Abstract
Bi-allelic inactivation of the VHL gene on chromosome 3p is the characteristic feature in most clear cell renal cell carcinomas (ccRCC). Frequent gene alterations were also identified in SETD2, BAP1 and PBRM1, all of which are situated on chromosome 3p and encode histone/chromatin regulators. The relationship between gene mutation, loss of protein expression and the correlations with clinicopathological parameters is important for the understanding of renal cancer progression. We analyzed PBRM1 and BAP1 protein expression as well as the tri-methylation state of H3K36 as a surrogate marker for SETD2 activity in more than 700 RCC samples. In ccRCC loss of nuclear PBRM1 (68%), BAP1 (40%) and H3K36me3 (47%) expression was significantly correlated with each other, advanced tumor stage, poor tumor differentiation (P < .0001 each), and necrosis (P < .005) Targeted next generation sequencing of 83 ccRCC samples demonstrated a significant association of genetic mutations in PBRM1, BAP1, and SETD2 with absence of PBRM1, BAP1, and HEK36me3 protein expression (P < .05, each). By assigning the protein expression patterns to evolutionary subtypes, we revealed similar clinical phenotypes as suggested by TRACERx Renal. Given their important contribution to tumor suppression, we conclude that combined functional inactivation of PBRM1, BAP1, SETD2 and pVHL is critical for ccRCC progression.
Introduction
Bi-allelic inactivation of the tumor suppressor gene von Hippel–Lindau (VHL) due to chromosome 3p deletion and gene mutation is a hallmark of clear cell RCC (ccRCC), the most common subtype of RCC [1]. Despite its prominent role as multi-adaptor protein that interacts with more than 30 different binding partners involved in many oncogenic processes, functional inactivation of pVHL is not sufficient for tumorigenesis [2]. Recent data suggest that additional genetic and epigenetic events in driver genes act cooperatively with a loss of pVHL function to promote ccRCC progression [3].
Systematic sequencing identified a broad spectrum of genetic lesions, some of which are closely linked to the clinical behavior of ccRCC [4]. Interestingly, three of those genes map, like VHL, to chromosome 3p and encode histone and chromatin regulators SETD2, BAP1, and PBRM1 [5]. PBRM1 encodes a subunit of the SWI/SNF (SWItch/Sucrose Non-Fermentable) transcription-modulating chromatin remodeling complex [6], BAP1 encodes the histone deubiquitinating enzyme BRCA1-associated protein 1 [7], and SETD2 encodes a methyltransferase that specifically trimethylates lysine-36 of histone H3 (H3K36me3) [8]. Mutations in PBRM1 have been detected in 40% of ccRCC, whereas 12 and 19% have sequence alterations in BAP1 and SETD2, respectively [3], [9].
All three proteins are involved in cellular pathways related to tumorigenesis and the high frequency of mutations in their genes support their role as tumor suppressors in ccRCC. Inactivation of PBRM1 has been shown to promote ccRCC cancer cell proliferation and migration [10] as well as to enhance the HIF-response [11]. BAP1 also contributes to chromosome stability by binding the microspherule protein 1 (MCRS1) which plays an essential role in spindle assembly [12]. Mice deficient for either VHL or VHL together with one allele of BAP1 developed multiple lesions spanning from benign cysts to cystic and solid ccRCC [13] suggesting a tumor suppressive cooperation between BAP1 and pVHL. Notably, a cooperative function was also reported for BAP1 and PBRM1 by demonstrating that combined loss of BAP1 and PBRM1 drive ccRCC in mice [14]. SETD2 depletion in ccRCC cells suggests a role in maintaining genome integrity through nucleosome stabilization, suppression of replication stress and the coordination of DNA repair [15]. Finally, SETD2 knockdown in renal primary tubular epithelial cells led to bypass the senescence barrier, facilitating a malignant transformation toward ccRCC [16].
In previous studies the clinical relevance of PBRM1, BAP1 and SETD2 expression was analyzed separately [17], [18], [19], [20] or only two of the three proteins were investigated in parallel [21], [22], [23] with partly controversial results. Here we aimed at investigating PBRM1 and BAP1 expression as well as methylation of H3K36me3 as surrogate marker for SETD2 activity simultaneously by immunohistochemistry (IHC) using tissue microarrays with more than 700 RCC and at correlating the expression data with gene mutation status, prognostic parameters and evolutionary subtypes of ccRCC as recently described [9].
Material and Methods
Cell lines, Cell and Tissue Microarrays
The cell lines used for cell microarrays were the same as previously described [24]. Wild type and transfected cell lines were authenticated by short tandem repeat profiling by IdentiCell (Department of Molecular Medicine, Aarhus University, Hospital Skejby, Aarhus, Denmark) or Microsynth (Balgach, Switzerland). Cell and tissue microarrays (CMA and TMA) were generated using a tissue microarrayer (Beecher Instruments Inc., Sun Prairie, WI, USA).
To analyze the expression of PBRM1, BAP1 and H3K36me3 in RCC, two tissue microarrays (TMA) containing 721 RCC and 44 normal kidney tissue (one punch per sample) were constructed as described [25]. Clinico-pathological information about RCC tissues and patients is listed in Suppl. Table 1. The samples were retrieved from the archives of the Department of Pathology and Molecular Pathology, University Hospital Zurich (Zurich, Switzerland) between the years 1993 to 2013. For each tumor, one representative tumor tissue block, with a minimum of 1 cm tumor diameter, was re-evaluated using hematoxylin and eosin-stained sections. Tumor samples with necrosis and high content of inflammatory cells were excluded. Only those cases with representative tumor regions that contained at least 70% tumor cells were selected for the TMA construction. All tumors were reviewed by two pathologists specialized in uropathology (R.O. and H.M.). Classification, grading and staging was performed according to TNM (8th ed.) [26] and 2016 WHO classification [27]. This study was approved by the local commission of ethics (KEK-ZH-No.2013–0629).
Immunohistochemistry Analysis
CMA and TMA sections (2.5 μm) on glass slides were subjected to immunohistochemical analysis according to the conditions listed in Suppl. Table 2. Tumors were considered positive if tumor cells showed unequivocal strong or weak nuclear expression. The intensity of staining detected in normal proximal tubules and glomeruli as well as in non-tumorous HEK and HK-2 cells was considered as reference for normal/strong staining. A conventional cut-off >5% positive tumor cells was used to prevent false positivity. For staining evaluation, stained slides were scanned using a NanoZoomer (Hamamatsu Photonics, Shizuoka, Japan). Analysis of the immunostainings were done by S.B., R.O. and J.R.
