Abstract
Genetic manipulation of genes to upregulate specific branches of metabolic pathways is a method that is commonly used to improve fruit quality. However, the use of a single gene to impact several metabolic pathways is difficult. Here, we show that overexpression of the single gene SlMYB75 (SlMYB75-OE) is effective at improving multiple fruit quality traits. In these engineered fruits, the anthocyanin content reached 1.86 mg g−1 fresh weight at the red-ripe stage, and these SlMYB75-OE tomatoes displayed a series of physiological changes, including delayed ripening and increased ethylene production. In addition to anthocyanin, the total contents of phenolics, flavonoids and soluble solids in SlMYB75-OE fruits were enhanced by 2.6, 4, and 1.2 times, respectively, compared to those of wild-type (WT) fruits. Interestingly, a number of aroma volatiles, such as aldehyde, phenylpropanoid-derived and terpene volatiles, were significantly increased in SlMYB75-OE fruits, with some terpene volatiles showing more than 10 times higher levels than those in WT fruits. Consistent with the metabolic assessment, transcriptomic profiling indicated that the genes involved in the ethylene signaling, phenylpropanoid and isoprenoid pathways were greatly upregulated in SlMYB75-OE fruits. Yeast one-hybrid and transactivation assays revealed that SlMYB75 is able to directly bind to the MYBPLANT and MYBPZM cis-regulatory elements and to activate the promoters of the LOXC, AADC2 and TPS genes. The identification of SlMYB75 as a key regulator of fruit quality attributes through the transcriptional regulation of downstream genes involved in several metabolic pathways opens new avenues towards engineering fruits with a higher sensory and nutritional quality.
Subject terms: Secondary metabolism, Plant molecular biology, Plant breeding, Plant stress responses, Plant physiology
Genetics: one small tweak, one giant leap for tomatoes
A single gene has been identified in tomatoes that regulates multiple aspects of fruit quality, including levels of health-promoting anthocyanins, opening the door to engineering more nutritious and better-tasting tomatoes. Fruit breeders have long-manipulated genes to enhance traits like yield or disease resistance, but it is rare to find a single gene that can improve multiple aspects of fruit quality. In this study, Zhengguo Li at Chongquing University in China and colleagues show that over-expressing a transcription factor called SIMYB75 results in striking deep purple tomatoes enriched in anthrocyanins–antioxidants thought to be protective against various diseases - which tomatoes are usually devoid of. They also had higher levels of other health-promoting phytochemicals and enhanced production of aroma volatiles, which can influence the taste and flavor of fruit.
Introduction
As an indispensable daily supplier of nutrition to humans, fruit offers abundant nutrients, including vitamins, minerals, antioxidants and fiber1. Therefore, improving the nutritional and sensory qualities of fruit is becoming a primary goal to meet consumer requirements for healthier food. Nevertheless, breeding efforts to date have primarily concentrated on yield and disease resistance traits, whereas sensory and nutritional aspects such as the pigment, aroma, taste and health-promoting compounds have not received sufficient attention. In recent years, tremendous advances in functional genomics and biotechnology tools have presented new prospects for producing high-quality fruits that can better meet consumer expectations.
Anthocyanins are water-soluble pigments that are widely distributed in many plants, where they confer their typical appealing color to various tissues and organs in a number of species. With regards to their impact on human health, numerous studies have indicated that anthocyanins have high antioxidant properties, with the scavenging of active oxygen species2. In addition, anthocyanins have been reported to play a role in protecting against age-related degenerative diseases, especially by inhibiting tumor cell growth3. Anthocyanins are synthesized via the flavonoid pathway, which is a branch of the phenylalanine pathway4. The accumulation of anthocyanins in plants is regulated by a series of environmental conditions and developmental signals5,6. Two types of genes have been suggested to participate in the anthocyanin pathway, (i) those encoding key enzymes for anthocyanin biosynthesis4 and (ii) those coding for the transcription factors (TFs) that regulate the spatio-temporal expression of functional genes7. Three types of TFs, R2R3-MYB, bHLH and WD40, are known to control the anthocyanin biosynthetic pathway, and most likely, these TFs work together through the formation of a transcription complex named MBW8,9. However, several studies have indicated that MYB TFs could also function independently of the MBW complex to regulate the biosynthesis of anthocyanins, thus assigning a crucial role to MYB factors per se in controlling anthocyanin biosynthesis7,10,11.
Tomatoes are one of the most widely consumed fruit crops in the world, which motivates their frequent use as a model species to study nutritional metabolism. Moreover, the tomato is an excellent candidate for the transgenic enhancement of its flavonoid content because of its low basal content of these compounds3,12. Although tomato plants with both Aft and atv alleles could accumulate small amounts of anthocyanin, the mechanism and key genes controlling this variation have not yet been deciphered13,14. The silencing of the SlDET1 gene in tomatoes has resulted in altered light-mediated developmental processes along with an enhanced flavonoid content15. Similarly, overexpression of the chalcone isomerase gene in tomatoes could increase the flavonoid content up to 78-fold exclusively in the peel, but the overall level in the fruits remained low12. In contrast to the functional genes, manipulating the expression of TFs proved more effective at modulating the secondary metabolism. Lc and C1 are two TFs in maize that control anthocyanin biosynthesis; however, overexpression of these two TFs in tomatoes greatly increased the flavonoid content, but not the anthocyanin content16. Overexpression of AtMYB12 under the control of the fruit-specific E8 promoter resulted in orange tomatoes that contained significantly high contents of flavonoid and caffeoyl quinic acid17. However, the most significant increase in the anthocyanin content of tomato fruits was first obtained by expressing Del and Ros1, two snapdragon genes encoding bHLH and MYB TFs, respectively, which were under the control of the E8 promoter, resulting in fully purple tomato fruits3. More recently, crossing the AtMYB12-overexpression line with the Del/Ros1 line was reported to enhance anthocyanin accumulation18. Interestingly, this study indicated that AtMYB12 not only promoted flavonoid biosynthesis, but it also improved the carbon supply from the primary metabolism, energy and reducing power, which resulted in a larger aromatic amino acid supply for secondary metabolism18. It is important to mention that the purple tomato fruits obtained in these studies always resulted from the cooperative action of two or more TFs. This result causes difficulty for traditional breeders, since it is challenging to find a wild species that could highly express these two or three key regulatory genes at the same time.
Recently, the MYB TF gene Cs6g17570 was identified to play a critical role in regulating anthocyanin biosynthesis in blood oranges19. Interestingly, the upregulation of its homologous gene AtMYB113-like resulted in the increased accumulation of anthocyanin in Arabidopsis, whereas its downregulation led to a reduced amount of this pigment. Moreover, this process was reported to be dependent on the MBW complex20. Through a homology alignment, we identified a tomato gene, named SlMYB75 (SGN, https://solgenomics.net/search/locus) or SlAN2, which showed the highest sequence similarity with Cs6g17570 (Supplementary Fig. S1). Overexpression of SlAN2 was previously reported to improve tomato plant resistance to high temperature, cold and oxidative stresses21,22. A recent study showed that overexpression of SlANT1 or SlAN2 induced accumulation of anthocyanin that was unevenly distributed in tomato fruit, but only SlAN2 was able to act as a positive regulator of anthocyanin biosynthesis in vegetative tomato tissues under stress conditions23. At the same time, SlAN2-overexpression fruits displayed an orange color, fast softening and higher ethylene content24. In addition, several other MYB TFs are also involved in some fruit quality attributes, such as primary metabolism, secondary metabolism and organic acid metabolism18,25, and a consumer sensory analysis showed that there is a consumer preference for purple tomato fruits, for their better perceived flavor26. Thus, the potential role of SlMYB75 in tomato fruit quality remains to be investigated.
