1-g dose (21); therefore, it is above the MICs of most EAEC (89%) and ETEC (95%) strains with an MIC of azithromycin less than 256 μg/ml. The activity of rifaximin against EAEC and ETEC remained unchanged compared to previous studies (22, 23). Rifaximin is a nonabsorbable antibiotic, reaching a fecal concentration of 7,961 μg/g with a dose of 800 mg daily for 3 days (24), which is far above the MIC90 that we found for EAEC and ETEC.
The main mechanisms of resistance to cefotaxime are ESBLs. Different ESBLs have been described to date, with the main types being TEM-type, SHV-type, and CTX-M-type, with the CTX-M-type being the most currently extended ESBL at a global level (25–28). Travelers have been shown to be potential carriers of the ESBL-producing Enterobacteriaceae in the intestinal tract, facilitating the dissemination of these microorganisms between countries (8, 28–33). The two most prevalent STs detected were ST38 carrying CTX-M-15 and ST131 carrying CTX-M-27. CTX-M-15-producing EAEC strains belonging to ST38 from India causing TD have been described previously, demonstrating that ST38 is a successful EAEC group (34, 35); however, in our study, this ST was also found in ETEC strains. The ST5584 ETEC strain carrying the blaCTX-M-15 gene isolated from a traveler to China was not found among a collection of E. coli strains causing diarrhea in China, reported in a recent study (36). Different replicon type profiles (FIB/FII, K, and Y) were found in these three ST38 EAEC strains. In addition, CTX-M-15 and other types of CTX-M-producing E. coli ST38 clones, mainly ExPEC, have been detected in Saudi Arabia. In fact, some uropathogenic E. coli (UPEC) ST38 strains were also described as carrying the aggR gene, a main feature of EAEC (37). In China, the UK, Bangladesh, and Nigeria, ST38 was found among the most frequent STs in a collection of EAEC strains (38, 39).
CTX-M-27-producing E. coli ST131 strains have been described in several countries (40), and the blaCTX-M-27 gene has been detected in EPEC and EIEC strains isolated in China (36). This blaCTX-M-27 gene has also been detected in one EAEC strain isolated from surface water (41). However, as far as we know, its presence in ETEC strains has not been reported. In this study, both ETEC strains came from Southeast Asia/India, and the blaCTX-M-27 gene was located in an IncF plasmid, as expected (42–44).
The main enzymatic mechanisms of E. coli associated with the acquisition of resistance to AMC include (i) hyperproduction of plasmid-mediated class A β–lactamases, such as TEM-1 and SHV-1; (ii) plasmid-mediated AmpC-type β–lactamase (p-AmpC); (iii) chromosomal AmpC β–lactamase (c-AmpC); (iv) production of inhibitor-resistant TEM (IRT) β–lactamases; and (v) plasmid-mediated β–lactamase OXA-1 (45). Among the EAEC and ETEC strains resistant to AMC, a plasmid-mediated AmpC (ACT-20) was detected only in the two strains with the highest MIC. This type of p-AmpC has previously been found in a strain of Enterobacter hormaechei isolated from dog feces (46), but so far, it has not been described in bacteria causing infections in humans. The EAEC and ETEC strains with moderate resistance to AMC presented an OXA-1 enzyme that is currently the most frequently found mechanism of resistance to AMC (47).
The acquisition of resistance to quinolones in E. coli can be either chromosome or plasmid mediated. Chromosomal mutations generating resistance to quinolones are those associated mainly with the gyrA and parC genes encoding the A subunits of DNA gyrase and topoisomerase IV, respectively, which are the protein targets of these antibacterial agents. In addition, mutations that produce an overproduction of an efflux pump or a decreased expression of a gene encoding a porin can also reduce the accumulation of the quinolone, and hence, increase resistance. The plasmid-mediated mechanisms of resistance to quinolones are related to the presence of the following three genes/gene types: (i) the qnr genes, which protect the protein target of the binding of the quinolones; (ii) the aac(6′)-Ib-cr gene, which produces the acetylation of a radical group of some quinolones generating a decrease in activity; and (iii) the qepA or opxAB genes, which are quinolone efflux pumps (48).