Next Generation Sequencing
DNA extracted from 2–3 punched (0.6 mm diameter) and matched tumor-normal FFPE samples from 83 ccRCC patients was analyzed. These tumors were also part of the TMA cohort. Total DNA was extracted as previously described [28]. The high coverage next generation sequencing data of these biopsies was generated and analyzed in a recent ccRCC study, where all details about the sequencing and computational analysis pipeline can be found [29].
Statistics
Contingency table analysis and Pearson's chi-square tests were used to analyze the associations between PBRM1, BAP1 and SETD2 expression patterns, gene mutations and pathological parameters. Overall survival rates were determined according to the Kaplan–Meier method and analyzed for statistical differences using a log-rank test. A Cox proportional hazard analysis was used to test for independent prognostic information. The statistics were performed with SAS Institute's StatView 5.0 statistical package (Cary, NC, USA) and SPSS Statistics 25 (IBM).
Results
PBRM1, BAP1, SETD2 and H3K36me3 Antibody Testing in RCC Cell Lines
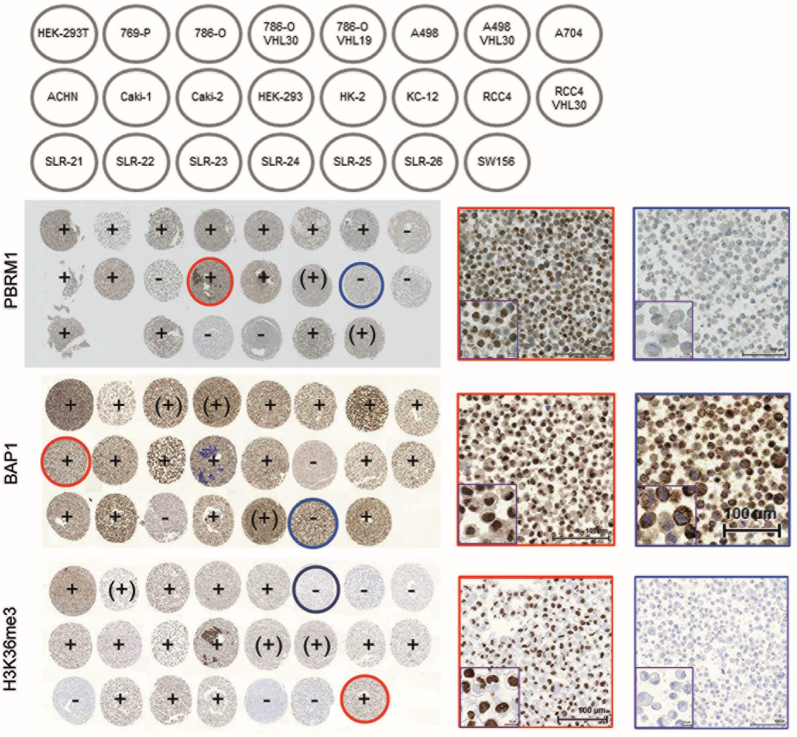
The suitability and specificity of the antibodies against PBRM1, BAP1, SETD2 and H3K36me3 on formalin-fixed material were optimized by testing several immunostaining protocols on a CMA consisting of 20 RCC and three kidney cell lines (Fig. 1). As expected, strong nuclear expression of the four proteins was seen in non-tumorous HEK and HK-2 cells. PBRM1 was not expressed in A704, Caki-2, RCC4, SLR-24, SLR-25 and confirmed previous data [20], [30]. Nuclear and cytoplasmic BAP1 positivity was seen in most of the RCC cell lines including 769-P known to be BAP1 mutated [31] but still expresses the protein. KC-12 and SLR-23 were BAP1 negative in both cellular compartments, whereas SLR-26 was strongly positive only in the cytoplasm. Although SETD2 mutations have been reported for A-498, A-704, and Caki-1 [32], all RCC cell lines were SETD2 positive. An antibody, which specifically binds methylated lysine at amino acid position 36 of histone 3 was used for immunostaining to test SETD2’s activity in our RCC cell lines. H3K36me3 was absent in SETD2 mutated A-498 and A-704 but not in Caki-1 suggesting different effects of SETD2 mutations on SETD2 function in these cell lines. Notably, H3K36me3 was also negative in ccRCC cell lines SLR21, SLR25, and SLR26 with yet unknown SETD2 mutation status. Based on these results we decided to use H3K36me3-specific antibody as surrogate marker for SETD2 activity for TMA analysis.
Figure 1.
Schematic illustration of the cell microarray (CMA) with different RCC and kidney cell lines (top). Immunohistochemical staining of PBRM1, BAP1 and H3K36me3 with representative examples of nuclear positive (circled red) and negative (circled blue) stained cell lines (20- and 60-fold magnification). Core diameter: 0.6 mm.
Expression Frequencies of PBRM1, BAP1 and SETD2 in RCC Subtypes
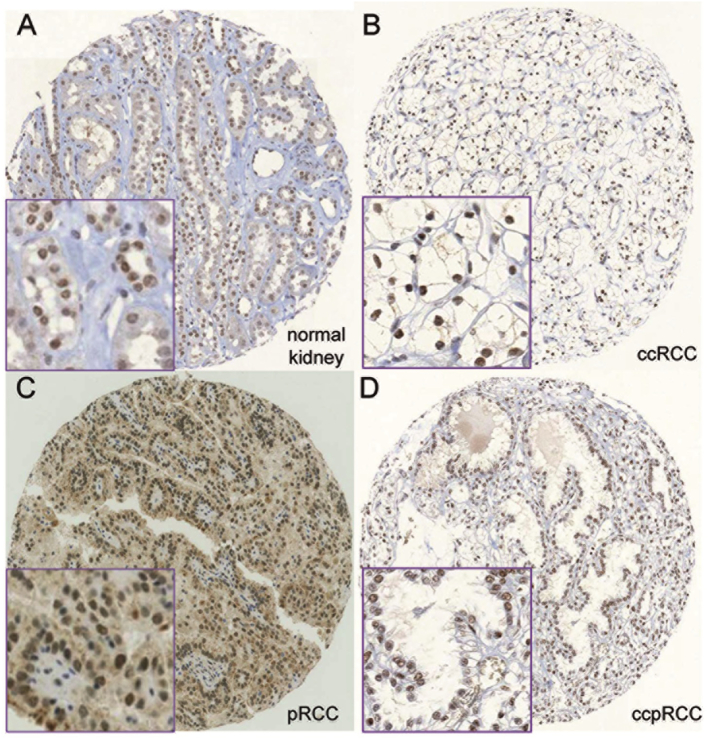
Normal tubular and glomerular cells showed strong staining similar to the expression patterns observed in cell lines without mutations. For all antibodies, we observed renal tumors with strong, weak or absent staining. In most previous studies, calculations were performed by grouping tumors in either negative versus positive or low versus high expression, although different criteria were used for interpreting immunohistochemical staining [17], [18], [19], [21], [33], [34]. Weak staining intensity may be caused by mutations and chromosomal loss of 3p, which may influence protein expression of PBRM1, BAP1, and H3K36me3, but also by the age of paraffin blocks, tissue fixation, and IHC detection protocols, making it difficult to distinguish between genomic driven and artificial reduction of protein expression. Therefore, we decided to keep tumors with weak and strong staining separated for the calculations and used a 3-tierd scoring system (negative, weak, strong staining) for all antibodies. As in our ccRCC set the portion of pT2 and pT4 tumors was very low (9% and 1.6%, respectively), we grouped pT1 and pT2 ccRCC (organ-confined) as well as pT3 and pT4 ccRCC (advanced tumors) for further calculations. Expression frequencies of PBRM1, BAP1 and H3K36me3 in different RCC subtypes are listed in Table 1. Examples of immunostained RCCs as well as normal kidney are illustrated in Fig. 2.