In the present study, we attempt to describe the physiological and metabolic changes in purple tomatoes produced by overexpressing a single SlMYB75 TF (SlMYB75-OE). The data indicated that SlMYB75 can effectively induce the accumulation of anthocyanin in various tissues, and its ectopic expression leads to increased ethylene production and enhanced phenolic, flavonoid and volatile aroma contents. Moreover, transcriptomic profiling showed that many genes involved in the ethylene response, phenylpropanoid and isoprenoid pathways were greatly affected in SlMYB75-OE fruits. Using yeast one-hybrid and dual-luciferase assays, we found that the conserved MYBPLANT (AAACCAACCC) and MYBPZM (ACCTACCC) elements were the core binding sites of SlMYB75, and we revealed the ability of SlMYB75 to activate the promoters of the LOXC, AADC2 and TPS genes. This study demonstrates that a single SlMYB75 TF is able to increase the anthocyanin content to 1.86 mg g−1 fresh weight at the red ripening stage and promote volatile aroma accumulation partly through the transcriptional regulation of downstream genes involved in the corresponding metabolic pathways.
Materials and methods
Plant materials, growth conditions and chemicals
Solanum lycopersicum cv. Micro-Tom was selected as the wild type (WT) in this study. All the tomato seedlings were grown in an intelligent greenhouse under standard conditions (16/8 h and 25 °C/18 °C day/night cycle, 80% humidity and 250 µmol m−2 s−1 light intensity). For the hormone and stress treatments, the leaves of 1-month-old tomato plants treated with different reagents were harvested. For each tissue/organ type, samples were collected from at least six healthy plants. All the samples were mixed and frozen under liquid nitrogen immediately. The sample experiments were conducted three independent times.
For the chemicals, Folin-Ciocalteu phenol reagent was purchased from Sigma (St. Louis, MO, USA). HPLC-grade methanol was purchased from Merck KgaA (Darmstadt, Germany). The other reagents were all of analytical grade and were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China).
Vector construction and plant transformation
The full length of the SlMYB75 coding sequence was amplified from tomato cDNA and then cloned into the K303 expression vector, which contain two 35s promoters from Gateway technology. The final vector was transferred into GV3101, and Agrobacterium-mediated transformation was performed as described previously27,28. The positive transgenic plants were identified by PCR, and homozygous plants from T2 or later generations were used for the experiments. All the primers used in this study are listed in Supplementary Table S2.
RNA-Seq analysis
Total RNA was extracted from the fruits at the mature green (MG) and breaker (BR + 0) stages using an RNeasy Plant Mini Kit (Tiangen, China) according to the manufacturer’s protocol. Samples from both WT and SlMYB75-OE fruits were collected with two biological replicates. The concentration and quality of the RNA were assayed using a NanoDrop Lite spectrophotometer (Thermo Scientific). The cDNA libraries were constructed and then sequenced using a BGISEQ-500 System (BGI Inc.). Clean data were obtained after they were filtered and then mapped to the reference genome of S. lycopersicum in the Tomato SGN database (http://solgenomics.net/) with Bowtie2. The homogenized data were used to calculate the gene expression levels with RSEM. The criteria for defining differentially expressed genes (DEGs) were fold change ≥2.00 and P value ≤ 0.05. The raw transcriptome reads of this study have been deposited in the NCBI Short Read Archive under accession number SRP158557.
Real time quantitative PCR
One microgram of total RNA (RNeasy Plant Mini Kit, Tiangen) was used to synthesize first strand cDNA with PrimeScriptTM RT reagent Kit using gDNA Eraser (Perfect Real Time) (TAKARA, Japan). Quantitative real-time PCR was performed with a Bio-Rad CFX system (Bio-Rad, USA) using SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (TAKARA, Japan). Each sample was collected as three independent biological replicates, and the relative fold differences were calculated using a comparative Ct method. SlUBI was used as the internal reference over the entire experiment.
Measurement of metabolites
For the total anthocyanin extraction, 0.1 g of lyophilized powder samples was incubated in 1 mL of pH 1.0 buffer solution (50 mM KCL, 150 mM HCL) and 1 mL of pH 4.5 buffer solution (400 mM CH3COONa, 240 mM HCL) for 24 h in the dark at 4 °C with gentle shaking, respectively. The samples were centrifuged at 14,000×g for 20 min. The total anthocyanin content in the supernatant was measured spectrophotometrically and expressed in mg of petunidin-3-(p-coumaroyl rutinoside)-5-glucoside (extinction coefficient 17000, molecular weight 934) per gram fresh weight3,23.
For the ethylene measurement, fruits at different development stages were harvested and placed in an open 50 mL jar for 2 h to minimize the wound ethylene caused by picking. The jars were sealed and incubated at room temperature for 1 h, and 1 mL of headspace gas was collected and then injected into a VARIAN CP3800 gas chromatograph equipped with a flame ionization detector (USA). The samples were compared with reagent-grade ethylene standards of known concentrations and normalized to the fruit weight29. At least 10 individual fruits were measured for each sample.
The total soluble solids and titratable acids were determined using a digital refractometer (Atago PR-101R, Tokyo, Japan) and titration method, respectively30. Each replicate contained 20 fresh tomato fruits and all the determinations were performed in triplicate.
For the total phenolic and flavonoid extractions, 1 g of lyophilized powder samples were incubated with methanol (80%, 24 mL) at 25 °C for 12 h with shaking and then centrifuged at 3000×g for 10 min at 4 °C. The residue was extracted twice more using the same procedure, and then the supernatants were collected and finally diluted to 50 mL with methanol to detect the total phenolics and flavonoids. The total phenolic content was determined using the Folin-Ciocalteu method. The total flavonoid content was measured as described previously31.
To measure the primary component of phenolics and flavonoids, 0.5 g of lyophilized powder was extracted with methanol (80%, 12 mL) and dimethyl sulfoxide (1:1, v/v) using the same procedures as the procedure used with the total phenolics and flavonoids. After the samples were filtered through a 0.22 μm syringe filter, the phenolics and flavonoids were measured using the HPLC method described previously31.
The concentration of aroma volatiles was determined as described previously, with some modifications30. Three grams of lyophilized powder were homogenized with 5 mL of saturated sodium chloride solution, and then 5 μL of ethyl nonanoate was added as an internal reference. The solution was incubated at 40 °C for 30 min, and a solid-phase microextraction (SPME) needle with a 1 cm-long fiber coated with 65 μm divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) fibers (Supelco Co., Bellefonte PA, USA) was used to extract the volatiles. A GCMS-QP2010 gas chromatograph-mass spectrometer system (Shimadzu Corporation, Kyoto, Japan) with an Rtx-5MS (Restek)-fused silica capillary column (5% diphenyl, 95% dimethyl polysiloxane) (0.32 mm, 30 m, 0.5 lm, J&W Scientific, Folsom CA, USA) was used for the compound confirmation. GC-MS Postrun Analysis software (SHIMADZU, GC-MS-QP2010, Japan) was used to evaluate the chromatograms and mass spectra. The compounds were identified by comparing their mass spectra with the data system library (NIST08). The concentrations of the volatile components were expressed as ng g−1 FW h−1.
Yeast one-hybrid assay
Yeast one-hybrid (Y1H) assays were performed using a Matchmaker Gold Yeast One Hybrid System (Clontech). To construct the prey and bait vectors, the full length of the SlMYB75 open reading frame (ORF) sequence was cloned and inserted into the pGADT7 vector, and the eight conserved cis-elements were cloned into the pAbAi vector. The bait plasmids were transformed into the Y1H Gold strain according to the manufacturer’s instructions. Aureobasidin A (AbA) was used to screen the minimal inhibitory concentration for the bait strains. The prey plasmid was transformed into a bait yeast strain to determine the DNA-protein interaction by screening them on SD medium with AbA and without leucine.
Dual-luciferase transient expression assay
For the dual-luciferase assay, the full-length ORF of SlMYB75 was cloned and inserted into a pGreenII 62-SK vector (effector), and the promoter sequence of the specific different expression gene (DEG) was cloned and inserted into pGreenII 0800 LUC vector (reporter). After their transformation into GV3101, the effector and reporter strains were cultured and then resuspended with infiltration buffer (10 mM MES, 10 mM MgCl2, 200 mM acetosyringone, pH 5.6) to an OD 600 of 1.0–1.5. The mixtures of effector and reporter cultures were infiltrated into Nicotiana benthamiana leaves with needleless syringes. Firefly luciferase and Renilla luciferase were assayed at 3 days after infiltration using Dual Luciferase Reporter Assay System reagents (Promega). The binding activity of SlMYB75 to the promoter of each specific DEG was calculated by finding the LUC to REN ratio. At least six biological replicates were conducted for each combination32.