In this study, the NALr CIPi-s phenotype shown by both ETEC and EAEC strains was mainly associated with a mutation in the gyrA gene, with the exception of one ETEC strain showing a NALi CIPi phenotype that did not have any mutation in the gyrA gene but presented the qnrS gene. The qnr gene was not detected in a previous study performed with ETEC and EAEC strains resistant to quinolones (20). In addition, in the present study, the NALr CIPr phenotype was related to a double mutation in the gyrA and parC genes alone or together with the qnrS or aac(6′)Ib-cr gene. In a study performed in India, the main mechanisms of resistance to quinolone in ETEC were also amino acid changes in GyrA and ParC. They did not find any Qnr determinant, but 65% of the strains presented the aac(6′)Ib-cr gene (49).
In summary, our results strengthen the message that resistance to quinolones and third-generation cephalosporins has increased in EAEC and ETEC strains causing of TD, mainly in patients traveling to India and Africa, and especially sub-Saharan Africa. In addition, the ST38 and ST131 carrying the blaCTX-M-15 and blaCTX-M-27 genes, respectively, are highly prevalent in ESBL-producing EAEC and ETEC strains. The cephamycinase ACT-20 has also been described for the first time in EAEC and ETEC strains causing TD in patients who had traveled to Central America. The percentages of resistance to azithromycin in EAEC and ETEC isolates from patients to Southeast Asia/India and Africa are above 25%; however, the high concentration of azithromycin reached in the intestinal tract can surpass the MIC of most of azithromycin-resistant strains. Meanwhile, rifaximin is still active against EAEC and ETEC strains, and strains with an MIC of >32 μg/ml were not found. However, it is not recommended as empirical treatment for inflammatory febrile diarrhea due to its nonabsorbable nature.
The preliminary data obtained regarding the prevalence of resistance to quinolones challenge the recommendation of use of this antibiotic in the treatment of TD in patients visiting or coming from the geographical areas studied, especially India and Africa, although further studies must be done in order to elucidate the prevalence of resistance to fluoroquinolones in larger collections of EAEC and ETEC strains causing TD, as well as in other etiological agents of this infectious disease. In addition, it must be taken into account that in vitro susceptibility testing does not always correlate with lack of success in clinical practice (50).
MATERIALS AND METHODS
Bacterial isolates.
EAEC and ETEC clinical isolates causing TD were investigated in this study. The bacterial isolates were collected from 2011 to 2017. These strains were isolated from patients who were travelers and had diarrhea at the time they visited at the tropical medicine unit in our hospital. None of the patients required hospital admission. The stool samples were collected during the acute phase of diarrhea and were processed within 2 h of collection. The stool specimens were cultured for E. coli and other bacterial enteropathogens using conventional methods. Single-colony subcultures of all different colonial morphotypes growing on MacConkey agar were identified by conventional criteria (51). These colonies were tested by PCR to detect EAEC and ETEC, as described elsewhere (52).
Antimicrobial susceptibility testing.
The susceptibilities of EAEC and ETEC against ampicillin, amoxicillin-clavulanic acid, cefotaxime, imipenem, chloramphenicol, tetracycline, co-trimoxazole, nalidixic acid, and ciprofloxacin were determined by disk diffusion following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations. Meanwhile, the MICs of amoxicillin-clavulanic acid, cefotaxime, cefoxitin, and azithromycin were determined by the Etest method, and the MIC of rifaximin was obtained using the microdilution method according to EUCAST guidelines (53). E. coli ATCC 25922 and E. coli ATCC 35218 were used as controls. The Clinical and Laboratory Standards Institute (CLSI) and EUCAST breakpoints were used to define resistance to nalidixic acid and ciprofloxacin, respectively. The breakpoints of azithromycin considered were those described by EUCAST for Salmonella enterica serovar Typhi (MIC, ≤16 μg/ml for wild-type isolates).
Detection of β–lactam and quinolone resistance mechanisms.
A double-disk synergy test was carried out in the cefotaxime-resistant isolates in order to confirm the extended-spectrum β-lactamase (ESBL) carriage (54). The detection of ESBL genes (blaCTX-M, blaSHV, blaOXA, and blaTEM genes) was carried out by PCR and DNA sequencing under previously described conditions (55). In addition, strains resistant to amoxicillin-clavulanic acid were tested for cefoxitin susceptibility by Etest in order to confirm the presence of cephamycinases, which were also detected by PCR and DNA sequencing using specific primers, as described previously (56).
