Abstract
Background
We previously found that loss of lncRNA-AZIN2 splice variant (AZIN2-sv) increases cardiomyocyte (CM) proliferation and attenuates adverse ventricular remodelling post-myocardial infarction (MI). However, whether inhibition of AZIN2-sv can simultaneously induce angiogenesis and thus improve prognosis after MI is unclear.
Methods
We used in situ hybridization and quantitative PCR to determine AZIN2-sv expression in endothelial cells. Knockdown and overexpression were performed to detect the role of AZIN2-sv in endothelial cell function, angiogenesis and prognosis after MI. RNA pulldown, RNA immunoprecipitation and luciferase reporter assays were used to determine the interaction with talin1 (Tln1) protein and miRNA-214 (miR-214). DNA pulldown and chromatin immunoprecipitation (ChIP) assays were used to study AZIN2-sv binding to upstream transcription factors.
Findings
AZIN2-sv was enriched in cardiac endothelial cells. The loss of AZIN2-sv reduced endothelial cell apoptosis and promoted endothelial sprouting and capillary network formation in vitro. Moreover, in vivo, the loss of AZIN2-sv induced angiogenesis and improved cardiac function after MI. Mechanistically, AZIN2-sv reduced Tln1 and integrin β1 (ITGB1) protein levels to inhibit neovascularization. AZIN2-sv activated the ubiquitination-dependent degradation of Tln1 mediated by proteasome 26S subunit ATPase 5 (PSMC5). In addition, AZIN2-sv could bind to miR-214 and suppress the phosphatase and tensin homologue (PTEN)/Akt pathway to inhibit angiogenesis. With regard to the upstream mechanism, Bach1, a negative regulator of angiogenesis, bound to the promoter of AZIN2-sv and increased its expression.
Interpretation
Bach1-activated AZIN2-sv could participate in angiogenesis by promoting the PSMC5-mediated ubiquitination-dependent degradation of Tln1 and blocking the miR-214/PTEN/Akt pathway. Inhibition of AZIN2-sv induced angiogenesis and myocardial regeneration simultaneously, thus, AZIN2-sv could be an ideal therapeutic target for improving myocardial repair after MI.
Fund
National Natural Science Foundations of China.
Keywords: AZIN2-sv, Angiogenesis, Tln1, Ubiquitination
Graphical abstract
Research in context.
Evidence before this study
We previously reported that loss of lncRNA-AZIN2 splice variant (AZIN2-sv) could induce cardiomyocyte (CM) proliferation and attenuate adverse ventricular remodelling post-myocardial infarction (MI). Notably, our preliminary results found that the inhibition of AZIN2-sv caused an increase in cardiac capillary density. These evidences suggested a multifaceted regulatory role of AZIN2-sv loss in post-MI repair through coordinated cardiac angiogenesis induction and CM regeneration promotion.
Added value of this study
Here, we demonstrate that AZIN2-sv was abundant in cardiac endothelial cells (ECs). The loss of AZIN2-sv facilitated EC sprouting and capillary network formation and induced angiogenesis after MI, improving subsequent prognosis after MI. Mechanically, AZIN2-sv promotes the ubiquitination-dependent degradation of talin1 (Tln1) mediated by PSMC5, which reduces integrin β1 (ITGB1) protein levels to inhibit neovascularization. Meanwhile, AZIN2-sv binds to miR-214 and suppresses the phosphatase and tensin homologue (PTEN)/Akt pathway to inhibit angiogenesis. Bach1, a negative regulator of angiogenesis, was found to bind to the AZIN2-sv promoter and increase its expression.
Implication of all the available evidence
Our study sheds light on an important myocardial repair effect of AZIN2-sv in regulating both CM regeneration and angiogenesis after MI. AZIN2-sv involvement in angiogenesis and CM regeneration may be an ideal target for facilitating myocardial repair after MI.
Alt-text: Unlabelled Box
1. Introduction
An orchestrated interaction between myocardial hyperplasia and endothelial activation to replenish cardiomyocytes (CMs) and induce angiogenesis is required for functional repair and improved prognosis after myocardial injury, including myocardial infarction (MI), viral myocarditis and primary cardiomyopathy, etc. [[1], [2], [3]]. Currently, studies of myocardial regeneration in the repair process after myocardial injury mainly focus on gene-targeted interventions [4]. Emerging evidence indicates that non-coding RNAs (ncRNAs) are responsible for specialized biological processes during cardiac regeneration and repair, which may provide a new opportunity for therapeutic interventions via regenerative medicine in the heart [4,5]. Many studies have explored ncRNAs that promote CM regeneration in terms of cell cycle re-entry [6], CM apoptosis reduction [7] and heart ageing delay [8]. Indeed, several miRNAs have been shown to have potential to improve myocardial repair by inducing angiogenesis after MI [9,10]. However, except for miRNAs, ncRNAs that could facilitate the improvement of myocardial injury by allowing cardiomyogenesis and angiogenesis together, as well as the subsequent angiogenic signals with cardiac regenerative capacity, have not been explored.
Among the numerous non-coding genes, long non-coding RNAs (lncRNAs) are increasingly showing the potential to promote both myocardial regeneration and endothelial activation according to their role in cell proliferation [11], cardiac development [12,13], and blood vessel formation [14]. The role of certain lncRNAs in improving endothelial function and inducing angiogenesis has been identified, such as lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and lncRNA MANTIS in regulating endothelial migration and vascular sprouting [15,16]. In our previous work [17], we found that loss of the cardiac-specific lncRNA-AZIN2 splice variant (AZIN2-sv) promoted CM regeneration through the activation of the PI3K/Akt pathway, and intriguingly, that the inhibition of AZIN2-sv caused an increase in capillary density. Mechanistically, mass spectrometry results have showed that talin1 (Tln1) may be a downstream binding protein of AZIN2-sv, which is considered as a key regulator in mediating endothelial migration and spreading. In the light of these findings, we hypothesized that loss of AZIN2-sv may have the potential to induce endothelial activation and angiogenesis to improve the prognosis of myocardial injury while regulating CM regeneration.
In the present study, we aimed to verify the role of AZIN2-sv in CM regeneration via endothelial activation, angiogenesis and subsequent prognosis improvement after MI, as well as the detailed underlying mechanism. Our results shed light on a novel molecular target that participates in myocardial repair by promoting both CM regeneration and angiogenesis after MI.
2. Materials and methods
Human heart samples were collected from patients undergoing open surgical repair according to protocols approved by the Research Ethics Committees of Zhongshan People's Hospital. All procedures complied with the Declaration of Helsinki. The animal experiments in this study were approved by the Animal Research Committee at Southern Medical University, and all procedures were in accordance with the Institutional Guidelines for Animal Research and the Guide for the Care and Use of Laboratory Animals published by the US NIH (2011) [18]. All methods are described in detail in the supplemental material.
2.1. Cell culture and transfection
AC16 human CM-like cells, human cardiac fibroblasts (HCFBs) and human umbilical vein endothelial cells (HUVECs) were donated by Mingkong Bio Co. Ltd. (Guangzhou, China). The cells were incubated in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% foetal bovine serum (Gibco, Life Technologies, Australia) at 37 °C with 5% CO2. Adenoviral vectors harbouring plasmids or 4-in-1 shRNA (Vigene Biosciences, Shandong, China) were added to cells at a multiplicity of infection (MOI) of 100 to overexpress or inhibit AZIN2-sv. The RNA interference target sequences for AZIN2-sv are in Supplemental Table S1. Scrambled RNA was used as the control. After 24 h, cells were maintained in normal fresh medium for 48 h before experiments and subsequent analyses. Small interfering RNAs (siRNAs) against PSMC5 and pcDNA 3.1-Bach1 were generated by Kidan Bio Co. Ltd. (Guangzhou, China). The interference sequences are provided in Supplemental Table S1. Transfections were performed with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Bridgewater, NJ, USA) according to the manufacturer's instructions.
2.2. Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed as previously described [20]. Total RNA was extracted from CMs using an E.Z.N.A.® Total RNA Kit II (Norcross, GA, USA). Related mRNAs were quantified with the SYBR Green PCR Kit (Takara) and the LightCycler 480 II System (Roche Diagnostics, Basel, Switzerland). The primer sequences are provided in Supplemental Table S2.
2.3. In situ hybridization
AZIN2-sv expression in heart and other organs was detected by in situ hybridization. Tissue sections were dewaxed in xylene, rehydrated through an ethanol gradient and then treated with 3% H2O2 for 10 min. Subsequently, the sections were treated with pepsin diluted in 3% fresh citrate buffer at 37 °C for 30 min and then washed. Then, hybridization with DIG-labelled probes (probe sense: Bis-P22758; Bersin Bioscience Co. Ltd., Guangzhou, China) was performed overnight at 37 °C after pre-hybridization using 20 μL of pre-hybridization solution for 2 h at 37 °C. The sections were subjected to high-stringency washes with 2× SSC, 0.5× SSC and 0.2× SSC for 5, 15 and 15 min, respectively, at 37 °C. Afterwards, the sections were incubated in blocking solution for 30 min at 37 °C and then incubated with alkaline phosphatase (AP)-conjugated sheep anti-DIG Fab fragments for 60 min at room temperature. Positive staining of AZIN2-sv was observed by adding BM purple AP substrate (Roche, Basel, Switzerland) according to the manufacturer's instructions and was measured digitally using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD).
2.4. RNA fluorescence in situ hybridization
RNA fluorescence in situ hybridization (FISH) was performed as previously described [20]. Slides were mounted and analysed using a Carl Zeiss LSM880 confocal microscope (Zeiss Corporation, Germany).
