Abstract
MicroRNAs are small regulatory noncoding RNAs that repress gene expression at the posttranscriptional level. Previous studies have reported that the expression of miR-23, miR-27, and miR-24, driven from two miR-23–27–24 clusters, is altered by various pathophysiological conditions. However, their functions in skeletal muscle have not been clarified. To define the roles of the miR-23–27–24 clusters in skeletal muscle, we generated double-knockout (dKO) mice muscle-specifically lacking the miR-23–27–24 clusters. The dKO mice were viable and showed normal growth. The contractile and metabolic features of the muscles, represented by the expression of the myosin heavy chain and the oxidative markers PGC1-α and COX IV, were not altered in the dKO mice compared with wild-type mice. The dKO mice showed increased cross-sectional areas of the oxidative fibers. However, this dKO did not induce functional changes in the muscles. The dKO mice also showed normal adaptation to voluntary wheel running for 4 weeks, including the glycolytic-to-oxidative fiber type switch, and increases in mitochondrial markers, succinate dehydrogenase activity, and angiogenesis. In conclusion, our data demonstrate that the miR-23–27–24 clusters have subtle effects on skeletal muscle development and endurance-exercise-induced muscle adaptation.
Introduction
MicroRNAs (miRNAs) are short noncoding RNAs that negatively regulate gene expression at the posttranscriptional level1. This repressive regulation predominantly relies on the seed regions in the 5′ regions of the miRNAs, which bind to their complementary sequences, usually in the 3′ untranslated regions (UTRs) of the target mRNAs2. A single miRNA has hundreds of mRNA targets and a single mRNA is targeted by multiple miRNAs2. Because a miRNA modestly represses the expression of a number of its target genes3,4 and most human mRNAs are predicted to be conserved targets of miRNAs5, miRNAs are considered to be critical regulatory molecules that fine tune global gene expression.
The capacity of miRNAs to repress their target mRNAs largely depends on their expression levels6,7. Therefore, miRNAs highly expressed in a specific tissue may have significant effects on gene expression in that tissue. For example, several miRNAs, including miR-1, miR-133, and miR-206, have been identified as specifically and highly expressed in striated muscle8 and their functions have been extensively studied. A number of loss-of-function studies have reported that the conventional knockout of miR-1 and miR-133a impaired heart development, causing neonatal and embryonic lethality9–14, although less than 25% of miR-133a-1/miR-133a-2 double KO (miR-133a dKO) mice survived until adulthood, with dilated cardiomyopathy10. The surviving miR-133a dKO mice displayed abnormalities in their skeletal muscle after 4 weeks of age, characterized by progressive centronuclear myopathy in the fast-twitch myofibers, mitochondrial dysfunction, and a glycolytic-to-oxidative muscle type switch11. Furthermore, at 3 months of age, the miR-133a dKO mice displayed a reduced capacity for endurance exercise and lower mitochondrial biogenesis after 6 weeks of treadmill exercise11,15.
We have previously demonstrated that miRNAs produced from the miR-23–27–24 clusters are also highly expressed in skeletal muscle16. There are two paralogous miR-23–27–24 clusters: miR-23a–27a–24-2 (miR-23a cluster) and miR-23b–27b–24-1 (miR-23b cluster) located on chromosomes 8 and 13, respectively, in the mouse genome and on chromosomes 19 and 9, respectively, in the human genome. A number of studies have reported that the miRNAs in the miR-23–27–24 clusters are altered in response to physiological and/or pathological changes in the skeletal muscle. A recent study reported that miR-24 is downregulated in response to acute contusion muscle injury17, and other studies have reported that muscle wasting conditions, such as diabetes and limb immobilization, are associated with the downregulation of miR-23 and miR-27 in the skeletal muscle18–20. Numerous studies have reported that miRNA expression in skeletal muscle is also altered by various types of exercise21, and have suggested that these changes in miRNAs contribute to the beneficial effects of exercise22,23. It has been reported that miR-23 is downregulated in the skeletal muscle by a single bout of acute endurance exercise, in both mice and humans24,25.
Because the miR-23–27–24 clusters are highly expressed in skeletal muscle and their expression is associated with many pathophysiological conditions, we speculated that the miR-23–27–24 clusters play important roles in the skeletal muscle biology. However, the functions of the miR-23–27–24 clusters in the skeletal muscle remain unclear. In this study, to investigate their functions, we generated mice in which the miR-23–27–24 clusters were muscle-specifically knocked out and determined their muscle phenotypes. We also examined the changes in the skeletal muscle phenotypes of the knockout mice in response to endurance exercise. This is the first study to report the muscle-specific loss of function of the miR-23–27–24 clusters in vivo.
Results
miRNA expression in the muscle-specific miR-23a/b-cluster-knockout mice
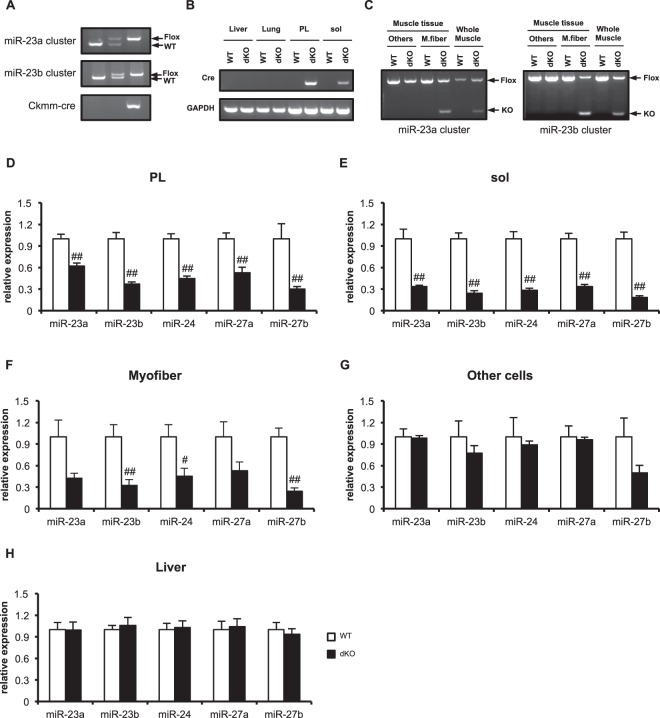
To generate the muscle-specific-miR-23a/b-cluster KO (dKO) mice, we produced miR-23a/b-cluster-floxed mice (Fig. 1) and cross-bred the miR-23a/b-cluster-floxed mice with Ckmm-cre mice (Fig. 2A). The dKO mice were born at the normal Mendelian rate, with no abnormalities in their appearance. The body, heart, and muscle weights did not differ between the dKO mice and their littermate controls (wild type) (Table 1). To confirm the Ckmm-cre (also called Mck-cre)-induced recombination, we first determined expression of Cre recombinase in muscle and non-muscle tissues. The Cre mRNA was detected in the dKO mouse skeletal muscle (both in plantaris (PL) and soleus (sol)), but not in non-muscle tissues (Fig. 2B). Since the Ckmm promoter is activated in fully differentiated muscle fibers26, we examined the degree of the Ckmm-cre-induced recombination in isolated myofibers from adult skeletal muscle. We observed that the recombination was occurred only in myofibers but not in other cells in muscle tissue of the dKO mice (Fig. 2C).
