Abstract
Prunus mume is the only plant in the genus Prunus of the Rosaceae family with a characteristic floral scent, and the main component of this scent is benzyl acetate. By contrast, benzyl acetate is not synthesized in Prunus persica flowers. Here, we searched for benzyl alcohol acetyltransferase (BEAT) genes based on genomic data from P. mume and P. persica and found 44 unique PmBEATs in P. mume. These genes, which were mainly detected in clusters on chromosomes, originated from gene duplication events during the species evolution of P. mume, and retroduplication and tandem duplication were the two dominant duplication patterns. The genes PmBEAT34, PmBEAT36 and PmBEAT37, which were generated by tandem duplication, were highly expressed in flowers, and their highest levels were detected during the blooming stage. In vitro, PmBEAT34, PmBEAT3, and PmBEAT37 all had benzyl alcohol acetyltransferase activity that was localized in the cytoplasm. Overexpression of the PmBEAT36 or PmBEAT37 genes increased benzyl acetate production in the petal protoplasts of P. mume, and interference in the expression of these genes slightly decreased the benzyl acetate content. In addition, light and temperature regulated the expression of the PmBEAT34, PmBEAT36 and PmBEAT37 genes. According to these results, we hypothesize that the expansion of the PmBEAT genes in the genome induce the characteristic floral scent of P. mume.
Subject terms: Plant evolution, Plant molecular biology
Chinese plum: Unraveling the genetics of scent
Prunus mume, Chinese plum, owes its unique scent to extra gene copies. Researchers knew the main component of P. mume blooms’ prized scent, but the details of its production were unknown. Qixiang Zhang at the Beijing Forestry University and co-workers investigated the genetic basis of the scent, and uncovered an interesting evolutionary tale. A search for the scent production genes showed that P. mume has 44 copies; closely related P. persica, which lacks the special scent, has only two. Further analysis showed that the genes are clustered together, indicating they originated from duplication events, the evolutionary equivalent of accidental copying. The superfluous copies, released from their usual responsibilities, were free to evolve new functions, including scent production. These results illuminate how new plant traits can evolve, and unravel the mystery of this special scent.
Introduction
Prunus mume (mei) is a traditional flower in China that was domesticated more than 3000 years ago as an ornamental and fruit plant. Mei belongs to the Prunus species of the Rosaceae family, and the flowers open in early spring. Mei flowers have a unique aroma compared to other plants in Prunus, such as peach, apricot and plum. P. mume originates from the southwestern part of China and the Yangtze River basin. The Yangtze River basin, which is in the subtropical monsoon climate zone, is the main P. mume cultivation area, and the flowering time is usually between January and February, when the daily average temperature ranges from 4 to 10 °C. The flowers are believed to have a richer fragrance in the evening and in slightly cold weather. However, no studies have rigorously examined how light or temperature affects the synthesis of flower scent in P. mume.
Throughout the long cultivation history of P. mume in China, breeders have produced many different varieties. Although these varieties have differences in scent that are detectable by the human nose, benzyl acetate has been reported to be the main component of the fragrance of P. mume1. Benzyl acetate is synthesized by the phenylpropanoid pathway. The terminal reaction utilizes benzyl alcohol and acetyl coenzyme A (acetyl-CoA) as substrates. During catalysis by acetyl-CoA/benzyl alcohol acetyltransferase (BEAT), the acetyl group of acetyl-CoA is transferred to the carbonyl group of benzyl alcohol to generate benzyl acetate. The first benzyl alcohol acetyltransferase was reported in Clarkia breweri (CbBEAT), in which the major floral scent constituent is also benzyl acetate. Some other alcohols have been shown to be catalyzed by CbBEAT; however, the highest catalytic activity occurred when benzyl alcohol was the substrate2. Aharoni et al.3 identified an alcohol acyltransferase gene in strawberry (SAAT) and showed that SAAT could utilize multiple alcohols as substrates, including benzyl alcohol. An alcohol acetyltransferase 1 (RhAAT1) gene has also been characterized in roses. Additionally, overexpression of this RhAAT1 gene in transgenic petunia plants was shown to produce higher levels of benzyl acetate, although RhAAT1 displayed higher acetyltransferase activity when geraniol was the substrate than when aromatic alcohols were the substrate in vitro4,5. Several types of alcohol acyltransferases have been reported in different species of plants, and they all can use a broad range of acyl-CoAs and alcohols as substrates. Some examples are anthraniloyl-CoA:methanol acyltransferase (AMAT), which is responsible for the formation of methyl anthranilate in Washington Concord grapes; acetyl-CoA:(Z)−3-hexen-1-ol acetyltransferase (CHAT), which is responsible for the production of (Z)−3-hexen-1-yl acetate in Arabidopsis; and coniferyl alcohol acyltransferase (PhCFAT), which is responsible for the synthesis of coniferyl acetate in petunia6–12. The motifs HXXXD and DFGWG are considered to be highly conserved in CoA-dependent acyltransferases13. The HXXXD motif is located in the center of the reaction channel of the protein structure, and the DFGWG motif is considered indispensable for catalysis. Another conserved motif, LSXTLXXXYXXXG, plays an important role in reactions that use acetyl-CoA as a cosubstrate3.
Gene duplication is a form of mutation in which a genomic region is replicated. Gene duplication plays an important role in species evolution because it provides raw materials for the evolution of new genes and new genetic functions. Multiple mechanisms contribute to gene duplication, including tandem duplication, segmental duplication, transposon-mediated duplication and retroduplication. Tandem duplication results from unequal crossing-over events and generates tandemly arrayed paralogous genes14. Segmental duplication originates from rearranged genomic regions after whole-genome duplications and occurs most frequently in plants15. In transposon-mediated duplication, genes are usually captured by Mutator-like elements16,17. Retroduplication is another transposable element (TE)-associated mechanism that occurs when the messenger RNA (mRNA) of an expressed gene is reverse transcribed to DNA and then inserted into the genome18. These duplicate genes are referred to as retrogenes and usually contain no introns. Because the regulatory sequences in the promoter are usually not duplicated, most retrogenes are believed not to be functional. However, some evidence suggests that retrogenes are functional in plants. Wang et al.19 found some retrogenes in rice to be under selection19. Retrogene transcripts and coding proteins have been detected in the rice and Arabidopsis SKP1 gene family20,21. In addition, approximately one-fourth and one-third of retrogenes in rice and Arabidopsis, respectively, exhibit expression patterns similar to their source genes22,23.
We sequenced and assembled the genome of P. mume in 2012. Based on genomic data, we showed that the BEAT gene family had expanded notably in P. mume, compared with Malus×domestics and Fragaria vesca in Rosaceae; additionally, most of the BEAT genes were clustered, suggesting that these BEAT genes originated from serial duplication events24. Here, we have further characterized the BEAT genes in P. mume. We identified a group of PmBEAT genes that was expanded in the P. mume genome compared to that of Prunus persica, which does not exhibit the benzyl acetate synthetic pathway in flowers. We selected the PmBEAT genes with high expression levels in flowers and analyzed their spatiotemporal expression patterns and benzyl alcohol acetyltransferase activities, and the results demonstrate the important role of PmBEAT genes in the synthesis of benzoyl acetate. Finally, we explored how light or temperature can affect the benzyl acetate synthesis pathway.