2.5. Flow cytometry
Cells in single-cell suspensions at a density of 1 × 106/mL after digestion in trypsin without EDTA were transfected for 24 h. Cell apoptosis was analysed by flow cytometry according to the instructions of the Annexin V/PI kit, and the percentage of apoptotic cells was determined in triplicate (%). Median Fluorescence Intensity (MFI) was used to represent EdU activity.
2.6. Tube-formation and spheroid-formation assays
Transfection of cultured cells was performed as previously mentioned. Matrigel Growth Factor Reduced (BD) Basement Membrane Matrix was prepared and incubated with 1.5 × 104 HUVECs in EBM and 1% foetal calf serum (FCS) for 4 h. After fixing the cells with 4% PFA, images of tube formation were obtained on a Zeiss AxioVision microscope (Jena, Germany). The cumulative length of all sprouts of each spheroid or the maximal distance of the migrated cells were used to quantify HUVEC spheroids. Approximately 10 spheroids were analysed for each experiment.
2.7. Scratch wound (migration) assay
Transfected HUVECs were cultured in EBM-2 at 37 °C and 5% CO2. Scratches in the cell monolayer were generated with a 200-μL tip, and the cells were imaged at 0, 6, 24 and 48 h with a Zeiss Axiovert 135 microscope. Subsequently, the distance between cell fronts was measured with an AxioVision documentation system (Zeiss).
2.8. Rat aortic ring assay
Rat aortic ring assays were performed as previously described [21]. Aortas were isolated from Sprague-Dawley rats and cut into approximately 1-mm-long rings after cleaning the fibroadipose tissue. The aortic rings were placed into Growth Factor Reduced Matrigel-coated wells and then sealed with a 100-μL overlay of Matrigel Medium 200 containing with and without VEGF. The wells were transfected with AZIN2-sv and incubated at 37 °C/5% CO2 for 6 days. At the end of the experiment, the formed microvessel sprouting were fixed and imaged under a dissecting microscope. Three independent experiments were performed.
2.9. Western blot analysis
Western blot analysis was performed as previously reported [22]. The primary antibodies used were against Tln1 (1:100, ab157808; Abcam), integrin β1 (ITGB1) (1:200, sc-374429; Santa Cruz) and PSMC5 (1:50, ab178681; Abcam). GAPDH (1:200, sc-25778; Santa Cruz) was used as the loading control. Alexa Fluor 480 (1:10,000, ab175772; Abcam) was used as the secondary antibody. Immunoreactive bands were visualized with Odyssey Software (version 1.2; LI-COR, Lincoln, NE, USA). Protein expression was measured using ImageJ Analysis software (NIH, Bethesda, MD, USA).
2.10. Immunoprecipitation
Cells were homogenized in 1× RIPA buffer supplemented with protease/phosphatase inhibitors. The lysates were cleared by centrifugation, and the supernatants were prepared for immunoblotting or immunoprecipitation with the antibodies described above. Immunoblotting signals were detected using Odyssey Software (version 1.2; LI-COR, Lincoln, NE, USA).
2.11. Pulldown assay
The probes of AZIN2-sv and its antisense RNA for RNA pulldown were designed as previously described [20]. For DNA pulldown, the probes were synthesized by Saicheng Bio Co. Ltd., Guangzhou, China. Biotinylated probes were mixed with proteins isolated from HUVECs and incubated with streptavidin-coated magnetic beads (SA10004; Invitrogen). RNase-free BSA and yeast tRNA (Sigma, Shanghai, China) were used to prevent non-specific binding of protein complexes. Finally, proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and the specific bands were extracted, followed by western blot analysis.
2.12. RNA immunoprecipitation (RIP)
RIP experiments were performed with the Magna RIP™ RNA-binding protein Immunoprecipitation Kit (Millipore, Stafford, VA, USA) according to the manufacturer's protocol. Anti-Tln1 antibody (1:100, ab157808; Abcam) and anti-PSMC5 antibody (1:50, ab178681; Abcam) were used to coprecipitate RNA. RNA quantity was detected by qRT-PCR.
2.13. Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed according to the EZ-CHIP kit (Millipore, Temecula, CA, USA). Anti-Bach1 (1:100, ab49657; Abcam) antibodies were used to precipitate the DNA-protein complexes. The immunoprecipitated DNA was examined by PCR. Primers specific for the AZIN2-sv promoter containing the E-box were 5′-GAGATGAGTGAGCAG-3′(forward) and 5′- CTGCTGGGCTGGCGGGGCGCA GG-3′ (reverse).
2.14. Luciferase activity assay
The pGL3-WT luciferase reporter plasmid was generated by ligating oligonucleotides.
containing a wild-type (WT) AZIN2-sv promoter into the vector pGL3-basic (Promega). The pGL3-Mut plasmid with a mutant target site in the E-box was synthesized by Kidan Bio Co. Ltd. (Guangzhou, China). Cells were seeded in 24-well plates (1 × 105/well) and transfected with pGL3-basic, pGL3-WT, or pGL3-Mut using Lipofectamine 2000 (Invitrogen). The pcDNA 3.1-Bach1 was co-transfected into HUVECs with the luciferase constructs as described above. The miR-214 mimic was co-transfected into HUVECs with AZIN2-sv-wt and AZIN2-sv-mut as previously described [17]. After 48 h, firefly and Renilla luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) and each well had three replicates.
2.15. Immunofluorescence
Immunofluorescence staining was performed as previously described [23]. Frozen sections of cardiac tissues (3 mm) were fixed with acetone, permeabilized with 0.2% Triton X-100, blocked in 3% BSA, and incubated with the desired primary antibodies against cTnT (1:200, ab45932; Abcam) and CD105 (1:200, ab11414;Abcam). After incubation at 4 °C overnight, the sections were incubated with Alexa Fluor 488 and 647 dye-conjugated secondary antibodies (1:200, ab150117 and ab150075, respectively; Abcam) for 2 h at room temperature. For terminal transferase-mediated dUTP nick end-labelling (TUNEL) assays, the sections were stained using fluorescein-conjugated TUNEL reagent in the In Situ Cell Death Detection Kit (Roche). DAPI was used to stain the nuclei. Positive cells were counted from five randomly selected fields in non-consecutive sections using confocal microscopy (Carl Zeiss, LSM880).
2.16. Histology and immunohistochemistry
Pathological examination was performed as previously described [24]. Fourteen days after MI, samples of skin, brain, liver, spleen, muscle and lung were fixed with 10% formalin, dehydrated and embedded in paraffin. Then, 4-um-thick sections were cut and stained with haematoxylin and eosin. For immunohistochemical staining, the slides were incubated at 4 °C overnight with primary antibodies and for 30 min with a biotinylated secondary antibody, followed by incubation in a horseradish peroxidase-labelled streptavidin solution. Then, the slides were stained with diaminobenzidine and counterstained with haematoxylin. The primary antibodies used were aSMA (1:1000, ab179467, Abcam), CD34 (1:1000, ab25124, Abcam), and vWF (1:1000, ab6994, Abcam).
2.17. MI model establishment and cardiac function evaluation
MI was induced in rats by ligation of the left anterior descending coronary artery as previously described [20]. Adult Sprague-Dawley (SD) rats (180–200 g) were anaesthetized with intraperitoneal injections of 3% pentobarbital sodium (40 mg/kg). For sham operation, an analogous surgical operation was performed without occlusion of the coronary artery. Immediately post-ligation, AAV9 vectors (1 × 1011 viral genome particles per animal) containing shAZIN2-sv or shITGB1 were injected into the myocardium bordering the infarct zone (single injection) using an insulin syringe with a 30-gauge needle. The RNA interference target sequences are provided in Supplemental Table S1. The animal chests were closed, and the rats were kept warm for several minutes until recovery. According to previous study [25], the perfusion score was assessed by PET-CT (Siemens, Munich, Germany) 8 weeks after MI. The left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) were measured by M-mode echocardiography. All rats were sacrificed by an overdose of pentobarbital sodium (150 mg/kg intraperitoneal injection) or cervical dislocation at the specified time points.
2.18. Statistical analysis
Quantitative data are expressed as the mean ± SEM. Differences between two groups were evaluated using two-tailed unpaired Student's t-tests, and multiple comparisons were performed by one-way analysis of variance (ANOVA), followed by Bonferroni tests in the case of equal variances and Dunnett's C test in the case of unequal variances (SPSS 16.0; SPSS Inc., Chicago, IL, USA). The Kaplan-Meier method was used to calculate survival, and significance was determined by log-rank test. For all tests, p < .05 was considered significant.