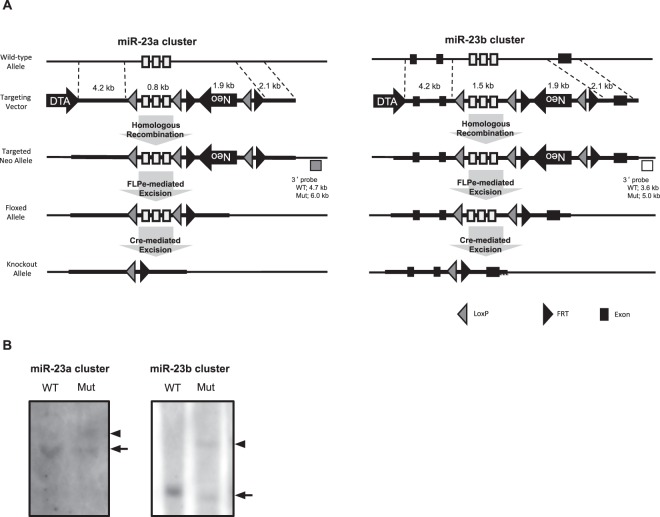
Figure 1.
Schematic representation of the targeted deletion of miR-23–27–24 clusters. (A) Targeted deletion of miR-23a and miR-23b clusters. (B) Representative images of Southern blotting analysis with 3′ external probes specific for the WT (arrows) and mutant alleles (arrowheads) of miR-23a and miR-23b clusters.
Figure 2.
Muscle-specific knockout of miR-23–27–24 clusters. (A) Representative images of the genotyping of the floxed and WT alleles of miR-23–27–24 clusters and the Ckmm-cre allele from mouse genomic DNA. (B) mRNA expression of Cre in liver, lung, plantaris (PL) and soleus (sol) of the WT and dKO mice. (C) PCR with genomic DNA to detect Cre-induced recombination in skeletal muscle. Other cells include myoblasts and fibroblasts, etc., M. fiber: isolated myofibers, Whole muscle: whole EDL muscle, Flox: floxed allele. KO: knockout allele. (D–H) Expression of mature miRNAs generated from miR-23–27–24 clusters; miR-23a, miR-23b, miR-24, miR-27a, and miR-27b in plantaris muscle (D) soleus muscle (E) myofibers (F) other cells in muscle tissue (G) and liver (H) of WT and dKO mice. Data are means ± SEM (n = 5–6). ##P < 0.01 compared with WT.
Table 1.
Bodyweight, heart and skeletal muscle weight.
| Genotype | Body weight (g) | Heart (%) | PL (%) | Sol (%) |
|---|---|---|---|---|
| WT | 26.01 ± 0.47 | 0.430 ± 0.008 | 0.062 ± 0.001 | 0.032 ± 0.001 |
| dKO | 26.89 ± 0.52 | 0.428 ± 0.007 | 0.064 ± 0.001 | 0.032 ± 0.001 |
WT, control littermate mice. dKO, Ckmm-cre induced muscle-specific miR-23a/b clusters double-KO mice. Heart (n = 8 for each group), plantaris (n = 6 for each group), and soleus (n = 6 for each group) weights (n = 7 for each group) per bodyweight were presented as percentage values. Data are presented as means ± SEM.
We next quantified the mature miRNA expression from the miR-23–27–24 clusters in the dKO mice skeletal muscles. These mature miRNAs were all significantly reduced in the plantaris (Fig. 2D) and soleus muscles (Fig. 2E) of the dKO mice compared with their levels in the control mice. We also confirmed that the mature miRNAs were decreased only in myofibers (Fig. 2F) but not in other cells of muscle tissue (Fig. 2G) such as myoblasts and fibroblasts of the dKO mice. To confirm that the deletion of the miRNAs was muscle specific, we quantified the expression of these miRNAs in the liver. The expression of the mature miRNAs from the miR-23–27–24 clusters in the liver was not altered by cross-breeding with the Ckmm-cre mice (Fig. 2H).
Skeletal muscle phenotype of the muscle-specific miR-23–27–24-cluster-knockout mice
Skeletal muscle is a complex of muscle fibers that express distinct contractile proteins. Rodent skeletal muscle fibers express four different subtypes of MyHC: type I (slow-twitch and oxidative), type IIa (fast-twitch and oxidative), type IId/x (fast-twitch and glycolytic), and type IIb (fast-twitch and glycolytic). Because the combination of these fiber types determines the contractile and metabolic features of the whole muscle tissue, we first determined the expression of the MyHC isoforms and oxidative markers in the skeletal muscle. Two different muscles with different compositions of the MyHC types were analyzed: the soleus, which is mainly composed of oxidative type I and IIa fibers, and the plantaris, which is mainly composed of glycolytic type IIa and IIb fibers.
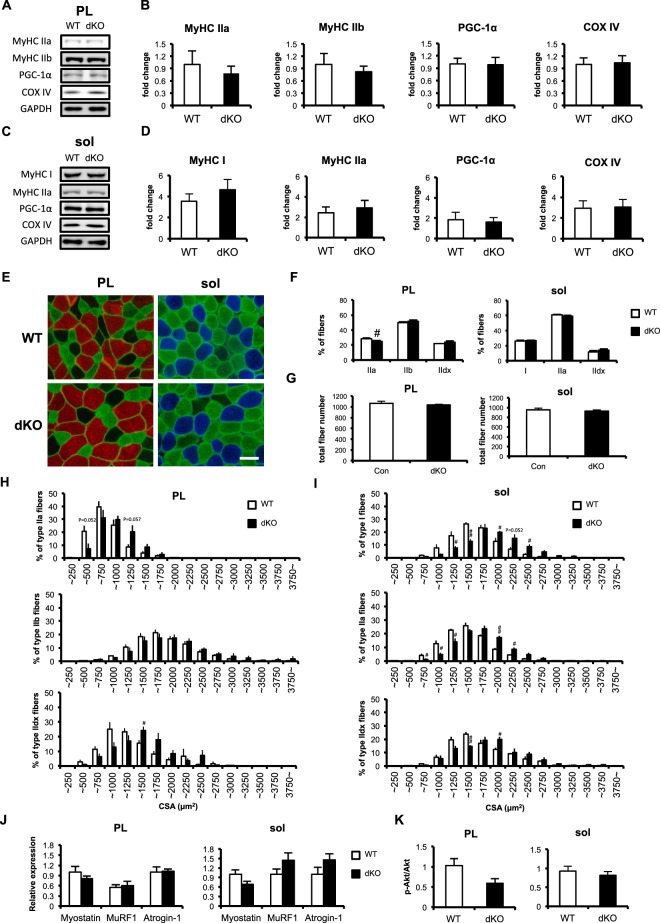
The expression of MyHC IIa and IIb proteins in the plantaris muscle was not altered by the muscle-specific dKO of the miR-23–27–24 clusters (Fig. 3A,B). The expression of PGC-1α, a master regulator of mitochondrial biogenesis, and COX IV, an enzyme complex involved in the mitochondrial respiratory electron transfer chain, was also unchanged by the dKO. Consistent with the observations in the fast plantaris muscle, there was no significant difference between the dKO and WT mice in the protein expression of MyHC I or IIa, PGC-1α, or COX IV in the slow soleus muscle.
