Abstract
Background
DNA mismatch repair (MMR) defects are a major factor in colorectal tumorigenesis in Lynch syndrome (LS) and 15% of sporadic cases. Some adenomas from carriers of inherited MMR gene mutations have intact MMR protein expression implying other mechanisms accelerating tumorigenesis. We determined roles of DNA methylation changes and somatic mutations in cancer-associated genes as tumorigenic events in LS-associated colorectal adenomas with intact MMR.
Methods
We investigated 122 archival colorectal specimens of normal mucosae, adenomas and carcinomas from 57 LS patients. MMR-deficient (MMR-D, n = 49) and MMR-proficient (MMR-P, n = 18) adenomas were of particular interest and were interrogated by methylation-specific multiplex ligation-dependent probe amplification and Ion Torrent sequencing.
Findings
Promoter methylation of CpG island methylator phenotype (CIMP)-associated marker genes and selected colorectal cancer (CRC)-associated tumor suppressor genes (TSGs) increased and LINE-1 methylation decreased from normal mucosa to MMR-P adenomas to MMR-D adenomas. Methylation differences were statistically significant when either adenoma group was compared with normal mucosa, but not between MMR-P and MMR-D adenomas. Significantly increased methylation was found in multiple CIMP marker genes (IGF2, NEUROG1, CRABP1, and CDKN2A) and TSGs (SFRP1 and SFRP2) in MMR-P adenomas already. Furthermore, certain CRC-associated somatic mutations, such as KRAS, were prevalent in MMR-P adenomas.
Interpretation
We conclude that DNA methylation changes and somatic mutations of cancer-associated genes might serve as an alternative pathway accelerating LS-associated tumorigenesis in the presence of proficient MMR.
Fund
Jane and Aatos Erkko Foundation, Academy of Finland, Cancer Foundation Finland, Sigrid Juselius Foundation, and HiLIFE.
Keywords: Lynch syndrome, Colorectal adenoma, DNA methylation, DNA mismatch repair, LINE-1 methylation, Mutation, Tumorigenesis, Tumor suppressor
Research in context.
Evidence before this study
Functional DNA mismatch repair is critical for cancer avoidance. DNA mismatch repair genes comply with the Knudson's two-hit paradigm for tumor suppressor genes. Lynch syndrome individuals have inherited a mutant copy of a given DNA mismatch repair gene which makes them susceptible to cancer, but as the product of the remaining normal copy of the same gene is available, tumor development is prevented. Tissues with an intact allele present show normal expression of the respective mismatch repair protein. However, recent evidence reveals that 10–46% of colorectal adenomas from Lynch syndrome patients retain mismatch repair protein expression, yet a tumor has developed, implying that proficient mismatch repair alone is insufficient to prevent tumor initiation. Molecular events needed in addition to or alternative to deficient DNA mismatch repair to initiate tumorigenesis, as well as the chronological sequence of events, is obscure. Deficient mismatch repair is associated with a characteristic mutational signature consisting of instability at repeat sequences and a designated pattern of base substitutions. Utilization of such mutational preferences to determine if a given alteration has occurred before or after mismatch repair deficiency, together with other approaches, has resulted in varying conclusions of early or late appearance of mismatch repair defects in Lynch syndrome tumorigenesis. Based on the above cited and other (recent) literature retrieved from PubMed, together with our previous observation that a mismatch repair-proficient subset exists among adenomas from a nation-wide registry of Lynch syndrome individuals, this study was undertaken to explore the tumorigenic events in Lynch syndrome-associated colorectal adenomas retaining MMR protein expression.
Added value of this study
Our investigation is one of the very few existing studies focusing on mismatch repair-proficient colorectal adenomas from Lynch syndrome mutation carriers, with the aim to determine the roles of DNA methylation changes and somatic mutations in cancer-associated genes in such neoplastic lesions. We show that tumor initiation may precede the loss of mismatch repair protein and identify molecular alterations present in adenomas with retained mismatch repair protein expression. While many of these changes were more prevalent in mismatch repair deficient adenomas, some were less frequent or even absent in the latter. Specifically, we found that hypermethylation of IGF2, NEUROG1, CDKN2A, CRABP1, SFRP1, and SFRP2, and especially KRAS mutations may accelerate, or possibly initiate, Lynch syndrome-associated tumorigenesis when the wild-type allele of the predisposing mismatch repair gene is still present as judged from retained protein expression. The emerging differences between mismatch repair deficient and proficient adenomas suggest that tumorigenesis in Lynch syndrome may be driven by different pathways, as supported by recent findings from Lynch syndrome-associated colorectal carcinomas.
Implications of all the available evidence
Recent studies, including this study, have shed light on the neoplastic process associated with colon tumorigenesis in Lynch syndrome. The fact that adenoma development is possible in the absence of biallelic inactivation of the predisposing mismatch repair gene shows that the traditional Knudson's two-hit mechanism is not the only one that applies to mismatch repair gene mutation-associated tumorigenesis. Genetic and epigenetic alterations observed in mismatch repair proficient adenomas may qualify for alternative tumor-initiating or promoting events. The findings have important clinical implications. As 27% of Lynch syndrome adenomas from our investigation and up to 46% of adenomas from published studies express the mismatch repair protein corresponding to the gene mutant in the germline, retained mismatch repair protein expression in a colorectal adenoma cannot be used to rule out Lynch syndrome in diagnostics. Genetic and epigenetic changes detected in mismatch repair proficient adenomas may provide potential biomarkers of increased tumor risk. Genetic heterogeneity in Lynch syndrome adenomas and carcinomas may be relevant for the design of targeted therapies or preventive measures. Larger sample sizes are required to confirm the recent findings described above and the specific contributions of the observed alterations to tumorigenesis. Further investigations are also needed to explore the extent to which the findings from Lynch syndrome may apply to sporadic colorectal tumorigenesis.
Alt-text: Unlabelled Box
1. Introduction
Lynch syndrome (LS) is a hereditary cancer predisposing syndrome caused by germline defects in DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS2 [1], or rarely deletions in the 3′ end of EPCAM gene that lead to hypermethylation of MSH2 gene promoter [2]. These germline defects cause a reduced level of MMR protein, i.e. haploinsufficiency, which accelerates the occurrence of de novo somatic mutations [3] or compromises other functions of MMR genes, such as apoptosis signaling [4], thereby increasing the risk for early onset malignancies. Those defects together with epigenetic events cause an increased risk of cancer [5], primarily colorectal cancer (CRC) and endometrial cancer [6].
The “second hit” leading to a loss of the remaining functional allele of a MMR gene is typically caused by loss of heterozygosity (LOH) or somatic mutations [[7], [8], [9]]. However, MMR deficiency is thought to appear as a secondary event in LS tumorigenesis, supported by multiple studies observing that 10–46% of adenomas show retained expression of MMR protein [[9], [10], [11], [12], [13]], and MMR deficiency is associated with larger polyps and higher grade [10,14,15]. The nature, timing and order of other molecular hits driving the malignant transformation are yet to be identified.
Most LS CRCs exhibit microsatellite instability (MSI) caused by defects in the MMR system [16]. CpG island methylation phenotype (CIMP) characterized by aberrant CpG island methylation in promoters of various tumor suppressor genes (TSGs) [17], is prevalent in LS CRC, and occurs already at early stages of tumor development, but clearly increases along with dysplasia [9]. The molecular drivers behind CIMP, and its role in hereditary cancer remain obscure. Accumulation of somatic mutations involving the epigenetic regulatory genes may serve as a mechanism for CIMP [8].
Long interspersed elements (LINEs) form a class of retrotransposons that constitute approximately 17% of the human genome, and are normally heavily methylated [18]. In cancer, LINEs can be activated due to DNA hypomethylation. LINE-1 hypomethylation serves as a surrogate marker for global hypomethylation that can lead to chromosomal instability (CIN) [19]. LINE-1 is a 6 kb long retrotransposon that can, once activated, induce transcription of other genes [18]. Moreover, activated LINE-1 can change the epigenetic regulation of adjacent genes [20], and can occur already at early stages in malignant transformation of CRC [21,22].