3. Results
3.1. Cardiac endothelium-enriched AZIN2-sv inhibits HUVEC migration
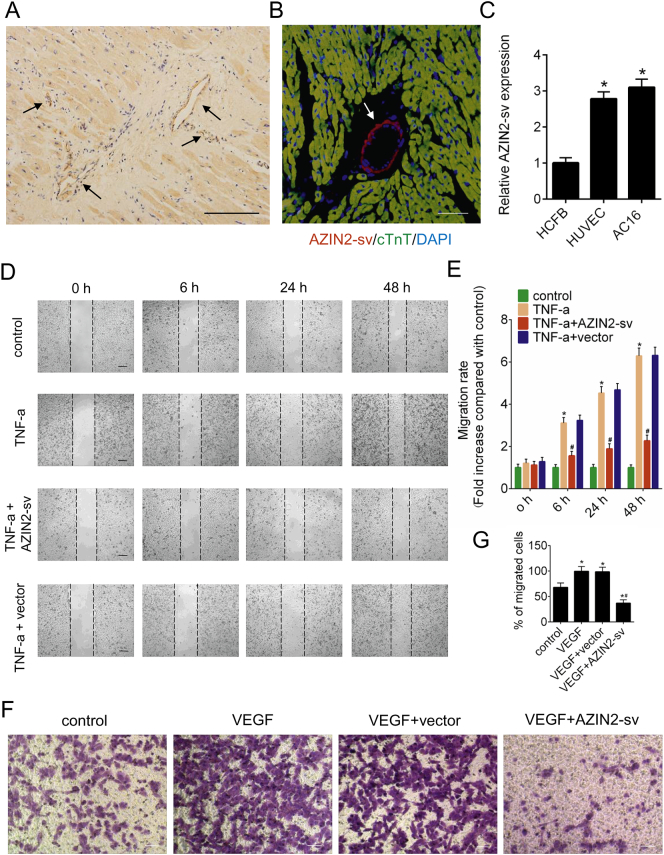
In situ hybridization was performed to determine AZIN2-sv localization, and the results showed that, in addition to being expressed in CM, AZIN2-sv was enriched in the vascular endothelium of human (Fig. 1A) and rat (Fig. 1B, Supplemental Fig. 1A) hearts. We compared AZIN2-sv expression in AC16 human CMs, HUVECs and HCFBs. The qRT-PCR results suggested that AZIN2-sv expression in AC16 cells and HUVECs was higher than that in HCFBs (p < .05; Fig. 1C). The effect of AZIN2-sv on endothelial cell migration was also investigated. Adenoviral vectors harbouring AZIN2-sv or shAZIN2-sv were transfected to overexpress and reduce AZIN2-sv expression, respectively (p < .05; Supplemental Fig. 1B, C). Increased expression of tumour necrosis factor (TNF)-α contributes to endothelial cell migration, thereby affecting vascular stability. We found that AZIN2-sv significantly decreased the number of migrating cells induced by TNF-α and AZIN2-sv knockdown promoted cells migration (p < .05; Fig. 1D, E, Supplemental Fig. 1D, E). Similar results were obtained by transwell assays, which indicated that AZIN2-sv could inhibit the invasion of HUVECs treated with VEGF and AZIN2-sv knockdown induced the invasion through Matrigel-coated polycarbonate membranes (p < .05; Fig. 1F, G, Supplemental Fig. 1H, I). These data show that myocardial-specific AZIN2-sv is enriched in cardiac endothelial cells and is a negative regulator of endothelial function.
Fig. 1.
Endothelial cell-enriched AZIN2-sv inhibits HUVEC migration in vitro.
(A) Representative image of in situ hybridization to confirm AZIN2-sv localization in the human heart (bars, 500 μm). Arrows indicate vessels. (B) Representative image of RNA FISH and immunofluorescence for AZIN2-sv and cTnT in the rat heart (bars, 40 μm). Red indicates the location of AZIN2-sv, green indicates the cardiomyocytes, and blue indicates the nucleus. Arrows indicate vessels. (C) AZIN2-sv expression in HCFBs, HUVECs and AC16 cells by qRT-PCR. ⁎p < .05 vs. HCFB; n = 6 per group. (D) Cell migration was assessed using wound-healing assays. Images were captured at 0, 6, 24, and 48 h after TNF-α treatment (10 ng/mL). The horizontal line indicates the wound edge. Migration was estimated by measuring cell numbers within the wounded region (bars, 500 μm). (E) Quantification of the endothelial cell migration rate. ⁎p < .05 vs. control. #p < .05 vs. TNF-a; n = 6 per group. (F) HUVEC invasion assessed by transwell assay (bars, 500 μm). (G) Quantification of invaded cells. ⁎p < .05 vs. control. #p < .05 vs. VEGF; n = 6 per group.
3.2. AZIN2-sv induces endothelial apoptosis and inhibits vessel formation
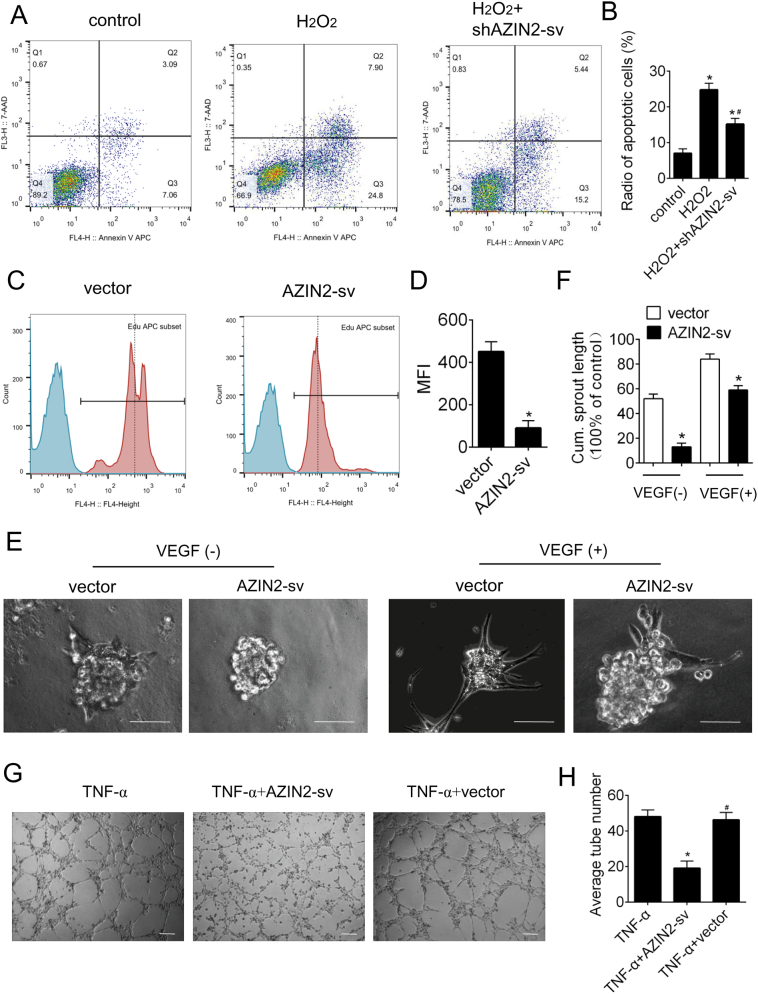
We further evaluated the effect of AZIN2-sv on endothelial cell apoptosis. Knockdown of AZIN2-sv prevented apoptosis in endothelial cells exposed to H2O2 and enhanced AZIN2-sv increased the radio of apoptotic cells (p < .05; Fig. 2A, B, Supplemental Fig. 1 J, K). Moreover, flow cytometry results showed that compared with that of control cells, proliferation was reduced in endothelial cells that overexpressed AZIN2-sv (p < .05; Fig. 2C, D). VEGF was used to promote endothelial spheroid formation. AZIN2-sv overexpression obviously suppressed vascular sprouting stimulated by VEGF (p < .05; Fig. 2E, F). The role of AZIN2-sv in vessel formation was also investigated. AZIN2-sv overexpression significantly decreased TNF-α-induced tube formation (p < .05; Fig. 2G, H). In addition, the presence of AZIN2-sv significantly antagonized VEGF-induced sprouting in rat aortic rings as shown in Supplemental Fig. 1C, D (p < .05). Thus, AZIN2-sv induces apoptosis and reduces proliferation in endothelial cells and inhibits endothelial vessel formation in vitro.
Fig. 2.
AZIN2-sv induces endothelial apoptosis and inhibits vessel formation.
(A) Flow cytometric analysis of HUVEC apoptosis was performed forty-eight hours after transfection with shAZIN2-sv. H2O2 (400 μM) was used to induce apoptosis of endothelial cells. (B) Quantification of apoptotic endothelial cells. ⁎p < .05 vs. control. #p < .05 vs. H2O2; n = 6 per group. (C) HUVEC proliferation was analysed by flow cytometry. (D) Quantification of Median Fluorescence Intensity (MFI). ⁎p < .05 vs. vector; n = 6 per group. (E) Representative images of vascular sprouting. Cells were treated with or without VEGF (10 ng/mL). Forty-eight hours after transfection with control vector or AZIN2-sv, HUVEC spheroids were allowed to undergo sprouting in a 3-dimensional (3D) matrix for 24 h (bars, 500 μm). (F) Quantification of the cumulative length of sprouts per spheroid. ⁎p < .05 vs. control. (G) HUVECs were seeded on Matrigel matrix and stimulated with TNF-α (10 ng/mL). Formation of tube-like structures was observed 24 h after TNF-α treatment. The average number of tubes formed per field was statistically analysed (bars, 500 μm). (H) Quantification of the average number of tubes. ⁎p < .05 vs. TNF-a. #p < .05 vs. TNF-a + AZIN2-sv; n = 6 per group.