Results
Benzyl alcohol acetyltransferase activity in the flowers of P. mume
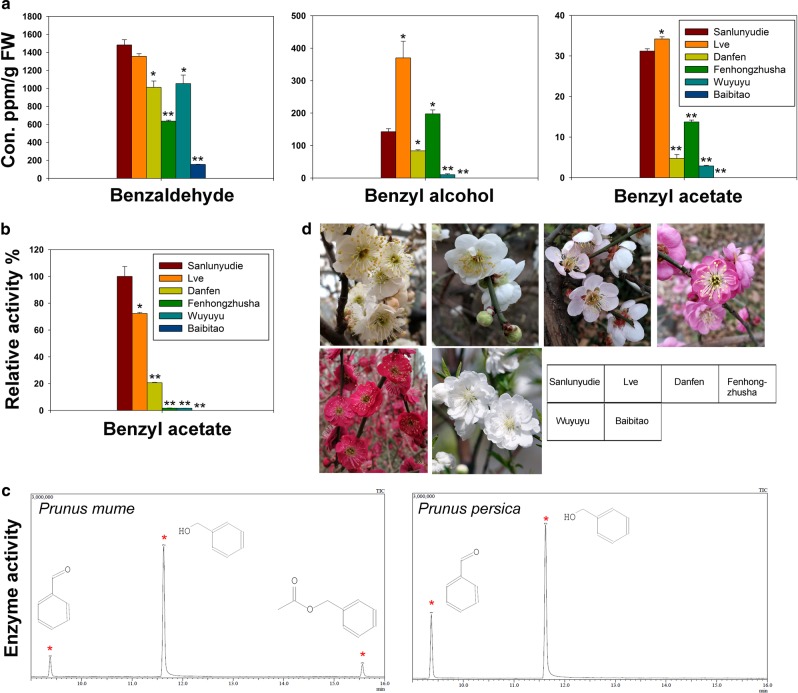
In earlier research, we found that the dominant compound responsible for the fragrance of P. mume flowers was ester benzyl acetate25, which is synthesized by the benzenoid/phenylpropanoid pathway. CbBEAT in C. breweri has been reported to catalyze the synthesis of benzyl acetate using benzyl alcohol and acetyl-CoA as substrates, and the substrate benzyl alcohol has been shown to be synthesized from the reduction of benzaldehyde in plants2,10. Differences exist in the content of endogenous benzyl acetate in the flowers of different P. mume varieties. The content in the white flower varieties ‘Sanlunyudie’ and ‘Lve’ is significantly higher than that in the red flower varieties ‘Danfen’, ‘Fenhongzhusha’ and ‘Wuyuyu’ (Fig. 1a, d). In addition, the contents of benzaldehyde and benzyl alcohol differ among varieties, but the differences do not correspond to the flower colors. We extracted the total protein of the flower for analysis of the benzyl alcohol acetyltransferase activity in vitro. The results showed that enzyme activity in the white flower varieties was significantly higher than that in the red flower varieties (Fig. 1b). These results suggest that the content of benzyl alcohol and the activity of benzyl alcohol acetyltransferase are two important factors that determine the benzyl acetate content in different varieties. P. mume and P. persica both belong to the Prunus genus of the Rosaceae family. However, no benzyl alcohol or benzyl acetate was detected in the flowers of P. persica ‘Baibitao’ (Fig. 1a, d). Additionally, benzyl acetate could not be synthesized in ‘Baibitao’ when benzyl alcohol was supplied for enzyme activity analysis (Fig. 1b, c). These results indicate that the synthesis efficiency differs among the varieties of P. mume and that the pathway for the synthesis of benzyl acetate with benzyl alcohol as the substrate does not exist in the flowers of P. persica.
Fig. 1. Analyses of endogenous benzyl acetate content and benzyl alcohol acetyltransferase activity in P. mume and P. persica.
a Analysis of the contents of endogenous benzaldehyde, benzyl alcohol and benzyl acetate in the blooming flowers of different varieties of P. mume and P. persica using GC-MS. Benzyl propionate was used as the internal standard for quantitative analysis. The data are presented as the mean values of three replicates ± SD. b The relative activities of benzyl alcohol acetyltransferase in the blooming flowers of P. mume and P. persica. Data are presented as the mean values of three replicates ± SD. *P < 0.05 and **P < 0.01 (Student’s t-test). Three independent experiments were performed with similar results. c The gas chromatograms of P. mume (‘Sanlunyudie’) and P. persica (‘Baibitao’) from enzyme activity analysis. Benzyl alcohol was supplied in excess in the experiment. The peaks corresponding to benzaldehyde, benzyl alcohol and benzyl acetate are indicated with red asterisks. d Flowers of different varieties of P. mume and P. persica, including ‘Sanlunyudie’, ‘Lve’, ‘Danfen’, ‘Fenhongzhusha’ and ‘Wuyuyu’ (varieties of P. mume), and ‘Baibitao’ (a variety of P. persica)
Identification of the BEAT genes in the Prunus mume genome
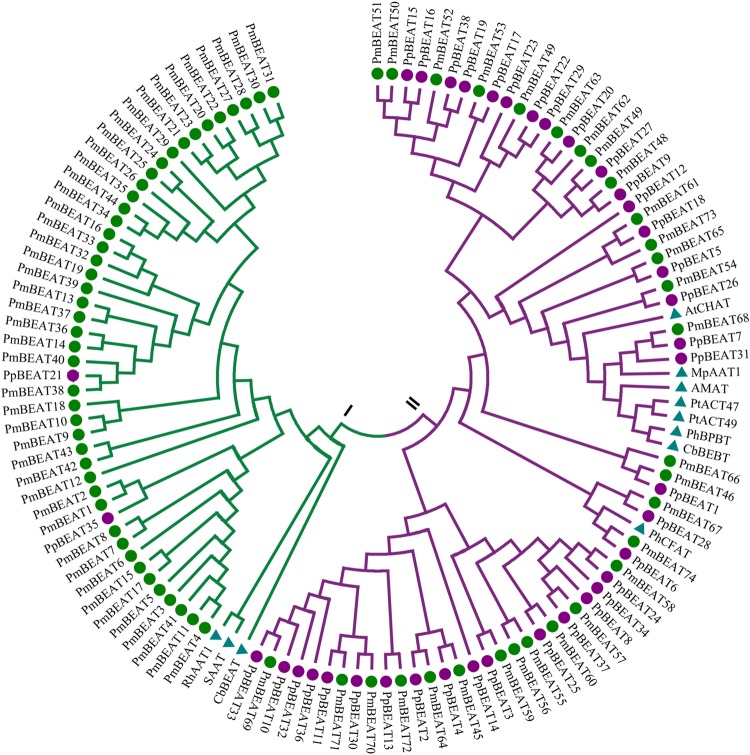
To identify the BEAT genes in the genome, BLASTP was performed using the amino acid sequence of CbBEAT as a query. In total, this method yielded 74 genes from the P. mume genome and 38 genes from the P. persica genome. The evolutionary history was inferred, and a phylogenetic tree was constructed that included other reported alcohol acetyltransferases (Fig. 2). Cluster analysis showed that CbBEAT, SAAT, RhAAT1 and the 44 PmBEATs were all in group I, which also included 2 PpBEATs from P. persica. PpBEAT35 has the highest sequence similarity with PmBEAT1 and PmBEAT2, both with 48% identity. PpBEAT21 has the highest sequence similarity with PmBEAT38, with differences in only 24 amino acids (95% identity) (Fig. S1). In group II, 30 PmBEATs and 36 PpBEATs were distributed evenly throughout the tree, and other acetyltransferases were clustered with them. This result indicates that the BEAT genes in group I have far more unique copies in the P. mume genome than in P. persica. We inferred that the multiple copies of PmBEATs in group I may contribute to the production of benzyl acetate in the flowers of P. mume. Thus, we selected the 44 PmBEATs in group I for further study.
Fig. 2. Phylogenetic tree of benzyl alcohol acetyltransferases obtained from P. mume and P. persica genomes and previously reported alcohol acetyltransferases.
The tree was generated using MEGA 6 software with the neighbor-joining method. The green solid points denote benzyl alcohol acetyltransferases from P. mume, the purple solid points denote benzyl alcohol acetyltransferases from P. persica and the blue triangles denote reported alcohol acetyltransferases from other species
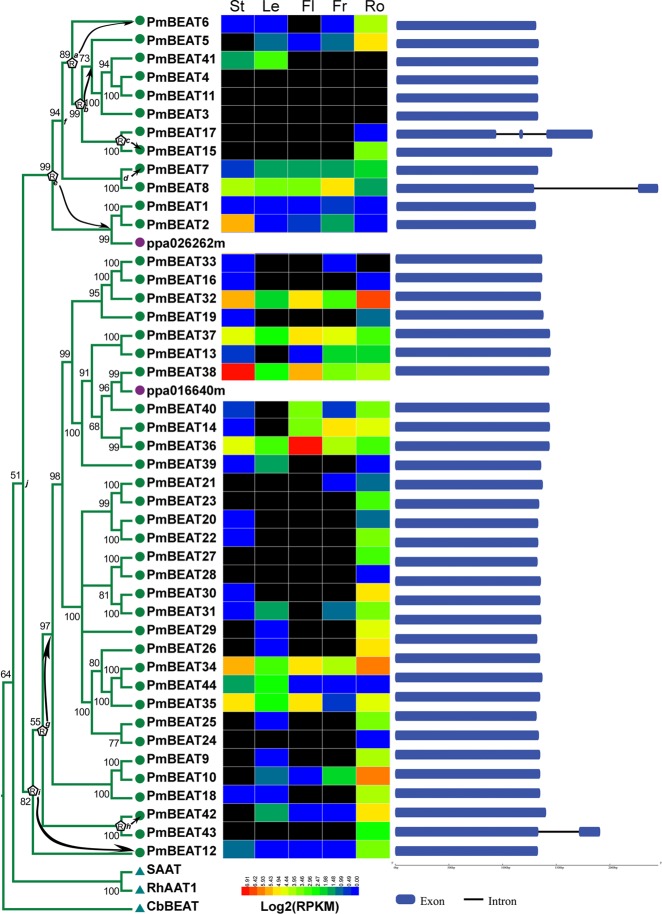
Based on the transcriptome data of P. mume24, gene expression heatmaps were drawn to indicate the expression patterns of PmBEATs in different organs. The results show that different expression levels can be detected for 18 PmBEAT genes in the buds, 22 genes in the fruits, 26 genes in the stems, 24 genes in the leaves and 36 genes in the roots (Fig. 3). The PmBEAT family may play multiple roles in different organs. Gene structure analysis showed that the PmBEAT genes do not contain introns, with the exceptions of PmBEAT17, PmBEAT8 and PmBEAT43.