Figure 3.
Skeletal muscle phenotype of miR-23–27–24-cluster-dKO mice. (A) Representative images of western blotting analysis of plantaris muscle. (B) Quantitative data for the protein expression of MyHC type IIa, MyHC type IIb, PGC-1α, and COX IV. Data are means ± SEM (n = 6). (C) Representative images of western blotting analysis of soleus muscle. (D) Quantitative data for the protein expression of MyHC type I, MyHC type IIa, PGC-1α, and COX IV. Data are means ± SEM (n = 5–6). (E) Representative images of plantaris and soleus muscles immunofluorescently stained for MyHC type IIa fibers and dystrophin in green, and type IIb fibers in red. MyHC type IId/x fibers are unstained (black). Scale bar, 50 μm. (F) Proportions of muscle fiber types in plantaris and soleus muscles. Proportion of each fiber type is presented as a percentage. Muscle fibers in entire cross-sections were counted. I, MyHC type I; IIa, MyHC type IIa; IIb, MyHC type IIb; IId/x, MyHC type IId/x. Data are means ± SEM (n = 4–5). #P < 0.05 compared with WT. (G) Total fiber numbers in plantaris and soleus muscles. Muscle fibers in entire cross-sections were counted. Data are means ± SEM (n = 4–5). (H) Distributions of MyHC type IIa fibers (top) MyHC type IIb fibers (middle), and MyHC type IId/x fibers (bottom) in plantaris muscle. Graphs show percentages of fibers per cross-sectional area (CSA). Data are means ± SEM (n = 5). (I) CSA distributions of MyHC type I fibers (top), MyHC type IIa (middle), and MyHC type IId/x fibers (bottom) in soleus muscle. Data are means ± SEM (n = 5). (J) mRNA expression of Myostatin, MurF1 and Atrogin-1 in WT and dKO mice skeletal muscle. Data are means ± SEM (n = 6). K, Quantitative data for the phospho-Akt/Akt by western blotting in WT and dKO mice skeletal muscle. Data are means ± SEM (n = 6).
To further understand the effects of the miR-23–27–24 clusters on the contractile properties of the skeletal muscle, we determined fiber-type compositions and cross sectional areas (CSAs) of the plantaris and soleus muscles (Fig. 3E). There were no differences in the muscle-fiber compositions of the plantaris or soleus muscles between the dKO and WT mice, except that the proportion of MyHC IIa fibers was reduced in the dKO plantaris muscle (Fig. 3F). The numbers of muscle fibers in both the plantaris and soleus were similar in the WT and dKO mice (Fig. 3G). The CSA analysis of each MyHC fiber type revealed that the CSAs of the type IIa and IId/x fibers were larger in the plantaris muscles of the dKO mice than in the WT, whereas the CSA of the type IIb fiber was not altered (Fig. 3H). The CSAs of the type I, IIa, and IId/x fibers in the soleus muscles of the dKO mice were also larger than those in the WT (Fig. 3I). We then examined expressions of muscle hypertrophy and atrophy-related genes including MuRF1, Atrogin1 and Myostatin that are reported to be targets of miR-23a/b cluster miRNAs16,27. We observed that these mRNAs were not changed in the dKO mice (Fig. 3J). We also examined whether the signaling pathway for protein synthesis during muscle hypertrophy28 was increased in the dKO mice and observed that phosphorylated Akt (p-Akt) was not changed in the dKO mice (Fig. 3K). Indeed, there was no significant difference in the muscle strengths of the WT and dKO mice (Table 2), suggesting that the increased CSAs of the muscle fibers in the dKO mice did not influence their muscle strength.
Table 2.
Grip test.
| Genotype | Strength (N/g) |
|---|---|
| WT | 0.06 ± 0.007 |
| dKO | 0.07 ± 0.003 |
Grip strength per bodyweight (N/g). WT (n = 3), control littermate mice. dKO (n = 3), Ckmm-cre induced muscle-specific miR-23a/b clusters double-KO mice. Data are presented as means ± SEM.
miR-23–27–24 clusters are not essential for the adaptation of adult skeletal muscle to 4 weeks of voluntary running
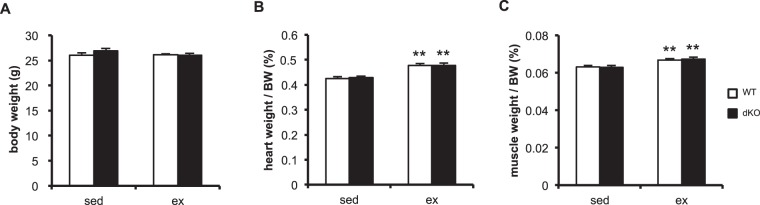
Next, we exposed the mice to 4 weeks of voluntary wheel running to induce skeletal muscle adaptation because skeletal muscle is a highly adaptive tissue that undergoes various physiological changes in response to endurance exercise. During the experimental period, the WT and dKO mice ran similar distances (Table 3). There was no significant difference in the bodyweights of the WT and dKO mice (Fig. 4A). The voluntary running increased the heart and plantaris muscle weights (Fig. 4B,C) in both the WT and dKO mice.
Table 3.
Running distance.
| Genotype | Distance (km) |
|---|---|
| WT | 11.07 ± 0.87 |
| dKO | 10.12 ± 0.63 |
Mean running distance per day during 4-week voluntary running. WT (n = 10), control littermate mice. dKO (n = 7), Ckmm-cre induced muscle-specific miR-23a/b clusters double-KO mice. Data are presented as means ± SEM.
Figure 4.
Heart and muscle weights before and after 4 weeks of voluntary wheel running. (A) Bodyweights of WT and KO mice. Heart (B) and plantaris muscle weights (C) per bodyweight (BW) of WT and dKO mice. Data are means ± SEM (n = 6–8). **P < 0.01 main effect of exercise.