3.3. AZIN2-sv knockdown promotes angiogenesis and thus improves prognosis after MI
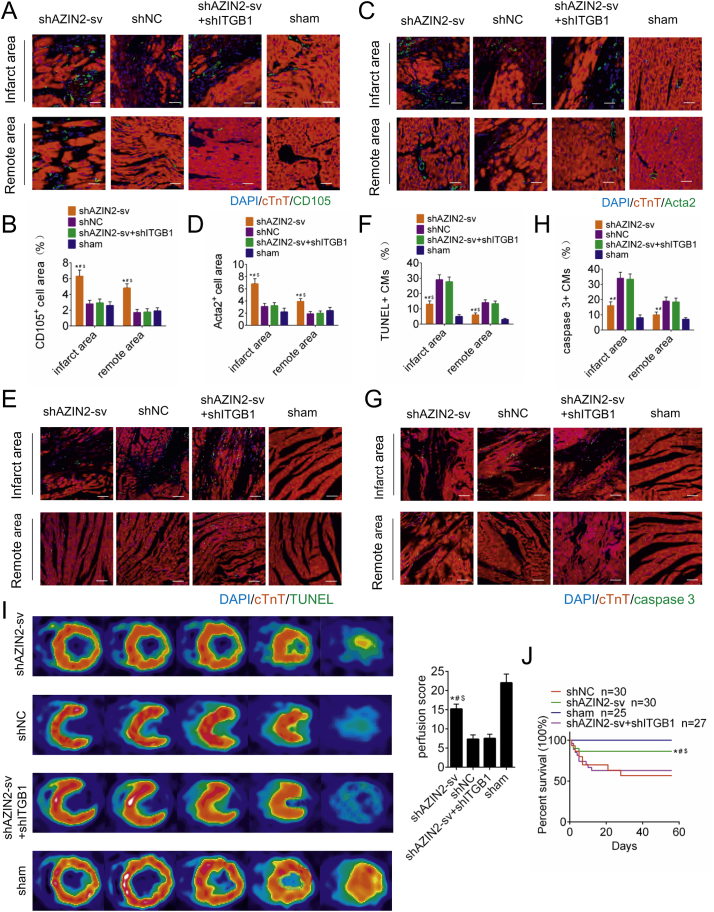
Neovascularization after MI was evaluated, and ITGB1 was knocked down to inhibit neovascularization. The transfection effect of AAV9 vectors is shown in Supplemental Fig. 2A, B (p < .05). Endoglin (CD105) is expressed on immature blood vessels and is a marker of angiogenesis. At 14 days after MI, compared with shNC- and sham-treated rats, the shAZIN2-sv -treated rats displayed increased CD105 staining in the peri-infarct and distant areas, while ITGB1 knockdown further reduced the number of CD105-positive cells (p < .05; Fig. 3A, B). Moreover, Acta2-positive arteriolar density was higher in the shAZIN2-sv group than in the shNC and sham groups, and this increase in arteriolar density could be inhibited by ITGB1 knockdown (p < .05; Fig. 3C, D). In addition, CM apoptosis was assessed. Compared with those in rats in the shNC group, TUNEL- and caspase-3-positive CMs in the peri-infarct and distant areas were markedly reduced in rats in the shAZIN2-sv group at 14 days after MI (p < .05; Fig. 3E-H). Increased neovascularization can improve prognosis after MI. At day 30 post-MI, myocardial perfusion score identified by PET/CT improved significantly in the shAZIN2-sv group and worsened when ITGB1 was knocked down (p < .05; Fig. 3I). The loss of AZIN2-sv brought the improvement of left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS), which were offset by ITGB1 knockdown at day 30 and 60 after MI (p < .05; Supplemental Fig. 2C). The same trend was also reflected in the survival rate. AZIN2-sv knockdown improved the survival rate after MI, which was aggravated when ITGB1 was knocked down (p < .05; Fig. 3J). In addition, the effect of AZIN2-sv knockdown on angiogenesis of other organs was also evaluated. Immunohistochemistry results showed that reduced AZIN2-sv did not cause changes in the vascular density of brain, muscle, lung, liver, skin and spleen (Supplemental Fig. 3A, B). These data suggest that the loss of AZIN2-sv can specifically promote myocardial angiogenesis and improve prognosis after MI.
Fig. 3.
AZIN2-sv knockdown promotes angiogenesis and improves prognosis after MI.
(A) Representative image of immunofluorescence staining for CD105 in the infarct area and distant myocardium (bars, 40 μm). Blue indicates the nucleus, red indicates the myocardium, and green indicates the neovascularization. (B) Quantification of neovascular density. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB1. $p < .05 vs. sham; n = 6 per group. (C) Representative image of immunofluorescence staining for Acta2 in the infarct area and distant myocardium (bars, 40 μm). Blue indicates the nucleus, red indicates the myocardium, and green indicates the arteriole. (D) Quantification of arteriolar density. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB1. $p < .05 vs. sham; n = 6 per group. (E) Representative image of TUNEL positivity in apoptotic CMs (bars, 40 μm). Blue indicates the nucleus, red indicates the myocardium, and green indicates the apoptotic nucleus. (F) Quantification of TUNEL-positive cells. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB1. $p < .05 vs. sham; n = 6 per group. (G) Representative image of immunofluorescence staining for caspase-3 (bars, 40 μm). Blue indicates the nucleus, red indicates the myocardium and green indicates the apoptotic nucleus. (H) Quantification of caspase-3-positive CMs. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB1. $p < .05 vs. sham; n = 6 per group. (I) Perfusion score determined by PET-CT. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB1. $p < .05 vs. sham; n = 6 per group. (J) Rat survival curves. ⁎p < .05 vs. shNC. #p < .05 vs. shAZIN2-sv + shITGB11. $p < .05 vs. sham; shAZIN2-sv n = 30, shNC n = 30, shAZIN2-sv + shITGB1 n = 27, sham n = 25.
3.4. AZIN2-sv binds to Tln1 protein and combines with miR-214 directly
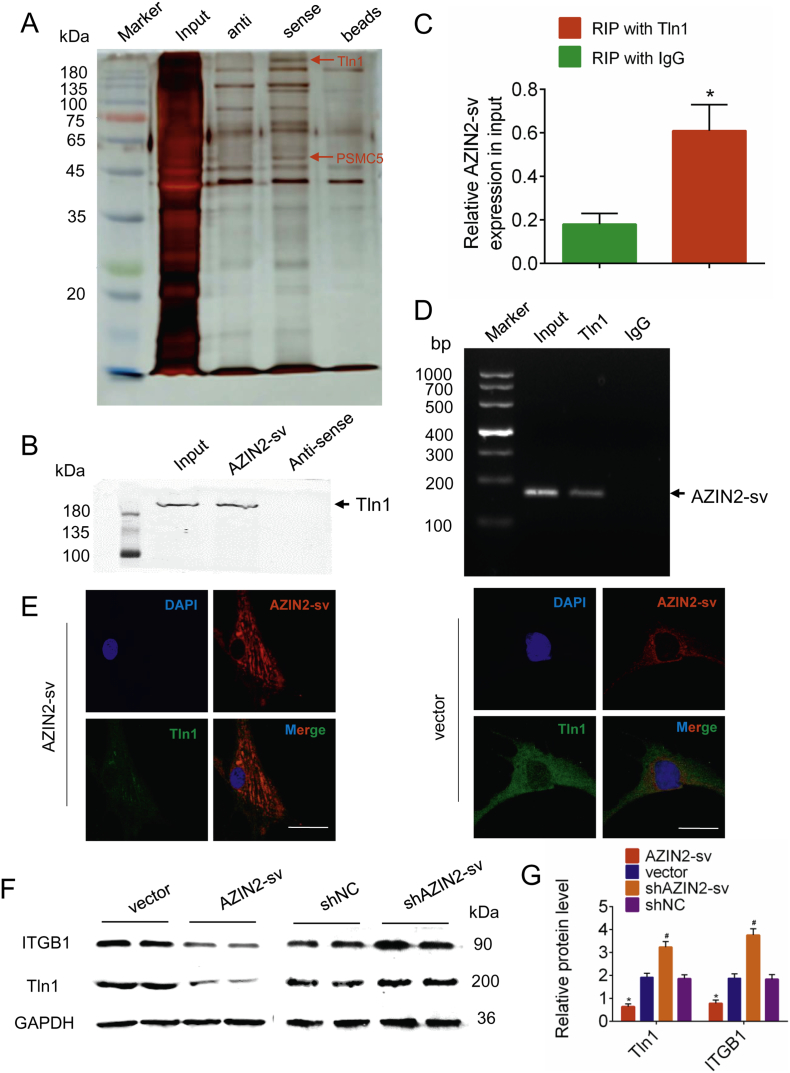
Next, the specific interaction protein of AZIN2-sv was identified. Tln1, which is expressed in CMs and endothelial cells, was pulled down by AZIN2-sv in CMs in our previous study (Supplemental Fig. 4A, B, Supplemental Table 3). We thus speculated that AZIN2-sv interacts with Tln1 in HUVECs. In line with this hypothesis, RNA pulldown assays were performed to confirm the interaction between AZIN2-sv and Tln1 in HUVECs (Fig. 4A, B). Moreover, RIP was used to demonstrate whether Tln1 could bind to AZIN2-sv in vivo. Indeed, compared with a non-specific IgG antibody, the anti-Tln1 antibody could enrich AZIN2-sv (p < .05; Fig. 4C, D). Furthermore, FISH and immunofluorescence staining for Tln1 showed the colocalization of Tln1 and AZIN2-sv in the cytoplasm when AZIN2-sv or control vector was transfected in HUVECs, which showed a decrease in Tln1 level when AZIN2-sv was overexpressed (Fig. 4E). Moreover, western blotting suggested that, compared with the respective control, AZIN2-sv overexpression decreased the expression of Tln1 and ITGB1, while AZIN2-sv knockdown had the opposite effect (p < .05; Fig. 4F, G). In addition, we also investigated the regulatory effect of AZIN2-sv on the miR-214/PTEN/Akt pathway in endothelial cells. The results indicated that in HUVECs, AZIN2-sv could act as a miR-214 sponge that releases PTEN and blocks Akt phosphorylation to inhibit angiogenesis (Supplemental Fig. 5A–E). These data show that AZIN2-sv can not only directly act on Tln1 protein but also block the miR-214/PTEN/Akt pathway.