The aim of this study was to investigate molecular mechanisms, DNA methylation changes and somatic mutations, as events contributing to LS-associated colorectal tumorigenesis. Our focus was on precursors of cancer, adenomas, based on the observation by Valo et al. [9], that many LS-associated adenomas retain the expression of the MMR protein corresponding to the germline mutation. DNA methylation changes were investigated in eight CIMP marker genes, LINE-1, and seven candidate TSGs associated with early colon tumorigenesis [23]. Moreover, adenomas were studied for somatic alterations in mutational hotspots of 22 cancer-associated genes. DNA methylation changes and mutations were compared between MMR-proficient (MMR-P) and MMR-deficient (MMR-D) adenomas to identify possible initiating or promoting molecular changes in adenomas in which the “second hit” leading to the loss of MMR function had not (yet) occurred.
2. Materials and methods
2.1. Patient samples
Study material consisted of formalin-fixed, paraffin-embedded (FFPE) specimens of normal mucosae, adenomas and carcinomas from 57 LS patients (Table 1). Specimens were collected at the Helsinki University Central Hospital and the Jyväskylä Central Hospital during 2013–2016. All the patients were represented in the nationwide Hereditary Colorectal Cancer Registry of Finland, and the information of verified germline mutations is available in Supplementary Table S1. In this study, we used normal mucosae and adenoma specimens, extending the sample series from our previous study [9] with 23 specimens including 13 adenomas with low-grade dysplasia, seven adenomas with high-grade dysplasia and three carcinomas. Normal mucosae collected for this study were supplemented with 13 older archival specimens of normal mucosae of Lynch syndrome patients to increase a total number of the reference group (n = 29). Tumor material was evaluated for histology and tumor content by the pathologist. The detailed characteristics of adenoma specimens are presented in Supplementary Table S2. Patients whose adenoma specimens were used in this study were between 26 and 74 years old (mean 52·4 ± 2·9 CI 95%) at the time of biopsy. This study was approved by the Institutional Review Board of the Central Finland Health Care District (K—S shp Dnro4/2011) and the collection of archival specimens by the National Authority for Medicolegal Affairs (Dnro 1272/04/044/07) and the National Supervisory Authority for Welfare and Health (Valvira, Dnro 10,741/06.01.03.01/2015).
Table 1.
Lynch syndrome individuals and eligible specimens.
| No of patients or specimens | |
|---|---|
| Patients with germline mutation in | |
| MLH1 | 44 |
| MSH2 | 8 |
| MSH6 | 5 |
| Total | 57 |
| Colorectal specimens | |
| Normal colonic mucosa | 29 |
| Low dysplasia adenoma | 47 |
| High dysplasia adenoma | 21 |
| Carcinoma | 25 |
| Total | 122 |
2.2. DNA extraction
DNA extraction from FFPE samples was performed by the phenyl-chlorophorm method [24]. DNA concentrations were measured by the Nanodrop spectrophotometer (Thermo Fisher Scientific). Additionally, DNA samples from adenoma specimens were quantified by the Qubit™ Fluorometer (Thermo Fisher Scientific), and these values were used in the next generation sequencing (NGS) protocol.
2.3. Immunohistochemical (IHC) staining for MMR protein expression
IHC was done according to the standard procedures using the primary antibodies described previously [9,25].
2.4. Microsatellite instability (MSI) analysis
Mononucleotide repeat markers BAT25 and BAT26, which have been shown to sensitive and specific markers of high-degree MSI (MSI-H) [26,27], were used to study MSI status. When either one of the markers was unstable, a tumor was considered MSI, and if both markers were normal a tumor was considered microsatellite-stable (MSS).
2.5. Methylation assay by methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)
The MS-MLPA was used to study DNA methylation changes in the promoters of eight CIMP-associated genes, seven selected candidate TSGs, and LINE-1, according to the manufacturer's instructions (MRC Holland, Amsterdam, The Netherlands). The method is described in more detail in Valo et al. [9] Briefly, 150–250 ng of DNA was used for each reaction. MS-MLPA is based on the hybridizing probes including the recognition sequence of GCGC for the methylation-sensitive endonuclease HhaI. Methylation prevents digestion reaction to occur, and after the PCR amplification a signal peak appears if template DNA is methylated [28]. Methylation dosage ratios (Dm) were calculated by the following formula: Dm = (Px/Pctrl)Dig / (Px/Pctrl)Undig, where Dm is a methylation dosage ratio (degree of methylation), Px is a peak area of the given probe, Pctrl is a sum of the peak area of all control probes, Dig stands for sample digested with HhaI, and Undig stands for the undigested sample. The average degree of methylation (Dm) plus one standard deviation or two standard deviations (i.e. stringency level II that was used for the CIMP analysis for the more accurate classification) calculated for normal mucosae samples determined a threshold value for the hypermethylation for each probe. In tumor samples, probes were defined methylated, if the degree of methylation exceeded the threshold value. The results from individual tumor samples were combined to define the relative frequency of hypermethylation.
CIMP status was investigated using the SALSA MLPA probemix ME042-B2 (MRC Holland) including 3–6 probes for the promoter of each marker gene: CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1, CDKN2A, MLH1 and CRABP1. The gene was considered methylated when one fourth or more of the probes were methylated [29]. The tumor sample was classified CIMP(+) using the Weisenberger panel: three or more genes out of CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1 should be methylated [30].
LINE-1 hypomethylation was studied by the custom-made LINE-1-MS-MLPA assay described in Pavicic et al. [31] Briefly, the assay included in total of ten probes, of those three were methylation-sensitive probes targeting regions with the HhaI restriction site inside the LINE-1 promoter sequence. The remaining seven probes provided a set of control probes targeting other regions without the restriction site in the LINE-1 sequence. LINE-1 hypomethylation was calculated based on L1-M2 probe [31].
Similarly, the custom-made MS-MLPA panel was designed to study promoter methylation of the selected candidate TSGs involved in early colon tumorigenesis in a mouse model for LS: DKK1, SFRP2, CDH1, HOXD1, SFRP5, SLC5A8, and SFRP1 [23]. Detailed design is described in Valo et al. [9] The custom-made probes were used together with the reference probe mix of the SALSA MLPA P300-A2 Human DNA Reference-2 (MRC Holland).
2.6. Amplicon-based next generation sequencing (NGS)
In total of 63 DNA samples of FFPE adenoma specimens were eligible for the sequencing study. Libraries were prepared from 4 to 15 ng of input DNA according to the manufacturer's protocol of the Ion AmpliSeq Library Kit 2.0 (Rev. C.0) (Thermo Fisher Scientific) using the Ion AmpliSeq Colon and Lung Cancer Panel (Thermo Fisher Scientific). The panel targets mutational hotspots in 22 genes totaling 14.6 kb: AKT1, ALK, BRAF, CTNNB1, DDR2, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NOTCH1, NRAS, PIK3CA, PTEN, SMAD4, STK11, and TP53. The libraries were purified using the AMPure XP magnetic beads (Beckman Coulter, Krefeld, Germany), and the quantification was performed with the Qubit Fluorometer with the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Library amplification and enrichment steps were performed according to the manufacturer's protocol of the Ion PGM Hi-Q View OT2 Kit (Rev.C.0) (Thermo Fisher Scientific) on the Ion OneTouch2 and Ion OneTouch ES instruments, respectively (Thermo Fisher Scientific). Sequencing was performed using the Ion PGM Hi-Q View Sequencing Kit (Rev.C.0) on the Ion PGM 316v2 Chip (Thermo Fisher Scientific). The barcoded (Ion Xpress Barcode Adapters, Thermo Fisher Scientific) libraries were pooled in the sets of 8–16 libraries.