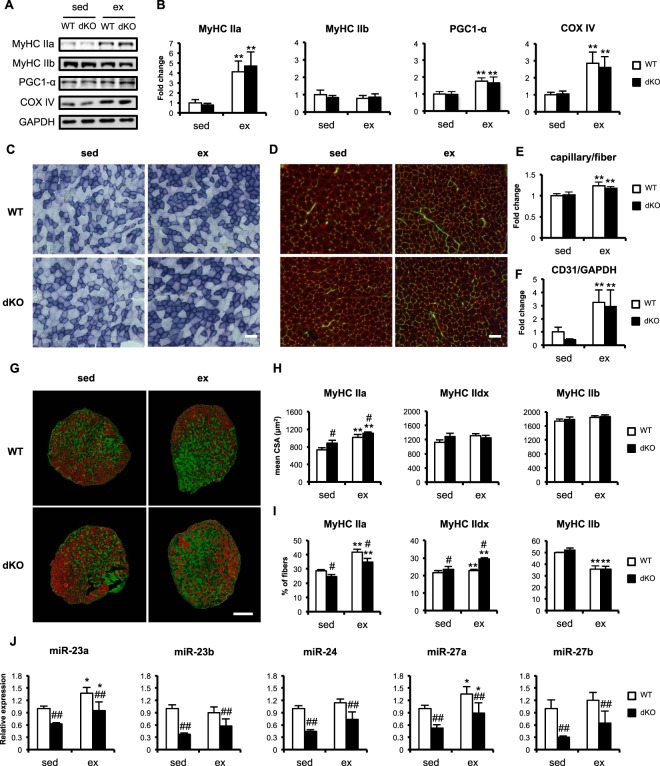
Generally, long-term endurance exercise increases the oxidative capacity of the skeletal muscle, represented by an increase in oxidative fibers, increased levels of mitochondrial marker proteins, such as PGC-1α and COX IV, and increased SDH activity29–32. Consistent with the previous studies33–36, MyHC IIa, PGC1-α, and COX IV increased after the 4 weeks of voluntary wheel running in both the WT and dKO mice, whereas MyHC IIb expression was not affected by either the mouse genotype or the voluntary running (Fig. 5A,B). SDH activity was also elevated by the voluntary running in both the WT and dKO mice (Fig. 5C).
Figure 5.
miR-23–27–24 clusters are not essential for endurance-exercise-induced muscle adaptation. (A) Representative images of western blotting analysis. (B) Quantitative data for the protein expression of MyHC type IIa, MyHC type IIb, PGC-1α, and COX IV. Data are means ± SEM (n = 6). **P < 0.01 main effect of exercise. (C) Representative images of SDH staining in plantaris muscle. Scale bar, 100 μm. (D) Representative immunofluorescent images of capillaries in plantaris muscle. Dystrophin is shown in red, CD31 in green. Scale bar, 100 μm. Quantitative data for the capillary-to-fiber ratio (E) and CD31 expression (F) in plantaris muscle. Data are means ± SEM (n = 5). **P < 0.01 main effect of exercise. (G) Representative images of plantaris muscle immunofluorescently stained for MyHC type IIa fibers and dystrophin in green, type IIb fibers in red. MyHC type IId/x fibers are unstained (black). Scale bar, 500 μm. (H) Mean cross-sectional areas (CSAs) of MyHC type IIa, MyHC type Id/x, and MyHC type IIb fibers in plantaris muscle. Data are means ± SEM (n = 4–5). #P < 0.05 main effect of genotype. **P < 0.01 main effect of exercise. (I) Proportions of MyHC IIa, MyHC IId/x, and MyHC IIb fibers in plantaris muscle. Data are means ± SEM (n = 4). #P < 0.05 main effect of genotype. **P < 0.01 main effect of exercise. (J) Expressions of the mature miRNAs in miR-23a/b clusters after 4 weeks of voluntary running. Data are means ± SEM (n = 5). ##P < 0.01 main effect of genotype. *P < 0.05 main effect of exercise. sed, sedentary group; ex, exercised group.
We also examined whether miR-23–27–24 deletion influenced exercise-induced angiogenesis34. Although the capillary density and CD31 expression in the plantaris muscle was elevated after exercise training, there was no significant difference between the WT and dKO mice (Fig. 5D–F).
The 4 weeks of voluntary running also induced increases in the CSAs and numbers of glycolytic-to-oxidative fibers in the plantaris muscles (Fig. 5G). The CSA of type IIa fibers were increased by voluntary running in both the WT and dKO mice (Fig. 5H). The CSAs of type IId/x and IIb fibers were not altered by either voluntary running or the genotype (Fig. 5H). The amounts of type IIa and IId/x fibers were increased by voluntary running, whereas the amounts of type IIb fibers decreased (Fig. 5I). The muscle-specific deletion of miR-23–27–24 reduced the proportion of type IIa fibers in the plantaris muscle, but increased the proportion of type IId/x fibers, regardless of exercise (Fig. 5I).
To determine whether the voluntary running influences expression of the miR-23a/b cluster miRNAs, we quantified the post-exercise level of these miRNAs. The miR-23a and miR-27a expressions were mildly increased in both the WT and dKO mice after the 4 weeks of voluntary running. However, each miRNA in miR-23a/b clusters was still significantly reduced in the dKO mice regardless of the voluntary running (Fig. 5J,K). Because exercise-induced muscle adaptation is occurred by a consequence of cumulative bouts of exercise, we also examined whether an acute bout of exercise has an influence on expression of the miRNAs. The acute bout of wheel running decreased miR-23a and miR-24 in the WT mice. The miR-23b and miR-27a were also decreased by the acute exercise both in the WT and dKO mice while they were still significantly reduced in the dKO mice compared to WT mice (Supplemental Fig. S1). The miR-27b did not show significant changes in response to the acute exercise both in the WT and dKO mice (Supplemental Fig. S1).
Taken together, these results indicate that 4 weeks of voluntary running was sufficient to induce skeletal muscle adaptation and that the muscle-specific deletion of the miR-23–27–24 clusters did not affect those changes.
Discussion
In this study, we have demonstrated that Ckmm-cre induced the ablation of the miR-23a/b clusters, which did not produce a significant skeletal muscle phenotype, either in the resting or exercised state. The only change observed in the dKO mice was the enlarged CSA of the oxidative muscle fibers (Fig. 3H,I). The distributions of CSA of the type IIa and type IId/x fibers in the dKO plantaris muscle were larger than in the WT (Fig. 3H), although the mean CSA only increased in the type IIA fibers (Fig. 5H). In the soleus muscle, the distributions of CSA of the type I, type IIa, and type IId/x fibers were also larger than in the WT (Fig. 3I). This suggests that the absence of the miR-23a/b clusters more strongly affected the phenotype of the oxidative fibers than that of the glycolytic fibers because the expression of the miR-23a/b clusters is greater in the soleus (where slow-twitch and oxidative fibers are dominant) than in the plantaris (where fast-twitch and glycolytic fibers are dominant)16,27.