Fig. 4.
Tln1 is a downstream interactor protein of AZIN2-sv.
(A) Silver-stained SDS-PAGE gel image representing proteins immunoprecipitated by AZIN2-sv and its antisense RNA in endothelial cells. The arrow indicates the region of the gel excised for western blotting. (B) Tln1 protein was detected in the specific band by western blotting. (C) RIP assays were performed using an antibody against Tln1 or negative control IgG. The purified RNA was used for qRT-PCR analysis, and enrichment of the AZIN2-sv was normalized against the input. ⁎p < .05 vs. RIP with IgG; n = 6 per group. (D) Agarose gel electrophoresis of RIP products. Arrow indicates enriched AZIN2-sv. (E) FISH and immunofluorescence for AZIN2-sv and Tln1 forty-eight hours after transfection with AZIN2-sv or control vector (bars, 40 μm). Blue indicates the nucleus, and red and green indicate the location of AZIN2-sv and Tln1, respectively. (F) ITGB1 and Tln1 protein levels detected by western blotting when AZIN2-sv was knocked down or overexpressed (GAPDH is the internal reference). (G) Quantification of protein levels in (F). ⁎p < .05 vs. vector. #p < .05 vs. shNC; n = 6 per group.
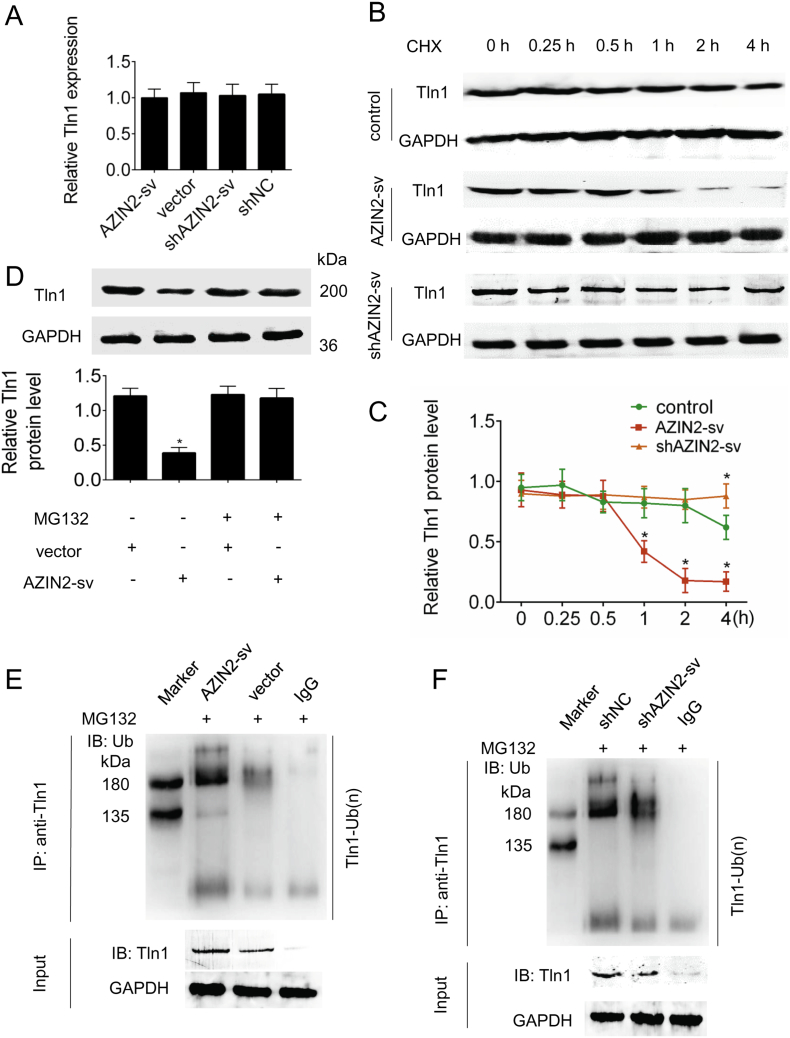
3.5. AZIN2-sv promotes Tln1 ubiquitin-mediated degradation at the translational level
We further explored the molecular consequences of the interaction between AZIN2-sv and Tln1. qRT-PCR showed that the levels of Tln1 mRNA were not altered when AZIN2-sv was overexpressed or knocked down (Fig. 5A). Treatment of AZIN2-sv-overexpressing cells with the protein synthesis inhibitor cycloheximide resulted in a notably shorter half-life of Tln1 than that in control cells, while AZIN2-sv knockdown delayed this decrease (p < .05; Fig. 5B, C). Furthermore, treatment of AZIN2-sv-overexpressing cells with the proteasome inhibitor MG132 resulted in increased levels of endogenous Tln1 protein compared with those in control cells, suggesting that the ubiquitin-proteasome pathway may play a critical role in the AZIN2-sv-mediated degradation of the Tln1 protein (p < .05; Fig. 5D). Indeed, Tln1 ubiquitination was dramatically increased in cells overexpressing AZIN2-sv compared with that in control cells but was substantially decreased in cells with AZIN2-sv knockdown (Fig. 5E, F). Collectively, these results indicate that the interaction of AZIN2-sv with Tln1 promotes Tln1 degradation through ubiquitination.
Fig. 5.
AZIN2-sv promotes Tln1 degradation through the ubiquitin-proteasome pathway.
(A) Tln1 mRNA level was detected by qRT-PCR when AZIN2-sv was knocked down or overexpressed. (B) AZIN2-sv was overexpressed and silenced in HUVECs, and the cells were then treated with cycloheximide (CHX; 20 μg/mL) for the indicated time. Tln1 protein levels were analysed by immunoblotting (GAPDH is the internal reference). (C) Quantification of the protein level in (B). ⁎p < .05 vs. control; n = 6 per group. (D) AZIN2-sv or control vector was transfected into HUVECs, and the cells were then treated with MG132 (5 μM) or vehicle for 24 h. Cell lysates were analysed by immunoblotting. ⁎p < .05 vs. vector; n = 6 per group. (E, F) HUVECs with AZIN2-sv overexpression (E) or knockdown (F) were treated with MG132 (5 μM) for 24 h. Cell lysates were immunoprecipitated with either control IgG or an antibody against Tln1 and analysed by immunoblotting with a ubiquitin (Ub)-specific antibody. Bottom, input from cell lysates.
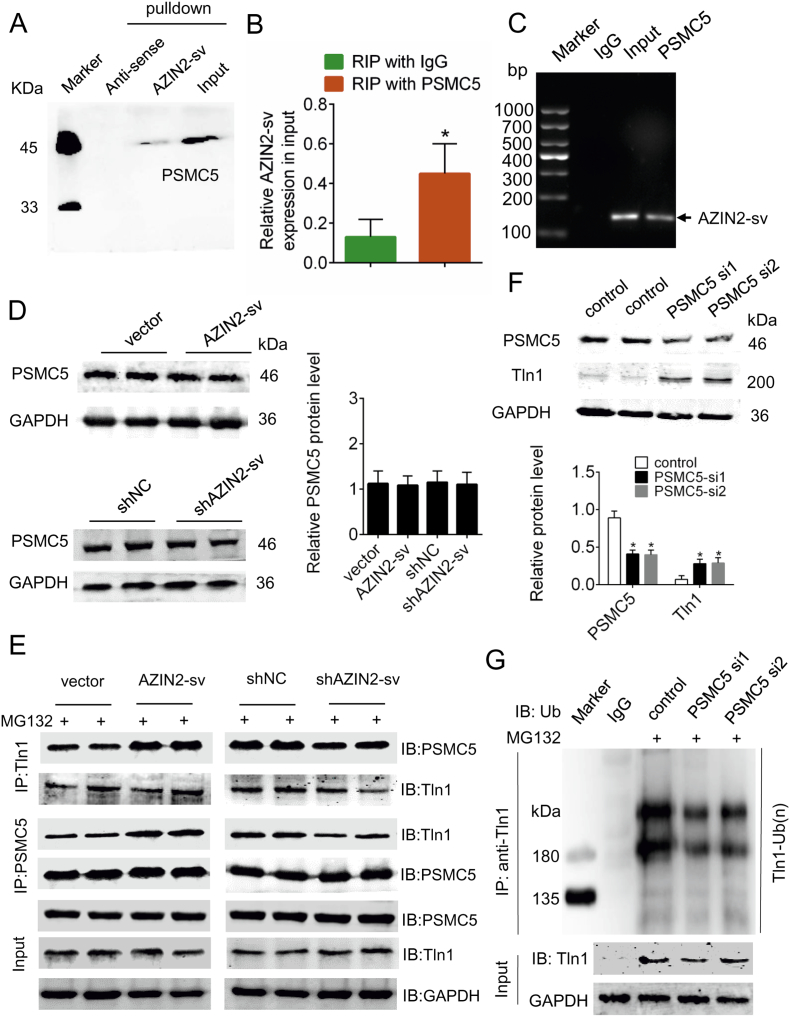
3.6. The ubiquitin-proteasome degradation pathway of Tln1 is mediated by PSMC5
Previous studies have suggested that the ubiquitin-proteasome pathway requires the participation of key enzymes. Intriguingly, among the AZIN2-sv-interacting proteins, we identified proteasome 26S subunit ATPase 5 (PSMC5), a core component of the 26S proteasome, which is the major non-lysosomal ATP-dependent proteolytic machinery that degrades polyubiquitinated proteins (Supplemental Fig. 4A, C, Supplemental Table 3). Using western blotting, we verified the presence of PSMC5 in the protein complex (Fig. 6A). RIP assays were used to demonstrate that PSMC5 could bind to AZIN2-sv in vivo (p < .05; Fig. 6B, C). Overexpression or knockdown of AZIN2-sv did not affect PSMC5 expression (Fig. 6D). However, AZIN2-sv knockdown had a certain impact on the interaction of PSMC5 with Tln1. Coimmunoprecipitation assays showed that enhanced AZIN2-sv expression increased the binding between Tln1 and PSMC5, while AZIN2-sv knockdown decreased the mutual binding of Tln1 with PSMC5 (Fig. 6E). We thus speculated that Tln1 is a substrate of PSMC5. Indeed, we observed an increase in Tln1 protein levels when PSMC5 expression was silenced (p < .05; Fig. 6F). Moreover, immunoprecipitation assays indicated that PSMC5 silencing led to diminished ubiquitination (Fig. 6G). Together, these results indicate that AZIN2-sv may function as a mediator that strengthens the PSMC5-Tln1 interaction, thereby promoting PSMC5-mediated ubiquitination and degradation of Tln1.