The sequencing data were analyzed utilizing the Torrent Suite software (v5.2.2). Coverage analysis was performed by using the plug-in of coverageAnalysis (v5.2.1.2), and variant calling was conducted with the variantCaller (v5.2.1.39). Default analysis parameters were applied on signal processing, base calling, read alignment and variant calling. Reads were aligned at the human genome reference assembly of h19. Variant calling files were imported in the VarSeq software (GoldenHelix) and annotated with the databases of COSMIC Cancer Gene Census 71 [32], ExAC Variant Frequencies 0.3, dbSNP 149 (NCBI), and RefSeq Genes 105 Interim v1 (NCBI), and in silico functional prediction tools (SIFT, PolyPhen2 HVAR, MutationTaster, MutationAssessor, FATHMM, and FATHMM MKL). For the final step of variant analysis, only variants with quality score ≥ 30, read depth ≥ 50, and VAF ≥ 0·05 were considered true positives. Moreover, all variants mapped outside the target regions, producing synonymous variant, or variants reported in the 1000 Genomes project (NCBI dbSNP 149) and/or in the ExaC database with minor allele frequency (MAF) of 0·01 or more, were filtered out.
2.7. Statistical analysis
Statistical analyses were performed using the SPSS software, version 24.0 (IBM SPSS Inc., Chicago, IL, USA). Group-wise comparisons for categorical variables were calculated by using the Fisher's exact-test. For continuous variables, normal distribution was tested by Shapiro-Wilk test, and a parametric (Student's t-test or one-way ANOVA with Tukey's post hoc test, homogeneity of variances tested by Levene's test) or non-parametric (Mann-Whitney U test/Kruskal-Wallis one-way ANOVA) test was used, as appropriate. Two-sided p values <0·05 were considered significant. The Bonferroni correction method was applied on p values obtained from multiple comparisons.
2.8. Data sharing statement
All relevant data supporting this study is published along the paper. Case-specific DNA methylation data is available upon request.
3. Results
3.1. MMR protein expression and MSI status
The colorectal specimen collection described in Table 1 was used for IHC investigations to evaluate if the remaining wild-type allele of the germline mutated MMR gene had undergone somatic inactivation. Protein expression of the relevant MMR genes was lost in all carcinomas (Table 2). Among adenomas, 35% (16/46) of low dysplasia cases and 10% (2/21) of high dysplasia cases retained the protein expression of the predisposing MMR gene (Table 2). Normal mucosae (n = 43) were tested previously and all showed normal MMR protein expression [9].
Table 2.
The proportions of retained MMR protein expression in adenomas and carcinomas by IHC (extended series of the previous study [9]).
| MLH1 | MSH2 | MSH6 | Totala | |
|---|---|---|---|---|
| Adenoma with low-grade dysplasia | 11/34 (32%) | 3/8 (38%) | 2/4 (50%) | 16/46 (35%) |
| Adenoma with high-grade dysplasia | 1/17 (6%) | 0/2 (0%) | 1/2 (50%) | 2/21 (10%) |
| Carcinoma | 0/15 (0%) | 0/3 (0%) | 0/5 (0%) | 0/23 (0%) |
MMR IHC could not be performed on two carcinoma and one adenoma with low-grade dysplasia specimens, and thus, they are missing from this table compared to the numbers presented in Table 1.
MSI status was determined by the mononucleotide markers BAT25 and BAT26. We found a clear association between IHC and MSI status (p < 0·0001). All tested carcinomas and all adenomas with absent MMR protein showed MSI, with one exception of adenoma with low dysplasia having stable microsatellites. All adenomas with intact MMR protein expression for which enough DNA was available for MSI analysis (n = 11) were MSS. Throughout this paper, adenomas with absent MMR protein or presence of MSI or both are considered MMR-deficient (MMR-D, n = 49) and those with neither abnormality MMR-proficient (MMR-P, n = 18).
The existence of significant MMR-proficient subsets of LS-adenomas indicates the necessity of other somatic events to initiate malignant transformation, leading us to investigate DNA methylation changes and somatic mutations in LS-associated adenomas (see below).
3.2. DNA methylation changes
3.2.1. CIMP markers
Methylation analysis of eight genes (CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1, CDKN2A, MLH1 and CRABP1) linked to CIMP was used to assess the frequency of CIMP, a high-degree coordinated methylation of CpG islands that are normally unmethylated. CIMP(+) was defined according to the Weisenberger panel [30]. The frequency of CIMP(+) increased along with the dysplasia grade, occurring in 0% (0/29), 11% (4/37), 17% (3/18), and 45% (10/22) of normal mucosae, adenomas with low-grade dysplasia, adenomas with high-grade dysplasia, and carcinomas, respectively.
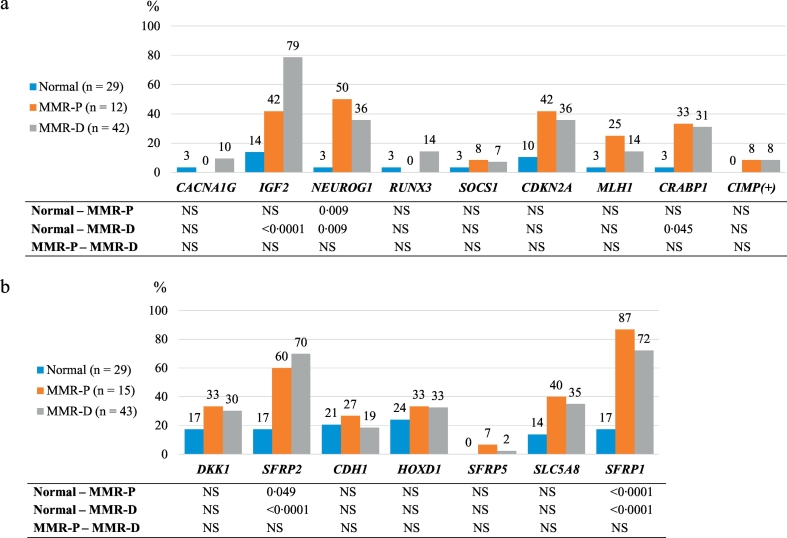
Altered methylation was evaluated in two ways, by determining the frequencies of hypermethylated samples based on defined cut-off methylation dosage (Dm) values for hypermethylation for each marker gene with normal mucosa as a reference (see Materials and Methods) and alternatively, by treating the degree of methylation (by Dm values) as a continuous variable in the samples. Fig. 1a shows the frequencies of hypermethylation for eight CIMP markers in adenomas stratified by MMR status, with normal mucosae as a reference. Hypermethylation frequency of NEUROG1 was increased already in MMR-P adenomas compared to normal colon (p = 0·009), and methylation frequency was higher than in MMR-D adenomas, although statistical significance was not obtained between MMR-P and MMR-D cases. MMR-D adenomas showed significantly elevated frequencies of hypermethylation relative to normal mucosa at IGF2 (p < 0·0001), NEUROG1 (p = 0·009) and CRABP1 (p = 0·045) loci. Among adenomas with MLH1 methylation depicted in Figs. 1a and 2a, only one, a MMR-D adenoma with high-grade dysplasia, revealed hypermethylation with the probe targeting the promoter region “C” of MLH1 (MLH1-I probe); hypermethylation of this region is associated with loss of MLH1 protein [33]. In the remaining cases, other regions of the MLH1 promoter (MLH1-II, III, and IV probes), not associated with protein expression, were involved.
Fig. 1.
Frequencies of methylated genes in normal colon, and MMR-P and MMR-D adenoma specimens. a. Hypermethylation frequencies of CIMP marker genes and frequencies of CIMP positive phenotype. b. Hypermethylation frequencies of candidate tumor suppressor genes. Bonferroni corrected two-sided p values are presented for pairwise comparisons. Abbreviations: MMR-D, MMR deficient; MMR-P, MMR proficient; NS, non-significant.