However, the muscle weights did not increase in the dKO mice despite the increase in CSAs (Table 1). The enlargement of the type IIa fibers might not have been sufficient to increase the total plantaris weight, because the predominant fiber type in this muscle is MyHC type IIb, the CSA of which was not altered in the dKO mice. The reduced percentage of type IIa fibers in the dKO plantaris muscle might also have offset the increased CSA, so that the total proportion of type IIa fibers did not differ in the WT and dKO plantaris muscles. However, we observed no significant changes in the muscle weight, proportions of fiber types, or total fiber numbers in the dKO soleus muscle, despite the increased CSA (Fig. 3F,G). It can be assumed that the decreasing trend in the proportion of type IIa fibers, which is the dominant fiber type in the soleus (Fig. 3F, P = 0.244) and in the total fiber numbers (Fig. 3G, P = 0.28), although it was not statistically significant, may have, at least partly, cancelled out the CSA-enlargement-induced increase in muscle mass. The skeletal muscle strength was unchanged in the dKO mice (Table 2), suggesting that the changes in the oxidative fibers caused by the ablation of the miR-23a/b clusters did not induce functional changes in the skeletal muscle.
We have previously reported that miR-23 suppresses PGC-1α by binding to the 3′ UTR of its transcript37. PGC-1α is a critical transcriptional coactivator that regulates exercise-induced skeletal muscle adaptation, including mitochondrial biogenesis38, energy metabolism32, and angiogenesis34. A negative correlation between miR-23 and PGC-1α expression in response to an acute bout of endurance exercise has also been reported24. In this study, we demonstrated that PGC-1α, oxidative MyHC type IIa, and COX IV expression and SDH activity in the skeletal muscle were unchanged in the dKO mice (Fig. 3B,D), even in response to voluntary running (Fig. 5B,C). This is consistent with our previous study, in which miR-23a transgenic mice did not differ from the WT in response to exercise39. In the dKO mice, miR-23a and miR-27a were elevated after the 4 weeks of voluntary running although their expression levels were still lower than those in the WT mice (Fig. 5J). It might be possible that the elevated expressions of miR-23a and miR-27a compensate the exercise-induced muscle adaptation in the dKO mice. Taken together, our results suggest that the miR-23a/b clusters are not essential for endurance-exercise-induced muscle adaptation.
Angiogenic adaptation in response to exercise is important because it allows the management of the increased the expenditure of energy and oxygen that occurs in skeletal muscle. It has been reported that skeletal-muscle-derived angiogenic signals, such as vascular endothelial growth factor (VEGF), are important in exercise-induced angiogenesis40. Previous studies have demonstrated that miR-23 and miR-27 exert proangiogenic effects by targeting sprouty 2 and semaphorin 6A, which inhibit angiogenesis41, whereas miR-24 promotes endothelial cell apoptosis and inhibits vascularization by targeting the endothelial transcription factor GATA2 and the P21-activated kinase PAK4, which inhibits proapoptotic signals in endothelial cells42. In this study, we have demonstrated that the deletion of the miR-23a/b clusters in skeletal muscle did not inhibit the increase in capillary formation in response to endurance exercise (Fig. 5D–F). This result suggests that muscle miR-23a/b clusters are not essential for exercise-induced angiogenesis. Because each miRNA expressed from the miR-23a/b clusters has either a pro- or antiangiogenic function, the overall angiogenic effects of the clusters might be neutral.
We generated muscle-specific miR-23a/b-cluster-dKO mice using the Ckmm-cre mouse. The Ckmm-cre mouse has been used in a number of studies of gene functions in skeletal muscle26,43. Muscle creatinine kinase promoter is activated in the postdifferentiated stage of skeletal muscle26, which first emerges on embryonic day 13.544 when multinucleated primary myofibers are generated35,45. Also, it has been reported that Ckmm-cre is activated in entire myonuclei within myofibers at 1.5 months of age when myonuclear accretion is completed for mice muscle growth26. Therefore, the phenotypes of the dKO mice are attributable to the absence of the miR-23a/b clusters in multinucleated skeletal muscle.
Each miRNA expressed from the miR-23a/b clusters in the dKO was significantly reduced in the skeletal muscle, but we still detected some residual miRNA expression (Fig. 2D,E). Because mononucleated muscle stem cells (also called ‘satellite cells’) are resident in the adult skeletal muscle and other cell types, such as endothelial cells, neuronal cells, and fibroblasts, which are not affected by Ckmm-cre (Fig. 2C)43, it can be assumed that the miRNAs expressed from the miR-23a/b clusters in these cells might be present in the dKO mice. We also noted that there were more residual miR-23a cluster miRNAs than miRNAs from the miR-23b cluster in the dKO skeletal muscles (Fig. 2B,C), whereas the miRNAs expressed from the miR-23a cluster were more reduced in the lungs of endothelial-cell-specific miR-23a/b-cluster dKO mice46. Although the transcriptional regulation of the miR-23a/b clusters has not been clarified, it has been reported that the promoter regions of the miR-23a cluster and miR-23b cluster are differently activated by MEF-2C and PGC-1α27, and that the promoter activity of the miR-23a cluster may differ in fibroblasts and myocytes47. The different transcriptional regulation of the miR-23a/b clusters in each cell type and the different transcriptional regulation of the miR-23a and miR-23b clusters may contribute to the different patterns of the residual miRNAs expressed from the miR-23a/b clusters in the dKO mice tissues.
Because the clustered miRNAs are simultaneously transcribed in the same polycistronic transcript48,49 and have targets that are involved in similar biological processes4,50, it can be assumed that miR-23, miR-27, and miR-24 function coordinately in response to physiological changes. Several studies have reported that miR-2351,52, miR-2736, and miR-2453 are involved in myoblast differentiation. Most of these studies reported that miR-23, miR-27, and miR-24 promote myoblast differentiation36,52,53, whereas the other study reported that miR-23a exerts the opposite effect51. Wang et al. reported that miR-23a is downregulated in the early stages of myogenesis and upregulated dramatically in the late stages to inhibit muscle differentiation by targeting the muscle structural proteins MYH1, MYH2, and MYH451. In contrast, Mercatelli et al. reported that both miR-23a and miR-23b are upregulated gradually from the early stages of myoblast differentiation, to promote myoblast differentiation by targeting the expression of thioredoxin reductase 1 (TRXR1). TRXR1 is part of the thioredoxin system, which controls redox homeostasis but also appears to repress the myogenic genes encoding myogenin and MyHC in the later stages myoblast differentiation52. miR-27 is upregulated in the early stages of myoblast differentiation and promotes myoblast differentiation by inhibiting paired-box transcription factor 3 (PAX3), which maintains the undifferentiated state36. miR-24 expression is upregulated gradually during myoblast differentiation53, and promotes myoblast differentiation by increasing the expression of both the early and late myogenic genes, MEF2, myogenin, and MyHC53. These findings indicate that individual miRNAs from the miR-23a/b clusters regulate myoblast differentiation by controlling the expression of myogenic genes. Because the Ckmm-cre-induced miR-23a/b-cluster dKO mice displayed no significant abnormalities in their skeletal muscle, it is conceivable that the functions of the miR-23a/b clusters more strongly affect myoblasts than fully differentiated muscle fibers. Further genetic studies, especially targeting miR-23a/b clusters in the early stage of muscle development, should demonstrate the precise roles of the miR-23a/b clusters in skeletal muscle biology.