Fig. 3. Gene retroduplication, structure and expression profiling of PmBEAT genes in different organs.
The genes belonging to group I are listed. The evolutionary distances were computed using the Poisson correction method. The letter R on the nodes of the tree indicates the positions where retroduplication has occurred. Hypothesized donor genes and retrogenes in retroduplication events are connected by arrows. The colored boxes following the genes indicate their expression profiling. The color gradient from red to blue denotes expression levels from high to low. The genes with no transcription detected are in black. Five different organs are stems (St), leaves (Le), flowers (Fl), fruits (Fr), roots (Ro). In the gene structure analysis, the exons and introns are indicated by blue boxes and thin lines, respectively
Location and duplication of PmBEAT genes in linkage groups
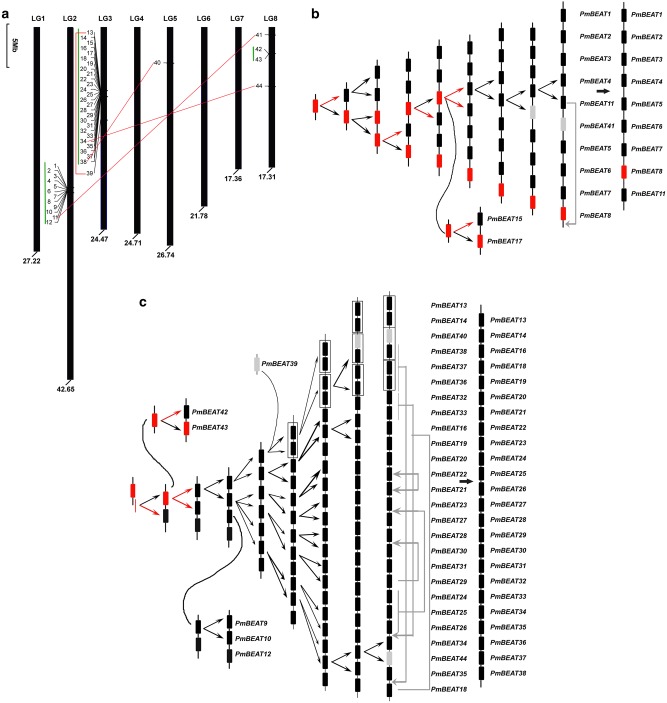
Based on the genome database, we located 44 PmBEAT genes in group I in eight linkage groups, which correspond to eight chromosomes in P. mume (Fig. 4a). Tandem and segmental duplication events were analyzed to investigate the evolutionary processes of the PmBEAT family. Four segmental duplication events were identified in the PmBEAT family on the basis of the sequence similarity (Fig. 4a). Tandem duplication is the main evolutionary cause of expansion in the PmBEAT gene family. The distribution of the three clusters in LG2, LG3 and LG8 suggest that they originated from tandem duplication. We speculated on these tandem duplication processes based on the phylogenetic relationships among PmBEATs (Fig. 3). Phylogenetically, the genes PmBEAT1, 2, 3, 4, 5, 6, 7, 8 and 11 are in a single clade, suggesting that they may have resulted from tandem duplication and thus share the same ancestral gene (Fig. 4b). PmBEAT15 and PmBEAT17 in LG3 likely also originated from this ancestral gene. The largest PmBEAT gene cluster is located on LG3 and contains 24 tandem-arrayed members: PmBEAT13, 14, 16, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37 and 38. However, these genes may be the result of more complex tandem duplication events. The clade they form also contains genes from other locations: PmBEAT9, 10 and 12 are located in the LG2 cluster, whereas PmBEAT42 and 43 are located in LG8 (Fig. 4c). In addition, the ectopic recombination of chromosomes may have changed the locations of some genes in this gene cluster, including PmBEAT36/37/38, 32/33, 21/22, 24/25/26, 29 and 18. A similar event may have also occurred in PmBEAT11 on LG2 (Fig. 4b). In addition, chromosome exchange between LG2 and LG3 may have changed the positions of PmBEAT9/10/12 and PmBEAT15/17. Interestingly, we also found that the PmBEAT12/13/36/27/38 cluster may have originated from a two-gene cluster through two tandem duplication events (Fig. 4c). The segmental duplication origins of PmBEAT39, 40, 41 and 44 are also indicated by the putative duplication processes.
Fig. 4. Linkage group locations and evolution of the PmBEAT genes in the genome of P. mume.
a The 8 linkage groups correspond to 8 chromosomes of P. mume. The locations of PmBEATs are marked in the picture. The length of each chromosome in Mb is indicated below. The scale refers to a 5 Mb chromosomal distance. The segmentally duplicated genes are represented by red lines and tandemly duplicated genes by green vertical lines. The hypothetical origins of the PmBEAT genes in the clusters LG2 (b) and LG3 (c) formed by tandem duplications are shown. The retroduplication events and intron-containing genes are indicated by red arrows and red boxes, respectively. The segmentally duplicated genes are indicated in gray. Changes in gene positions caused by putative gene transposition or chromosome inversion events are indicated by gray arrows
Since most of the PmBEAT genes lack introns, retroduplication may have occurred during gene expansion. Because a retrogene originates from the mRNA of a donor gene, the two have identical sequences except for the introns. Therefore, the retrogene will cluster together with the donor gene in the phylogenetic tree. Since PmBEAT8, PmBEAT17 and PmBEAT43 contain introns, possibly, nodes d, c and h of their recent ancestors also contain introns, as do nodes a, b, e, f, g, i and j of their elder ancestors. According to this speculation, the most likely locations of retroduplication events are in nodes a, b, c, d, e, g, h and i of the tree shown in Fig. 3, and the intron-containing genes are indicated in Fig. 4b, c. Additional evidence for the occurrence of retroduplication is that 12 of the PmBEAT genes have poly(A)-like tails in their 3′ flanking sequences; however, direct repeats cannot be identified in the 5′ and 3′ flanking sequences (Fig. S4). The poly(A) and short directed repeats of retrogenes very likely can be easily masked by base substitutions and/or insertions and deletions during the evolutionary process and thus may not be recognized26.
Expression patterns of PmBEAT genes
To further study the spatiotemporal expression patterns of PmBEAT genes in the variety ‘Sanlunyudie’, all PmBEAT genes except PmBEAT1, 3, 15, 28, 30, 42 and 43 were cloned from the experimental variety. The primers were designed according to the coding DNA sequences (CDS) from the genome database (Table S1). Some differences were observed at the nucleic acid and amino acid levels from the published sequences of PmBEATs from the wild plant used for genome sequencing. Remarkably, a fragment deletion in the coding sequence of PmBEAT19 resulted in frameshifts with premature termination by introducing a stop codon (Fig. S2).