To determine the quinolone resistance mechanisms, mutations in the quinolone resistance-determining region of the gyrA and parC genes were detected by PCR, and sequencing was performed as described elsewhere (57, 58). The purified PCR products visualized in gels were processed for DNA sequencing and analyzed in an automatic DNA sequencer (ABI 377; PerkinElmer, Emeryville, CA, USA) using the BigDye Terminator cycle sequencing kit (version 3.1; PerkinElmer). Detection of the qnr genes screening for the qnrA, qnrB, qnrC, qnrD, and qnrS genes was performed by multiplex PCR using a combination of specific primers (59). Bacterial strains positive for each qnr gene were used as positive controls and were run in each batch of samples tested. Detection of the aac(6′)-Ib-cr gene was performed using specific primers described previously (60).
Plasmid typing and MLST.
Replicon typing was then performed in the strains carrying the blaCTXM genes to determine the potential plasmids carrying this resistance gene; the primers used were designed by Carattoli et al. in 2006 (42) but adapted amplification protocols for commensal and pathogenic E. coli isolates described by Johnson et al. were employed (61), or the PCR-based replicon typing kit was used (Diatheva, Cartoceto, Italy). In the same set of strains, the multilocus sequence typing (MLST) was determined analyzing by amplification seven housekeeping genes (adk, fumC, icd, purA, gyrB, recA, and mdh) (62). The database available at https://enterobase.warwick.ac.uk/ was used for assigning sequence types (STs) and clonal complexes (CCs). Strains carrying plasmids from incompatibility group IncF were further characterized following the plasmid MLST website (https://pubmlst.org/plasmid/) protocol, developed by Keith Jolley and sited at the University of Oxford (63).
Statistical analysis.
Data of the present study are presented as frequencies. The prevalence of resistance was compared to previous data using the binomial test (64). Proportions were compared using a chi-square test or Fisher’s exact test if the application conditions of the chi-square test were not met. Significance was set at P < 0.05. The analysis was carried out using Stata (65).
Supplementary Material
ACKNOWLEDGMENTS
This study received funding from the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (grant REIPI RD16/0016/0010), and was cofinanced by the European Development Regional Fund “A way to achieve Europe.” This work was also supported by award 2017 SGR 0809 from the Agència de Gestió d’Ajuts Universitaris i de Recerca of the Generalitat de Catalunya.
We thank ALFASIGMA S.p.A. for providing us with the rifaximin-α.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01745-18.
REFERENCES
- 1.Leder K, Torresi J, Libman MD, Cramer JP, Castelli F, Schlagenhauf P, Wilder-Smith A, Wilson ME, Keystone JS, Schwartz E, Barnett ED, von Sonnenburg F, Brownstein JS, Cheng AC, Sotir MJ, Esposito DH, Freedman DO, GeoSentinel Surveillance Network. 2013. GeoSentinel surveillance of illness in returned travelers, 2007–2011. Ann Intern Med 158:456–468. doi: 10.7326/0003-4819-158-6-201303190-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagmann SHF, Han PV, Stauffer WM, Miller AO, Connor BA, Hale DC, Coyle CM, Cahill JD, Marano C, Esposito DH, Kozarsky PE, GeoSentinel Surveillance Network. 2014. Travel-associated disease among US residents visiting US GeoSentinel clinics after return from international travel. Fam Pract 31:678–687. doi: 10.1093/fampra/cmu063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steffen R, Hill DR, DuPont HL. 2015. Traveler’s diarrhea: a clinical review. JAMA 313:71–80. doi: 10.1001/jama.2014.17006. [DOI] [PubMed] [Google Scholar]
- 4.Zboromyrska Y, Hurtado JC, Salvador P, Alvarez-Martínez MJ, Valls ME, Mas J, Marcos MA, Gascón J, Vila J. 2014. Aetiology of traveler’s diarrhea: evaluation of a multiplex PCR tool to detect different enteropathogens. Clin Microbiol Infect 20:O753–O759. doi: 10.1111/1469-0691.12621. [DOI] [PubMed] [Google Scholar]
- 5.Jiang ZD, DuPont HL. 2017. Etiology of travelers’ diarrhea. J Travel Med 24:S13–S16. 2017. doi: 10.1093/jtm/tax003. [DOI] [PubMed] [Google Scholar]
- 6.Government of Canada. 2015. Statement on travelers’ diarrhea. Canada.ca https://www.canada.ca/en/public-health/services/travel-health/about-catmat/statement-travellers-diarrhea.html#a55.