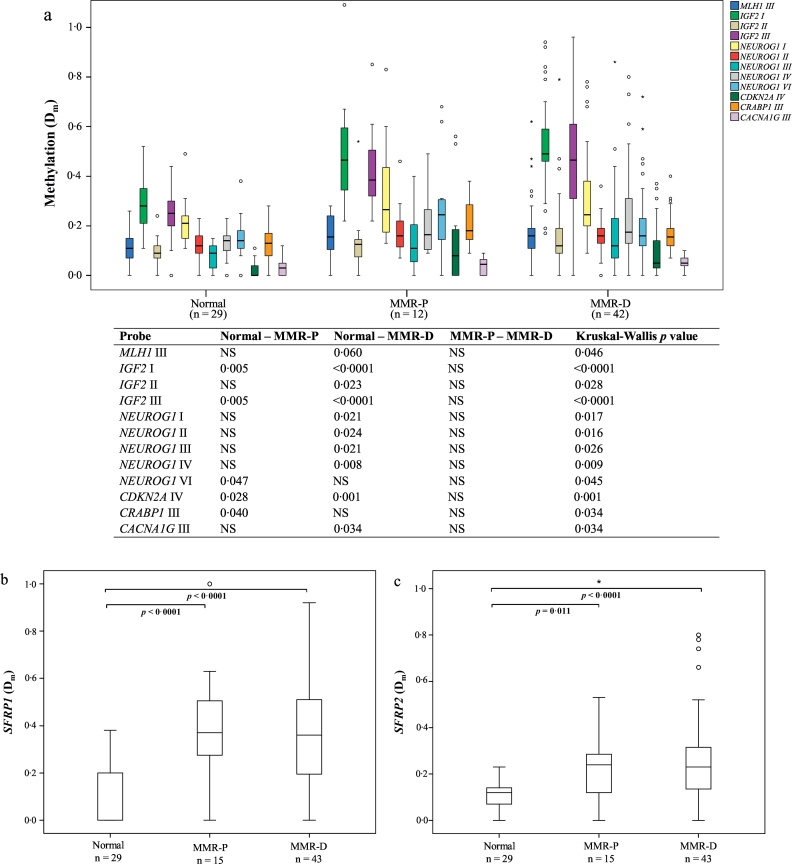
Fig. 2.
Comparison of the average degrees of methylation in normal colon, and MMR-P and MMR-D adenoma specimens. a. The CIMP probes showing statistical significance (p < 0·05) with Kruskal-Wallis test. b. Average degree of methylation of SFRP1. c. Average degree of methylation of SFRP2. Bonferroni corrected two-sided p values <0·10 are presented for the pairwise comparisons. The upper and lower edges of the boxes are the 75th and 25th percentiles, the horizontal line inside the box denotes the median, the whiskers indicate the highest and lowest values, and the asterisks and open circles stand for outliers. Abbreviations: Dm, methylation dosage; MMR-D, MMR deficient; MMR-P, MMR proficient.
When examining the average Dm values for the individual probes contributing to each CIMP marker gene (Fig. 2a), at least one probe of the following marker genes revealed a significant increase already in MMR-P adenomas vs. normal colon: IGF2, NEUROG1, CDKN2A, and CRABP1. Of those, almost all probes showed significantly increased methylation also in MMR-D adenomas vs. normal colon. There were two probes, one for CRABP1 and one for NEUROG1, which showed significantly elevated methylation solely in MMR-P adenomas (vs. normal colon).
3.2.2. Candidate tumor suppressor genes
We studied seven candidate TSGs (DKK1, SFRP2, CDH1, HOXD1, SFRP5, SLC5A8, and SFRP1) associated with early colon oncogenesis and being downregulated by promoter methylation in an experimental mouse model for LS [23]. Similarly to CIMP marker analysis, we first compared the hypermethylation frequencies of each gene in normal colon, MMR-P and MMR-D adenomas. Here, both SFRP1 and SFRP2 showed significantly increased frequency of hypermethylation in both MMR-P and MMR-D tumors compared to normal colon (Fig. 1b). Likewise, in the analysis of average Dm values, SFRP1 and SFRP2 showed increased methylation levels in both MMR-P and MMR-D adenomas compared to normal colon (Fig. 2b, c).
3.2.3. LINE-1
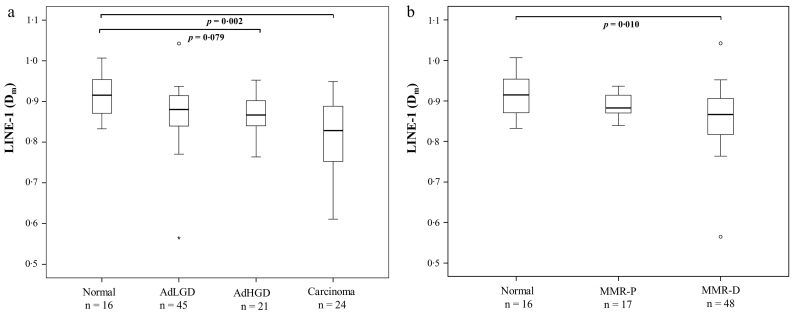
The average degree of methylation of LINE-1, a marker for global hypomethylation, decreased along with dysplasia (p = 0·005) (Fig. 3a). Statistically significant difference relative to normal colon was observed for carcinoma, but not for adenomas with high- or low-grade dysplasia, suggesting that LINE-1 methylation is a rather late event in LS-associated tumorigenesis. When comparing adenomas stratified by MMR status with normal colon, significantly decreased methylation level was observed for MMR-D adenomas (Fig. 3b), indicating that LINE-1 hypomethylation is likely to occur after the loss of MMR protein.
Fig. 3.
Average degree of LINE-1 methylation. a. LINE-1 methylation levels in all the specimens sorted by histology. b. LINE-1 methylation in normal colon, and MMR-P and MMR-D adenoma specimens. Bonferroni corrected two-sided p values <0·10 are presented in the figure. The upper and lower edges of the boxes are the 75th and 25th percentiles, the horizontal line inside the box denotes the median, the whiskers indicate the highest and lowest values, and the asterisks and open circles stand for outliers. Abbreviations: AdHGD, adenoma with high-grade dysplasia; AdLGD, adenoma with low-grade dysplasia; Dm, methylation dosage; MMR-D, MMR deficient; MMR-P, MMR proficient.
4. Effect of dysplasia grade on methylation results
To produce the results given in Fig. 1, Fig. 2, Fig. 3, adenomas with high- and low-grade dysplasia were combined. The same analyses were performed on adenomas with low-grade dysplasia only and are available in Supplementary Materials (Supplementary Fig. S1–S3). Adenomas with high-grade dysplasia included too few MMR-P cases (Table 2) and could not be studied as an independent group. As evident from Supplementary Figs. S1–S3, the results remained essentially similar to those derived from adenomas not stratified by dysplasia grade. Moreover, to exclude the effect of dysplasia grade on the results, we compared adenomas with high-grade dysplasia to those with low-grade dysplasia regardless of MMR status, and no significant differences were observed (data not shown).
4.1. Somatic mutations
Adenomas were studied for somatic mutations targeting the mutational hotspots in 22 cancer-associated genes (see Materials and Methods). As the amount of sample material of adenoma specimens was limited, this panel served as an excellent option to study somatic mutations even in those samples with a very low DNA content. In total, 59 out of initial 63 samples that were eligible for the sequencing study produced good quality sequencing library and sequencing results, including 16 MMR-P adenomas, out of which a majority (n = 14) were adenomas with low-grade dysplasia. For the obtained sequencing data, mean values for the read depth, the mapping uniformity between the amplicons, and the percentage of reads mapped on targets were 1379 (± 176 CI 95%), 94% (± 2 CI 95%), and 92% (± 1 CI 95%) respectively.