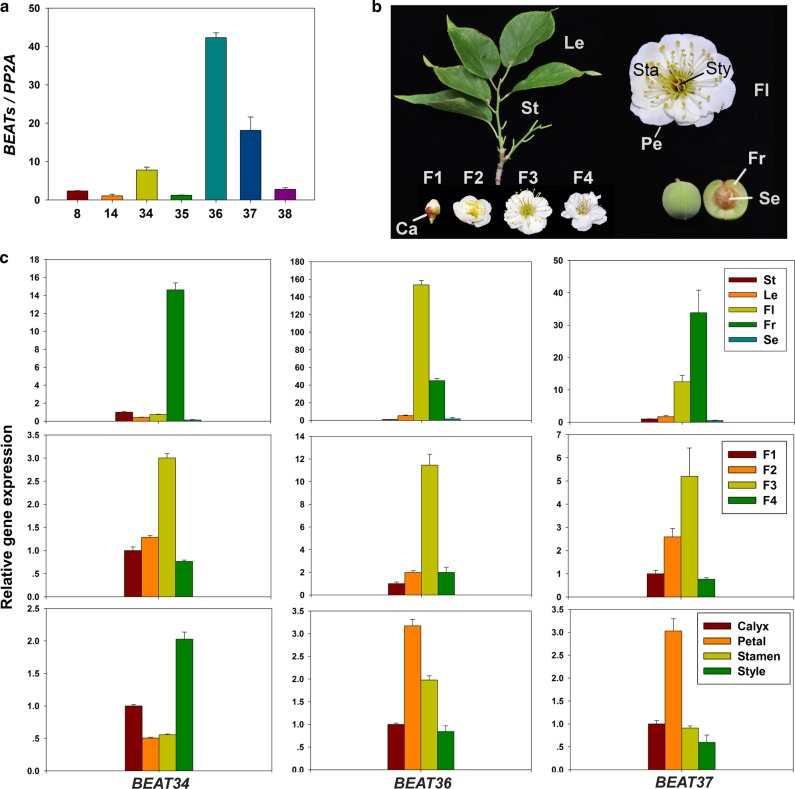
The endogenous synthesis of benzyl acetate has been reported to occur mainly in petals and stamens, with the benzyl acetate content reaching its peak during the blooming stage. We asked whether the expression patterns of PmBEATs are consistent with this pattern of endogenous benzyl acetate synthesis. To identify the key genes that control fragrance synthesis in the flowers, we determined the expression levels of PmBEATs in five different tissues of P. mume using semiquantitative reverse transcription polymerase chain reaction (RT-PCR; Fig. S3). The primers were designed according to the coding sequences of PmBEATs in the ‘Sanlunyudie’ variety (Table S2). We detected PmBEAT8, PmBEAT14, PmBEAT34, PmBEAT35, PmBEAT36, PmBEAT37 and PmBEAT38 in the flower tissues. This result is generally consistent with the transcriptome data. Next, the expression levels of these PmBEAT genes in the flowers were compared using real-time PCR (the primers are listed in Table S3). As shown in Fig. 5a, the expression levels of PmBEAT34, PmBEAT36 and PmBEAT37 were more than three times greater than the expression levels of other genes in the flowers of P. mume. We speculate that PmBEAT34, PmBEAT36 and PmBEAT37 may play the most important role in benzyl acetate synthesis. The results of an analysis of the temporal and spatial expression patterns of these genes (Fig. 5b) show that PmBEAT34 was mainly expressed in fruits and had relatively high expression levels in the styles. By contrast, PmBEAT36 was mostly expressed in flowers and fruits, but the highest levels of expression were in the flowers, particularly in the petals and stamens. Finally, PmBEAT37 was also mainly expressed in flowers and fruits, with high expression levels in fruits and the highest expression level in the petals (Fig. 5c). The flowering process was divided into four stages: F1 (the budding phase, before the flowers opened), F2 (the early flowering phase, when the flowers began to open but were not fully expanded), F3 (the blooming phase, when the flowers were unfolded with yellow stamens) and F4 (the late flowering phase, when the petals began to shed, and the stamens faded) (Fig. 5b). The expression levels of PmBEAT34, PmBEAT36 and PmBEAT37 all increased throughout the process of flowering, peaked during the blooming stage and then decreased during the late flowering stage (Fig. 5c). In addition, the expression levels of PpBEAT21 and PpBEAT35 in three tissues of P. persica were analyzed. As shown in Fig. S3, they were mainly expressed in leaves and stems and could not be detected in flowers.
Fig. 5. Expression patterns of PmBEATs.
a The expression levels of PmBEATs in blooming flowers determined using real-time PCR. b The plant materials used for gene expression pattern analysis. St stems, Le leaves, Fl flowers, Fr fruits, Se seeds, F1 budding phase, F2 early flowering phase, F3 blooming phase, F4 late flowering phase. A blooming flower was dissected into four different tissues, the calyx (Ca), petal (Pe), stamen (Sta) and style (Sty) to determine the gene expression. c The expression patterns of PmBEAT34, PmBEAT36 and PmBEAT37 in different organs, flowering stages and flower parts. The gene PmPP2A was used as a reference gene. The data are presented as the mean values of three replicates ± SD. Three independent experiments were performed with similar results
Conserved motif analysis of the PmBEATs
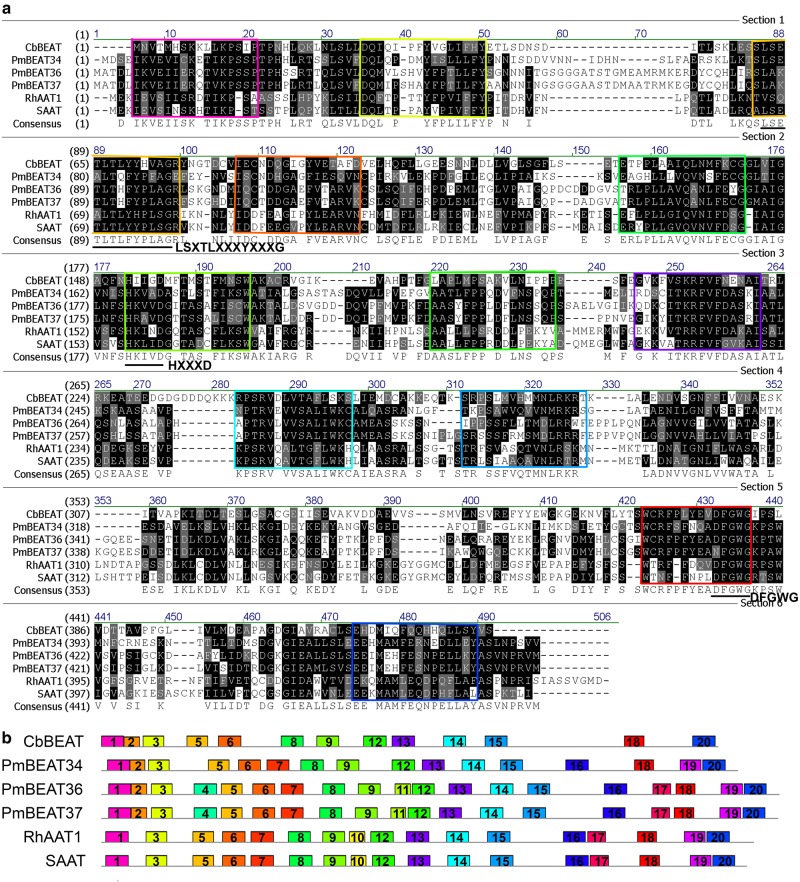
We then analyzed the conserved motifs of the PmBEAT34, PmBEAT36 and PmBEAT37 proteins. Their amino acid sequences were compared to those of the CbBEAT, RhAAT1 and SAAT proteins. The results show that the three PmBEAT proteins shared sequence identities of 24–28% with CbBEAT. Although the homology of the amino acid sequences was low, the protein motif analysis showed 12 consensus sequences, including the published HXXXD, DFGWG and LSXTLXXXYXXXG motifs (X is a variable-identity amino acid) (Fig. 6a). HXXXD and DFGWG have been reported to be the two most highly conserved motifs in CoA-dependent acyltransferases13. The HXXXD motif is located in the center of the reaction channel of the protein. The DFGWG motif near the C terminus of the protein is considered to be indispensable for catalysis. The LSXTLXXXYXXXG motif, located near the N terminus, may play an important role in reactions that use acetyl-CoA as a substrate3. To identify the conserved motifs and evaluate the structural divergences of the PmBEATs, the MEME online tool was used to analyze the motif compositions compared to those of other reported alcohol acetyltransferases. Figure 6b shows a model diagram of the distribution of conserved motifs. In total, 12 conserved motifs were identified in all the sequences: motifs 1, 3, 5, 6, 8, 9, 12, 13, 14, 15, 18 and 20. Motifs 5, 9 and 18 correspond to the LSXTLXXXYXXXG, HXXXD and DFGWG motifs, respectively (Fig. 6a). Other motifs were found in some sequences but not in others. Three motifs (7, 16 and 19) were shared in all sequences except that of the CbBEAT protein, whereas motif 2 was specifically identified in CbBEAT and the PmBEATs. Two motifs, 4 and 11, were present only in PmBEAT36 and PmBEAT37, whereas motif 10 was unique to the RhAAT1 and SAAT proteins. In addition, motif 17 was identified only in the PmBEAT36, PmBEAT37, RhAAT1 and SAAT proteins.
Fig. 6. Comparison of the amino acid sequences and motifs of PmBEATs with those of closely related alcohol acetyltransferases from other species.