- 7.Riddle MS, Connor BA, Beeching NJ, DuPont HL, Hamer DH, Kozarsky P, Libman M, Steffen R, Taylor D, Tribble DR, Vila J, Zanger P, Ericsson CD. 2017. Guidelines for the prevention and treatment of travelers’ diarrhea: a graded expert panel report. J Travel Med 24:S57–S74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kantele A, Laaveri T, Mero S, Vilkman K, Pakkanen SH, Ollgren J, Antikainen J, Kirveskari J. 2015. Antimicrobials increase travelers’ risk of colonization by extended-spectrum beta-lactamase-producing Enterobacteriaceae. Clin Infect Dis 60:837–846. doi: 10.1093/cid/ciu957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrheal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 10.Vila J, Vargas M, Ruiz J, Corachan M, Jimenez De Anta MT, Gascon J. 2000. Quinolone resistance in enterotoxigenic Escherichia coli causing diarrhea in travelers to India in comparison with other geographical areas. Antimicrob Agents Chemother 44:1731–1733. doi: 10.1128/AAC.44.6.1731-1733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila J, Vargas M, Ruiz J, Espasa M, Pujol M, Corachán M, Jiménez de Anta MT, Gascón J. 2001. Susceptibility patterns of enteroaggregative Escherichia coli associated with traveler’s diarrhea: emergence of quinolone resistance. J Med Microbiol 50:996–1000. doi: 10.1099/0022-1317-50-11-996. [DOI] [PubMed] [Google Scholar]
- 12.Zboromyrska Y, Vila J. 2016. Advanced PCR-based molecular diagnosis of gastrointestinal infections: challenges and opportunities. Expert Rev Mol Diagn 16:631–640. doi: 10.1586/14737159.2016.1167599. [DOI] [PubMed] [Google Scholar]
- 13.Aslani MM, Alikhani MY, Zavari A, Yousefi R, Zamani AR. 2011. Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int J Infect Dis 15:e136–e139. doi: 10.1016/j.ijid.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Ali MM, Ahmed SF, Klena JD, Mohamed ZK, Moussa TA, Ghenghesh KS. 2014. Enteroaggregative Escherichia coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. J Infect Dev Ctries 8:589–596. [DOI] [PubMed] [Google Scholar]
- 15.Guerra JA, Romero-Herazo YC, Arzuza O, Gómez-Duarte OG. 2014. Phenotypic and genotypic characterization of enterotoxigenic Escherichia coli clinical isolates from northern Colombia, South America. Biomed Res Int 2014:236260. doi: 10.1155/2014/236260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. 2005. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol 43:755–760. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina AM, Rivera FP, Pons MJ, Riveros M, Gomes C, Bernal M, Meza R, Maves RC, Huicho L, Chea-Woo E, Lanata CF, Gil AI, Ochoa TJ, Ruiz J. 2015. Comparative analysis of antimicrobial resistance in enterotoxigenic Escherichia coli isolates from two paediatric cohort studies in Lima, Peru. Trans R Soc Trop Med Hyg 109:493–502. doi: 10.1093/trstmh/trv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begum YA, Talukder KA, Azmi IJ, Shahnaij M, Sheikh A, Sharmin S, Svennerholm AM, Qadri F. 2016. Resistance pattern and molecular characterization of enterotoxigenic Escherichia coli (ETEC) strains isolated in Bangladesh. PLoS One 11:e0157415. doi: 10.1371/journal.pone.0157415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odetoyin BW, Hofmann J, Aboderin AO, Okeke IN. 2016. Diarrheagenic Escherichia coli in mother-child pairs in Ile-Ife, south western Nigeria. BMC Infect Dis 16:28. doi: 10.1186/s12879-016-1365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendez Arancibia E, Pitart C, Ruiz J, Marco F, Gascon J, Vila J. 2009. Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveler’s diarrhea. J Antimicrob Chemother 64:343–347. doi: 10.1093/jac/dkp178. [DOI] [PubMed] [Google Scholar]
- 21.Kong FYS, Rupasinghe TW, Simpson JA, Vodstrcil LA, Fairley CK, McConville MJ, Hocking JS. 2017. Pharmacokinetics of a single 1g dose of azithromycin in rectal tissue in men. PLoS One 12:e0174372. doi: 10.1371/journal.pone.0174372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sierra JM, Ruiz J, Navia MM, Vargas M, Gascon J, Vila J. 2001. In vitro activity of rifaximin against enteropathogens producing traveler’s diarrhea. Antimicrob Agents Chemother 45:643–644. doi: 10.1128/AAC.45.2.643-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz J, Mensa L, O’Callaghan C, Pons MJ, González A, Vila J, Gascón J. 2007. In vitro antimicrobial activity of rifaximin against enteropathogens causing traveler’s diarrhea. Diagn Microbiol Infect Dis 59:473–475. doi: 10.1016/j.diagmicrobio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Jiang ZD, Ke S, Palazzini E, Riopel L, Dupont H. 2000. In vitro activity and fecal concentration of rifaximin after oral administration. Antimicrob Agents Chemother 44:2205–2206. doi: 10.1128/AAC.44.8.2205-2206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 26.