Table 1.
PBRM1, BAP1 and H3K36me3 Expression in RCC Subtypes
| Clear Cell n (%) |
Papillary Type 1 n (%) |
Papillary Type 2 n (%) |
Chromophobe n (%) |
Oncocytoma n (%) |
Clear Cell Papillary n (%) |
|
|---|---|---|---|---|---|---|
| PBRM1 | ||||||
| Negative | 306 (68) | 27 (43.5) | 22 (41.5) | 32 (66.7) | 1 (8.3) | 1 (20) |
| Weak | 96 (21.3) | 19 (30.6) | 17 (32.1) | 10 (20.8) | 2 (16.7) | 3 (60) |
| Strong | 48 (10.7) | 16 (25.8) | 14 (26.4) | 6 (12.5) | 9 (75) | 1 (20) |
| Total | 450 | 62 | 53 | 48 | 12 | 5 |
| BAP1 | ||||||
| Negative | 193 (40.4) | 5 (7.5) | 9 (15.8) | 21 (45.7) | 0 (0) | 1 (20) |
| Weak | 205 (42.9) | 32 (47.8) | 31 (54.4) | 21 (45.7) | 5 (38.5) | 2 (40) |
| Strong | 80 (16.7) | 30 (44.8) | 17 (29.8) | 4 (8.6) | 8 (61.5) | 2 (40) |
| Total | 478 | 67 | 57 | 46 | 13 | 5 |
| H3K36me3 | ||||||
| Negative | 220 (47) | 1 (1.5) | 11 (19.6) | 6 (11.8) | 1 (7.7) | 0 (0) |
| Weak | 124 (26.5) | 3 (4.5) | 9 (16.1) | 11 (21.6) | 2 (15.4) | 2 (40) |
| Strong | 124 (26.5) | 62 (93.9) | 36 (64.3) | 34 (66.6) | 10 (76.9) | 3 (60) |
| Total | 468 | 66 | 56 | 51 | 13 | 5 |
Figure 2.
Representative images of immunohistochemically stained tissue samples: strong nuclear PBRM1 expression in normal kidney (a) and one ccRCC (b); strong nuclear BAP1 expression in one pRCC and strong nuclear H3K36me3 expression in one pccRCC (20- and 60-fold magnification; core diameter: 0.6 mm).
Absence of PBRM1 expression was observed in about two-thirds of ccRCCs and chromophobe RCCs (chRCCs). In contrast, only about 40% of both type 1 and type 2 of the papillary RCC (pRCC), 20% of the clear cell papillary RCCs and 8% of the oncocytomas were PBRM1-negative. Compared to PBRM1, the frequency of BAP1 and H3K36me3 negative tumors was considerably lower in ccRCC (40% and 47%), pRCC (type 1: 7.5% and 1.5%; type 2: 16% and 20%) as well as in chRCC (46% and 12%). Only one of 13 (8%) oncocytomas was H3K36me3 negative and one of 5 clear cell papillary RCC was BAP1 negative.
Correlation of Protein Expression with Tumor Stage and ISUP Grade
By linking our TMA IHC data to pathological parameters we found that loss of BAP1, H3K36me3 and PBRM1 was highly associated with late tumor stage as well as high nuclear differentiation grade (P < .0001, each; Table 2). In advanced stage ccRCC the portion of tumors which lost expression of the three proteins ranged between 50% and 81.5%. Similarly, the majority of grade 4 tumors were negative for BAP1, H3K36me3, and PBRM1 (51.1–83.7%).
Table 2.
Expression frequencies of PBRM1, BAP1 and H3K36me3 in relation to tumor stage and ISUP grade in ccRCC.
| pT1/pT2 n (%) |
pT3/pT4 n (%) |
P-value | Grade 1/2 n (%) |
Grade 3 n (%) |
Grade 4 n (%) |
P-value | |
|---|---|---|---|---|---|---|---|
| BAP1 | |||||||
| Negative | 93 (34.2) | 98 (50) | <0.0001 | 55 (29.1) | 66 (45.2) | 71 (51.1) | <0.0001 |
| Weak | 119 (43.7) | 81 (41.3) | 77 (40.7) | 65 (44.5) | 61 (43.9) | ||
| Strong | 60 (22.1) | 17 (8.7) | 57 (30.2) | 15 (10.3) | 7 (5) | ||
| Total | 272 | 196 | 189 | 146 | 139 | ||
| H3K36me3 | |||||||
| Negative | 104 (39.4) | 115 (59) | <0.0001 | 55 (30.5) | 66 (52.1) | 89 (64) | <0.0001 |
| Weak | 71 (26.9) | 47 (24.1) | 52 (28.9) | 44 (30.1) | 25 (18) | ||
| Strong | 89 (33.7) | 33 (16.9) | 73 (40.6) | 26 (17.8) | 25 (18) | ||
| Total | 264 | 195 | 180 | 146 | 139 | ||
| PBRM1 | |||||||
| Negative | 151 (59) | 150 (81.5) | <0.0001 | 92 (53.2) | 98 (71) | 113 (83.7) | <0.0001 |
| Weak | 67 (26.2) | 25 (13.6) | 48 (27.7) | 30 (21.7) | 18 (13.3) | ||
| Strong | 38 (14.8) | 9 (4.9) | 33 (19.1) | 10 (7.2) | 4 (3) | ||
| Total | 256 | 184 | 173 | 138 | 135 |
Also, loss of PBRM1, H3K36me3, and BAP1 expression was significantly associated with presence of necrosis and, excepting H3K36me3, a high density of tumor infiltrating lymphocytes (Table 3). Although tumor stage and WHO/ISUP grade were highly linked to patient survival (P < .0001, each) (Suppl. Fig. 1 A and B), only loss of PBRM1 and H3K36me3 expression showed a trend to worse outcome, but this did not reach significance (Suppl. Fig. 1 C-E). Multivariate analysis of PBRM1, BAP1 and H3K36me3 expression did not add independent prognostic information (Suppl. Table 3).