a Comparison of the amino acid sequences of alcohol acetyltransferases. The conserved motifs are shown in colored squares; different colors reflect the different motifs in (b), and the reported conserved motifs are indicated with black lines. b Comparison of the motifs of alcohol acetyltransferases. Each motif is represented by a colored block with a number. The lengths and positions of the blocks correspond to the lengths and positions of the motifs in the individual protein sequences
Enzyme activities of the PmBEAT proteins
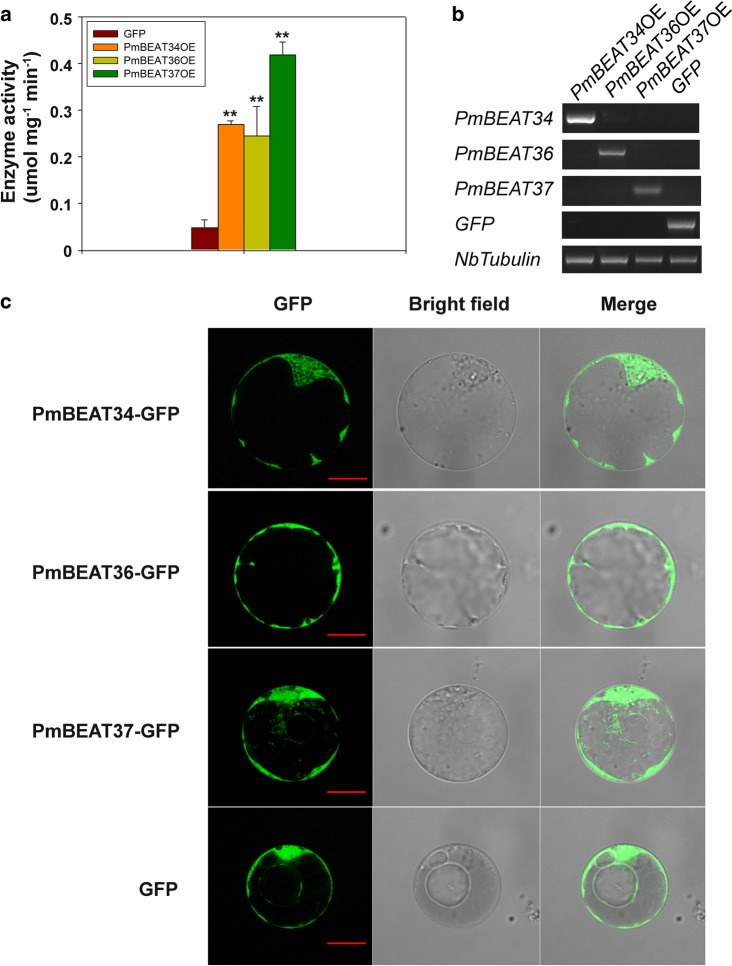
To verify the benzyl alcohol acetyltransferase activities of PmBEATs, PmBEAT34, PmBEAT36 and PmBEAT37 were heterologously expressed in the leaves of tobacco plants, and the crude proteins were extracted to determine the enzyme activities of PmBEATs in in vitro systems. The hydrosulfonyl produced from acetyl-CoA was used to calculate the enzyme activity using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) with a spectrophotometer at a wavelength of 412 nm. As shown in Fig. 7a, significant enzyme activities were detected in the leaves, which overexpressed PmBEAT34, PmBEAT36 and PmBEAT37 compared with the control. In addition, the activity of PmBEAT37 was higher than that of PmBEAT34 and PmBEAT36. The gene expression levels in tobacco leaves were detected using RT-PCR (Fig. 7b). To further assess the location at which benzyl acetate was synthesized in the cell, constructs with PmBEATs infused with green fluorescent protein (GFP) were introduced into the petal protoplasts of P. mume to determine the subcellular localizations. As shown in Fig. 7c, PmBEAT34, PmBEAT36 and PmBEAT37 were all localized in the cytoplasm. These results indicate that benzyl acetate was synthesized in the cytoplasm of petal cells.
Fig. 7. Enzyme activity and subcellular localization analysis of PmBEATs.
a The benzyl alcohol acetyltransferase activity of PmBEAT34, PmBEAT36 and PmBEAT37. The PmBEAT genes were expressed in tobacco leaves by Agrobacterium injection, and the crude protein was extracted for enzyme activity analysis. The data are presented as the mean values of three replicates ± SD. **P < 0.01 (Student’s t-test). Three independent experiments were performed with similar results. b Detection of the expression levels of PmBEAT34, PmBEAT36 and PmBEAT37 in tobacco leaves by sq-RT-PCR. NbTubulin was used as the reference gene. c Confocal images of petal protoplasts expressing PmBEAT proteins containing the GFP tag. The scale bars are 20 μM
PmBEATs affect the synthesis of benzyl acetate in the petal cells of P. mume
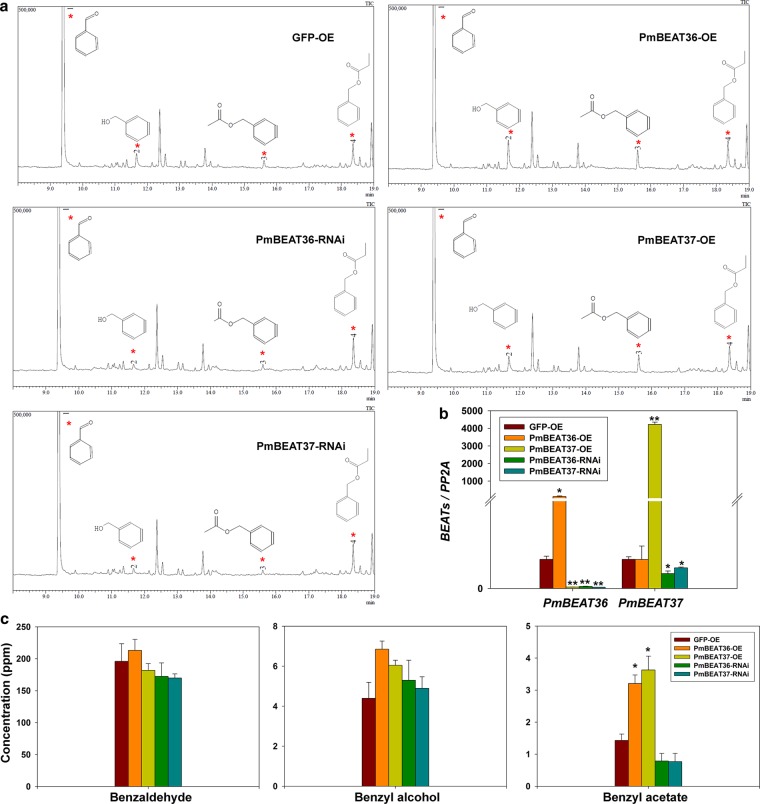
The petals were the main tissue in which benzyl acetate synthesis occurred in P. mume. Because mature genetic transformation and regeneration systems were not available, the protoplast transient transformation system was used to verify the functioning of PmBEATs in vivo. Since PmBEAT34 was mainly expressed in the fruits, and PmBEAT36 and PmBEAT37 were mainly expressed in the flowers and fruits and had the highest expression levels in petal tissue, PmBEAT36 and PmBEAT37 were selected for functional verification in the petal protoplasts. The petal protoplasts were prepared and introduced with overexpressing or interfering gene constructions. After incubation at 20 °C for 30 h, the protoplasts were collected for endogenous component extraction, and the contents of benzaldehyde, benzyl alcohol and benzyl acetate were then detected using gas chromatography-mass spectrometry (GC-MS; Fig. 8a). The RNA was also extracted for gene expression analysis. As shown in Fig. 8b, the expression levels of PmBEAT36 and PmBEAT37 were significantly increased in the protoplasts of samples overexpressing those PmBEATs and were reduced in the interfering expression samples. We note that the overexpression of PmBEAT37 also inhibited the expression of PmBEAT36 in protoplasts. As shown in Fig. 8c, the contents of benzaldehyde, benzyl alcohol and benzyl acetate were calibrated using an internal standard. The levels of benzyl acetate increased significantly when PmBEAT36 or PmBEAT37 was overexpressed, and decreased slightly in the interfering expression samples compared with the control. These results indicate that the expression levels of PmBEAT36 or PmBEAT37 can affect the synthesis of benzyl acetate in the flower cells of P. mume. In addition, we found that the content of benzyl alcohol was slightly elevated when PmBEAT36 or PmBEAT37 was overexpressed in cells, whereas no obvious differences were observed in the benzaldehyde content.
Fig. 8. Expression levels of PmBEATs affect the benzyl acetate content in the petal protoplasts of P. mume.