Statistical analysis
All the experiments were repeated at least three times, and the results were presented with the standard deviations. Student’s t-tests were used to analyze the data, and a difference was considered to be statistically significant when P < 0.05.
Results
Overexpression of a single SlMYB75 induces anthocyanin accumulation
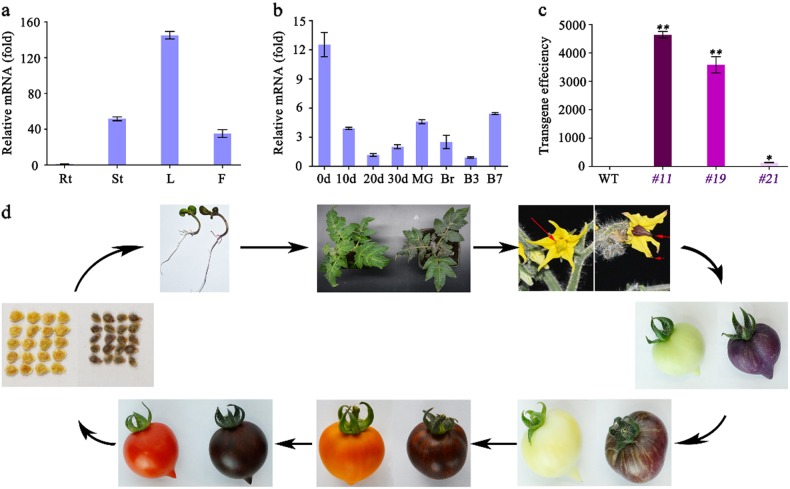
A qRT-PCR investigation of the SlMYB75 expression pattern indicated that transcripts corresponding to this gene are detected in all tissues, but they display higher expression levels in vegetative tissues, especially in leaf organs (Fig. 1a and b). However, unlike the Cs6g17570 gene19, the expression level of SlMYB75 in tomato fruit organs is very low (Fig. 1a, b). To address the functional significance of SlMYB75, a tomato “Micro-Tom” cultivar was transformed with a sense construct of SlMYB75, leading to the generation of three independent transgenic homozygous lines. The expression of SlMYB75 in SlMYB75-OE plants was assessed by qRT-PCR, indicating a dramatic upregulation in line #11 and line #19, while the #21 line only showed a slight upregulation (Fig. 1c), and then lines #11 and 19 were selected to perform further experiments. A phenotypic evaluation revealed that the SlMYB75-OE lines accumulated abundant amounts of anthocyanins in both vegetative and reproductive organs, especially in the stamen, in which the color turned totally purple (Fig. 1d). Furthermore, the SlMYB75-OE lines displayed significantly smaller seeds than the WT (Fig. 1d). To further investigate the expression characteristics of SlMYB75, we checked the expression level of this TF in WT seedlings treated with different hormones or ones that were subjected to stress conditions. The data showed that SlMYB75 is responsive to all the treatments applied here, consistent with the presence of the corresponding cis-elements identified in the promoter of SlMYB75 (Supplementary Fig. S2). These results indicated that overexpressing a single SlMYB75 TF can lead to abundant anthocyanin accumulation in both vegetative and reproductive organs, and this TF could be induced by various hormones or stress conditions.
Fig. 1. SlMYB75 expression pattern and generation of transgenic plants.
a, b, qRT-PCR analysis of SlMYB75 expression level. Rt, root; St, stem; L, leaf; and F, flower; 0, 10, 20, and 30 d represent the fruits at 0, 10, 20, and 30 d after flowering; MG, mature green; and Br, B3, and B7 represent the fruits at 0, 3, and 7 d after the breaker stage. c, The SlMYB75 mRNA level in fruits from the #11, #19 and #21 transgenic lines. d, Phenotypic characterization of WT (left) and #11 (right) plants at different growth stages. The data represent the means ± SD of three biological replicates. “*” and “**” indicate significant differences between SlMYB75-OE tomato and WT with P < 0.05 and P < 0.01, respectively, as determined by the t-test
Physiological features of the SlMYB75-OE tomato plants
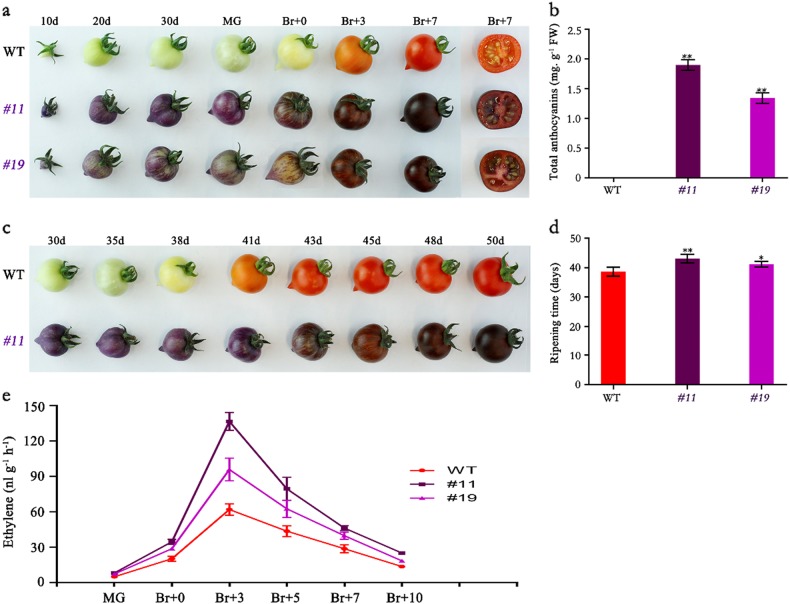
One of the striking features of the SlMYB75-OE lines is the purple color exhibited by several organs, including the seeds, stamens and fruits. An assessment of the anthocyanin accumulation revealed high levels in the SlMYB75-OE fruits, and these compounds were not detected in the WT (Fig. 2a, b). Despite their deep purple color, it was easy to identify the different ripening stages of the transgenic fruits by checking the color of the low-anthocyanin region (Fig. 2a). Moreover, SlMYB75-OE plants exhibited 3–5-day delays in the occurrence of fruit ripening onset compared to the WT (Fig. 2c, d). Strikingly, ripening-associated ethylene production was significantly higher in SlMYB75-OE tomatoes at all ripening stages (Fig. 2e), which contrasted with the delayed ripening. These data suggested that overexpression of a single SlMYB75 TF can result in a series of physiological changes.
Fig. 2. Phenotypic and physiological characteristics of WT and SlMYB75-OE plants.