Table 3.
PBRM1, BAP1 and H3K36me3 Expression and Presence of Necrosis and Tumor Infiltrating Lymphocytes in ccRCC
| Necrosis n (%) |
P-value | Tumor Infiltrating Lymphocytes n (%) |
|||||
|---|---|---|---|---|---|---|---|
| Absent | Present | Sparse | Moderate | Dense | P-value | ||
| BAP1 | |||||||
| Negative | 130 (38.6) | 61 (44.5) | 0.0023 | 25 (36.8) | 93 (39.7) | 73 (42.4) | 0.0162 |
| Weak | 138 (40.9) | 66 (48.2) | 23 (33.8) | 101 (43.2) | 80 (46.5) | ||
| Strong | 69 (20.5) | 10 (7.3) | 20 (29.4) | 40 (17.1) | 19 (11) | ||
| Total | 337 | 137 | 68 | 234 | 172 | ||
| H3K36me3 | |||||||
| Negative | 139 (42) | 81 (60.5) | 0.0014 | 27 (40.3) | 113 (47.2) | 88 (50.3) | ns |
| Weak | 94 (28.4) | 27 (20.1) | 18 (26.9) | 89 (29.3) | 71 (21.3) | ||
| Strong | 98 (29.6) | 26 (19.4) | 22 (32.8) | 28 (23.6) | 12 (28.4) | ||
| Total | 331 | 134 | 67 | 229 | 169 | ||
| PBRM1 | |||||||
| Negative | 198 (62.7) | 105 (80.8) | 0.0009 | 31 (51.7) | 148 (67.6) | 124 (74.3) | 0.001 |
| Weak | 80 (25.3) | 16 (12.3) | 16 (26.7) | 44 (20.1) | 36 (21.6) | ||
| Strong | 38 (12) | 9 (6.9) | 13 (21.7) | 27 (12.3) | 7 (4.2) | ||
| Total | 316 | 130 | 60 | 219 | 167 | ||
Relationship Among PBRM1, BAP1 and H3K36me3 Expression Patterns in ccRCC
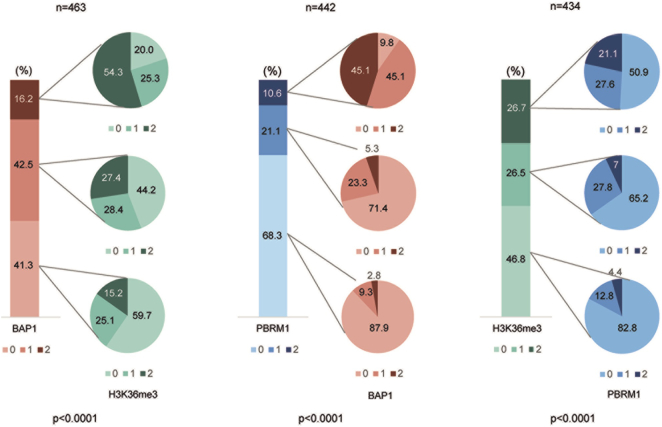
Next, we investigated potential associations of the expression data yielded for BAP1, H3K36me3 and PBRM1 in ccRCC. As illustrated in Fig. 3, the correlations of BAP1, H3K36me3 and PBRM1 expression patterns were highly significant among each other (P < .0001). For example, 88% of PBRM1 negative tumors were also negative for BAP1. Similarly, 83% of tumors with H3K36me3 loss were also deficient for PBRM1, and 60% of BAP1 negative ccRCC had no methylated H3K36.
Figure 3.
Relationships between BAP1, PBRM1 and H3K36me3 expression patterns in ccRCC. 0, 1, 2 refers to negative, weak and strong nuclear expression.
Association of Mutations in PBRM1, BAP1, and SETD2 with Loss of Protein Expression
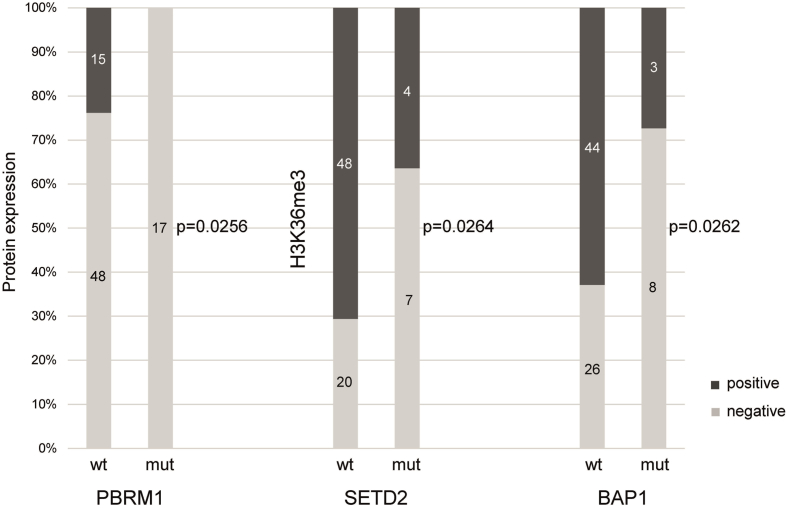
DNA sequencing data including non-silent mutations of PBRM1, BAP1 and SETD2 are summarized in Suppl. Table 4. Seventeen of 83 (20%) ccRCC had PBRM1 mutations and 11 tumors had BAP1 and SETD2 mutations (13%, each). In 4 ccRCC (5%) both PBRM1 and BAP1 were affected, 2 tumors had mutations in PBRM1 and SETD2 (2%), and 1 tumor in BAP1 and SETD2 (1%). None of the analyzed tumors had non-silent mutations in all 3 genes. Sequence alterations that highly likely lead to truncation of the proteins (frameshift, nonsense, splice site) were seen in 15 of 17 (88%; PBRM1), 6 of 11 (56%; BAP1), and 9 of 11 (82%; SETD2) mutated ccRCC. There was no association with tumor grade and stage. We compared also our sequence data with protein expression. As shown in Fig. 4, PBRM1, BAP1, and SETD2 mutations correlated with loss of PBRM1, BAP1 expression and H3K36me3 methylation.
Figure 4.
Correlation of PBRM1, SETD2, and BAP1 mutation and protein expression.