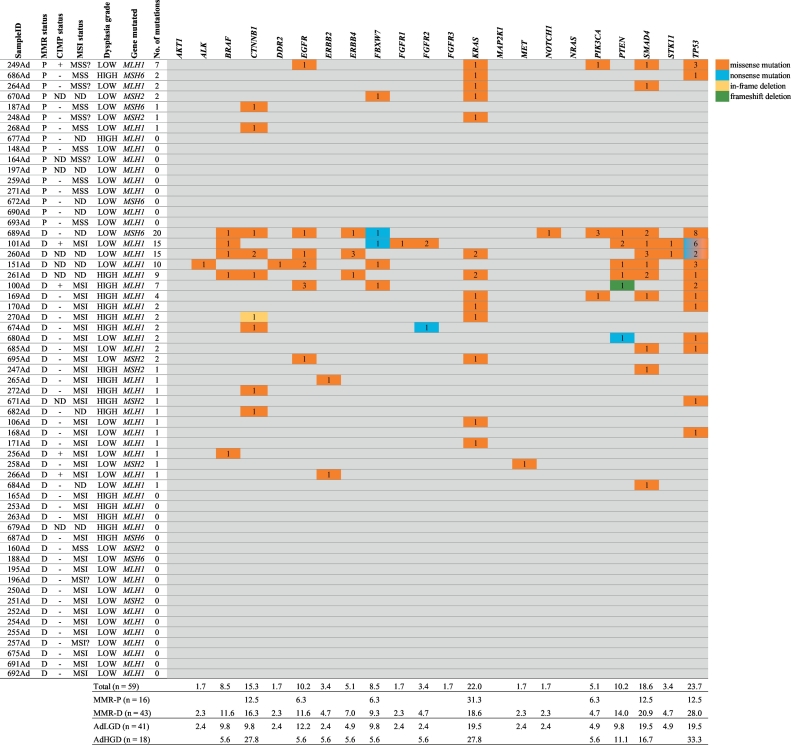
Here, we present non-synonymous mutations that have previously been reported in the COSMIC database [32], emphasizing their cancer-associated nature. The most frequently mutated genes were TP53, followed by KRAS, SMAD4, and CTNNB1, found in 24% (n = 14), 22% (n = 13), 19% (n = 11), and 15% (n = 9) of the adenomas, respectively. Overall, 54% (32/59) of adenomas harbored at least one mutation. One MMR-P sample (249Ad) included clearly more mutations (n = 7) than other MMR-P adenomas (Fig. 4). This particular case was the only CIMP(+) MMR-P adenoma. In total, there were five CIMP(+) adenomas and three of those harbored several mutations in multiple genes (Fig. 4). A vast majority of the mutations were missense mutations (Fig. 4). The full list of our COSMIC mutations is available in Supplementary Table S3.
Fig. 4.
Mutation status of adenomas stratified by their MMR status and number of mutations. Mutations presented in this table are reported in the COSMIC database (v71). Numbers in the colored cells indicate the number of mutations affecting a given gene in the sample in question, and color distinguishes the type of mutation. Table below shows mutation frequencies in adenomas stratified by MMR status or dysplasia grade. Note: question mark after MSI or MSS status indicates that only one probe produced an interpretable result (for MSS?) or due to poor quality, the result was weakly interpretable (MSI?). Abbreviations: AdHGD, adenoma with high-grade dysplasia; AdLGD, adenoma with low-grade dysplasia; CIMP, CpG island methylator phenotype; D, deficient; MMR, DNA mismatch repair; MMR-D, MMR deficient; MMR-P, MMR proficient; ND, not done; P, proficient.
We observed a somewhat higher average number of mutations in MMR-D adenomas compared to MMR-P adenomas (2·4 vs. 1·0), although no statistical difference was found (p = 0·323). As the most intriguing finding, KRAS mutations were clearly more prevalent in MMR-P adenomas (Fig. 4) (5/16, 31% vs. 8/43, 19%), although the difference is statistically non-significant. Considering only KRAS mutations in the codons 12, 13, 61, or 146 regarded pathogenic [[34], [35], [36]], these were clearly more prevalent in MMR-P tumors (5/16, 31% vs. 7/43, 16%). The observation remained the same when only adenomas with low-grade dysplasia were included: the frequency of pathogenic KRAS mutations was 29% (4/14) in MMR-P cases vs. 15% (4/27) in MMR-D cases. Detailed molecular characteristics of MMR-P adenomas are presented case by case in Table 3.
Table 3.
Molecular characteristics of Lynch syndrome adenoma specimens with retained MMR protein expression, undergone somatic mutation analysis.
| Patient ID | Sample ID | Dysplasia grade | Germline mutation | Hypermethylated CIMP markers | Hypermethylated candidate TSGs | Somatic mutations (VAF %) | MSI status | CIMP status |
|---|---|---|---|---|---|---|---|---|
| 73 | 148Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | IGF2 | None | None | MSS | (−) |
| 3 | 164Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | ND | ND | None | MSSa | ND |
| 16 | 187Ad | LOW | MSH6 c.900dup | NEUROG1 | SFRP1 | CTNBB1 p.Ser45Phe (37) | MSS | (−) |
| 95 | 197Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | ND | SFRP1 | None | ND | ND |
| 13 | 248Ad | LOW | MSH2 c.1667_1671del | CDKN2A | SFRP2, SLC5A8, SFRP1 | KRAS p.Gly12Asp (7) | MSSa | (−) |
| 17 | 249Ad | LOW | MLH1 c.546-2A > G | MLH1, IGF2, NEUROG1, CDKN2A, CRABP1 | SFRP2, HOXD1, SFRP1 | EGFR p.Ala767Val (5), KRAS p.Gly12Val (8), PIK3CA p.Arg693His (5), SMAD4 p.Pro514Ser (5), TP53 p.Arg282Trp (6); p.Pro250Leu (5); p.Gly244Ser (5) | MSSa | (+) |
| 45 | 259Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | NEUROG1 | SFRP2, SFRP1 | None | MSS | (−) |
| 76 | 264Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | NEUROG1, CDKN2A, CRABP1 | SFRP1 | KRAS p.Gly12Val (29), SMAD4 p.Pro102Leu (18) | MSSa | (−) |
| 95 | 268Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | MLH1, IGF2, NEUROG1, CRABP1 | none | CTNNB1 p.Thr41Ala (5) | MSS | (−) |
| 101 | 271Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | none | SFRP1 | None | MSS | (−) |
| 12 | 670Ad | LOW | MSH2 c.1552_1553_del | ND | SFRP2, CDH1, HOXD1, SLC5A8, SFRP1 | FBXW7 p.Arg465His (7), KRAS p.Ala146Thr (10) | ND | ND |
| 16 | 672Ad | LOW | MSH6 c.900dup | ND | SFRP2, CDH1, HOXD1, SLC5A8, SFRP1 | None | ND | ND |
| 73 | 677Ad | HIGH | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | None | SFRP2, SFRP1 | None | ND | (−) |
| 112 | 686Ad | HIGH | MSH6 c.3195_3199del | IGF2, NEUROG1, CDKN2A, CRABP1 | SFRP2, SLC5A8, SFRP1 | KRAS p.Ala146Thr (45), TP53 p.Glu180Lys (5) | MSS | (−) |
| 120 | 690Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | None | SFRP2, CDH1, HOXD1, SOCS1, SLC5A8, SFRP1 | None | ND | (−) |
| 123 | 693Ad | LOW | MLH1 c.1731 + 2247_1897-402del (Mutation 1) | IGF2, NEUROG1, CDKN2A | SFRP2, CDH1, HOXD1, SFRP5, SOCS1, SLC5A8, SFRP1 | None | MSS | (−) |
Abbreviations: CIMP, CpG island methylator phenotype; MSS, microsatellite stable; MSI, microsatellite instable; ND, not done; TSGs, tumor suppressor genes; VAF, variant allele frequency.
Only one of the two microsatellite markers produced an interpretable result.
Mutational frequencies between adenomas with low-grade and high-grade dysplasia were mostly similar, with no statistically significant differences observed (Fig. 4). However, multiple genes, such as BRAF (4/41, 10% vs. 1/18, 6%) and SMAD4 (8/41, 20% vs. 3/18, 17%) were slightly more frequently mutated in adenomas with low-grade dysplasia than in tumors with high-grade dysplasia. Instead, mutations in CTNNB1 (5/18, 28% vs. 4/41, 10%), KRAS (5/18, 28% vs. 8/41, 20%), and TP53 (6/18, 33% vs. 8/41, 20%) occurred more often in advanced stage adenomas.
One adenoma with low-grade dysplasia (256Ad) carried the BRAF V600E mutation (VAF 5%). The BRAF mutation was the only mutation detected in this specimen. Other BRAF missense mutations that were found in four cases occurred in the codons 455, 467, and 469. The BRAF V600E mutant tumor was characterized by MMR-D, CIMP(+), MSI, and germline point mutation in MLH1 (c.543C > G). Moreover, this tumor harbored hypermethylation in multiple promoters, including all three probes of IGF2, two probes of NEUROG1, one CRABP1, all three probes of RUNX3, SFRP1, HOXD1, and SFRP2.