Fig. 6.
PSMC5 mediates Tln1 ubiquitination and degradation.
(A) HUVEC lysates were incubated with in vitro-synthesized biotin-labelled sense or antisense AZIN2-sv for biotin pulldown followed by immunoblot analysis. (B) RIP assays were performed using an antibody against PSMC5 or negative control IgG. The purified RNA was used for qRT-PCR analysis, and the enrichment of AZIN2-sv was normalized against the input. ⁎p < .05 vs. RIP with IgG; n = 6 per group. (C) Agarose gel electrophoresis of RIP products. Arrow indicates the enriched AZIN2-sv. (D) PSMC5 protein levels detected by western blotting when AZIN2-sv was overexpressed and knocked down (GAPDH is the internal reference). (E) HUVECs with AZIN2-sv overexpression or knockdown were treated with MG132 (5 μM) for 24 h. Cell lysates were immunoprecipitated with an antibody against Tln1 or PSMC5. The precipitates and input were analysed by immunoblotting. (F) Immunoblots for PSMC5 and Tln1 in HUVECs transfected with PSMC5-targeting siRNAs or scrambled control siRNAs (GAPDH is the internal reference). ⁎p < .05 vs. control; n = 6 per group. (G) HUVECs transfected with PSMC5-targeting siRNAs or scrambled control siRNAs were treated with MG132 (5 μM) for 24 h. Cells lysates were immunoprecipitated with either control IgG or antibody against Tln1. The precipitates and input were analysed by immunoblotting.
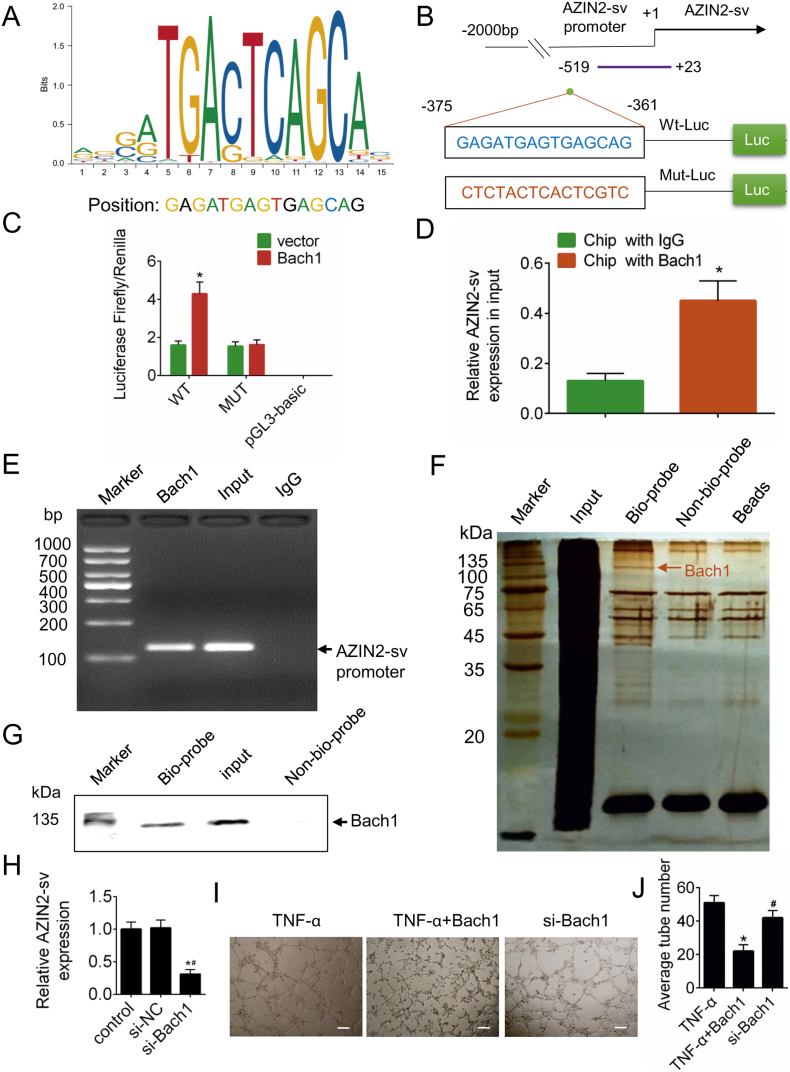
3.7. Bach1 directly binds to the promoter region of AZIN2-sv and enhances its expression
To examine the mechanisms underlying the transcriptional regulation of AZIN2-sv, we predicted the potential transcription factor that binds to the promoter region of AZIN2-sv and identified Bach1 as a likely candidate binding to the AZIN2-sv promoter (−375 bp to −361 bp) (Fig. 7A). Indeed, luciferase activity increased when Luc-Bach1-WT was transfected but decreased when Luc-Bach1-MUT was transfected (p < .05; Fig. 7B, C). Additionally, ChIP assays showed that Bach1 directly binds to the AZIN2-sv promoter (p < .05; Fig. 7D, E). In addition, DNA pulldown was performed to identify the specific protein that could bind to the promoter of AZIN2-sv (Fig. 7F). Bach1 could be detected in the specific bands (Fig. 7G). When Bach1 was silenced, AZIN2-sv expression decreased (p < .05; Fig. 7H). The effect of Bach1 on vessel formation was also investigated. TNF-α was used to induce the formation of tubes, and Bach1 overexpression suppressed tube formation, while Bach1 silencing had the opposite effect (p < .05; Fig. 7I, J). These results indicate that Bach1, an angiogenesis repressor, can promote AZIN2-sv expression.
Fig. 7.
AZIN2-sv is regulated by the transcription factor Bach1.
(A) Predicted binding sites for Bach1 on the promoter of AZIN2-sv. (B) Construction of WT-Luc and Mut-Luc promoter sequences of AZIN2-sv. (C) Luciferase activity in HUVECs transfected with WT-Luc or Mut-Luc after transfection with Bach1 overexpression constructs and scrambled control. ⁎p < .05 vs. vector; n = 6 per group. (D) ChIP was performed using an antibody against Bach1 or negative control IgG. Purified RNA was used for qRT-PCR analysis, and the enrichment of the AZIN2-sv promoter was normalized against the input. ⁎p < .05 vs. ChIP with IgG; n = 6 per group. (E) Agarose gel electrophoresis of ChIP products. Arrow indicates enriched AZIN2-sv promoter. (F) Silver-stained SDS-PAGE gel image representing proteins immunoprecipitated by the AZIN2-sv promoter and control probes. The arrow indicates the region of the gel excised for western blotting. (G) Bach1 protein was detected in the specific band by western blotting. (H) AZIN2-sv expression by qRT-PCR after silencing Bach1. ⁎p < .05 vs. control. #p < .05 vs. si-NC; n = 6 per group. (I) Representative tube-like structures were observed 24 h after TNF-α (10 ng/mL) treatment and Bach1 intervention (bars, 500 μm). (J) Quantification of the average number of tubes. ⁎p < .05 vs. TNF-a. #p < .05 vs. TNF-a + Bach1; n = 6 per group.