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:9. doi: 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doi Y, Iovleva A, Bonomo RA. 2017. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J Travel Med 24:S44–S51. doi: 10.1093/jtm/taw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woerther P-L, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sole M, Pitart C, Oliveira I, Fàbrega A, Muñoz L, Campo I, Salvador P, Alvarez-Martínez MJ, Gascón J, Marco F, Vila J. 2014. Extended spectrum β-lactamase-producing Escherichia coli faecal carriage in Spanish travelers returning from tropical and subtropical countries. Clin Microbiol Infect 20:O636–O639. doi: 10.1111/1469-0691.12592. [DOI] [PubMed] [Google Scholar]
- 30.Kuenzli E, Jaeger VK, Frei R, Neumayr A, DeCrom S, Haller S, Blum J, Widmer AF, Furrer H, Battegay M, Endimiani A, Hatz C. 2014. High colonization rates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli in Swiss travelers to South Asia–a prospective observational multicentre cohort study looking at epidemiology, microbiology and risk factors. BMC Infect Dis 14:528. doi: 10.1186/1471-2334-14-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaita K, Aoki K, Suzuki T, Nakaharai K, Yoshimura Y, Harada S, Ishii Y, Tachikawa N. 2014. Epidemiology of extended-spectrum β-lactamase producing Escherichia coli in the stools of returning Japanese travelers, and the risk factors for colonization. PLoS One 9:e98000. doi: 10.1371/journal.pone.0098000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lääveri T, Vlot JA, van Dam AP, Häkkinen HK, Sonder GJB, Visser LG, Kantele A. 2018. Extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-PE) among travellers to Africa: destination-specific data pooled from three European prospective studies. BMC Infect Dis 18:341. doi: 10.1186/s12879-018-3245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, Grobusch MP, Lashof AMO, Molhoek N, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. 2017. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travelers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 17:78–85. doi: 10.1016/S1473-3099(16)30319-X. [DOI] [PubMed] [Google Scholar]
- 34.Guiral E, Mendez-Arancibia E, Soto SM, Salvador P, Fabrega A, Gascon J, Vila J. 2011. CTX-M-15-producing enteroaggregative Escherichia coli as cause of travelers’ diarrhea. Emerg Infect Dis 17:1950–1953. doi: 10.3201/eid1710.110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattaway MA, Jenkins C, Ciesielczuk H, Day M, DoNascimento V, Day M, Rodríguez I, van Essen-Zandbergen A, Schink AK, Wu G, Threlfall J, Woodward MJ, Coldham N, Kadlec K, Schwarz S, Dierikx C, Guerra B, Helmuth R, Mevius D, Woodford N, Wain J. 2014. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis 20:1935–1937. doi: 10.3201/eid2011.131845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai L, Wang L, Yang X, Wang J, Gan X, Wang W, Xu J, Chen Q, Lan R, Fanning S, Li F. 2017. Prevalence and molecular characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from diarrheic patients in China. Front Microbiol 8:144. doi: 10.3389/fmicb.2017.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. 2015. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother 70:2757–2762. doi: 10.1093/jac/dkv188. [DOI] [PubMed] [Google Scholar]
- 38.Okeke IN, Wallace-Gadsden F, Simons HR, Matthews N, Labar AS, Hwang J, Wain J. 2010. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One 5:e14093. doi: 10.1371/journal.pone.0014093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chattaway MA, Jenkins C, Rajendram D, Cravioto A, Talukder KA, Dallman T, Underwood A, Platt S, Okeke IN, Wain J. 2014. Enteroaggregative Escherichia coli have evolved independently as distinct complexes within the Escherichia coli population with varying ability to cause disease. PLoS One 9:e112967. doi: 10.1371/journal.pone.0112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumura Y, Pitout JD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, Peirano G, DeVinney R, Bradford PA, Motyl MR, Tanaka M, Nagao M, Takakura S, Ichiyama S. 2016. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis 22:1900–1907. doi: 10.3201/eid2211.160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franz E, Veenman C, van Hoek AH, Husman A de R, Blaak H. 2015. Pathogenic Escherichia coli producing extended-spectrum β-lactamases isolated from surface water and wastewater. Sci Rep 5:14372. doi: 10.1038/srep14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, Whichard JM, Rossolini GM. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg Infect Dis 12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coque TM, Novais Â, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mshana SE, Imirzalioglu C, Hossain H, Hain T, Domann E, Chakraborty T. 2009. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a university hospital in Germany. BMC Infect Dis 9:97. doi: 10.1186/1471-2334-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smet A, Vaes R, Praud K, Doublet B, Daminet S, Cloeckaert A, Haesebrouck F. 2014. New broad-spectrum β-lactamases emerging among Enterobacteriaceae from healthy cats and dogs: a public health concern? Int J Antimicrob Agents 44:81–82. doi: 10.1016/j.ijantimicag.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Ortega A, Oteo J, Aranzamendi-Zaldumbide M, Bartolomé RM, Bou G, Cercenado E, Conejo MC, González-López JJ, Marín M, Martínez-Martínez L, Merino M, Navarro F, Oliver A, Pascual A, Rivera A, Rodríguez-Baño J, Weber I, Aracil B, Campos J. 2012. Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 56:3576–3581. doi: 10.1128/AAC.06393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fàbrega A, Madurga S, Giralt E, Vila J. 2009. Mechanism of action of and resistance to quinolones. Microb Biotechnol 2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pazhani GP, Chakraborty S, Fujihara K, Yamasaki S, Ghosh A, Nair GB, Ramamurthy T. 2011. QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhea in Ahmedabad, India. Indian J Med Res 134:214–223. [PMC free article] [PubMed] [Google Scholar]
- 50.Cavallo J, Mérens A. 2008. Spectre d’activité antibactérien d’un antibiotique et catégorisation clinique. Pathol Biol (Paris) 56:300–304. doi: 10.1016/j.patbio.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 51.Álvarez Martínez M, Buesa Gómez J, Castillo García J, Vila Estape J. 2008. Diagnóstico microbiológico de las infecciones gastrointestinales. Procedimientos en microbiología clínica. Sociedad de Enfermedades Infecciosas y Microbiología Clínica (SEIMC), Barcelona, Spain: https://www.seimc.org/contenidos/documentoscientificos/procedimientosmicrobiologia/seimc-procedimientomicrobiologia30.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-Blázquez J, Vargas M, Nataro JP, Vila J, Gascón J. 2005. Validation of a PCR technique for the detection of enteroagreggative Escherichia coli causing traveler’s diarrhea. Enferm Infecc Microbiol Clin 23:479–481. doi: 10.1157/13078826. [DOI] [PubMed] [Google Scholar]
- 53.EUCAST. 2018. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf. [Google Scholar]
- 54.Calvo J, Cantón R, Fernández-Cuenca F, Mirelis B, Navarro F. 2011. Detección fenotípica de mecanismos de resistencia en gramnegativos. Procedimientos en microbiología clínica. Sociedad de Enfermedades Infecciosas y Microbiología Clínica (SEIMC), Barcelona, Spain. [DOI] [PubMed] [Google Scholar]
- 55.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 56.Chuong L, Van Prachayasittikul V, Isarankura Na Ayudhya C, Lawung R. 2018. Multiplex PCR scheme for variant plasmid mediated class C β-lactamase typing. J Clin Lab Anal 32:e22298. doi: 10.1002/jcla.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vila J, Ruiz J, Marco F, Barcelo A, Goñi P, Giralt E, Jimenez de Anta T. 1994. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother 38:2477–2479. doi: 10.1128/AAC.38.10.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vila J, Ruiz J, Goñi P, De Anta MT. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother 40:491–493. doi: 10.1128/AAC.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 60.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoel PG. 1984. Introduction to mathematical statistics, 5th ed Wiley, New York, NY. [Google Scholar]
- 65.StataCorp. 2017. Stata statistical software: release 15. StataCorp LLC, College Station, TX. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.