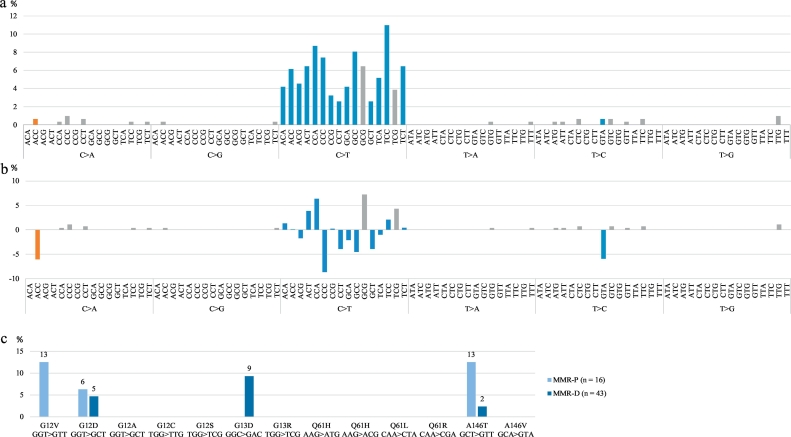
We also addressed the mutation signatures by Alexandrov et al. [37], taking the MMR deficiency related signature 6 as a reference. The adjacent bases for single base pair substitutions were determined and mutation patterns constructed for adenomas based on the relative frequencies of mutation types among all mutations found (Fig. 5a, b). In this analysis, in addition to the COSMIC mutations, we also included other variants fulfilling the filtering criteria (see Materials and Methods), although not found in the COSMIC database. The full list of the mutations used in this analysis is available in Supplementary Table S3. Based on the total of 310 missense or nonsense mutations, the following observations were made. C > T transitions were clearly the most prevalent substitutions in both MMR-P and MMR-D adenomas. However, there were some differences in base triplets harboring the transitions. Signature 6 is characterized by C > T transitions at NpCpG sites [37], thus, we investigated the frequencies of these mutations among all mutations found in MMR-P and MMR-D adenomas. Indeed, they were more prevalent in MMR-D adenomas (53/277, 19% vs. 3/33, 9%), although statistical significance was not reached. Intriguingly, the mutation with the highest probability to occur in MMR-D tumors according to the mutation signature, C > T at GpCpG, was missing in MMR-P adenomas (0/16), but was present (12/43, 28%) in MMR-D adenomas (p = 0·025). Moreover, we observed ApCpC, CpCpC, and GpTpA sites to be enriched with mutations in MMR-P adenomas compared to MMR-D adenomas with >5% difference in the relative mutation frequency. Signature 6 is characterized also by small indels, but in our study, only three deletions (1–3 bp) were found, all in MMR-D adenomas.
Fig. 5.
Distributions of different mutation types. a. Relative frequencies of different mutation types among all mutations found in all adenomas. b. The difference of relative mutation frequencies between MMR-D and MMR-P adenomas (MMR-D – MMR-P). In a. and b., orange color indicates that a mutation type occurs exclusively in MMR-P adenomas, whereas gray color indicates exclusive occurrence in MMR-D adenomas, and blue color indicates mutation types present in both MMR-P and MMR-D tumors. c. Frequency of KRAS mutations and mutation patterns in MMR-P and MMR-D adenomas. Abbreviations: MMR-D, MMR deficient; MMR-P, MMR proficient.
In addition, we examined pathogenic KRAS mutations (codons 12, 13, 61, and 146) that can cause constitutive activation of KRAS protein [34,35], and all possible mutations based on the flanking nucleotides (Fig. 5c). KRAS G13D mutations (n = 4) occurred exclusively in MMR-D adenomas, in agreement with published findings of this mutation being associated with MSI tumors [38,39]. Codon 12 and 146 mutations were more prevalent in MMR-P adenomas (see Discussion).
5. Discussion
In the majority of colorectal cancers, the adenoma-carcinoma sequence is a well-established path, where adenomatous polyps have a high risk of developing into carcinomas. However, in LS, recent findings such as frequent development of colorectal cancer despite regular colonoscopic surveillance to remove adenomas has challenged the classical view [40]. Furthermore, the tumorigenic stage at which MMR deficiency appears is controversial. Observations of MMR protein loss associated with high-grade dysplastic and larger polyps supports deficient MMR being a late event (this study and [10,[13], [14], [15],41]). However, the discovery of histologically normal colonic crypts with a loss of MMR protein expression, i.e. MMR-deficient crypt foci (MMR-DCF) [42,43], as well as a recent finding of MMR-deficiency preceding adenoma formation in LS suggest early appearance of MMR deficiency [44]. In our investigation, all LS carcinomas showed a complete loss of the MMR protein expression, whereas 10% of adenomas with high-grade dysplasia and 35% adenomas with low-grade dysplasia maintained the expression, implying that MMR protein loss occurs relatively late. Ahadova et al. [38] proposed a model of three different pathways driving LS-associated tumorigenesis, which could reconcile the contradictory findings of the adenoma-carcinoma sequence and the timing of MMR-deficiency. The model covers late appearance of deficient MMR (MMR-P adenoma with secondary MMR inactivation) as well as early MMR deficiency (MMR-DCF evolving into carcinoma either via MMR-D adenoma or directly without any polyp precursor).
Only limited information is available of molecular alterations present in LS adenomas lacking MMR deficiency as an obvious driver of tumorigenesis (MMR-P adenomas). We addressed DNA methylation changes and somatic mutations of cancer-associated genes as possible cancer-initiating or promoting events. Methylation was studied for eight CIMP markers, seven selected candidate TSGs [23], and LINE-1. The frequency of CIMP(+) and the average degree of LINE-1 hypomethylation increased with the degree of dysplasia. In general, they were rather rare events in adenomas with low-grade dysplasia or in MMR-P adenomas, indicating that wide-spread hypo- and hypermethylation occurs at later stages of tumorigenesis. Nevertheless, MMR-P adenomas revealed significantly elevated methylation levels (expressed as average Dm values) relative to normal mucosa in four CIMP marker genes: IGF2, CRABP1, NEUROG1, and CDKN2A (Fig. 2a) and the candidate TSGs SFRP1 (Fig. 2b) and SFRP2 (Fig. 2c) Our findings from LS adenomas are supported in the sporadic context by Ibrahim et al. [21] They recognized hypermethylation of NEUROG1 and SFRP2 as early events in sporadic colorectal carcinogenesis, with NEUROG1 being hypermethylated in hyperplastic polyps and SFRP2 in adenomatous polyps. Moreover, in our previous study comparing LS individuals without vs. with colorectal cancer, SFRP1 hypermethylation occurred as a “field defect” in high-risk normal mucosa from the latter group [9]. The baseline level of LINE-1 methylation may vary according to the clinical category of colorectal cancer [31,45]. Progressive LINE-1 hypomethylation from normal mucosa to adenoma to carcinoma accompanies both LS-associated (Fig. 3a) and sporadic colon tumorigenesis [22].
We used a sensitive technique (amplicon-based next-generation sequencing) to screen the hotspot regions of 22 established cancer-associated genes for somatic mutations in LS-adenomas. Variant alleles occurring with 5% or higher frequencies were considered. Mutations were somewhat more prevalent in MMR-D than MMR-P adenomas (average 2·4 vs. 1·0 mutations per sample, statistically non-significant). Additionally, MMR status of the adenomas was reflected in the base substitution and sequence context-based mutational signatures defined by Alexandrov et al. [37] so that the MMR-deficiency-associated signature 6 was overrepresented among MMR-D adenomas, especially C > T at GpCpG with a statistically significant difference, whereas ApCpC, CpCpC, and GpTpA sites were enriched with mutations in MMR-P adenomas (Fig. 5b). Previous studies have shown that targeted capture and sequencing data can successfully be used to determine mutational signatures [46]. However, the differences we report in mutation spectra must be interpreted with caution, as we targeted solely mutational hotspots in a limited number of genes, and the number of MMR-P cases was relatively modest. Chang et al. [47] used RNA-seq data to determine mutation profiles in 19 LS-associated (adenomatous) polyps. 16 (84%) were non-hypermutated and MMR-proficient except one, whereas three (16%) were hypermutated, MMR-deficient and exhibited signature 6 by Alexandrov et al. [37] Regardless of mutation rates, all LS polyps displayed immune activation, the mechanism of which remained unknown. A tumorigenic model was hypothesized in which immune deregulation constitutes one of the earliest steps and precedes the accumulation of genomic alterations.