4. Discussion
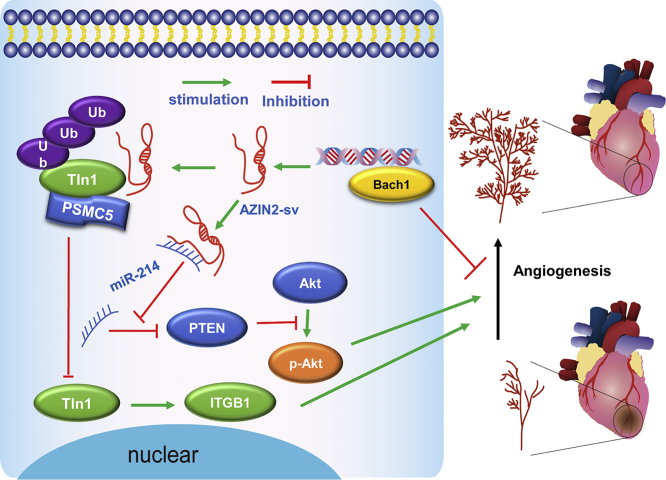
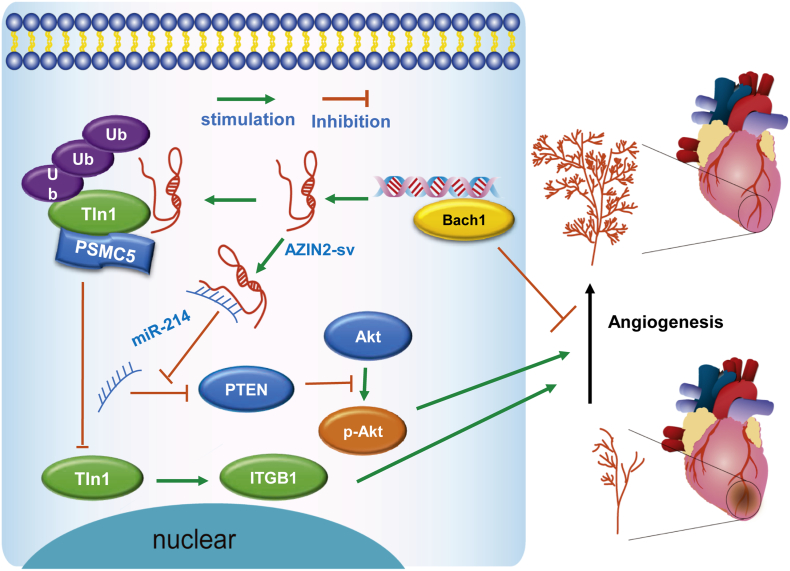
In this study, we identified that the loss of cardiac-specific AZIN2-sv promoted endothelial activation and induced angiogenesis in vitro and in vivo, thereby improving prognosis post-MI. AZIN2-sv knockdown increased Tln1 protein levels by blocking its ubiquitination mediated by PSMC5 and activated the miR-214/PTEN/Akt pathway to promote neovascularization. In addition, the transcription factor Bach1, which inhibits angiogenesis, directly bound to the AZIN2-sv promoter and increased its expression via an upstream mechanism. These findings suggest that AZIN2-sv, which participates in CM regeneration and angiogenesis, could be used to improve myocardial repair after MI (Fig. 8).
Fig. 8.
A schematic representing the role of AZIN2-sv in angiogenesis.
AZIN2-sv is an anti-angiogenesis lncRNA that acts by regulating Tln1 degradation through ubiquitination and binding to miR-214, resulting in the suppression of ITGB1 and inhibition of the Akt pathway and poor prognosis after MI. The transcription factor Bach1, a negative regulator of angiogenesis, can bind to the AZIN2-sv promoter and enhance its expression.
It is equally important to promote CM regeneration and induce angiogenesis for repair after MI [26]. Our present work indicated that the loss of AZIN2-sv, a lncRNA involved in CM proliferation, as previously reported by us, can enhance migration, vascular sprouting and tube formation in endothelial cells and further improve prognosis by increasing angiogenesis after MI. Previous studies have suggested that increased capillary density significantly correlated with reduced apoptosis of CMs and better prognosis post-MI [[27], [28], [29]]. The current results showed that with AZIN2-sv knockdown, neovascularization and arteriolar density labelled by CD105 and Acta2 in the peri-infarct region and distant myocardium markedly increased, which significantly reduced the apoptotic CMs, decreased the infarct area and improved the survival rate after MI. Moreover, ITGB1 knockdown served as an angiogenesis inhibitor and offset the benefits of AZIN2-sv knockdown, which further indicated the role of angiogenesis in myocardial repair after MI. Based on the regulation of CM proliferation demonstrated in our previous study and the currently described effects of AZIN2-sv on angiogenesis, we suggest that AZIN2-sv may serve as an ideal target for cardiac repair after MI. In addition, AZIN2-sv is a cardiac-specific lncRNA, which localizes its effect on angiogenic regulation to the heart. Previous studies have reported that lncRNAs are tissue specific, which allows them to be more precisely employed in the regulation of development and pathological processes. The muscle-specific lncRNA linc-MD1 has been shown to regulate the timing of muscle differentiation and play a crucial role in the control of factors involved in the myogenic programme [30]. Another cardiac-specific lncRNA, Bvht, was shown to be required for the commitment of nascent mesoderm towards a cardiac fate and was found to be critical in the establishment of the cardiovascular lineage during mammalian development [13]. Our present results suggest that AZIN2-sv could promote myocardial angiogenesis but did not affect vessel formation in other organs, including skeletal muscle, liver, spleen, lung, brain and skin. Thus, AZIN2-sv can induce CM regeneration and specifically promote myocardial angiogenesis, and these mechanisms may be valuable for future clinical applications.
This study revealed that the anti-angiogenic effect of AZIN2-sv was achieved by acting on the downstream protein Tln1 and miR-214. Previous studies have reported that lncRNAs can interact with proteins in various forms and function as protein decoys, protein scaffolds, protein target guides, and intracellular signals [31,32]. In our study, using RNA pulldown and RIP assays, we determined that AZIN2-sv could interact with Tln1 and downregulate its protein level. LncRNAs mainly regulate the expression of downstream proteins at the transcriptional or translational level [33]. Our current results showed that AZIN2-sv promoted the degradation of Tln1 protein without affecting its gene expression, which showed that the AZIN2-sv regulated Tln1 level at the translation level. Tln1, which functions as a regulator of integrin activity by binding to β-integrin tails, is known to be important for cell spreading and flattening required for angiogenesis [34,35]. The present results showed that AZIN2-sv could inhibit Tln1 protein levels and reduce the expression of β-integrin, both of which are essential for endothelial development and vessel formation. Further, we identified that the reduction of Tln1 by AZIN2-sv occurred through the ubiquitin degradation pathway. The inhibition of lncRNA binding to proteins can be achieved in a variety of ways including blockage of ATP synthesis, inhibition of phosphorylation and induction of acetylation [[36], [37], [38]]. Our present study showed that MG132, a protease inhibitor, could prevent AZIN2-sv-mediated Tln1 degradation. Moreover, we observed an increase in the binding of a ubiquitin ligase to Tln1 when AZIN2-sv was overexpressed, while AZIN2-sv knockdown resulted in the opposite effect. These results deepen our understanding of Tln1 regulation and provide insights into new approaches for future interventions. A recent study reported that LINC00673 can reinforce the interaction of PTPN11 with PRPF19, an E3 ubiquitin ligase, and promote PTPN11 degradation through ubiquitination [39]. In the present study, we found that the ubiquitin-mediated degradation of Tln1 by AZIN2-sv was dependent on PSMC5, the regulatory subunit of the 26S proteasome. PSMC5 has been reported to interact with the N-terminal region of Prp19 to recruit its target proteins to the proteasome and activate the ubiquitin proteasome degradation system [40,41]. In line with these findings, our results indicated the important role of PSMC5 in the ubiquitin proteasome pathway. In addition, our present study suggested that AZIN2-sv could act as a miR-214 sponge to release PTEN, which blocked activation of the Akt pathway to restrain angiogenesis. The PTEN/Akt signalling pathway has been shown to play an important role in angiogenesis [42,43], and our previous study reported its activation in CM and activation in myocardial regeneration. The current results indicated that regulatory effect of AZIN2-sv on miR-214/PTEN/Akt signalling could be achieved in ECs, which suggested that this mechanism was not cell specific. Thus, AZIN2-sv can promote Tln1 downregulation through the ubiquitinated protease degradation pathway dependent on PSMC5 and block the activation of miR-214/PTEN/Akt pathway to suppress angiogenesis.
In this study, we also found that AZIN2-sv expression could be regulated by the transcription factor Bach1. Recent studies have shown that transcription factors can bind to the promoter regions and regulate the transcription of lncRNAs [44,45]. In the present study, using ChIP and luciferase assays, we found that Bach1 bound to the promoter of AZIN2-sv at specific sites, and moreover, Bach1 could increase AZIN2-sv expression. Bach1 has been previously reported to act as a transcriptional repressor [46], while our results indicated a positive correlation between Bach1 levels and AZIN2-sv expression. A possible explanation for this phenomenon is that other molecules regulated by Bach1 are involved in the transcriptional activation of AZIN2-sv. A recent study showed that Bach1 could recruit histone deacetylase 1 to the interleukin-8 (IL-8) promoter in HUVECs [47]. In addition, Bach1 has been reported to induce endothelial cell apoptosis and inhibit tube formation [48]. Our present study showed that Bach1 overexpression could reduce tube formation; moreover, Bach1-activated AZIN2-sv could restrain endothelial function and angiogenesis. These findings were consistent with the results of previous reports and indicated that Bach1 may also be an intervention target for vascular diseases. Thus, AZIN2-sv transcription can be activated by Bach1, which is also a negative regulator of angiogenesis.
Nevertheless, our study has some limitations. The current results show that the transcriptional repressor Bach1 could promote AZIN2-sv expression, but the underlying mechanism needs to be investigated in the future. Although Bach1 is involved in vessel formation, its role in myocardial repair after MI needs to be further explored. Because Tln1 protein also exists in CMs, although we have demonstrated that AZIN2-sv could suppress CM proliferation by targeting the PTEN/Akt pathway, it is still an interesting topic to explore the mechanism with Tln1 of AZIN2-sv in CMs.
In conclusion, inhibition of AZIN2-sv improves prognosis post-MI by inducing angiogenesis and CM regeneration; Bach1-activated AZIN2-sv regulates angiogenesis by promoting Tln1 degradation via the ubiquitination pathway and activating the miR-214/PTEN/Akt pathway. Thus, AZIN2-sv is involved in angiogenesis and CM regeneration and may be an ideal target for facilitating myocardial repair after MI.
The following are the supplementary data related to this article.
Supplementary figures
Supplementary material
Gene-interfering base sequences
RT-PCR primer sequences
Detail mass spectrometry data
Funding sources
This work was supported by grants to Jianping Bin from the National Natural Science Foundation of China (No. 81771857 and 81571698). The funders have no roles in the study design, data collection, data analysis, interpretation, or writing of the report.