Assigning PBRM1, BAP1 and SETD2 Expression Patterns to Evolutionary ccRCC Subtypes
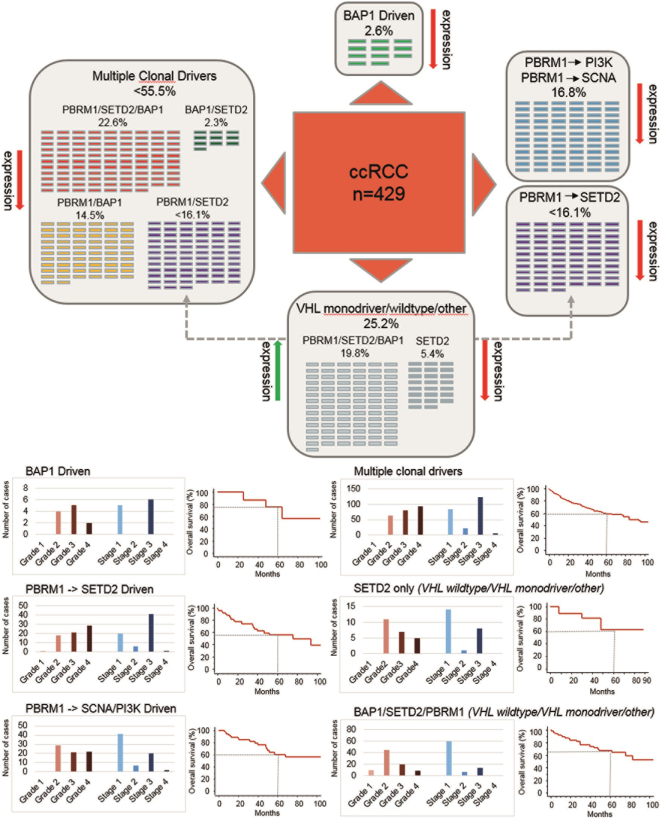
Comparisons of sequencing and expression data obtained from our ccRCC cohorts imply that loss of PBRM1, BAP1 and SETD2 expression is caused by gene mutations but also by additional, yet unknown, molecular and epigenetic mechanisms. As the three proteins are critical key players in five of seven deterministic evolutionary trajectories [9], we attempted to assign our TMA results to the PBRM1, SETD2 and BAP1 driven ccRCC subtypes (Fig. 5A and B). About half of the tumors belonged to the subtype “multiple clonal drivers” in which at least two of the three proteins were negative. This subtype consisted mainly of high grade and late stage ccRCC. The majority of BAP1 driven tumors, which represented the smallest group (2.6%), were characterized by elevated grade and a better survival rate (5-year survival rate: 75%) compared to the other subtypes (about 60%). As we were not able to distinguish between the subtypes “PBRM1- > PI3K” and “PBRM1- > SCNA”, the two subtypes both characterized by early PBRM1 inactivation were combined. This group was enriched for lower grade (40% grade 2) and early stage tumors (69%). The remaining cases consisted of only SETD2 negative ccRCC and tumors in which all three proteins were expressed. We therefore grouped these tumors into “VHL monodriver” and “VHL wildtype” subtype. Notably, the majority of tumors presented at lower grade and early stage.
Figure 5.
A. ccRCC grouped by evolutionary subtypes: BAP1 driven (only BAP1 negative), PBRM1- > SCNA and PBRM1- > PI3K (only PBRM1 negative), Multiple clonal drivers (≥2 BAP1, SETD2 or PBRM1 negative), VHL monodriver/wildtype and other (PBRM1, BAP1 and SETD2 positive as well as only SETD2 negative). PBRM1 and SETD2 negative tumors were assigned to both multiple clonal drivers and PBRM1- > SETD2 subtype (gray arrow). B. Distribution of tumor grade, tumor stage and overall survival per subtype.
Discussion
We provide a comprehensive expression analysis of BAP1, PBRM1 as well as the methylation state of H3K36me3 as surrogate marker for SETD2 activity in RCC cell lines and more than 600 clinically well characterized human RCC tumor specimens. Loss of expression of the three proteins was highest in ccRCC with about 70% for PBRM1, 40% for BAP1, and 50% for H3K36me3. Similar frequencies were obtained for PBRM1 and BAP1 in chRCC. In pRCC, in which 5–10% of the tumors have also PBRM1, SETD2, and BAP1 mutations [35], approximately 40% were PBRM1 negative in both main subtypes type 1 and type 2. Of note, compared to pRCC type a higher deficiency of BAP1 and H3K36me3 was observed in the more aggressive pRCC type 2 (type 1: 8% and 2%; type 2: 16% and 20%). In oncocytoma and clear cell papillary RCC almost all tumors were positive for the three proteins. Furthermore, our results demonstrate that the absence of the three proteins in ccRCC is highly related to each other suggesting overlapping down-regulatory mechanisms of their genes in many of the tumors. Mutations in PBRM1, BAP1 and SETD2 contribute to loss of protein expression. Finally, the significant association of loss of expression of BAP1, SETD2 and PBRM1 with advanced tumor stage and high tumor grade together with their critical role in recently identified evolutionary trajectories [9] emphasizes an important concerted tumor-suppressive role of the three proteins in ccRCC.
By doubling the number of RCCs from 300 to 600 we could confirm the results from a previous study [20] showing that loss of PBRM1 expression is strongly linked to a more aggressive tumor behavior. We further extended our study by analyzing the expression of BAP1 and H3K36me3 and obtained associations, which were very similar to those observed for PBRM1. The strong relationship among the expression profiles of PBRM1, BAP1, and H3K36me3 may be explained by the location of PBRM1, BAP1, and SETD2 on chromosome 3p. As chromosome 3p deletion occurs in about 90% of ccRCC [2], our data imply that in most early stage tumors the remaining alleles of PBRM1, BAP1 and SETD2 are still expressed, albeit to a lower dosage in comparison to normal tubular kidney cells. Following the classical two-hit hypothesis of tumor suppressor genes [36], gene inactivation by mechanisms such as truncating mutations occurs mainly in late stage and high grade tumors.
The fractions of non-expressing tumors in our tumor cohort was generally higher (40% BAP1, 50% H3K36me3, 70% PBRM1) compared to those described in several previous studies, which ranged between 10–50% for BAP1 [19], [21], [22], [37], 14–34% for SETD2 [18], [23], and 31–70% for PBRM1 [20], [21], [23], [38], [39]. The use of different scoring criteria, antibodies, immunostaining protocols, numbers and fixation of the tumors included in the analyses as well as large sections versus tissue microarrays may explain some of the discrepancies. However, regardless of the diverse patient cohorts and strategies used to examine ccRCC, the consistent finding of these studies is that loss of expression of PBRM1, SETD2 and BAP1 is closely linked to tumor progression [17], [18], [19], [23], [40].