The most frequently mutated genes in our study were TP53, KRAS, SMAD4, and CTNNB1, in decreasing order (Fig. 4). While TP53 mutations are considered rare in MSI LS tumors [38,39], such mutations occurred in up to one third of our LS adenomas (Fig. 4). The frequency of TP53 mutations in MMR-P adenomas or adenomas with low-grade dysplasia was 13% and 20%, respectively. Commonly, TP53 mutations have been associated with advanced CRC tumors [48], but recently, TP53 mutations were found already at early dysplastic stage in sporadic colorectal adenomas [49]. Intriguingly, we found a higher prevalence of pathogenic KRAS mutations in MMR-P than MMR-D adenomas (Fig. 4). Codon 12 mutations (especially G12V) are proposed to be associated with MSS CRC [38,39] and MMR-P tumors [38], which is concordant with our finding (Fig. 5c). Besides G12V, KRAS A146T mutation occurred more often in MMR-P than MMR-D adenomas in our investigation, whereas in the study by Kloth et al. [39] focusing on verified or suspected LS patients, A146T was associated with MSI tumors.
Our finding of a BRAF V600E mutation positive (MMR-D) adenoma (case 256Ad in Fig. 4) is significant since BRAF V600E mutation generally argues against LS [50]. We note that the variant allele frequency was low (5%), and confirmation by immunohistochemical methods [25] was not possible since there was no adenoma tissue left. At least four additional tumors from germline carriers of MMR gene mutations have been reported to carry BRAF V600E to date [50], together with our observation emphasizing that BRAF V600E mutation in a colorectal tumor is not an absolute indicator to exclude LS in a diagnostic practice.
The suggestion that LS tumorigenesis may proceed along different pathways is supported by a recent study by Binder et al. [51] who found two major molecular subgroups of LS-CRCs, one resembling sporadic MSI CRC and another one sporadic MSS CRC, with respect to mutation spectra and microsatellite length distributions. Furthermore, the mutation spectra suggested differences in DNA methylation between the two groups [51]. Integrated analysis of genomic and transcriptomic alterations has revealed extensive pathway sharing between hypermutated and non-hypermutated colorectal carcinomas; for example, Wnt signaling is overactive in above 90% of all tumors via APC inactivation or CTNNB1 activation [52]. Sekine et al. [44] compared APC mutation profiles in MMR-P and MMR-D adenomas from LS patients and found that frameshift mutations affecting repeat sequences accounted for a major share of all APC mutations in MMR-D adenomas but were rare in MMR-P adenomas, suggesting that MMR deficiency generally precedes APC inactivation and adenoma formation. On the other hand, Homfray et al. [53] compared APC mutation profiles in sporadic colorectal cancers with vs. without MSI, and the lack of essential differences led the authors to conclude that APC mutations occur prior to MMR inactivation. The gene panel we used did not include APC but did include the CTNNB1 gene, mutations of which were recently linked to immediate invasive growth, possibly explaining interval cancers in LS [54]. Mutation frequencies of CTNNB1 did not differ between MMR-P (13%) and MMR-D adenomas (16%) (Fig. 4).
A possibility remains that some MMR-P adenomas arising in LS patients are sporadic. Mutation frequencies in our MMR-P adenomas for the four commonly affected genes discussed above (TP53, KRAS, SMAD4, and CTNNB1) were comparable to those reported for sporadic colorectal adenomas in the literature [49,55]. However, MMR-P and MMR-D adenomas were diagnosed at similar ages, between 26 and 70 (mean 51·4 ± 3·0 CI 95%) years for MMR-P adenomas (n = 18) and between 31 and 74 (mean 52·8 ± 2·8 CI 95%) years for MMR-D adenomas (n = 47, age information unavailable for two cases), providing no obvious reason to suspect a higher proportion of sporadic cases in the MMR-P group. In addition, it is important to note that MMR genes have many other anti-carcinogenic functions beyond MMR and these functions may be even more sensitive to reduced levels of MMR gene product than the repair function. Cell line studies by Cejka et al. [4] revealed that MMR proficiency was restored at low concentrations of MLH1, whereas checkpoint activity required a full complement of MLH1 protein. Our recent results from Mlh1+/− mice showed normal Mlh1 protein expression and absence of MSI in colorectal carcinomas that developed in these animals [56]. Instead of MSI, the tumors revealed signs of chromosomal instability, consistent with decreased mRNA expression of Mlh1 and several other chromosomal segregation-associated genes in normal mucosa.
In summary, the presence of a strong predisposing defect (inherited MMR gene mutation) in every cell but the absence of tumor development until additional hits (to the predisposing MMR gene and other genes) occur in a target tissue, makes LS a valuable model to dissect the multistep tumorigenesis of colon and other organs. Our finding of MMR-P adenomas shows, first, that tumor formation in LS is possible even in the absence of a second hit to inactivate the wild-type allele of the predisposing MMR gene. Second, we identify possible alternative events prevalent in MMR-P adenomas that may serve as early alterations accelerating, or possibly initiating, LS-associated tumorigenesis when MMR protein expression is still intact. Hypermethylation of IGF2, NEUROG1, CDKN2A, CRABP1, SFRP1, and SFRP2, and especially KRAS mutations may represent such events. Third, the emerging epigenetic and genetic differences between MMR-P and MMR-D adenomas imply the existence of different alternative routes to cancer development in LS, consistent with observations of genetic heterogeneity in LS carcinomas as discussed above. Further studies with large sample sizes are necessary to deepen the understanding of tumorigenic sequences leading to LS cancers, thereby improving early recognition of increased tumor risk and cancer prevention in LS.
Acknowledgments
Acknowledgements
We thank the patients contributing to this study. We thank Beatriz Alcala-Repo and Kirsi Pylvänäinen for collecting the samples and patient information. Saila Saarinen and Omar Youssef are thanked for laboratory support. The work was funded by the Jane and Aatos Erkko Foundation (MN, J-PM, and PP), Academy of Finland (grant no. 294643) (PP), the Cancer Foundation Finland (J-PM and PP), the Sigrid Juselius Foundation (PP), and HiLIFE (PP). Funding sources had no involvement in the study design, in the collection, analysis, or interpretation of data, in the writing, nor in the decision to submit the paper for publication.
Declaration of interests
During the conduct of the study, Dr. Nyström received a grant from the Jane and Aatos Erkko Foundation, Dr. Mecklin received grants from the Jane and Erkko Foundation and the Cancer Foundation Finland, and Dr. Peltomäki received grants from the Jane and Aatos Erkko Foundation, the Academy of Finland, the Cancer Foundation Finland, the Sigrid Juselius Foundation, and the HiLIFE. Other authors declare no conflicts of interests.
Author contributions
SM-N, SV, and PP designed the study. LR-S, AL, and J-PM provided the patient data and samples. AR performed the histopathological analyses. VS and SK provided sequencing expertise and access to the facilities. MN shared epigenetic expertise and information of epigenetic markers derived from a mouse model of LS. SM-N and SV performed the laboratory experiments, and data analyses. SM-N and PP wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.018.