a, Phenotype of WT, #11 and #19 fruits at different developmental stages. b, The total anthocyanin content was measured in WT and SlMYB75-OE fruits at BR + 7. c, Fruit ripening process of a representative line (#11) compared with the WT. d, Quantification of fruit ripening by days to reach the breaker stage in the WT and SlMYB75-OE lines. The first and second branches of each plant from a total of 20–24 plants for each line were used to quantify the ripening time. e, Ethylene production of WT and SlMYB75-OE fruits was measured at different developmental stages. The values represent the means of at least 10 individual fruits. The statistical significance was based on Student’s t-test, *P < 0.05; and **P < 0.01, and the data are the means ± SD
Transcriptome profiling of the SlMYB75-OE tomato fruits
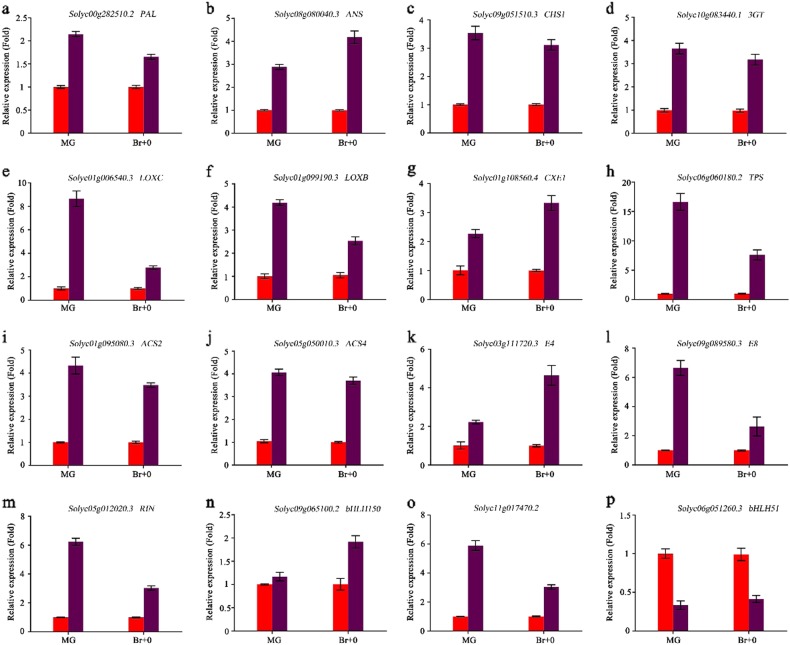
To gain insight into the extent of transcriptomic reprogramming induced by the ectopic expression of SlMYB75, we performed genome-wide transcriptomic profiling in WT and SlMYB75-OE fruits (line #11) at the MG and BR + 0 stages. Compared to the WT, a total of 1576 and 2843 genes were differently expressed (DEGs) in SlMYB75-OE tomatoes at the MG and BR + 0 stages, respectively (Files S1 and S2). Based on the Gene Ontology (GO) assignment, the common DEGs of the MG and BR + 0 stages (675 common DEGs) were assigned to three categories, namely, biological process, cellular component and molecular function. “Biological process” was primarily focused on metabolic and cellular processes. Alterations in the molecular function analysis were primarily involved in the binding and catalytic activities (Supplementary Fig. S3 and File S3). Furthermore, 675 common DEGs were enriched in 73 KEGG pathways. The top five enriched pathways were the biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, protein processing in the endoplasmic reticulum, plant hormone signal transduction and flavonoid biosynthesis (Supplementary Fig. S4 and File S4). These results indicated that SlMYB75 overexpression impacted multiple processes, including transcription, stress responses, secondary metabolism and phytohormone signaling pathways. Based on the phenotype properties and potential application values of the SlMYB75-OE tomato, we focused on genes involved in metabolic processes, ripening-related processes and TFs (Table 1). To validate the accuracy of the generated transcriptome data, a total of 16 genes related to the above-cited processes were selected for qRT-PCR analysis. The outcome of this targeted expression analysis was highly consistent with the transcriptome data for all the tested genes (Fig. 3).
Table 1.
List of different expression genes (DEGs) between WT and #11 tomato fruits at the MG and BR + 0 stages. Genes marked with asterisks were validated by qPCR
| ITAG 2.40 Tomato | Log2-fold (#11/WT; MG) | Log2-fold (#11/WT; BR + 0) | Functional annotation |
|---|---|---|---|
| Phenylalanine pathway | |||
| Solyc10g086180.2 | 1.66 | 1.07 | Phenylalanine ammonia-lyase |
| Solyc00g282510.2* | 1.68 | 1.03 | Phenylalanine ammonia-lyase-like |
| Solyc05g052240.3 | 1.34 | 1.71 | Probable chalcone-flavonone isomerase 3 |
| Solyc08g080040.3* | 1.51 | 1.68 | Leucoanthocyanidin dioxygenase-like |
| Solyc09g091510.3* | 1.87 | 1.60 | Chalcone synthase 1 |
| Solyc12g088170.2 | 8.96 | 10.01 | Anthocyanin acyltransferase |
| Solyc10g083440.1* | 1.85 | 1.46 | Anthocyanidin 3-O-glucosyltransferase-like |
| Flavor related | |||
| Solyc01g006540.3* | 2.86 | 1.19 | LOXC |
| Solyc01g099190.3* | 2.49 | 1.46 | LOXB |
| Solyc01g006560.3 | 2.25 | 1.38 | LOXF |
| Solyc08g006740.3 | 2.00 | 1.71 | AADC2 |
| Solyc01g108560.4* | 1.03 | 2.16 | CXE1 |
| Solyc06g060180.2* | 4.40 | 1.90 | TPS |
| Solyc10g011920.2 | 1.10 | 1.66 | Aromatic amino acid lyase |
| Ripening related pathway | |||
| Solyc01g095080.3* | 2.20 | 1.94 | ACS2 |
| Solyc05g050010.3* | 1.80 | 1.98 | ACS4 |
| Solyc03g111720.3* | 1.91 | 3.69 | E4 |
| Solyc09g089580.3* | 1.74 | 1.50 | E8 |
| Solyc09g075440.3 | 1.23 | 1.76 | ETR3(NR) |
| Solyc05g012020.3* | 3.09 | 1.68 | RIN |
| Solyc03g044300.3 | 2.39 | 1.40 | AP2a |
| Solyc03g031860.3 | 2.60 | 1.71 | PSY1 |
| Solyc10g080210.2 | 2.54 | 2.33 | PG2a |
| Other genes | |||
| Solyc09g065100.2* | 0.62 | 1.34 | bHLH150 |
| Solyc12g007070.2 | 2.62 | 1.28 | Heat stress transcription factor C-1 |
| Solyc11g017470.2* | 2.39 | 1.02 | NAC domain-containing protein 2-like |
| Solyc10g079050.2 | 1.46 | 1.71 | Transcription factor bHLH130-like |
| Solyc10g009550.3 | −2.50 | −1.06 | Probable WRKY transcription factor 30 |
| Solyc09g014990.3 | −1.93 | −1.18 | Probable WRKY transcription factor 26 |
| Solyc06g051260.3* | −1.46 | −1.25 | Transcription factor bHLH51 |
| Solyc03g115850.3 | −0.99 | −1.63 | NAC domain-containing protein 100 |
| Solyc10g080030.2 | −1.89 | −1.69 | MADS-box transcription factor 23-like |
| Solyc12g014140.2 | −1.93 | −1.81 | TCP transcription factor 3 |
| Solyc11g028020.2 | −1.35 | −2.09 | TAGL11 transcription factor |
| Solyc10g005010.3 | −2.31 | −3.38 | NAC domain-containing protein 43 |
| Solyc08g080490.3 | −1.52 | −1.53 | 2S sulfur-rich seed storage protein 2-like |
| Solyc09g025210.3 | −1.68 | −2.32 | 12S seed storage protein CRA1-like |
| Solyc09g072560.3 | −1.68 | −2.55 | 11S globulin seed storage protein 2-like |
Fig. 3. Genes selected from the transcriptome data were validated by qPCR.
Four phenylalanine metabolism-related, four aroma volatile-related, five ethylene signaling-related and three transcription factors were selected. The red column represents WT fruits, and the purple column represents SlMYB75-OE fruits. The data represent the means ± SD of three biological replicates
Change in the ripening-related metabolic processes in SlMYB75-OE tomatoes
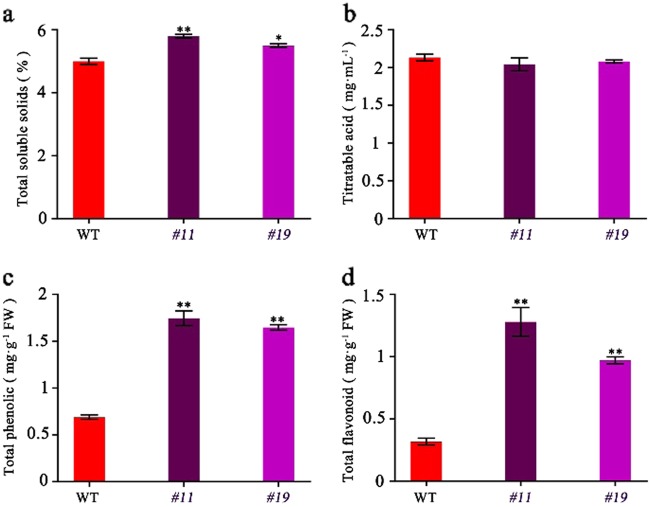
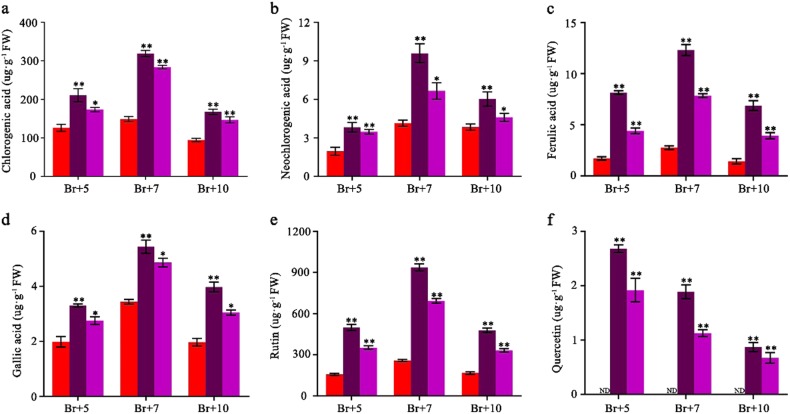
To further investigate the metabolic changes in the SlMYB75-OE tomatoes, we measured the metabolite contents relevant to the DEGs at the BR + 5, BR + 7 and BR + 10 ripening stages. Interestingly, the total soluble solids (TSS), which primarily reflect the sugar content, was significantly higher in the SlMYB75-OE tomatoes than in the WT (Fig. 4a) while the titratable acids showed no significant difference between WT and SlMYB75-OE tomatoes (Fig. 4b). Strikingly, the total contents of phenolics and flavonoids were dramatically increased in SlMYB75-OE tomatoes (Fig. 4c, d). A further characterization of the changes in phenolics and flavonoids contents revealed that the SlMYB75-OE fruits exhibited higher levels of chlorogenic acid, neochlorogenic acid, ferulic acid, gallic acid, rutin and quercetin than those in the WT (Fig. 5). These data indicated that in addition to increasing the anthocyanin content, overexpression of SlMYB75 also led to elevated levels of sugar, phenolic and flavonoid compounds.