a Petal protoplasts transfected with PmBEAT overexpression or RNA-interference constructs were collected to determine the three components of the floral fragrance using GC-MS. The peaks corresponding to benzaldehyde, benzyl alcohol and benzyl acetate are indicated with red asterisks. The sample overexpressing the GFP was used as a control. b The expression levels of PmBEAT36 and PmBEAT37 in protoplasts were analyzed by real-time PCR. c Quantitative analysis of benzaldehyde, benzyl alcohol and benzyl acetate in protoplasts. The data are presented as the mean values of three replicates ± SD. *P < 0.05 and **P < 0.01 (Student’s t-test). Three independent experiments were performed with similar results
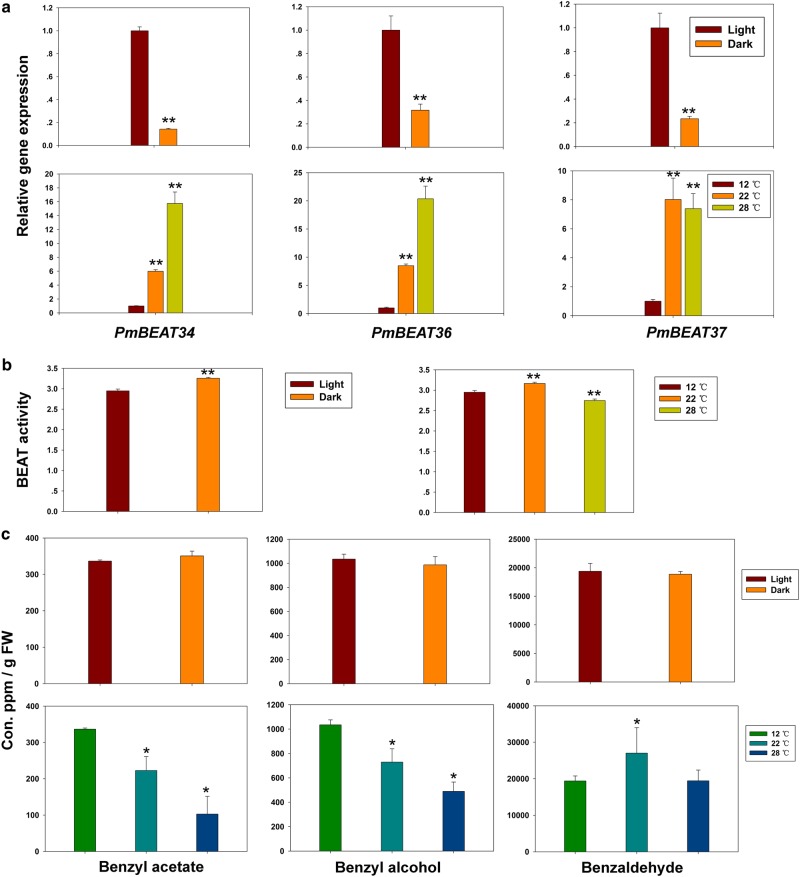
Light and temperature can affect the expression of the PmBEATs
To further study the regulatory mechanisms of PmBEAT gene expression, the promoter sequences of PmBEAT34, PmBEAT36 and PmBEAT37 were obtained from the genome database of P. mume, and a search for cis-acting regulatory elements was performed using the PlantCARE online tool. Several light- or temperature-responsive elements were identified in the promoter sequences of PmBEAT genes (Table 1). To examine how light affected the expression of the PmBEATs, detached branches with buds were dipped in water and cultured in a 12 °C incubator with or without light. When the buds were blooming, the flowers were collected for gene expression analysis. As shown in Fig. 9a, the gene expression levels of PmBEATs were higher in the flowers that opened in the light than in those that opened in the dark. These results indicate that light positively regulates the PmBEAT gene expression. The benzyl alcohol acetyltransferase activity and endogenous fragrance content were also analyzed in flowers that opened in the light versus the dark. Interestingly, the enzyme activity significantly increased when the flowers opened in the dark (Fig. 9b), but no differences were observed in the contents of benzyl acetate, benzyl alcohol and benzaldehyde in the light versus dark treatments (Fig. 9c). Possibly, a post-transcriptional regulation mechanism exists that affects the enzyme activity, and thus the benzyl alcohol content may be a more important limiting factor in the synthesis of benzyl acetate.
Table 1.
The light- and temperature-responsive elements in the promoters of PmBEAT genes
| Gene | Site name | Sequence | Number | Function |
| PmBEAT34 | 5' UTR Py-rich stretch | TTTCTTCTCT | 1 | Cis-acting element conferring high transcription levels |
| TTTCTCTCTCTCTC | 1 | |||
| AE-box | AGAAACAA | 1 | Part of a module for light response | |
| Box 4 | ATTAAT | 1 | Part of a conserved DNA module involved in light responsiveness | |
| G-box | CACGTG | 1 | Cis-acting regulatory element involved in light responsiveness | |
| CACACATGGAA | 1 | |||
| CACGTA | 1 | |||
| TACGTG | 1 | |||
| GA-motif | AAAGATGA | 1 | Part of a light-responsive element | |
| GT1-motif | GGTTAA | 1 | Light-responsive element | |
| AATCCACA | 1 | |||
| Gap-box | AAATGGAGA | 1 | Part of a light-responsive element | |
| L-box | ATCCCACCTAC | 1 | Part of a light-responsive element | |
| LTR | CCGAAA | 1 | Cis-acting element involved in low-temperature responsiveness | |
| Sp1 | CC(G/A)CCC | 1 | Light-responsive element | |
| TCT-motif | TCTTAC | 2 | Part of a light-responsive element | |
| chs-CMA1a | TTACTTAA | 1 | Part of a light-responsive element | |
| rbcS-CMA7a | GTCGATAAGG | 1 | Part of a light-responsive element | |
| PmBEAT36 | AE-box | AGAAACAA | 2 | Part of a module for light response |
| Box 4 | ATTAAT | 2 | Part of a conserved DNA module involved in light responsiveness | |
| G-box | CACGAC | 1 | Cis-acting regulatory element involved in light responsiveness | |
| GACACGTAGT | 1 | |||
| GTGGC-motif | GATTCTGTGGC | 1 | Part of a light-responsive element | |
| HSE | AAAAAATTTC | 3 | Cis-acting element involved in heat-stress responsiveness | |
| AGAAAATTCG | 1 | |||
| L-box | ATCCCACCTAC | 1 | Part of a light-responsive element | |
| Sp1 | GGGCGG | 1 | Light-responsive element | |
| TCT-motif | TCTTAC | 2 | Part of a light-responsive element | |
| PmBEAT37 | AT1-motif | AATTATTTTTTATT | 1 | Part of a light-responsive module |
| ATCT-motif | AATCTGATCG | 1 | Part of a conserved DNA module involved in light responsiveness | |
| Box 4 | ATTAAT | 2 | Part of a conserved DNA module involved in light responsiveness | |
| Box I | TTTCAAA | 2 | Light-responsive element | |
| CATT-motif | GCATTC | 1 | Part of a light-responsive element | |
| G-box | CACGAC | 1 | Cis-acting regulatory element involved in light responsiveness | |
| GAG-motif | GGAGATG | 1 | Part of a light-responsive element | |
| AGAGAGT | 2 | |||
| PmBEAT37 | GATA-motif | GATAGGG | 1 | Part of a light-responsive element |
| HSE | AAAAAATTTC | 1 | Cis-acting element involved in heat-stress responsiveness | |
| I-box | GATAGGG | 1 | Part of a light-responsive element | |
| L-box | ATCCCACCTAC | 1 | Part of a light-responsive element | |
| LTR | CCGAAA | 2 | Cis-acting element involved in low-temperature responsiveness | |
| Sp1 | CC(G/A)CCC | 4 | Light-responsive element | |
| TCT-motif | TCTTAC | 3 | Part of a light-responsive element | |
| chs-CMA1a | TTACTTAA | 2 | Part of a light-responsive element |
Fig. 9. Light and temperature affect the benzyl acetate synthesis pathway.
(a) The expression levels of PmBEAT34, PmBEAT36 and PmBEAT37, (b) the benzyl alcohol acetyltransferase activity and (c) the endogenous contents of benzaldehyde, benzyl alcohol and benzyl acetate in blooming flowers under different light or temperature treatments. The data are presented as the mean values of three replicates ± SD. * indicates P < 0.05, and ** indicates P < 0.01 (Student’s t-test). Three independent experiments were performed with similar results
To assess how temperature affects the synthesis of benzyl acetate, detached branches were treated in lighted incubators at 12 °C, 22 °C and 28 °C. Once the buds were blooming, the flowers were analyzed for gene expression, enzyme activity and endogenous fragrance content. As shown in Fig. 9a, the gene expression levels of PmBEATs increased significantly with temperature. However, the benzyl alcohol acetyltransferase activity was increased at 22 °C and decreased at 28 °C compared to that at 12 °C (Fig. 9b). The inconsistency between gene expression and enzyme activity also implies the existence of post-transcriptional regulation of enzyme activity. By contrast, the contents of benzyl acetate and benzyl alcohol decreased as the temperature increased (Fig. 9c). In addition, the benzaldehyde content increased at 22 °C, but no significant difference was observed at 28 °C compared to the treatment at 12 °C. The trend in endogenous benzyl acetate content was consistent with the benzyl alcohol substrate, which might suggest that the content of benzyl alcohol is the most important factor affecting benzyl acetate synthesis at different temperatures.