Appendix A. Supplementary data
Supplementary Tables S1-S3, and Supplementary Figures S1-S3
References
- 1.Thompson B.A., Spurdle A.B., Plazzer J.P. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ligtenberg M.J., Kuiper R.P., Chan T.L. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Tsutsumi S., Kawaguchi T. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012;22:208–219. doi: 10.1101/gr.123109.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cejka P., Stojic L., Mojas N. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lao V.V., Grady W.M. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moller P., Seppala T.T., Bernstein I. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the prospective Lynch Syndrome Database. Gut. 2018;67:1306–1316. doi: 10.1136/gutjnl-2017-314057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemminki A., Peltomaki P., Mecklin J.P. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat Genet. 1994;8:405–410. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 8.Porkka N., Valo S., Nieminen T.T. Sequencing of Lynch syndrome tumors reveals the importance of epigenetic alterations. Oncotarget. 2017;8:108020–108030. doi: 10.18632/oncotarget.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valo S., Kaur S., Ristimaki A. DNA hypermethylation appears early and shows increased frequency with dysplasia in Lynch syndrome-associated colorectal adenomas and carcinomas. Clin Epigenetics. 2015;7:71. doi: 10.1186/s13148-015-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halvarsson B., Lindblom A., Johansson L., Lagerstedt K., Nilbert M. Loss of mismatch repair protein immunostaining in colorectal adenomas from patients with hereditary nonpolyposis colorectal cancer. Mod Pathol. 2005;18:1095–1101. doi: 10.1038/modpathol.3800392. [DOI] [PubMed] [Google Scholar]
- 11.Muller A., Beckmann C., Westphal G. Prevalence of the mismatch-repair-deficient phenotype in colonic adenomas arising in HNPCC patients: results of a 5-year follow-up study. Int J Colorectal Dis. 2006;21:632–641. doi: 10.1007/s00384-005-0073-6. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M., Nakajima T., Sugano K. Mismatch repair deficiency in Lynch syndrome-associated colorectal adenomas is more prevalent in older patients. Histopathology. 2016;69:322–328. doi: 10.1111/his.12941. [DOI] [PubMed] [Google Scholar]
- 13.Yurgelun M.B., Goel A., Hornick J.L. Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev Res (Phila) 2012;5:574–582. doi: 10.1158/1940-6207.CAPR-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Jong A.E., Morreau H., Van Puijenbroek M. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology. 2004;126:42–48. doi: 10.1053/j.gastro.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 15.Meijer T.W., Hoogerbrugge N., Nagengast F.M., Ligtenberg M.J., van Krieken J.H. In Lynch syndrome adenomas, loss of mismatch repair proteins is related to an enhanced lymphocytic response. Histopathology. 2009;55:414–422. doi: 10.1111/j.1365-2559.2009.03403.x. [DOI] [PubMed] [Google Scholar]
- 16.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino S., Nosho K., Kirkner G.J. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard G., Eiges R., Gaudet F., Jaenisch R., Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 20.Belancio V.P., Deininger P.L., Roy-Engel A.M. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibrahim A.E., Arends M.J., Silva A.L. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60:499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 22.Jiang A.C., Buckingham L., Barbanera W., Korang A.Y., Bishesari F., Melson J. LINE-1 is preferentially hypomethylated within adenomatous polyps in the presence of synchronous colorectal cancer. Clin Epigenetics. 2017;9:25. doi: 10.1186/s13148-017-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pussila M., Sarantaus L., Dermadi Bebek D. Cancer-predicting gene expression changes in colonic mucosa of Western diet fed Mlh1+/− mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isola J., Devries S., Chu L., Ghazvini S., Waldman F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994;145:1301–1308. [PMC free article] [PubMed] [Google Scholar]
- 25.Thiel A., Heinonen M., Kantonen J. BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch. 2013;463:613–621. doi: 10.1007/s00428-013-1470-9. [DOI] [PubMed] [Google Scholar]
- 26.Esemuede I., Forslund A., Khan S.A. Improved testing for microsatellite instability in colorectal cancer using a simplified 3-marker assay. Ann Surg Oncol. 2010;17:3370–3378. doi: 10.1245/s10434-010-1147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loukola A., Eklin K., Laiho P. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC) Cancer Res. 2001;61:4545–4549. [PubMed] [Google Scholar]
- 28.Nygren A.O., Ameziane N., Duarte H.M. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg M., Hagland H.R., Soreide K. Comparison of CpG island methylator phenotype (CIMP) frequency in colon cancer using different probe- and gene-specific scoring alternatives on recommended multi-gene panels. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisenberger D.J., Siegmund K.D., Campan M. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 31.Pavicic W., Joensuu E.I., Nieminen T., Peltomaki P. LINE-1 hypomethylation in familial and sporadic cancer. J Mol Med (Berl) 2012;90:827–835. doi: 10.1007/s00109-011-0854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forbes S.A., Beare D., Boutselakis H. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1121. (D777–D83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng G., Peng E., Gum J., Terdiman J., Sleisenger M., Kim Y.S. Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer. 2002;86:574–579. doi: 10.1038/sj.bjc.6600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edkins S., O'Meara S., Parker A. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feig L.A., Cooper G.M. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loupakis F., Ruzzo A., Cremolini C. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrov L.B., Nik-Zainal S., Wedge D.C. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahadova A., Gallon R., Gebert J. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int J Cancer. 2018;143:139–150. doi: 10.1002/ijc.31300. [DOI] [PubMed] [Google Scholar]
- 39.Kloth M., Ruesseler V., Engel C. Activating ERBB2/HER2 mutations indicate susceptibility to pan-HER inhibitors in Lynch and Lynch-like colorectal cancer. Gut. 2016;65:1296–1305. doi: 10.1136/gutjnl-2014-309026. [DOI] [PubMed] [Google Scholar]
- 40.Moller P., Seppala T., Bernstein I. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iino H., Simms L., Young J. DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary non-polyposis colorectal cancer. Gut. 2000;47:37–42. doi: 10.1136/gut.47.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kloor M., Huth C., Voigt A.Y. Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: a pathological study. Lancet Oncol. 2012;13:598–606. doi: 10.1016/S1470-2045(12)70109-2. [DOI] [PubMed] [Google Scholar]
- 43.Shia J., Stadler Z.K., Weiser M.R. Mismatch repair deficient-crypts in non-neoplastic colonic mucosa in Lynch syndrome: insights from an illustrative case. Fam Cancer. 2015;14:61–68. doi: 10.1007/s10689-014-9751-2. [DOI] [PubMed] [Google Scholar]
- 44.Sekine S., Mori T., Ogawa R. Mismatch repair deficiency commonly precedes adenoma formation in Lynch Syndrome-Associated colorectal tumorigenesis. Mod Pathol. 2017;30:1144–1151. doi: 10.1038/modpathol.2017.39. [DOI] [PubMed] [Google Scholar]
- 45.Antelo M., Balaguer F., Shia J. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zehir A., Benayed R., Shah R.H. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang K., Taggart M.W., Reyes-Uribe L. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. 2018;4:1085–1092. doi: 10.1001/jamaoncol.2018.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolff R.K., Hoffman M.D., Wolff E.C. Mutation analysis of adenomas and carcinomas of the colon: early and late drivers. Genes Chromosomes Cancer. 2018;57:366–376. doi: 10.1002/gcc.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parsons M.T., Buchanan D.D., Thompson B., Young J.P., Spurdle A.B. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151–157. doi: 10.1136/jmedgenet-2011-100714. [DOI] [PubMed] [Google Scholar]
- 51.Binder H., Hopp L., Schweiger M.R. Genomic and transcriptomic heterogeneity of colorectal tumours arising in Lynch syndrome. J Pathol. 2017;243:242–254. doi: 10.1002/path.4948. [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Homfray T.F., Cottrell S.E., Ilyas M. Defects in mismatch repair occur after APC mutations in the pathogenesis of sporadic colorectal tumours. Hum Mutat. 1998;11:114–120. doi: 10.1002/(SICI)1098-1004(1998)11:2<114::AID-HUMU3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 54.Ahadova A., von Knebel Doeberitz M., Blaker H., Kloor M. CTNNB1-mutant colorectal carcinomas with immediate invasive growth: a model of interval cancers in Lynch syndrome. Fam Cancer. 2016;15:579–586. doi: 10.1007/s10689-016-9899-z. [DOI] [PubMed] [Google Scholar]
- 55.Chang P.Y., Chen J.S., Chang S.C. Acquired somatic TP53 or PIK3CA mutations are potential predictors of when polyps evolve into colorectal cancer. Oncotarget. 2017;8:72352–72362. doi: 10.18632/oncotarget.20376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pussila M., Toronen P., Einarsdottir E. Mlh1 deficiency in normal mouse colon mucosa associates with chromosomally unstable colon cancer. Carcinogenesis. 2018;39:788–797. doi: 10.1093/carcin/bgy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S3, and Supplementary Figures S1-S3