Fig. 4. Determination of the total soluble solids, titratable acids, total phenolics and total flavonoids in WT and SlMYB75-OE fruits at BR + 7.
Total soluble solids (a) and titratable acids (b) were measured using a digital refractometer and a titration method, respectively. The data are the means ± SD of at least 10 individual fruits for each line. The total phenolics (c) and total flavonoids (d) were calculated according to the gallic acid and rutin equivalents, respectively. All data are the means ± SD of three biological replicates, *P < 0.05; and **P < 0.01 (Student’s t-test)
Fig. 5. Phenolic and flavonoid accumulation in WT and SlMYB75-OE fruits at BR + 5, BR + 7, BR + 10. Chlorogenic acid.
(a), neochlorogenic acid (b), ferulic acid (c), gallic acid (d), rutin (e), and quercetin (d) were quantified according to their corresponding standards using an HPLC method. The data are the means ± SD of three biological replicates analyzed by Student’s t-test; *P < 0.05; and **P < 0.01
Overexpression of SlMYB75 affects volatile aroma metabolism
Considering that some of the DEGs such as LOX, AADC and CXE1 have been reported to play critical roles in aroma volatile production33,34, we performed a comparative analysis of aroma volatiles in WT and SlMYB75-OE tomatoes. Nearly 380 volatile compounds were identified in the Micro-Tom tomato fruits analyzed in this experiment, and only 38 characteristic volatiles were screened and quantified (Table 2). Based on the chemical nature of their precursors, tomato volatiles could be categorized into the following three groups: those derived from fatty acids, carotenoids and amino acids34. As shown in Table 2, the aroma compounds derived from fatty acids, especially aldehydes, accounted for a large part of the tomato aroma content. The amount of aroma volatiles in fruits at the BR + 5 stage was much lower than the levels at the BR + 7 and BR + 10 stages, and some of the aroma volatiles could not even be detected at the early ripening stage. Most of the volatiles showed significantly higher levels in SlMYB75-OE tomatoes than those in the WT, including trans-2-hexenal, hexanal, 3-hexenal, benzaldehyde, 3-nonanone, methyl salicylate and geranyl acetate. By contrast, some volatiles such as 2-propyl-1-pentanol and 2-ethylhexyl acrylate showed similar or lower contents in SlMYB75-OE tomatoes compared to the WT. Noticeably, the contents of terpenoids/terpenes, terpineol, (-)-4-terpineol, copaene, γ-terpinen and β-myrcene in the SlMYB75-OE fruits exhibited a dramatic increase, with some of them having more than 10 times higher levels than those in the WT, while the content in the WT stayed very low. Overall, the general trend is that volatiles in SlMYB75-OE tomatoes are significantly higher than they are in the WT, in particular those corresponding to aldehydes and terpenes.
Table 2.
Aroma volatiles emitted by WT and SlMYB75-OE fruits at the BR + 5, BR + 7 and BR + 10 stages. The data are expressed as the means ± standard deviation of triplicate samples
| Volatile compound | RT (min) | BR+5 Content (ng g−1 FW h−1) | BR+7 Content (ng g−1 FW h−1) | BR+10 Content (ng g−1 FW h−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | #11 | #19 | WT | #11 | #19 | WT | #11 | #19 | ||
| Aldehydes | ||||||||||
| trans-2-Hexenal | 4.90 | 259.65 ± 22.48 | 421.29 ± 31.71 | 331.96 ± 22.38 | 193.00 ± 32.87 | 368.42 ± 50.84 | 257.05 ± 36.45 | 219.73 ± 35.18 | 352.31 ± 25.68 | 303.40 ± 25.90 |
| Hexanal | 3.66 | 77.29 ± 11.74 | 116.96 ± 22.90 | 97.84 ± 7.12 | 44.35 ± 11.82 | 89.52 ± 21.54 | 77.92 ± 5.11 | 74.98 ± 8.37 | 196.91 ± 46.12 | 128.03 ± 19.14 |
| 3-Hexenal | 3.62 | 33.29 ± 2.98 | 56.18 ± 3.15 | 46.63 ± 2.37 | 12.07 ± 0.96 | 42.74 ± 2.73 | 28.51 ± 2.64 | 25.33 ± 2.89 | 48.25 ± 2.19 | 39.32 ± 3.28 |
| (Z)-2-Heptenal | 8.01 | 4.83 ± 0.54 | 11.46 ± 1.66 | 7.80 ± 0.61 | 6.38 ± 1.00 | 11.21 ± 1.80 | 8.86 ± 0.35 | 6.63 ± 0.52 | 14.43 ± 2.11 | 10.62 ± 0.74 |
| (E,E)-2,4-Heptadienal | 9.06 | 3.85 ± 0.24 | 7.64 ± 0.73 | 6.08 ± 0.13 | 2.95 ± 0.54 | 6.43 ± 1.76 | 4.77 ± 0.19 | 6.42 ± 1.72 | 16.48 ± 1.28 | 9.01 ± 1.47 |
| Heptanal | 6.55 | 2.59 ± 0.39 | 5.55 ± 0.70 | 4.97 ± 0.25 | 1.90 ± 0.52 | 4.82 ± 1.07 | 3.42 ± 0.43 | 3.16 ± 0.61 | 5.61 ± 1.75 | 4.38 ± 1.32 |
| Geranial | 15.05 | 12.27 ± 2.42 | 31.05 ± 2.99 | 21.22 ± 2.96 | 47.15 ± 7.63 | 276.86 ± 44.54 | 204.11 ± 41.07 | 66.92 ± 11.91 | 252.34 ± 15.73 | 179.60 ± 31.54 |
| Neral | 14.52 | ND | 6.33 ± 2.33 | 4.27 ± 1.03 | ND | 5.45 ± 0.46 | 3.18 ± 0.71 | 5.70 ± 1.37 | 22.40 ± 2.62 | 15.37 ± 1.96 |
| Benzaldehyde | 7.90 | 3.49 ± 0.89 | 6.38 ± 1.63 | 4.91 ± 1.10 | 3.73 ± 0.79 | 8.70 ± 1.13 | 6.70 ± 0.68 | 4.23 ± 0.92 | 7.63 ± 1.42 | 6.46 ± 0.45 |
| Alcohols | ||||||||||
| 6-Methyl-5-hepten-2-ol | 9.30 | 9.63 ± 2.62 | 11.04 ± 2.40 | 10.23 ± 0.43 | 19.38 ± 8.69 | 23.81 ± 2.63 | 21.54 ± 4.15 | 16.45 ± 3.52 | 27.56 ± 4.53 | 24.25 ± 2.73 |
| 2-Propyl-1-pentanol | 10.26 | 20.01 ± 5.13 | 10.04 ± 0.54 | 14.64 ± 2.68 | 13.82 ± 2.93 | 5.60 ± 3.16 | 8.75 ± 1.57 | 3.36 ± 1.25 | 1.91 ± 0.37 | 2.22 ± 0.75 |
| Eucalyptol | 14.91 | ND | ND | ND | ND | 108.95 ± 23.39 | 70.36 ± 9.40 | ND | 85.87 ± 7.43 | 53.91 ± 5.61 |
| 1-Octanol | 11.23 | 4.23 ± 0.82 | 9.57 ± 1.00 | 6.20 ± 0.34 | 4.71 ± 1.16 | 10.07 ± 2.78 | 7.21 ± 0.77 | 5.16 ± 0.49 | 9.60 ± 1.30 | 6.73 ± 0.72 |
| Matsutake alcohol | 8.98 | ND | ND | ND | ND | 21.52 ± 2.68 | 14.39 ± 2.82 | ND | 13.66 ± 1.27 | 7.89 ± 0.48 |
| Ketones | ||||||||||
| β-ionone | 18.81 | 1.66 ± 0.12 | 1.88 ± 0.43 | 1.79 ± 0.29 | 1.77 ± 0.39 | 2.86 ± 0.64 | 2.19 ± 0.91 | 1.89 ± 0.59 | 2.78 ± 0.52 | 2.32 ± 0.31 |
| 6-Methyl-5-h eptene-2-one | 8.96 | 64.57 ± 14.54 | 71.89 ± 18.57 | 69.16 ± 15.53 | 50.77 ± 7.89 | 58.18 ± 13.71 | 56.33 ± 12.84 | 161.80 ± 12.71 | 170.58 ± 36.07 | 165.23 ± 13.39 |