Mutations of PBRM1, BAP1 and SETD2 determined in 83 ccRCC revealed their important negative influence on protein expression, but in a considerable fraction of tumors loss or reduction of expression seem to follow mechanisms other than mutations and chromosome 3p loss. Hypermethylation of CpG islands in the gene promoter region were reported for several genes located on chromosome 3p, including VHL [41], [42]. However, no aberrant promoter hypermethylation of PBRM1, BAP1 and SETD2 were found in 50 ccRCC suggesting that silencing of the three genes by methylation is absent or rare in this tumor type [43]. Also, SETD2, BAP1 as well as PBRM1 mutations have been reported in pRCC, which has no chromosome 3p deletions [35]. Therefore, other mechanisms may drive down regulation on the transcriptional or (post-) translational level. Recently, PBRM1 has been reported to be functionally regulated by p53-induced protein degradation in RCC [44]. Whether or not this interaction promotes ccRCC is yet unclear.
Although gene mutations were significantly associated with loss of protein expression in our ccRCC cohort, BAP1 and H3K36me3 were expressed in a small subset of ccRCC despite BAP1 and SETD2 mutations. As shown in previous VHL studies [28], [45], [46], mutations can have different impact on expression, protein function and stabilization. It is therefore conceivable that PBRM1, BAP1, and SETD2 mutations identified in ccRCC, as well as in other cancer types, may also exert diverse effects on protein expression and function.
Inactivation of tumor suppressor genes by genetic or epigenetic events is crucial for tumor progression. In this context, PBRM1, BAP1, and SETD2 mutation and loss of expression was described to be associated with advanced tumor stage, high grade and patient outcome [17], [18], [19], [20], [21], [22], [23], [39], [40], [47], [48], [49]. Surprisingly, PBRM1-deficient tumor clones in mice were more sensitive to T-cell-mediated cytotoxicity compared to their wild-type counterparts [50]. Our study demonstrates a significant association between PBRM1 alterations and a high amount of TIL in ccRCC. This could in part explain a recent observation by Miao et al. showing an increased clinical benefit of immune checkpoint therapy in ccRCC patients with inactivated PBRM1 [51]. This suggests that PBRM1 loss induces changes resulting in an increased susceptibility to immunotherapy. PBRM1 loss-of-function mutations may thus serve as unfavorable prognostic marker but also as favorable predictive marker regarding PD-(L)1 blockade therapy. This finding also helps explain the conflicting data about the prognostic value of PBRM1 in ccRCC.
PBRM1, BAP1 and SETD2 are involved in chromatin remodeling by regulating transcription of genes and modifying histones [6], [7], [8]. Functional in vitro and in vivo analyses strongly suggest that their inactivation leads to increased cell proliferation and loss of genome integrity [10], [12], [15], [16]. It is therefore plausible that loss of PBRM1, BAP1 and SETD2 function is tightly linked to nuclear de-differentiation, which is accompanied by high mitotic rates of tumor cells, and larger tumor size. In this context, it was recently shown that deletion of VHL together with either BAP1 or PBRM1 drives tumor grade [11]. Notably, functional inactivation of the four tumor suppressors pVHL, PBRM1, BAP1 and SETD2 affects spindle orientation, spindle assembly, histone modification, nucleosome stabilization and chromatin remodeling, all of which strongly impair chromosomal stability [8], [12], [15], [52], [53]. The lack of concerted action of pVHL, PBRM1, BAP1 and SETD2 through chromosome 3p loss and additional molecular mechanisms may therefore increase the risk of chromosomal instability, which is characteristic for high-grade ccRCC [52].
Intratumor heterogeneity exists in most of ccRCC and can lead to confounding estimates of gene mutation and protein expression prevalence. Multiregion-NGS procedures demonstrated that in ccRCC the majority of identified driver aberrations derive from spatially separated subclones [4]. In addition, a deterministic nature of subclonal diversification has been identified, which allows ccRCC to be grouped into seven evolutionary subtypes each representing potential prognostic markers [9]. Notably, in five of these subtypes PBRM1, SETD2, and BAP1 mutations play a dominant role as driver genes. These subtypes were defined i) by mutations in PBRM1 followed by SETD2; ii) by PBRM1 mutations followed by alterations in the PI3K/AKT pathway; iii) by mutations in PBRM1 followed by a driver somatic copy number alteration event; iv) by BAP1 mutations as the single driver event with VHL, and v) by tumors in which at least two of the genes BAP1, SETD2, PBRM1 or PTEN where clonally mutated.
In almost all previous immunohistochemical and DNA sequencing studies usually one tissue section and one or very few punches per tumor sample were analyzed. Given the subclonal diversity and its different impact on patient outcome in ccRCC, it becomes evident why the frequencies and clinical relevance of gene mutations and protein expression of BAP1, SETD2, and PBRM1, let alone their subtyping, differ from each other [5], [17], [18], [19], [20], [21], [22], [23], [49]. The different combinations of BAP1, PBRM1 or SETD2 expression changes obtained from our TMA immunostainings support the proposed genetic and clonal evolution features [9] and suggest that the diverse clinical phenotypes of ccRCC may also be identified by protein expression data.
By grouping our tumors into the proposed evolutionary subtypes, we obtained clinical phenotypes that were quite similar to those revealed by comprehensive NGS analysis [9]. We observed on the protein level only 85 of 429 (19.8%) tumors without evidence of BAP1, PBRM1 or SETD2 expression changes. These tumors highly likely belong to “VHL wildtype” and “VHL monodriver” ccRCC as BAP1, PBRM1 and SETD2 are not affected in these subtypes. Notably, we identified one group of only SETD2 inactive ccRCC mainly consisting of lower grade and early stage tumors. This is in line with the hypothesis that a strong niche-specific selection of SETD2 mutant subclones induce a limited clonal growth [9]. Whether or not SETD2 only tumors belong to a different subtype is still unclear.
A BAP1 driven pattern with elevated higher grade tumors was only seen in 11 (2.6%) cases. PBRM1 driven subtypes followed by SETD2, as well as copy number alterations and alterations in the PI3K/AKT pathway were assigned to 69 (16.1%) and 72 (16.8%) tumors, respectively. Finally, the multiple clonal driver subtype represented the largest group consisting of 238 ccRCC (55.5%). In contrast to PBRM1- > SCNA/PI3K driven tumors, both multiple clonal driver and PBRM1- > SETD2 subtypes were enriched with higher grade and late stage ccRCC.