Declaration of interests
None.
Author contributions
Xinzhong Li wrote the paper, performed the experiments and analysed the data. Yili Sun, Senlin Huang, Yanmei Chen, Xiaoqiang Chen and Mengsha Li performed the experiments and analysed the data. Xiaoyun Si, Xiang He, Hao Zheng, Lintao Zhong and Yang Yang analysed the data and wrote the paper. Wangjun Liao, Yulin Liao designed the research. Guojun Chen, Jianping Bin designed the research and wrote the paper.
Contributor Information
Guojun Chen, Email: guojunchen1983@126.com.
Jianping Bin, Email: jianpingbin@126.com, jianpingbin@hotmail.com.
References
- 1.Lin Z., Pu W.T. Strategies for cardiac regeneration and repair. Sci Transl Med. 2014;6(239):239rv1. doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galdos F.X., Guo Y., Paige S.L., Vandusen N.J., Wu S.M., Pu W.T. Cardiac regeneration: lessons from development. Circ Res. 2017;120(6):941–959. doi: 10.1161/CIRCRESAHA.116.309040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uygur A., Lee R.T. Mechanisms of cardiac regeneration. Dev Cell. 2016;36(4):362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112(12):1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao X., Wang J., Wang Z., Du J., Yuan X., Huang W. MicroRNA profiling during rat ventricular maturation: a role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett. 2013;587(10):1548–1555. doi: 10.1016/j.febslet.2013.01.075. [DOI] [PubMed] [Google Scholar]
- 7.Yang X., Qin Y., Shao S., Yu Y., Zhang C., Dong H. MicroRNA-214 inhibits left ventricular remodeling in an acute myocardial infarction rat model by suppressing cellular apoptosis via the phosphatase and tensin homolog (PTEN) Int Heart J. 2016;57(2):247–250. doi: 10.1536/ihj.15-293. [DOI] [PubMed] [Google Scholar]
- 8.Boon R.A., Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495(7439):107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 9.Fiedler J., Jazbutyte V., Kirchmaier B.C., Gupta S.K., Lorenzen J., Hartmann D. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124(6):720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 10.Wen Z., Huang W., Feng Y., Cai W., Wang Y., Wang X. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0104666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D., Yang F., Yuan J.H., Zhang L., Bi H.S., Zhou C.C. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology (Baltimore, MD) 2013;58(2):739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 12.Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan S.X., Yang F., Yang Y., Tao Q.F., Zhang J., Huang G. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology (Baltimore, MD) 2012;56(6):2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 15.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zornig M., Braun T. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 16.Leisegang M.S., Fork C., Josipovic I., Richter F.M., Preussner J., Hu J. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136(1):65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., He X., Wang H., Li M., Huang S., Chen G. Loss of AZIN2 splice variant facilitates endogenous cardiac regeneration. Cardiovasc Res. 2018;114(12):1642–1655. doi: 10.1093/cvr/cvy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Research Council Committee for the Update of the Guide for the C, Use of Laboratory A. The National Academies Collection: Reports funded by National Institutes of Health. Guide for the Care and Use of Laboratory Animals. Washington (DC): National Academies Press (US) National Academy of Sciences; 2011.
- 20.Li X., He X., Wang H., Li M., Huang S., Chen G. Loss of long non-coding RNA ROCR facilitates endogenous cardiac regeneration. Cardiovasc Res. 2018;114(12):1642–1655. doi: 10.1093/cvr/cvy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicosia R.F., Ottinetti A. Modulation of microvascular growth and morphogenesis by reconstituted basement membrane gel in three-dimensional cultures of rat aorta: a comparative study of angiogenesis in matrigel, collagen, fibrin, and plasma clot. In vitro Cell Develop Biol. 1990;26(2):119–128. doi: 10.1007/BF02624102. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y., Zhang C., Shen S., Guo S., Zhong L., Li X. Ultrasound-targeted microbubble destruction enhances delayed BMC delivery and attenuates post-infarction cardiac remodelling by inducing engraftment signals. Clin Sci (Lond) 2016;130(23):2105–2120. doi: 10.1042/CS20160085. [DOI] [PubMed] [Google Scholar]
- 23.Guo S., Shen S., Wang J., Wang H., Li M., Liu Y. Detection of high-risk atherosclerotic plaques with ultrasound molecular imaging of glycoprotein IIb/IIIa receptor on activated platelets. Theranostics. 2015;5(4):418–430. doi: 10.7150/thno.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Zhao Z., Shen S., Zhang C., Guo S., Lu Y. Selective depletion of tumor neovasculature by microbubble destruction with appropriate ultrasound pressure. Int J Cancer. 2015;137(10):2478–2491. doi: 10.1002/ijc.29597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao Z., Chen B., Tan X., Zhao Y., Wang L., Zhu T. Coexpression of VEGF and angiopoietin-1 promotes angiogenesis and cardiomyocyte proliferation reduces apoptosis in porcine myocardial infarction (MI) heart. Proc Natl Acad Sci U S A. 2011;108(5):2064–2069. doi: 10.1073/pnas.1018925108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broughton K.M., Wang B.J., Firouzi F., Khalafalla F., Dimmeler S., Fernandez-Aviles F. Mechanisms of cardiac repair and regeneration. Circ Res. 2018;122(8):1151–1163. doi: 10.1161/CIRCRESAHA.117.312586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katare R., Riu F., Mitchell K., Gubernator M., Campagnolo P., Cui Y. Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circ Res. 2011;109(8):894–906. doi: 10.1161/CIRCRESAHA.111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silvestre J.S. Pro-angiogenic cell-based therapy for the treatment of ischemic cardiovascular diseases. Thromb Res. 2012;130(Suppl. 1):S90–S94. doi: 10.1016/j.thromres.2012.08.287. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X.L., Zhu R.R., Liu S., Xu H., Xu X., Wu Q.C. Notch signaling promotes angiogenesis and improves cardiac function after myocardial infarction. J Cell Biochem. 2018;119(8):7105–7112. doi: 10.1002/jcb.27032. [DOI] [PubMed] [Google Scholar]
- 30.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Kopp F., Chang T.C., Sataluri A., Chen B., Sivakumar S. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1–2):69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monkley S.J., Kostourou V., Spence L., Petrich B., Coleman S., Ginsberg M.H. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev Biol. 2011;349(2):494–502. doi: 10.1016/j.ydbio.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zovein A.C., Luque A., Turlo K.A., Hofmann J.J., Yee K.M., Becker M.S. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev Cell. 2010;18(1):39–51. doi: 10.1016/j.devcel.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcid E.A., Tsukiyama T. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014;28(21):2348–2360. doi: 10.1101/gad.250902.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.W P., X Y., H Y., L L., W C., X S. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 38.Chen R., Liu Y., Zhuang H., Yang B., Hei K., Xiao M. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45(17):9947–9959. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J., Huang X., Tan W., Yu D., Du Z., Chang J. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48(7):747–757. doi: 10.1038/ng.3568. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q., Yao J., Wani G., Chen J., Wang Q.E., Wani A.A. The ubiquitin-proteasome pathway is required for the function of the viral VP16 transcriptional activation domain. FEBS Lett. 2004;556(1–3):19–25. doi: 10.1016/s0014-5793(03)01347-4. [DOI] [PubMed] [Google Scholar]
- 41.Sihn C.R., Cho S.Y., Lee J.H., Lee T.R., Kim S.H. Mouse homologue of yeast Prp19 interacts with mouse SUG1, the regulatory subunit of 26S proteasome. Biochem Biophys Res Commun. 2007;356(1):175–180. doi: 10.1016/j.bbrc.2007.02.134. [DOI] [PubMed] [Google Scholar]
- 42.Xue M., Yao S., Hu M., Li W., Hao T., Zhou F. HIV-1 Nef and KSHV oncogene K1 synergistically promote angiogenesis by inducing cellular miR-718 to regulate the PTEN/AKT/mTOR signaling pathway. Nucleic Acids Res. 2014;42(15):9862–9879. doi: 10.1093/nar/gku583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang J., Ding M., Yang L., Liu L.Z., Jiang B.H. PI3K/PTEN/AKT signaling regulates prostate tumor angiogenesis. Cell Signal. 2007;19(12):2487–2497. doi: 10.1016/j.cellsig.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y., Jiang Y.Y., Xie J.J., Mayakonda A., Hazawa M., Chen L. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9(1):3619. doi: 10.1038/s41467-018-06081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H.T., Liu S., Liu L., Ma R.R., Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell cycle progression in gastric cancer. Cancer Res. 2018;78(20):5877–5890. doi: 10.1158/0008-5472.CAN-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mito S., Ozono R., Oshima T., Yano Y., Watari Y., Yamamoto Y. Myocardial protection against pressure overload in mice lacking Bach1, a transcriptional repressor of heme oxygenase-1. Hypertension (Dallas, Tex : 1979) 2008;51(6):1570–1577. doi: 10.1161/HYPERTENSIONAHA.107.102566. [DOI] [PubMed] [Google Scholar]
- 47.Jiang L., Yin M., Wei X., Liu J., Wang X., Niu C. Bach1 represses Wnt/beta-catenin signaling and angiogenesis. Circ Res. 2015;117(4):364–375. doi: 10.1161/CIRCRESAHA.115.306829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang L., Yin M., Xu J., Jia M., Sun S., Wang X. The transcription factor Bach1 suppresses the developmental angiogenesis of zebrafish. Oxid Med Cell Longev. 2017;2017:2143875. doi: 10.1155/2017/2143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary material
Gene-interfering base sequences
RT-PCR primer sequences
Detail mass spectrometry data