Discussion
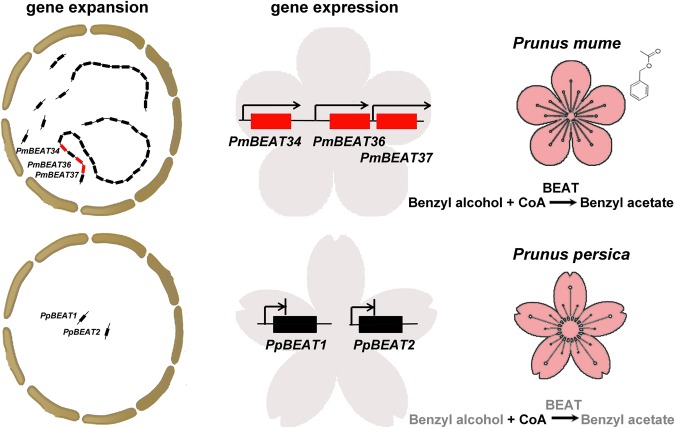
An examination of the distribution of the BEAT genes in the P. mume and P. persica genomes reveals many more PmBEAT genes in the P. mume genome. In group I, 44 PmBEAT genes were extracted from the P. mume genome, whereas 2 BEAT genes were extracted from the P. persica genome. This result indicates that the duplication and evolution events of the PmBEAT genes in the P. mume genome occurred after the differentiation of the two species (P. mume and P. persica). The 44 PmBEATs and 2 PpBEATs may share a common ancestral gene and a similar function. Our study showed that tandem duplication and retroduplication played dominant roles in the expansion of the PmBEAT gene family. The retroduplication events occurred earlier than the tandem duplication events. Most retrogenes are generally believed to be nonfunctional because of the absence of regulatory sequences, and retroduplication is believed to play a very minor role in the expansion of gene families. In the PmBEAT gene family, however, most of the members, except PmBEAT3, PmBEAT4 and PmBEAT11, have been detected in transcription data, indicating that they may be functional. PmBEAT34, PmBEAT36 and PmBEAT37 are generated by tandem duplication, and an examination of their enzyme activities in vitro and in vivo has indicated that the PmBEAT genes are functional. P. mume has higher benzyl alcohol acetyltransferase activity than P. persica and can synthesize benzyl acetate, which is manifested in the fragrance of the flowers and fruits of P. mume. The multiple copies of the PmBEAT genes play an important role in the characteristic aroma of P. mume. The expression of the two PpBEAT genes was detected only in leaves and stems but not in the peach flowers. We hypothesize that the expansion of the PmBEAT genes coupled with varied gene expression patterns resulted in specific expression in flowers and thereby induced the heightened activity of the benzyl alcohol acetyltransferase, which resulted in the synthesis of benzyl acetate in the flowers of P. mume (Fig. 10). The numerous repeats of the BEAT genes clustered in the genome are unique and have not been observed in other species. Our study indicates that the expansion of the PmBEAT genes in the genome resulted in the unique species characteristics of P. mume and presents new evidence that retroduplication can play an important role in the formation of new metabolic pathways.
Fig. 10. Model for PmBEAT gene evolution and function in the flowers of P. mume.
Two copies of PpBEAT genes exist in the genome of P. persica, but the expression of 2 PpBEATs genes was not observed in the flowers. Forty-four copies of PmBEAT genes were produced through gene expansion, and the expression of PmBEAT34, PmBEAT36 and PmBEAT37, which were generated by tandem duplications, were actively observed in the flowers and resulted in the high benzyl alcohol acetyltransferase activity observed in P. mume. In addition, benzyl alcohol can be synthesized in P. mume, but not in P. persica. Therefore, benzyl acetate, which is the main component of characteristic floral scent, can be synthesized in P. mume
In addition to the presence of the BEAT genes in the genome, the synthesis of benzyl acetate also requires sufficient benzyl alcohol and acetyl-CoA substrates. Acetyl-CoA is abundant in the cytoplast of plant cells. The characteristic fragrance of P. mume flowers is due to the benzyl alcohol synthesis pathway and to benzyl alcohol acetyltransferase. The benzyl alcohol acetyltransferase activity and the benzyl alcohol content limit the synthesis of benzyl acetate in P. mume. Our results suggest that benzyl alcohol acetyltransferase activity in flowers is regulated not only by the transcription levels of the PmBEAT genes but more importantly by post-transcriptional mechanisms. Although light can increase the expression of PmBEAT genes, it results in decreased benzyl alcohol acetyltransferase activity in the flowers and has no significant effect on the synthesis of endogenous benzyl acetate and benzyl alcohol. Temperature, in contrast, has considerable influence on the synthesis of endogenous benzyl alcohol and benzyl acetate. Higher temperatures can increase the expression levels of PmBEATs and the activity of benzyl alcohol acetyltransferase in flowers, but decrease the synthesis of the benzyl alcohol substrate, which restricts the synthesis of benzyl acetate and, ultimately, leads to a decrease in the content of endogenous benzyl acetate. In contrast to the results of the study on P. mume, neither benzyl alcohol nor benzyl alcohol acetyltransferase activity was detected in P. persica, indicating that no relevant metabolic pathway exists for the synthesis of benzyl acetate in the flowers of P. persica. Studies using the isotope tracing method in petunia reported that benzyl alcohol is produced by the reduction of benzaldehyde and is reversible10. Thus far, no enzyme has been identified that can synthesize benzyl alcohol in plants. We will focus on this question in future research.
The results of this study showed that PmBEAT34, PmBEAT36 and PmBEAT37 were highly expressed in floral organs, and they were verified to have benzyl alcohol acetyltransferase activities in vitro. However, the enzyme activities were tested using the crude extracts, which could only indirectly show that the candidate genes can enhance the benzyl alcohol acetyltransferase activity. To further study the specific enzyme activities of PmBEATs, the background interference should be eliminated. PmBEAT36 and PmBEAT37 are mainly expressed in petal tissues. Manipulating the expression levels of PmBEAT36 and PmBEAT37 in the petal protoplasts of P. mume affected the endogenous benzyl acetate content. We conclude that PmBEAT36 and PmBEAT37 are the major genes responsible for the synthesis of benzyl acetate in flowers. By contrast, PmBEAT34 is mainly expressed in fruits and may play a major role in fruit flavor. In addition, although some of the PmBEAT gene family members are not expressed as strongly as PmBEAT34, PmBEAT36 and PmBEAT37 in the floral organs, they may also participate in the synthesis of benzyl acetate in the flowers. The PmBEAT gene copy number in the P. mume genome is high, and the gene expression patterns are varied. Some genes were detected only in the roots, whereas some genes were not detected in the transcriptome data of the various organs at all. We speculate that their expression may be induced by certain conditions. Therefore, we suggest that the function of the PmBEAT gene family is not limited to the synthesis of benzyl acetate in flowers and fruits; rather, these genes may have additional functions in P. mume.
Materials and methods
Genomic data mining and PmBEAT identification
To identify PmBEATs in P. mume, local BLASTP searches in the P. mume genome database were performed using the CbBEAT full-length protein sequence as the reference sequence. The PpBEATs were obtained using the same method in the P. persica genome database. The gene IDs are shown in Table S4. The PmBEAT and PpBEAT protein sequences were aligned with those of published acyltransferase, namely CbBEAT (AAC18062), CbBEBT (AAN09796), RhAAT1 (AAW31948), SAAT (AAG13130), PhCFAT (DQ767969), PhBPBT (AAU06226), MpAAT1 (AAU14879), AMAT (AAW22989), PtACT47 (KP228018), PtACT49 (KP228019) and AtCHAT (AAN09797), using ClustalW. The evolutionary history was inferred using the neighbor-joining method27. The phylogenetic tree of these BEAT proteins and the alcohol acetyltransferases reported in other species was created in MEGA628. The evolutionary distances were computed using the Poisson correction method. A heat map was drawn according to the transcription data from the different organs of P. mume. The diagrams of gene structures were drawn based on P. mume sequencing data.
Physical locations and gene duplications of PmBEAT genes
To determine the physical locations of PmBEAT genes, the starting and ending positions of the PmBEAT genes in each linkage group were obtained from the genome database of P. mume. The tandem duplications were identified based on the close phylogenetic relationship between tandemly arrayed genes at the same chromosomal location20. To determine the gene segmental duplications, the CDSs of PmBEAT genes were blasted against each other (e value < 1e–10, identity >80%), and the genes with the highest identity value were found. To examine whether PmBEAT genes are retrogenes, 1 kb noncoding sequences in the 5′ and 3′ ends of the genes were extracted from the genome sequence to identify the poly(A) tails and direct repeats.
Gene expression pattern analysis
The primers were designed according to the CDS from the genome database to amplify the predicted PmBEAT genes from the genomic DNA and mRNA of ‘Sanlunyudie’. The predicted genes were then cloned into the pCloneEZ vector and then sequenced. The primers used in gene amplification are shown in Supplemental Table 1. The primers used for semiquantitative RT-PCR and real-time PCR were designed according to the coding sequences of the PmBEAT genes of ‘Sanlunyudie’. PmPP2A was used as a reference gene. The stems, leaves, flowers, fruits and seeds of ‘Sanlunyudie’ were collected for semiquantitative RT-PCR and real-time PCR to determine the gene expression in the different tissues. Flowers in four different stages during the open period and four different tissues from the blooming flowers were collected for real-time PCR to detect the temporal and spatial expression patterns of the genes. The primers used to detect PpBEAT gene expression are shown in Supplemental Table S2.
Amino acid sequencing and conserved motif analysis
The amino acid sequences of alcohol acetyltransferases were aligned using Vector NTI software. The MEME online tool (http://meme-suite.org) was used for conserved motif analysis with the following parameters: the maximum number of motifs was 20, the minimum motif width was 6 and the maximum motif width was 1629.