Despite of the different clinical phenotypes obtained from the evolutionary subtypes there was no significant correlation with overall survival. Only patients with BAP1 driven tumors showed a better outcome after 5 years (75% vs 60%).
In summary, we show a strong relationship between the expression profiles of PBRM1, BAP1 and SETD2 in ccRCC suggesting reciprocal synergy effects in the context of tumor suppression. Chromosome 3p deletion, which is very frequent and occurs early in ccRCC, may cause haploinsufficiency of PBRM1, BAP1, SETD2 as well as VHL, which may be a critical step toward tumor development [54]. Additional successive events affecting the second alleles of these genes might explain the formation of recently proposed evolutionary subtypes and the varying prognostic significance obtained for these tumor suppressors in previous studies [17], [18], [19], [20], [21], [23], [39], [40], [44], [47], [48], [55], [56].
The following are the supplementary data related to this article.
Supplementary figures
Supplementary tables 1-3.
Mutations in PBRM1, BAP1, and SETD2 identified in 83 ccRCC patients.
Acknowledgments
Acknowledgements
We thank Susanne Dettwiler and Fabiola Prutek from the Tissue Biobank and Christiane Mittmann from the in situ laboratory of the Department of Pathology and Molecular Pathology, University Hospital Zurich, for generating and immunostaining cell and tissue microarray sections.
Disclosure/Conflict of Interest
The authors report no conflict of interest and have nothing to disclose.
Footnotes
Funding: This study has been funded by the Swiss National Science Foundation (Grant No. 31003A_135792, the Zurich Cancer League (to H.M.), SystemsX.ch (Grant No. SXPHI0_142005) and the Swiss Cancer League (Grant No. KLS-2892-02-2012) (to N.B.).
References
- 1.Moch H. An overview of renal cell cancer: pathology and genetics. Semin Cancer Biol. 2013;23(1):3–9. doi: 10.1016/j.semcancer.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Frew IJ, Moch H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu Rev Pathol. 2015;10:263–289. doi: 10.1146/annurev-pathol-012414-040306. [DOI] [PubMed] [Google Scholar]
- 3.Riazalhosseini Y, Lathrop M. Precision medicine from the renal cancer genome. Nat Rev Nephrol. 2016;12(11):655–666. doi: 10.1038/nrneph.2016.133. [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46(3):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 6.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 7.Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun. 2016;7:10292. doi: 10.1038/ncomms10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70(11):4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 9.Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, O'Brien T, Lopez JI, Watkins TBK, Nicol D. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx Renal. Cell. 2018;173(3):595–610. doi: 10.1016/j.cell.2018.03.043. [e511] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao W, Li W, Xiao T, Liu XS, Kaelin WG., Jr. Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc Natl Acad Sci U S A. 2017;114(5):1027–1032. doi: 10.1073/pnas.1619726114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J, Ma J, Li W, Mo R, Zhang P, Gao K, Jin X, Xiao J, Wang C, Fan J. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett. 2015;369(1):167–174. doi: 10.1016/j.canlet.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang SS, Gu YF, Wolff N, Stefanius K, Christie A, Dey A, Hammer RE, Xie XJ, Rakheja D, Pedrosa I. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A. 2014;111(46):16538–16543. doi: 10.1073/pnas.1414789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu YF, Cohn S, Christie A, McKenzie T, Wolff N, Do QN, Madhuranthakam AJ, Pedrosa I, Wang T, Dey A. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 2017;7(8):900–917. doi: 10.1158/2159-8290.CD-17-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanu N, Gronroos E, Martinez P, Burrell RA, Yi Goh X, Bartkova J, Maya-Mendoza A, Mistrik M, Rowan AJ, Patel H. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34(46):5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Kluiver J, Osinga J, Westers H, van Werkhoven MB, Seelen MA, Sijmons RH, van den Berg A, Kok K. Functional studies on primary tubular epithelial cells indicate a tumor suppressor role of SETD2 in clear cell renal cell carcinoma. Neoplasia. 2016;18(6):339–346. doi: 10.1016/j.neo.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapur P, Christie A, Raman JD, Then MT, Nuhn P, Buchner A, Bastian P, Seitz C, Shariat SF, Bensalah K. BAP1 immunohistochemistry predicts outcomes in a multi-institutional cohort with clear cell renal cell carcinoma. J Urol. 2014;191(3):603–610. doi: 10.1016/j.juro.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Guo R, Zhang X, Liang Y, Kong F, Wang J, Xu Z. Loss of SETD2, but not H3K36me3, correlates with aggressive clinicopathological features of clear cell renal cell carcinoma patients. Biosci Trends. 2017;11(2):214–220. doi: 10.5582/bst.2016.01228. [DOI] [PubMed] [Google Scholar]
- 19.Minardi D, Lucarini G, Milanese G, Di Primio R, Montironi R, Muzzonigro G. Loss of nuclear BAP1 protein expression is a marker of poor prognosis in patients with clear cell renal cell carcinoma. Urol Oncol. 2016;34(8):338. doi: 10.1016/j.urolonc.2016.03.006. [e311–338] [DOI] [PubMed] [Google Scholar]
- 20.Pawlowski R, Muhl SM, Sulser T, Krek W, Moch H, Schraml P. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer. 2013;132(2):E11–E17. doi: 10.1002/ijc.27822. [DOI] [PubMed] [Google Scholar]
- 21.Joseph RW, Kapur P, Serie DJ, Parasramka M, Ho TH, Cheville JC, Frenkel E, Parker AS, Brugarolas J. Clear cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J Urol. 2016;195(1):180–187. doi: 10.1016/j.juro.2015.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur P, Pena-Llopis S, Christie A, Zhrebker L, Pavia-Jimenez A, Rathmell WK, Xie XJ, Brugarolas J. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14(2):159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Dulaimi E, Devarajan K, Parsons T, Wang Q, Liao L, Cho EA, O'Neill R, Solomides C, Peiper SC. Immunohistochemistry successfully uncovers intratumoral heterogeneity and widespread co-losses of chromatin regulators in clear cell renal cell carcinoma. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struckmann K, Mertz K, Steu S, Storz M, Staller P, Krek W, Schraml P, Moch H. pVHL co-ordinately regulates CXCR4/CXCL12 and MMP2/MMP9 expression in human clear-cell renal cell carcinoma. J Pathol. 2008;214(4):464–471. doi: 10.1002/path.2310. [DOI] [PubMed] [Google Scholar]