| Geranylacetone | 18.27 | 8.46 ± 1.52 | 15.84 ± 2.14 | 13.73 ± 1.92 | 13.82 ± 1.15 | 39.76 ± 5.49 | 27.06 ± 1.15 | 24.37 ± 4.34 | 40.88 ± 8.84 | 31.89 ± 5.01 |
| 3-Nonanone | 11.49 | 2.88 ± 0.73 | 13.47 ± 2.38 | 8.18 ± 0.97 | 91.51 ± 0.40 | 202.60 ± 37.01 | 151.44 ± 22.51 | 140.37 ± 13.72 | 279.56 ± 24.59 | 216.84 ± 24.57 |
| Farnesyl acetone | 24.82 | 3.27 ± 0.30 | 5.35 ± 0.28 | 4.42 ± 0.18 | 4.35 ± 0.76 | 8.42 ± 0.84 | 7.09 ± 0.92 | 4.32 ± 0.71 | 9.78 ± 2.36 | 7.47 ± 1.08 |
| Esters | ||||||||||
| Methyl salicylate | 13.59 | 4.75 ± 1.33 | 13.67 ± 1.07 | 11.19 ± 0.61 | 4.01 ± 1.69 | 20.95 ± 1.66 | 12.06 ± 1.06 | 2.88 ± 0.45 | 8.96 ± 0.89 | 6.97 ± 0.59 |
| Geranyl acetate | 17.15 | ND | ND | ND | 6.20 ± 1.04 | 31.49 ± 8.72 | 21.33 ± 3.07 | 6.78 ± 1.73 | 33.68 ± 4.80 | 21.09 ± 2.35 |
| Nerol acetate | 16.84 | ND | ND | ND | 4.65 ± 0.58 | 17.08 ± 2.55 | 10.57 ± 0.86 | 5.39 ± 1.33 | 16.32 ± 2.07 | 10.83 ± 1.61 |
| α-terpineol acetate | 16.66 | ND | ND | ND | 3.82 ± 0.43 | 21.86 ± 3.01 | 13.74 ± 2.75 | 4.95 ± 0.90 | 21.79 ± 5.01 | 15.64 ± 3.53 |
| 2-Ethylhexyl acrylate | 14.52 | 87.61 ± 16.45 | ND | ND | 61.36 ± 20.21 | ND | ND | ND | ND | ND |
| Isobutyl acetate | 4.13 | 0.34 ± 0.05 | 0.54 ± 0.08 | 0.50 ± 0.10 | 0.20 ± 0.08 | 0.78 ± 0.08 | 0.55 ± 0.07 | 0.18 ± 0.03 | 0.30 ± 0.08 | 0.29 ± 0.06 |
| Terpenoids/Terpenes | ||||||||||
| Linalool | 11.88 | 28.06 ± 6.20 | 134.50 ± 16.79 | 99.69 ± 6.14 | 34.47 ± 11.09 | 313.13 ± 40.92 | 234.12 ± 13.87 | 59.80 ± 13.92 | 370.44 ± 35.57 | 260.58 ± 38.45 |
| Terpineol | 13.68 | 4.48 ± 0.96 | 13.61 ± 3.12 | 11.10 ± 1.87 | 6.67 ± 3.57 | 27.87 ± 3.05 | 17.77 ± 5.06 | 8.62 ± 2.00 | 19.03 ± 4.04 | 12.59 ± 2.01 |
| (-)-4-Terpineol | 13.46 | 1.02 ± 0.46 | 11.08 ± 0.99 | 7.22 ± 0.47 | 16.37 ± 0.38 | 92.76 ± 14.58 | 69.05 ± 19.86 | 18.34 ± 3.24 | 63.52 ± 5.78 | 49.16 ± 8.37 |
| D-Limonene | 10.36 | 9.71 ± 1.38 | 19.62 ± 4.13 | 14.77 ± 1.53 | 19.79 ± 4.63 | 60.74 ± 14.19 | 16.18 ± 5.76 | 34.08 ± 6.72 | 79.74 ± 12.15 | 74.22 ± 9.23 |
| β-myrcene | 9.46 | 1.91 ± 1.05 | 4.38 ± 1.02 | 3.43 ± 1.19 | 12.53 ± 2.52 | 44.03 ± 12.17 | 25.94 ± 5.84 | 26.07 ± 2.44 | 50.97 ± 7.38 | 44.39 ± 4.12 |
| Copaene | 17.39 | 10.92 ± 2.15 | 29.06 ± 5.95 | 21.38 ± 2.57 | 4.14 ± 0.93 | 38.62 ± 9.59 | 23.16 ± 4.24 | 7.20 ± 1.94 | 9.03 ± 1.23 | 7.37 ± 0.57 |
| Ocimene | 10.83 | 0.46 ± 0.07 | 0.73 ± 0.50 | 0.58 ± 0.03 | 16.55 ± 2.42 | 24.04 ± 4.16 | 19.50 ± 3.43 | 14.59 ± 1.47 | 21.05 ± 3.46 | 16.23 ± 4.08 |
| β-trans-ocimene | 10.57 | 0.42 ± 0.07 | 1.24 ± 0.37 | 0.87 ± 0.13 | 5.41 ± 0.47 | 18.24 ± 1.73 | 12.02 ± 1.00 | 6.28 ± 0.90 | 15.26 ± 3.26 | 11.97 ± 2.47 |
| γ-terpinen | 11.05 | 0.56 ± 0.03 | 1.50 ± 0.25 | 0.92 ± 0.15 | 0.45 ± 0.25 | 7.28 ± 1.26 | 5.09 ± 0.96 | 1.45 ± 1.21 | 8.39 ± 0.60 | 6.26 ± 0.16 |
| Terpilene | 10.06 | ND | ND | ND | ND | 5.71 ± 0.50 | 3.09 ± 0.35 | ND | 6.18 ± 0.53 | 3.62 ± 0.45 |
| Terpinolen | 11.72 | ND | 1.12 ± 0.15 | ND | ND | 7.12 ± 2.15 | 3.21 ± 0.59 | 1.60 ± 0.23 | 11.88 ± 1.61 | 6.84 ± 1.81 |
| α-phellandrene | 9.74 | ND | ND | ND | ND | 5.10 ± 0.98 | 2.97 ± 0.69 | 3.82 ± 0.58 | 6.48 ± 0.93 | 4.96 ± 0.36 |
| β-cyclocitral | 14.15 | 1.02 ± 0.25 | 1.99 ± 0.23 | 1.56 ± 0.20 | 1.30 ± 0.30 | 1.84 ± 0.14 | 1.52 ± 0.35 | 1.35 ± 0.48 | 2.68 ± 0.51 | 2.08 ± 0.38 |
Note: The data are expressed as the means ± standard deviation of triplicate samples. RT retention time, ND not detectable
Analysis of the regulatory relationships between SlMYB75 TF and other genes
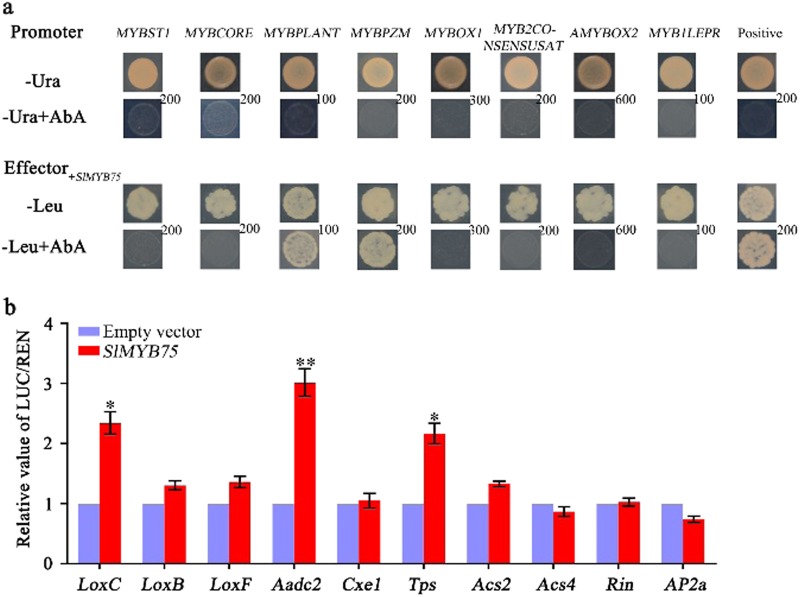
To explore whether DEGs could serve as direct targets of SlMYB75, we checked the ability of this MYB TF to bind the promoters of some of the DEGs directly using a yeast one-hybrid (Y1H) assay. Since many DEGs were identified in this study, we chose highly conserved elements rather than specific DEGs to verify which ones could be directly bound by SlMYB75. By analyzing the promoters with the New Place database tools, we identified eight conserved elements as the primary ones present in the DEG promoters (Supplementary Table S1). Tests on these eight conserved elements in Y1H assays indicated that only MYBPLANT (AAACCAACCC) and MYBPZM (ACCTACCC) could be recognized by SlMYB75 (Fig. 6a), suggesting they might be direct targets of this MYB gene. To verify whether SlMYB75 could interact with native promoters of some DEGs, we performed dual-luciferase assays with genes involved in ethylene signaling and volatile aroma metabolism. The data showed that SlMYB75 could only trans-activate the promoters of the LOXC, AADC2 and TPS genes (Fig. 6b).
Fig. 6. Regulatory roles of SlMYB75 on the conserved cis-elements and promoters of several DEGs.
a, The interactions between SlMYB75 and eight conserved cis-elements were determined by the yeast one-hybrid assay. No auto-activation ability was detected in the eight conserved cis-elements in yeast grown on SD/ura medium containing aureobasidin A (-Ura + AbA). The interactions were determined on SD/leu medium in the presence of AbA (-Leu + AbA). b, The transcription activation ability of SlMYB75 on the promoters of several DEGs was tested using dual luciferase assays. The LUC/REN ratio of the empty vector plus the promoter was used as the calibrator (set as 1). At least six biological replicates were conducted for each combination. The values are the means ± SD. *P < 0.05; and **P < 0.01 (Student’s t-test)
Discussion