Materials and Methods
Generation of mice with muscle-specific knockout of the miR-23~27~24 clusters
The plasmid pDT-loxP-loxPFRT-PGKneo-loxPFRT was kindly supplied by S. Takahashi (University of Tsukuba) and used to construct vectors targeting the miR-23a and miR-23b clusters (Fig. 1A). Each 5′ arm (4.2 kb), central domain (containing the miR-23a or miR-23b cluster, 1.5 kb), and 3′ arm (2.1 kb) of the miR-23a and miR-23b clusters were amplified from mouse genomic DNA and ligated into the targeting vector. The vector was then linearized and introduced into mouse embryonic stem (ES) cells derived from C57BL6/J mice by electroporation. The transfected ES cells were selected with Geneticin® (G418, GIBCO), and the homologously recombined cells were identified with a Southern blotting analysis using 5′ and 3′ external probes (Fig. 1A,B). The properly transfected ES clones were injected into blastocysts and transferred to pseudopregnant female mice to generate chimeric progeny mice. The chimeras were intercrossed to facilitate the germline transmission of the targeted allele, and then crossed with CAG-FLPe mice (Riken BRC, Ibaraki, Japan) to remove the neomycin-resistance cassette with flippase–flippase recognition target (Flp–FRT) recombination.
Muscle-specific miR-23a/b-cluster-knockout mice were generated by cross-breeding miR-23a/b-cluster-double-floxed mice and Ckmm-cre mice (Jackson Laboratory). The tail DNA of the mice was genotyped with PCR with the following primer pairs: miR-23a–27a–24-2, 5′-GCT CCA ACC TTC CTA CGG ATC GAT GC-3′ and 5′-CCT GAG GGG ACA TAA CTG GCT TT-3′; miR-23b–27b–24-1, 5′-TGC CCC CTG AGT GAG CAA ATC C-3′ and 5′-TCT ACA GAC AAG GCC CTT CAG ACA G-3′. The primers were also used to examine Cre-induced recombination. PCR product size for WT, floxed and KO alleles of miR-23a cluster are 1001 bp, 1107 bp and 197 bp, respectively, and those of miR-23b cluster are 1535 bp, 1670 bp and 170 bp, respectively. The Ckmm-cre mice were identified with PCR using the specific primers: 5′-ATC AGC TAC ACC AGA GAC GGA AA-3′ and 5′-GAG GTT CAC AGG GGG AGA AC-3′.
Animal experiments
All the mice were cage-housed in a temperature-controlled (21 °C) environment under a 12 h light/12 h dark cycle, with free access to food and water according to the Guideline for Experimental Animal Care issued by the Prime Minister’s Office of Japan. Eighteen-week-old male Ckmm-cre;miR-23a/b-cluster-double-floxed mice and their miR-23a/b-cluster-double-floxed littermates were used for the experiments. The mice in the exercise group were housed individually in cages equipped with running wheels54. They were familiarized with the cages for 2 days before commencing voluntary wheel running. The wheel rotations were counted with a computer system (Meltiest, Toyama, Japan) and the total running distance was calculated. The sedentary mice were also housed individually in the same-sized cages without running wheels during the experimental period. For long-term (4 weeks) voluntary running mice (22 weeks old of age), the muscle samples were harvested following a 24-hour resting period (with locked wheels) after the last bout of running. An acute bout of wheel running exercise was conducted as described previously55–57, and the mice were sacrificed 3 hours after the final session. Their skeletal muscles were harvested in the appropriate buffer or embedded in OCT compound. The animal protocols were approved by the Animal Care and Use Committee of the University of Tokyo and Waseda University.
Muscle strength
The grip strength of the mice in all four limbs was measured as described previously58,59. Briefly, each mouse was placed on the grid that attached to a force measurement device (DST-50N, Imada Co., Ltd) and pulled back horizontally until its grip failed. The procedure was conducted three times in succession as a set, and the strength in each session was recorded. Each mouse was subjected to a total of three sets, on 3 consecutive days. The mean strength of each mouse was adjusted to its bodyweight.
Muscle fiber isolation
The isolation of muscle fiber was performed as described previously60,61. Breifly, extensor digitorum longus (EDL) and flexor digitorum brevis (FDB) muscles were harvested and incubated with 0.2% collagenase II (Worthington Biochemical Corporation) in DMEM for 40 minutes. Isolated muscle fibers are collected with glass pipette.
RT-PCR
Total RNA was extracted from the animal tissues with Isogen® II (FujiFilm Wako Pure Chemical Corporation, Japan), according to the manufacturer’s instructions. 1 μg of total RNA was reverse transcribed using ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Ex Taq HS (Takara, Osaka, Japan) was used for PCR with the following gene-specific primers: Cre, 5′-GCC TGC ATT ACC GGT CGA TGC-3′ and 5′-CAG GGT GTT ATA AGC AAT CCC-3′; Gapdh, 5′-GAC CCC TTC ATT GAC CTC AAC-3′ and 5′-TAA GCA GTT GGT GGT GCA GGA-3′. Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) was used for real-time PCR with the following gene-specific primers: Myostatin, 5′-ACA GAG TCT GAC TTT CTA ATG CAA G-3′ and 5′-GGA GTC TTG ACG GGT CTG AG-3′; MuRF1, 5′-CTT CCA AGG ACA GAA GAC TGA GC-3′ and 5′-AGT CCT CCA GCT GAG AGA TGA T-3′; Atrogin1, 5′-CTC AGA GAG GCA GAT TCG CAA-3′ and 5′-AGG GTG ACC CCA TAC TGC T-3′; Gapdh, 5′-AAA TGG TGA AGG TCG GTG TG-3′ and 5′-TGA AGG GGT CGT TGA TGG-3′. Gapdh was used as internal standards.
miRNA analysis
TaqMan™ MicroRNA Assays (Applied Biosystems, Foster City, CA) for miR-23a (no. 000399), miR-23b (no. 000400), miR-27a (no. 000408), miR-27b (no. 000409), and miR-24 (no. 000402) were used to analyze the miRNAs, according to the manufacturer’s instructions. Briefly, 10 ng of total RNA was combined with reverse transcription (RT) reaction mixture containing RT primers, reverse transcription buffer, dNTPs with dTTP, reverse transcriptase, and RNase inhibitor, and the RT mixture was incubated with the following thermal protocol: 30 min at 16 °C, 30 min at 42 °C, and 5 min at 85 °C. Real-time PCR for each miRNA was performed with the Applied Biosystems StepOne Real-time PCR System in 96-well plates.