- 25.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 26.Paner GP, Stadler WM, Hansel DE, Montironi R, Lin DW, Amin MB. Updates in the eighth edition of the tumor-node-metastasis staging classification for urologic cancers. Eur Urol. 2018;73(4):560–569. doi: 10.1016/j.eururo.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol. 2016;70(1):93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Razafinjatovo C, Bihr S, Mischo A, Vogl U, Schmidinger M, Moch H, Schraml P. Characterization of VHL missense mutations in sporadic clear cell renal cell carcinoma: hotspots, affected binding domains, functional impact on pVHL and therapeutic relevance. BMC Cancer. 2016;16:638. doi: 10.1186/s12885-016-2688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore AL, Kuipers J, Singer J, Burcklen E, Schraml P, Beisel C, Moch H, Beerenwinkel N. Intra-tumor heterogeneity and clonal exclusivity in renal cell carcinoma. bioRxiv. 2018 [Google Scholar]
- 30.Murakami A, Wang L, Kalhorn S, Schraml P, Rathmell WK, Tan AC, Nemenoff R, Stenmark K, Jiang BH, Reyland ME. Context-dependent role for chromatin remodeling component PBRM1/BAF180 in clear cell renal cell carcinoma. Oncogene. 2017;6(1) doi: 10.1038/oncsis.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cell cancer research. Mol Cancer. 2016;15(1):83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckel-Passow JE, Serie DJ, Cheville JC, Ho TH, Kapur P, Brugarolas J, Thompson RH, Leibovich BC, Kwon ED, Joseph RW. BAP1 and PBRM1 in metastatic clear cell renal cell carcinoma: tumor heterogeneity and concordance with paired primary tumor. BMC Urol. 2017;17(1):19. doi: 10.1186/s12894-017-0209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Fu Q, An H, Chang Y, Zhang W, Zhu Y, Xu L, Xu J. Decreased expression of SETD2 predicts unfavorable prognosis in patients with nonmetastatic clear-cell renal cell carcinoma. Medicine (Baltimore) 2015;94(45) doi: 10.1097/MD.0000000000002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research N, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374(2):135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Park JY, Park MR, Hwang KC, Park KK, Park C, Cho SK, Lee HC, Song H, Park SB. Developmental arrest of scNT-derived fetuses by disruption of the developing endometrial gland as a result of impaired trophoblast migration and invasiveness. Dev Dyn. 2011;240(3):627–639. doi: 10.1002/dvdy.22568. [DOI] [PubMed] [Google Scholar]
- 38.Ho TH, Kapur P, Joseph RW, Serie DJ, Eckel-Passow JE, Parasramka M, Cheville JC, Wu KJ, Frenkel E, Rakheja D. Loss of PBRM1 and BAP1 expression is less common in non-clear cell renal cell carcinoma than in clear cell renal cell carcinoma. Urol Oncol. 2015;33(1):23. doi: 10.1016/j.urolonc.2014.10.014. [e29–23 e14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SH, Park WS, Park EY, Park B, Joo J, Joung JY, Seo HK, Lee KH, Chung J. The prognostic value of BAP1, PBRM1, pS6, PTEN, TGase2, PD-L1, CA9, PSMA, and Ki-67 tissue markers in localized renal cell carcinoma: A retrospective study of tissue microarrays using immunohistochemistry. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0179610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam SJ, Lee C, Park JH, Moon KC. Decreased PBRM1 expression predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Urol Oncol. 2015;33(8):340. doi: 10.1016/j.urolonc.2015.01.010. [e349–316] [DOI] [PubMed] [Google Scholar]
- 41.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91(21):9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21(45):6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 43.Ibragimova I, Maradeo ME, Dulaimi E, Cairns P. Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2, KDM6A and other chromatin-modifying genes is absent or rare in clear cell RCC. Epigenetics. 2013;8(5):486–493. doi: 10.4161/epi.24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macher-Goeppinger S, Keith M, Tagscherer KE, Singer S, Winkler J, Hofmann TG, Pahernik S, Duensing S, Hohenfellner M, Kopitz J. PBRM1 (BAF180) protein is functionally regulated by p53-induced protein degradation in renal cell carcinomas. J Pathol. 2015;237(4):460–471. doi: 10.1002/path.4592. [DOI] [PubMed] [Google Scholar]
- 45.Rechsteiner MP, von Teichman A, Nowicka A, Sulser T, Schraml P, Moch H. VHL gene mutations and their effects on hypoxia inducible factor HIFalpha: identification of potential driver and passenger mutations. Cancer Res. 2011;71(16):5500–5511. doi: 10.1158/0008-5472.CAN-11-0757. [DOI] [PubMed] [Google Scholar]
- 46.von Teichman A, Storz M, Dettwiler S, Moch H, Schraml P. Whole genome and transcriptome amplification: practicable tools for sustainable tissue biobanking? Virchows Arch. 2012;461(5):571–580. doi: 10.1007/s00428-012-1315-y. [DOI] [PubMed] [Google Scholar]
- 47.da Costa WH, Rezende M, Carneiro FC, Rocha RM, da Cunha IW, Carraro DM, Guimaraes GC, de Cassio Zequi S. Polybromo-1 (PBRM1), a SWI/SNF complex subunit is a prognostic marker in clear cell renal cell carcinoma. BJU Int. 2014;113(5b):E157–E163. doi: 10.1111/bju.12426. [DOI] [PubMed] [Google Scholar]
- 48.Kim JY, Lee SH, Moon KC, Kwak C, Kim HH, Keam B, Kim TM, Heo DS. The impact of PBRM1 expression as a prognostic and predictive marker in metastatic renal cell carcinoma. J Urol. 2015;194(4):1112–1119. doi: 10.1016/j.juro.2015.04.114. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research N Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrari de Andrade L., Tay R.E., Luoma A.M., Tsoucas D., Qiu X., Lim K., Rao P. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–775. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol. 2009;11(8):994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 53.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue K, Fry EA. Haploinsufficient tumor suppressor genes. Adv Med Biol. 2017;118:83–122. [PMC free article] [PubMed] [Google Scholar]
- 55.Hogner A, Krause H, Jandrig B, Kasim M, Fuller TF, Schostak M, Erbersdobler A, Patzak A, Kilic E. PBRM1 and VHL expression correlate in human clear cell renal cell carcinoma with differential association with patient's overall survival. Urol Oncol. 2018;36(3):94. doi: 10.1016/j.urolonc.2017.10.027. [e1–94.e14] [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Liu L, Qu Y, Xi W, Xia Y, Bai Q, Xiong Y, Long Q, Xu J, Guo J. Prognostic value of SETD2 expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. J Urol. 2016;196(5):1363–1370. doi: 10.1016/j.juro.2016.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables 1-3.
Mutations in PBRM1, BAP1, and SETD2 identified in 83 ccRCC patients.