In the last period, both classical and biotechnology-based breeding methods have made major progress that opened new prospects for crop improvement3,18,35. Anthocyanins are well known for their strong antioxidant and health-protection properties, but some of the most highly consumed fruits, such as tomatoes, are completely devoid of this compound. In the present study, we obtained remarkable purple tomatoes enriched with this health-promoting compound through the ectopic expression of a single SlMYB75 TF. Strikingly, SlMYB75-OE fruits also exhibited higher phenolics and flavonoids contents and enhanced the production of aroma volatiles. Our results indicated that SlMYB75 TF plays excellent roles in promoting fruit quality, and it can be directly used in genetic engineering or traditional breeding methods to produce high quality fruit.
The purple phenotype of SlMYB75-OE fruits in this study is remarkable, and its anthocyanin content could increase to 1.86 mg g−1 fresh weight (Fig. 2), which exceeded the results of earlier studies through overexpression of a single gene and previously required at least two genes to reach this level3,13,14,18,23,24. Given that it is difficult to find the parental lines that could highly express these two or three genes at the same time, together with its easy segregation characteristic, multi-gene strategies are not the best choice for breeders. Thus, expressing a single SlMYB75 provides a straightforward strategy towards improving the fruit nutritional and sensory quality. In addition, the content of phenolic and flavonoid compounds was also greatly enhanced in SlMYB75-OE fruits, which is similar to the results of a study on anthocyanin-producing grapes36. Phenolics, flavonoids, and anthocyains are all derived from phenylalanine, and the transcriptomic data showed that most of the genes involved in the phenylalanine pathway were upregulated in SlMYB75-OE fruits. Thus, SlMYB75 might participate in regulating different branches of the phenylpropanoid pathway.
It has been shown that there was no difference in the seed sizes between the WT and SlAN2-expression lines24. However, our results indicated the seeds of SlMYB75-OE tomato were significantly smaller than the WT ones (Fig. 1d). Previous studies indicated that plants that produce small seeds usually exhibit superior colonization abilities37, and thus SlMYB75-OE tomatoes may have a better dispersal capacity due to their abundant small seeds. In addition, it has been reported that SlAN2 overexpression confers enhanced tolerance to abiotic stresses such as cold, high temperature, high light, and oxidative stress21–23. This finding is consistent with our data showing that SlMYB75 is also responsive to NaCl, ABA, and SA (Supplementary Fig. S2). Collectively, these traits might confer a better capacity to survive under harsh environmental conditions to SlMYB75-OE tomatoes.
While ethylene is known to regulate fruit ripening positively through inducing ripening-associated genes and autocatalytic ethylene production38, two seemingly conflicting features of SlMYB75-OE tomatoes are their high level of ethylene production and delayed ripening (Fig. 2). As an attempt to explain the two seemingly inconsistent features, it may be speculated that the higher anti-oxidative properties of SlMYB75-OE tomatoes could mitigate the ethylene effect, since the ripening process comprises a series of physiological and biochemical changes, among which the anti-oxidative characteristics have been suggested to play a fundamental role39,40.
Aroma volatiles play a key role in the perception and acceptability of flowers, vegetables and fruits by consumers34. This property leads aroma towards being the focus of horticultural research, but little is known about improving the aroma content through the transcriptional regulation of fruits41. Consistent with the study in petunia flowers42, our results showed that the contents of the phenylalanine-derived aroma volatiles benzaldehyde and methyl salicylate were significantly higher in SlMYB75-OE fruits. Furthermore, the transcript level of fatty acid-related genes (LOXC, LOXB and LOXF)43 and the content of most fatty acid-derived aroma volatiles, especially trans-2-hexenal and (z)-2-heptenal, which are important contributors to flavor and consumer acceptance of tomato fruits44, were greatly increased in SlMYB75-OE fruits. The primary contributors to aroma volatiles in blood oranges ranked from high to low are as follows: terpenes, esters and then aldehydes45–47, but tomato fruits have very low levels of terpenes and contain only minute amounts of accumulated monoterpenes48. Interestingly, the terpene levels (Table 2) and transcript level of a terpene synthase gene (Solyc06g060180.2) (Table 1) were also greatly upregulated in SlMYB75-OE tomato fruits, suggesting that the isoprenoid pathway was also affected in SlMYB75-OE tomatoes. Studies on tomato mutants and rice indicated that there are interactions between the phenylpropanoid and isoprenoid pathways, but the mechanisms underlying the interactions responsible for the production of specific metabolites are still unclear49,50. To explore the molecular mechanism of SlMYB75 in promoting aroma volatiles, yeast one-hybrid and dual-luciferase assays were performed. The yeast one-hybrid assays revealed positive interactions between SlMYB75 and conserved MYBPLANT (AAACCAACCC) and MYBPZM (ACCTACCC) elements, supporting the idea that the CC(T/A)ACC sequence motif might be the core binding site of SlMYB75 (Fig. 6a), in a similar way with that of AtMYB12 as reported previously18. Consistent with this finding, the dual-luciferase assays showed that LOXC, AADC2 and TPS genes with promoters containing MYBPLANT (AAACCAACCC) and MYBPZM (ACCTACCC) elements (Supplementary Fig. S5) can be directly trans-activated by SlMYB75 (Fig. 6b). Thus, SlMYB75 has a regulatory function and promotes aroma volatile accumulation, probably by targeting downstream aroma volatile-related genes.