Extraction and analysis of the endogenous floral components of flowers
Fresh flowers in bloom were collected and ground into powder in liquid nitrogen. Approximately 0.2 g of powder was placed in a 2 mL centrifuge tube, and 1 mL ethyl acetate was added. The floral components were extracted on a vortex mixer for 15 min and then centrifuged at 12,000 rpm for 10 min. The supernatant was transferred to a new centrifuge tube, and anhydrous sodium sulfate was added to remove water. Benzyl propionate was added as an internal standard1. The components of the extracts were analyzed using GC-MS (GPC-GC, Shimadzu, Japan).
Plasmid construction
To detect the activities of PmBEAT proteins and verify their functions, the CDS sequences of PmBEAT34 (MH259759), PmBEAT36 (MH259760) and PmBEAT37 (MH259761) were cloned into the vector pSuper130030 with a Super promoter, using the In-Fusion method (In-Fusion HD Cloning Kit, PT5162-1). The PmBEAT34 complementary DN (cDNA) was amplified using the PmBEAT34OE-F (ACATTTAAATACTAGTatggattcagaaatcaaag) and PmBEAT34OE-R (TACCGGATCCACTAGTttaaacgacactcggattc) primers. The PmBEAT36 cDNA was amplified using the PmBEAT36OE-F (ACATTTAAATACTAGTatggccacagacttgatc) and PmBEAT36OE-R (TACCGGATCCACTAGTatgacccttggatttac) primers. The PmBEAT37 cDNA was amplified using the PmBEAT37OE-F (ACATTTAAATACTAGTatggccacagacttgatca) and PmBEAT37OE-R (TACCGGATCCACTAGTctacatgacccttggattta) primers.
To verify the functions of the PmBEAT36 and PmBEAT37 genes, specific fragments of the genes were cloned into the pFGC5941 vector. The two specific fragments of the PmBEAT36 gene were amplified using the PmBEAT36-RNAi-F1 (tttacaattaccatggcaaacataccaccatcttcg) and PmBEAT36-RNAi-R1 (tgggcgcgccccatggtgcaatacctttactgagtttgg) primers and the PmBEAT36-RNAi-F2 (gcaggtatttggatcctgcaatacctttactgagtttgg) and PmBEAT36-RNAi-R2 (gactcacctaggatcccaaacataccaccatcttcg) primers, respectively. The specific fragments of the PmBEAT37 gene were amplified using the PmBEAT37-RNAi-F1 (tttacaattaCCATGGgatgctgcgtcttattttcc) and PmBEAT37-RNAi-R1 (tgggcgcgccCCATGGtgagtgctgaaactacgagg) primers and the PmBEAT37-RNAi-F2 (gcaggtatttGGATCCtgagtgctgaaactacgagg) and PmBEAT37-RNAi-R2 (gactcacctaGGATCCgatgctgcgtcttattttcc) primers, respectively.
To study the subcellular localization of the PmBEAT proteins, the cDNA sequences of PmBEAT34, PmBEAT36 and PmBEAT37 were cloned into the pSuper1300-GFP vector using the In-Fusion method. The primers used were the same as for the pSuper1300 vector constructions.
Enzyme extraction and assay
For comparison of the activity of benzyl alcohol acetyltransferase in the flowers of P. persica and the different varieties of P. mume, the crude protein samples were extracted using a protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 0.1% NP-40 and a protease inhibitor cocktail (Roche). Then, 2 μg of crude protein was added to 1 mL of enzyme reaction buffer containing 50 mM Tris-HCl (pH 7.2), 3 mM 2-mercapto-ethanol, 0.5 mM acetyl-CoA and 1 mM benzyl alcohol. The assays were carried out in a 30 °C water bath for 1 h. The components were extracted with 0.5 mL ethyl acetate. Benzyl propionate was added as an internal standard, and the contents of benzaldehyde, benzyl alcohol and benzyl acetate were analyzed using GC-MS. The enzyme activity was calculated as the yield of benzyl acetate.
To detect the benzyl alcohol acetyltransferase activity of the PmBEATs, PmBEATs were transiently expressed in tobacco leaves by Agrobacterium tumefaciens-mediated transformation. The crude protein samples were extracted using a protein extraction buffer as described above. Then, 2 μg of crude protein was added to 1 mL of enzyme reaction buffer containing 50 mM Tris-HCl (pH 8.0), 0.5 mM acetyl-CoA, 1 mM benzyl alcohol, 0.01 mM MgCl2 and 1 mM DTNB. The assays were carried out in a 30 °C water bath, and the increase in absorbance at 412 nm was recorded using a spectrophotometer to detect the production of 2-nitro-5-thiobenzoic acid by the reaction of DTNB with free CoA31. The enzyme activity was calculated as the consumption of acetyl-CoA.
To verify the functions of PmBEAT36 and PmBEAT37 in floral synthesis, the purified plasmids were transformed into petal protoplasts of P. mume. After 30 h of incubation, the protoplasts were collected, and the endogenous components were extracted by vortexing in ethyl acetate for 15 min. Next, benzyl propionate was added as an internal standard, and the contents of benzaldehyde, benzyl alcohol and benzyl acetate were analyzed using GC-MS.
Electronic supplementary material
Acknowledgements
This work was supported by the Beijing Natural Science Foundation (6174044), National Natural Science Foundation of China (31471906), and Special Fund for Beijing Common Construction Project.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0104-4).
References
- 1.Hao RJ, et al. Emitted and endogenous floral scent compounds of Prunus mume and hybrids. Biochem. Syst. Ecol. 2014;54:23–30. doi: 10.1016/j.bse.2013.12.007. [DOI] [Google Scholar]
- 2.Dudareva N, D’Auria JC, Nam KH, Raguso RA, Pichersky E. Acetyl-CoA:benzylalcohol acetyltransferase--an enzyme involved in floral scent production in Clarkia breweri. Plant J. 1998;14:297–304. doi: 10.1046/j.1365-313X.1998.00121.x. [DOI] [PubMed] [Google Scholar]
- 3.Aharoni A, et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:647–662. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalit M, et al. Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/Citronellol acetyltransferase in developing rose petals. Plant Physiol. 2003;131:1868–1876. doi: 10.1104/pp.102.018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guterman I, et al. Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Mol. Biol. 2006;60:555–563. doi: 10.1007/s11103-005-4924-x. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, De Luca V. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 2005;44:606–619. doi: 10.1111/j.1365-313X.2005.02552.x. [DOI] [PubMed] [Google Scholar]
- 7.D’Auria JC, Pichersky E, Schaub A, Hansel A, Gershenzon J. Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 2007;49:194–207. doi: 10.1111/j.1365-313X.2006.02946.x. [DOI] [PubMed] [Google Scholar]
- 8.D’Auria JC, Chen F, Pichersky E. Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiol. 2002;130:466–476. doi: 10.1104/pp.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souleyre EJ, Greenwood DR, Friel EN, Karunairetnam S, Newcomb RD. An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J. 2005;272:3132–3144. doi: 10.1111/j.1742-4658.2005.04732.x. [DOI] [PubMed] [Google Scholar]
- 10.Boatright J, et al. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 2004;135:1993–2011. doi: 10.1104/pp.104.045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dexter R, et al. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 2007;49:265–275. doi: 10.1111/j.1365-313X.2006.02954.x. [DOI] [PubMed] [Google Scholar]
- 12.Chedgy RJ, Kollner TG, Constabel CP. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry. 2015;113:149–159. doi: 10.1016/j.phytochem.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Ma X, Koepke J, Panjikar S, Fritzsch G, Stockigt J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 2005;280:13576–13583. doi: 10.1074/jbc.M414508200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JZ. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 15.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005;15:621–627. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 18.Brosius J. Retroposons: seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong H, et al. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi N, et al. Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol. 2004;45:83–91. doi: 10.1093/pcp/pch009. [DOI] [PubMed] [Google Scholar]
- 22.Sakai H, et al. Retrogenes in rice (Oryza sativa L. ssp japonica) exhibit correlated expression with their source genes. Genome Biol. Evol. 2011;3:1357–1368. doi: 10.1093/gbe/evr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdelsamad A, Pecinka A. Pollen-specific activation of Arabidopsis retrogenes is associated with global transcriptional reprogramming. Plant Cell. 2014;26:3299–3313. doi: 10.1105/tpc.114.126011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. The genome of Prunus mume. Nat. Commun. 2012;3:1318. doi: 10.1038/ncomms2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao RJ, et al. A comparative analysis of characteristic floral scent compounds in Prunus mume and related species. Biosci. Biotechnol. Biochem. 2014;78:1640–1647. doi: 10.1080/09168451.2014.936346. [DOI] [PubMed] [Google Scholar]
- 26.Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 30.Bao F, et al. Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses. New Phytol. 2014;202:1320–1334. doi: 10.1111/nph.12760. [DOI] [PubMed] [Google Scholar]
- 31.Nimitkeatkai H, et al. Effect of jasmonates on ethylene biosynthesis and aroma volatile emission in Japanese apricot infected by a pathogen (Colletotrichum gloeosporioides) J. Agric. Food Chem. 2011;59:6423–6429. doi: 10.1021/jf2010996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.