Western blotting analysis
The dissected muscle tissues were immediately transferred to complete protein-loading buffer and homogenized with a glass homogenizer. The complete protein-loading buffer contained 50 mM Tris-HCl (pH 6.8), 1% SDS, 10% glycerol, 20 mM dithiothrietol, 127 mM 2-mercaptoethanol, 0.01% bromophenol blue, protease inhibitors (Roche), and phosphatase inhibitors (Sigma-Aldrich). The muscle homogenates were transferred to 1.5 ml microfuge tubes, denatured for 5 min at 95 °C, and centrifuged for 10 min at 12,000 × g. The protein concentrations of the homogenates were measured with the RC DC™ Protein Assay kit (Bio-Rad). The total proteins (10–40 μg) were loaded onto 6%, 7.5%, 10% and 12% gels and separated with SDS-PAGE for 1.5–2 h at 100 V. After SDS-PAGE, the proteins were transferred to a nitrocellulose membrane (Hybond ECL, GE Healthcare) at 100 V for 1 h in a chamber filled with transfer buffer. Nonspecific protein binding was blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 (TBST) for 1 h at room temperature. The membranes were incubated with the following primary antibodies: anti-myosin heavy chain (MyHC) type I (BA-F8, DSMZ, 1:100), anti-MyHC type IIa (SC-71, DSMZ, 1:100), anti-MyHC type IIb (BF-F3, DSMZ, 1:100), anti-PGC1-α (AB3242, Millipore, 1:500), anti-cytochrome c oxidase complex IV (anti-COX IV; #4844, Cell Signaling Technology, 1:1000), anti-CD31 (anti-PECAM-1; sc-46694, Santa Cruz Biotechnology, INC., 1:400), anti-Phospho-Akt (#4060, Cell signaling Technology, 1:1000), anti-Akt (#9272, Cell signaling Technology, 1:1000) and anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH; MAB374, Millipore, 1:1000). ECL™ Rabbit IgG, HRP-linked F(ab’)2 fragment (GE Healthcare) and Goat Anti-Mouse IgG (H + L)–HRP Conjugate (Bio-Rad) were used as the secondary antibodies. The signals were immunodetected with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) using the LAS-3000 Imaging System (Fuji Film, Japan). The signals were quantified with the ImageJ software (National Institutes of Health, Bethesda, MD).
Immunofluorescent staining
The OCT-embedded muscles were cut transversely into 10 μm sections at −20 °C and collected immediately on glass slides. The sections were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) on ice and permeabilized with 0.3% Triton X-100 in PBS. Monoclonal antibodies directed against MyHC I (BA.F8), MyHC IIa (SC.71), MyHC IIb (BF.F3), and CD31 (MCA2388, AbD Serotec) were diluted 1:25, 1:250, 1:25, and 1:100, respectively, in 5% normal goat serum (NGS) in PBS and used as the primary antibodies to identify the muscle fiber types and capillaries. Mouse anti-dystrophin antibody (D8043, Sigma) was diluted 1:100 in 5% NGS/PBS and used to determine the shapes of the myofibers. Alexa-Fluor-405-conjugated goat anti-mouse IgG2b, Alexa-Fluor-488-conjugated goat anti-mouse IgG1, Alexa-Fluor-549-conjugated goat anti-mouse IgM, and Dy549-conjugated goat anti-mouse IgG1 antibodies were all diluted 1:100 in 5% NGS/PBS and used as the secondary antibodies. All the secondary antibodies and NGS were purchased from Jackson ImmunoResearch Laboratories (PA, USA). Images of the immunofluorescently stained cross-sections were taken with an IX-70 fluorescence microscope (Olympus, Japan) equipped with a DS-Ri1 digital camera (Nikon, Japan). The subjects were blinded to investigators when the images were analyzed. The cross-sectional areas (CSAs) and capillary densities were calculated with the ImageJ software.
Enzymatic staining
The transverse sections of the OCT-embedded muscles were stained for succinate dehydrogenase (SDH), as described previously62. Images of the SDH-stained cross-sections were taken with an IX-70 fluorescence microscope equipped with a DS-Ri1 digital camera.
Statistics
All values are presented as means ± standard error of mean (SEM). Comparisons for two factors; genotype (WT vs dKO) and exercise (sed vs ex) were made with two-way factorial analysis of variance (ANOVA). Tukey’s HSD test was performed as post-hoc test. Student’s t test was used for comparisons of two groups. All statistical analyses were performed with IBM SPSS Statistics 24 (IBM, USA).
Supplementary information
Acknowledgements
This study was supported, in part, by Grants-in Aid for Young Investigators (A) (nos 18680047 and 21680049 to T.A.) and Grants-in Aid for Scientific Research (B) (nos 25282198 and 16H03239 to T.A.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. M.L. was supported by MEXT. S.W. and S.O. were supported by the Japan Society for the Promotion of Science.
Author Contributions
Individual author contributions were as follows: conception and design of the experiments (M.L., S.W., T.A.); data collection (M.L., S.W., S.O.); analysis and interpretation of data (M.L., S.W., T.A.); drafting the article and revising it critically for important intellectual content (M.L., T.A.); supervision of the study (T.U., K.S., T.A.). All authors approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Minjung Lee and Shogo Wada contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37765-3.