Overall, the outcome of the study indicates that overexpression of a single SlMYB75 TF can improve several tomato quality traits, including the sensory and nutritional aspects. This finding provides new opportunities for innovative breeding strategies aimed at generating tomato lines that better meet consumer requirement in terms of enriching health-promoting metabolites while better coping with a changing environment.
Electronic supplementary material
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0400101), the National Natural Science Foundation of China (31572175, 31772370), the Fundamental Research Funds for the Central Universities (2018CDXYSM0021) and the Committee of Science and Technology of Chongqing (cstckjcxljrc15).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0098-y).
References
- 1.Carrari F. Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 2006;57:1883–1897. doi: 10.1093/jxb/erj020. [DOI] [PubMed] [Google Scholar]
- 2.Yokozawa T, et al. Study on the inhibitory effect of tannins and flavonoids against the 1,1-diphenyl-2 picrylhydrazyl radical. Biochem. Pharmacol. 1998;56:213–222. doi: 10.1016/S0006-2952(98)00128-2. [DOI] [PubMed] [Google Scholar]
- 3.Butelli E, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotech. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- 4.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-Specific Induction of the Anthocyanin Biosynthetic Pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant. Sci. 2008;13:99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 8.Ramsay NA, Glover BJ. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant. Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Gao, Y. et al. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res.5, 10.1038/s41438-018-0032-3 (2018). [DOI] [PMC free article] [PubMed]
- 10.Mehrtens F. The Arabidopsis Transcription Factor MYB12 Is a Flavonol-Specific Regulator of Phenylpropanoid Biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takos AM, et al. Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muir SR, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotech. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- 13.Jones C, Mes P, Myers J. Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. J. Hered. 2003;94:449–456. doi: 10.1093/jhered/esg093. [DOI] [PubMed] [Google Scholar]
- 14.Sapir M, et al. Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J. Hered. 2008;99:292–303. doi: 10.1093/jhered/esm128. [DOI] [PubMed] [Google Scholar]
- 15.Davuluri GR, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotech. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bovy A. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes Lc and C1. Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008;56:316–326. doi: 10.1111/j.1365-313X.2008.03597.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015;6:8635. doi: 10.1038/ncomms9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, J. H. et al. Anthocyanin Biosynthesis Regulation in the Fruit of Citrus sinensis cv. Tarocco. Plant. Mol. Biol. Rep. 34, 1–13 (2016).
- 20.Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Meng X, et al. An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J. Plant. Physiol. 2015;175:1–8. doi: 10.1016/j.jplph.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Meng X, et al. Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol. Plant. 2014;58:121–130. doi: 10.1007/s10535-013-0376-3. [DOI] [Google Scholar]
- 23.Kiferle C, et al. Tomato R2R3-MYB Proteins SlANT1 and SlAN2: Same Protein Activity, Different Roles. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0136365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng X, et al. Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant. Physiol. Biochem. 2015;89:24–30. doi: 10.1016/j.plaphy.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Hu DG, et al. MdMYB1 Regulates Anthocyanin and Malate Accumulation by Directly Facilitating Their Transport into Vacuoles in Apples. Plant Physiol. 2016;170:1315–1330. doi: 10.1104/pp.15.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim W, Miller R, Park J, Park S. Consumer sensory analysis of high flavonoid transgenic tomatoes. J. Food Sci. 2014;79:S1212–S1217. doi: 10.1111/1750-3841.12478. [DOI] [PubMed] [Google Scholar]
- 27.Huang W, et al. Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol. 2017;15:472–488. doi: 10.1111/pbi.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, et al. Silencing of SlPL (Solyc03g111690), which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life, and reduced susceptibility to gray mold. Plant Biotechnol. 2017;15:1544–1555. doi: 10.1111/pbi.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, et al. The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl‐ERF. B3, shows contrasting effects on tomato fruit ripening. New Phytol. 2014;203:206–218. doi: 10.1111/nph.12771. [DOI] [PubMed] [Google Scholar]
- 30.Zheng H, Zhang Q, Quan J, Zheng Q, Xi W. Determination of sugars, organic acids, aroma components, and carotenoids in grapefruit pulps. Food Chem. 2016;205:112–121. doi: 10.1016/j.foodchem.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Phenolic compositions and antioxidant capacities of Chinese wild mandarin (Citrus reticulata Blanco) fruits. Food Chem. 2014;145:674–680. doi: 10.1016/j.foodchem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Ye YJ, et al. Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Sci. Rep. 2016;6:23632. doi: 10.1038/srep23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klee HJ, Tieman DM. Genetic challenges of flavor improvement in tomato. Trends Genet. 2013;29:257–262. doi: 10.1016/j.tig.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Baldwin EA, Bai J. Recent Advance in Aromatic Volatile Research in Tomato Fruit: The Metabolisms and Regulations. Food Bioprocess Technol. 2016;9:203–216. doi: 10.1007/s11947-015-1638-1. [DOI] [Google Scholar]
- 35.Sagor GHM, et al. A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant. Biotechnol. J. 2015;14:1116–1126. doi: 10.1111/pbi.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deluc L, et al. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006;140:499–511. doi: 10.1104/pp.105.067231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotta OC, Tanksley SD. Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet. 2009;5:1–12. doi: 10.1371/journal.pgen.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 2015;169:2380–2390. doi: 10.1104/pp.15.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conde C, Delrot S, Gerós H. Physiological, biochemical and molecular changes occurring during olive development and ripening. J. Plant. Physiol. 2008;165:1545–1562. doi: 10.1016/j.jplph.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Jimenez A, et al. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta. 2002;214:751–758. doi: 10.1007/s004250100667. [DOI] [PubMed] [Google Scholar]
- 41.Klee HJ. Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 2010;187:44–56. doi: 10.1111/j.1469-8137.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- 42.Spitzer-Rimon B, et al. EOBII, a Gene Encoding a Flower-Specific Regulator of Phenylpropanoid Volatiles’ Biosynthesis in Petunia. Plant Cell. 2010;22:1961–1976. doi: 10.1105/tpc.109.067280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J, et al. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 2014;65:419–428. doi: 10.1093/jxb/ert382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tieman D, et al. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- 45.Arena E, Guarrera N, Campisi S, Asmundo CN. Comparison of odour active compounds detected by gas-chromatography-olfactometry between hand-squeezed juices from different orange varieties. Food Chem. 2006;98:59–63. doi: 10.1016/j.foodchem.2005.04.035. [DOI] [Google Scholar]
- 46.Maccarone E, Campisi S, Fallico B, Rapisarda P, Sgarlata R. Flavor Components of Italian Orange Juices. J. Agric. Food Chem. 1998;46:2293–2298. doi: 10.1021/jf970949d. [DOI] [PubMed] [Google Scholar]
- 47.Moufida S. d. & Marzouk, B. Biochemical characterization of blood orange, sweet orange, lemon, bergamot and bitter orange. Phytochemistry. 2003;62:1283–1289. doi: 10.1016/S0031-9422(02)00631-3. [DOI] [PubMed] [Google Scholar]
- 48.Davidovich-Rikanati R, et al. Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nat. Biotech. 2007;25:899–901. doi: 10.1038/nbt1312. [DOI] [PubMed] [Google Scholar]
- 49.Enfissi EMA, et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell. 2010;22:1190–1215. doi: 10.1105/tpc.110.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JK, et al. Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 2010;58:12804–12809. doi: 10.1021/jf103277g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.