References
- 1.Bartel DP. MicroRNAs- Genomics, Review Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han YC, et al. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet. 2015;47:766–775. doi: 10.1038/ng.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherji S, et al. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N, et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J Clin Invest. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidersbach A, et al. microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development. PLoS Genetics. 2013;9:e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, et al. Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 2014;24:278–292. doi: 10.1038/cr.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Y, et al. Impaired exercise tolerance, mitochondrial biogenesis, and muscle fiber maintenance in miR-133a–deficient mice. The FASEB Journal. 2016;30:3745–3758. doi: 10.1096/fj.201600529R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wada S, et al. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286:38456–38465. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, et al. miR-24 and miR-122 Negatively Regulate the Transforming Growth Factor-beta/Smad Signaling Pathway in Skeletal Muscle Fibrosis. Mol Ther Nucleic Acids. 2018;11:528–537. doi: 10.1016/j.omtn.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39:219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson MB, et al. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol. 2014;306:C551–558. doi: 10.1152/ajpcell.00266.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, A. et al. miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia Muscle, 10.1002/jcsm.12296 (2018). [DOI] [PMC free article] [PubMed]
- 21.Silva GJJ, Bye A, El Azzouzi H, Wisloff U. MicroRNAs as Important Regulators of Exercise Adaptation. Prog Cardiovasc Dis. 2017;60:130–151. doi: 10.1016/j.pcad.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Aoi W, et al. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab. 2010;298:E799–806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto H, et al. MicroRNA-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and Forkhead box j3. Am J Physiol Endocrinol Metab. 2012;303:E1419–1427. doi: 10.1152/ajpendo.00097.2012. [DOI] [PubMed] [Google Scholar]
- 24.Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky M. A. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell AP, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol. 2013;591:4637–4653. doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi, P. et al. Stage-specific effects of Notch activation during skeletal myogenesis. Elife5, 10.7554/eLife.17355 (2016). [DOI] [PMC free article] [PubMed]
- 27.Allen DL, Loh AS. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am J Physiol Cell Physiol. 2011;300:C124–137. doi: 10.1152/ajpcell.00142.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skeletal Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holloszy JO. Biochemical Adaptations in Muscle. J Biol Chem. 1967;242:2278–2282. [PubMed] [Google Scholar]
- 30.Bengtsson J, Gustafsson T, Widegren U, Jansson E, Sundberg CJ. Mitochondrial transcription factor A and respiratory complex IV increase in response to exercise training in humans. Pflugers Arch. 2001;443:61–66. doi: 10.1007/s004240100628. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00936. [DOI] [PubMed] [Google Scholar]
- 32.Lira VA, Benton CR, Yan Z, Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;299:E145–161. doi: 10.1152/ajpendo.00755.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gulve EA, Rodnick KJ, Henriksen EJ, Holloszy JO. Effects of wheel running on glucose transporter (GLUT4) concentration in skeletal muscle of young adult and old rats. Mech Ageing Dev. 1993;67:187–200. doi: 10.1016/0047-6374(93)90122-8. [DOI] [PubMed] [Google Scholar]
- 34.Chinsomboon J, et al. The transcriptional coactivator PGC-1 mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci USA. 2009;106:21401–21406. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly AM, Zacks SI. The Histogenesis of Rat Intercostal Muscle. J Cell Biol. 1969;42:135–153. doi: 10.1083/jcb.42.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crist CG, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell AP, et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol Dis. 2013;49:107–117. doi: 10.1016/j.nbd.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Geng T, et al. PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. Am J Physiol Cell Physiol. 2010;298:C572–579. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada S, et al. MicroRNA-23a has minimal effect on endurance exercise-induced adaptation of mouse skeletal muscle. Pflugers Arch. 2015;467:389–398. doi: 10.1007/s00424-014-1517-z. [DOI] [PubMed] [Google Scholar]
- 40.Delavar H, et al. Skeletal myofiber VEGF is essential for the exercise training response in adult mice. Am J Physiol Regul Integr Comp Physiol. 2014;306:R586–595. doi: 10.1152/ajpregu.00522.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Q, et al. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA. 2011;108:8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiedler J, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 43.Brüning JC, et al. A Muscle-Specific Insulin Receptor Knockout Exhibits Features of the Metabolic Syndrome of NIDDM without Altering Glucose Tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/S1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 44.Lyons GE, et al. Developmental regulation of creatine kinase gene expression by myogenic factors in embryonic mouse and chick skeletal muscle. Development. 1991;113:1017–1029. doi: 10.1242/dev.113.3.1017. [DOI] [PubMed] [Google Scholar]
- 45.Harris AJ, Duxson MJ, Fitzsimons RB, Rieger F. Myonuclear birthdates distinguish the origins of primary and secondary myotubes in embryonic mammalian skeletal muscles. Development. 1989;107:771–784. doi: 10.1242/dev.107.4.771. [DOI] [PubMed] [Google Scholar]
- 46.Oikawa, S., Wada, S., Lee, M., Maeda, S. & Akimoto, T. Role of endothelial microRNA-23 clusters in angiogenesis in vivo. Am J Physiol Heart Circ Physiol., 10.1152/ajpheart.00742.2017 (2018). [DOI] [PubMed]
- 47.Hernandez-Torres F, Aranega AE, Franco D. Identification of regulatory elements directing miR-23a-miR-27a-miR-24-2 transcriptional regulation in response to muscle hypertrophic stimuli. Biochim Biophys Acta. 2014;1839:885–897. doi: 10.1016/j.bbagrm.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryazansky, S. S., Gvozdev, V. A. & Berezikov, E. Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics. 12, 10.1186/1471-2164-12-371 (2011). [DOI] [PMC free article] [PubMed]
- 50.Wang Y, Luo J, Zhang H, Lu J. microRNAs in the Same Clusters Evolve to Coordinately Regulate Functionally Related Genes. Mol Biol Evol. 2016;33:2232–2247. doi: 10.1093/molbev/msw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp Cell Res. 2012;318:2324–2334. doi: 10.1016/j.yexcr.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 52.Mercatelli, N. et al. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Scientific Reports7, 10.1038/s41598-017-07575-0 (2017). [DOI] [PMC free article] [PubMed]
- 53.Sun Q, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akimoto T, Ribar TJ, Williams RS, Yan Z. Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am J Physiol Cell Physiol. 2004;287:C1311–1319. doi: 10.1152/ajpcell.00248.2004. [DOI] [PubMed] [Google Scholar]
- 55.Wu H, et al. Activation of MEF2 by muscle activity is mediated though a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akimoto T, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 57.Choi S, et al. Transcriptional profiling in mouse skeletal muscle following a single bout of voluntary running: evidence of increased cell proliferation. Journal of Applied Physiology. 2005;99:2406–2415. doi: 10.1152/japplphysiol.00545.2005. [DOI] [PubMed] [Google Scholar]
- 58.Cabe PA, Tilson HA, Mitchell CL, Dennis R. A simple recording grip strength device. Pharmacol Biochem Behav. 1978;8:101–102. doi: 10.1016/0091-3057(78)90131-4. [DOI] [PubMed] [Google Scholar]
- 59.Maurissen JPJ, Marable BR, Andrus AK, Stebbins KE. Factors affecting grip strength testing. Neurotoxicology and Teratology. 2003;25:543–553. doi: 10.1016/s0892-0362(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 60.Pasut, A., Jones, A. E. & Rudnicki, M. A. Isolation and Culture of Individual Myofibers and their Satellite Cells from Adult Skeletal Muscle. Journal of Visualized Experiments, 10.3791/50074 (2013). [DOI] [PMC free article] [PubMed]
- 61.Gallot, Y., Hindi, S., Mann, A. & Kumar, A. Isolation, Culture, and Staining of Single Myofibers. Bio-Protocol6, 10.21769/BioProtoc.1942 (2016). [DOI] [PMC free article] [PubMed]
- 62.Nachlas MM, Tsou KC, de Souza E, Cheng CS, Seligman AM. Cytochemical demonstration of succinic dehydrogenase by the use of a new p-nitrophenyl substituted ditetrazole. J Histochem Cytochem. 1957;5:420–436. doi: 10.1177/5.4.420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.