Human-to-human transmission risk of avian influenza A(H7N9) virus has not changed over time and remains limited to date, although the highly-pathogenic avian influenza A(H7N9) virus emerged and the number of human cases increased dramatically during the 2016–17 epidemic wave.
Keywords: influenza A(H7N9), cluster, human-to-human transmissibility, genetic susceptibility
Abstract
Background
The 2016–17 epidemic of human infections with avian influenza A(H7N9) virus was alarming, due to the surge in reported cases across a wide geographic area and the emergence of highly-pathogenic A(H7N9) viruses. Our study aimed to assess whether the human-to-human transmission risk of A(H7N9) virus has changed across the 5 waves since 2013.
Methods
Data on human cases and clusters of A(H7N9) virus infection were collected from the World Health Organization, open access national and provincial reports, informal online sources, and published literature. We compared the epidemiological characteristics of sporadic and cluster cases, estimated the relative risk (RR) of infection in blood relatives and non–blood relatives, and estimated the bounds on the effective reproductive number (Re) across waves from 2013 through September 2017.
Results
We identified 40 human clusters of A(H7N9) virus infection, with a median cluster size of 2 (range 2–3). The overall RR of infection in blood relatives versus non–blood relatives was 1.65 (95% confidence interval [CI]: 0.88, 3.09), and was not significantly different across waves (χ2 = 2.66, P = .617). The upper limit of Re for A(H7N9) virus was 0.12 (95% CI: 0.10, 0.14) and was not significantly different across waves (χ2 = 1.52, P = .822).
Conclusions
The small cluster size and low Re suggest that human-to-human transmissibility of A(H7N9) virus has not changed over time and remains limited to date. Continuous assessment of A(H7N9) virus infections and human case clusters is of crucial importance for public health.
Asian lineage avian influenza A(H7N9) virus infections of humans were first identified in eastern China during spring 2013, and have caused subsequent annual epidemic waves of human infections with the A(H7N9) virus [1, 2]. A total of 1525 laboratory-confirmed cases of human infection with the A(H7N9) virus were reported in 27 of 31 provinces in mainland China as of 17 March 2018. A(H7N9) virus was determined to pose the highest risk of pandemic emergence and public health impact among novel influenza A virus subtypes to date [3].
The 2016–17 A(H7N9) epidemic wave (1 October 2016 to 30 September 2017) attracted global attention because of 3 developments. First, low-pathogenic avian influenza (LPAI) A(H7N9) virus evolved to highly-pathogenic avian influenza (HPAI) A(H7N9) virus following insertion of 4 basic amino acid sequences in the hemagglutinin cleavage site during circulation among poultry [4]. The World Health Organization first reported human infections with HPAI A(H7N9) virus in February 2017 [5]. Animal experiments indicated that an A(H7N9) virus bearing the hemagglutinin cleavage site basic amino acid insertion and the PB2 627K/701N mutation was highly lethal in mice and ferrets and was transmitted efficiently in ferrets by respiratory droplets, which may enhance its risk for humans [6, 7]. Second, A(H7N9) viruses diverged genetically into 2 separate lineages (Pearl River Delta lineage and Yangtze River Delta lineage), with the Yangtze River Delta lineage viruses differing antigenically from those of earlier epidemics [8]. Analyses of gene sequences revealed the predominance of Yangtze River Delta lineage viruses in the 2016–17 wave [8]. Third, the 2016–17 wave was the largest epidemic to date, with 749 laboratory-confirmed A(H7N9) virus infections. This surge was accompanied by an expansion in the geographic distribution of human infections with A(H7N9) virus from the eastern and southern provinces to the northern and western provinces, resulting in much wider coverage than during the previous 4 waves [9–11]. The evolving genetic features of A(H7N9) viruses, as well as the changing spatial-temporal pattern of A(H7N9) epidemics, raised concerns about the pandemic potential of A(H7H9) viruses circulating during the 2016–17 wave.
Our previous study showed that the epidemiological characteristics and clinical severities of laboratory-confirmed A(H7N9) cases had not changed substantially across waves [9]. HPAI A(H7N9) patients had similar epidemiological characteristics and disease severities as LPAI A(H7N9) patients [12, 13]. Clusters of 2 or more epidemiologically-linked cases of A(H7N9) virus infection have been reported infrequently, with limited human-to-human transmissibility during the 2013 spring, 2013–14, and 2014–15 waves [2, 14–16]. A recent study presented the characteristics of 40 clusters of case-patients during 5 waves in China, and found comparable cluster numbers, sizes, and proportions of clusters with probable human-to-human transmission across waves [17]. However, the study did not provide quantitative estimates of the transmission risk of the A(H7N9) virus across waves, and did not provide assessments of the clinical severity of and familial susceptibility to the A(H7N9) virus. Our objective was to describe the epidemiological characteristics and clinical severities of all clusters of human infection with the A(H7N9) virus in mainland China and to investigate whether human-to-human transmissibility has changed across 5 waves, with a focus on the 2016–17 wave, when the HPAI A(H7N9) virus emerged and the number of human cases increased dramatically.
METHODS
Sources of Data
Data on clusters of human infection with A(H7N9) virus from 2013 to September 2017 were collected from multiple data sources, including the World Health Organization (WHO) Disease Outbreak News, WHO Monthly Risk Assessment Summary reports, websites of national and provincial Health and Family Planning Commissions of China, Flu Trackers, HealthMap, avian influenza case reports from the Centre of Health Protection of Hong Kong, and a literature review of both English and Chinese databases. Our detailed literature search strategy is available in the Supplementary Data. Conflicting information was resolved based on the reports from the WHO. A detailed description of case definitions, surveillance for identification of cases, and laboratory testing for the A(H7N9) virus have been provided elsewhere [18]. Case definitions; detection methods; field investigations of laboratory-confirmed human infections with the A(H7N9) virus and 7-day monitoring of their close contacts; and data collection protocols remained the same across waves [12].
A cluster was defined as a group of 1 or more laboratory-confirmed cases of A(H7N9) virus infection that occurred in the same setting, such as a household, a hospital, a residential institution, a military barrack, a recreational camp, or a neighborhood, and in which the infected individuals had the onset of symptoms within 2 weeks of each other [15]. We included 1 clinically-suspected case with an A(H7N9) virus infection in the first cluster, as it had been described elsewhere [2]. Cluster size was defined as the total number of laboratory-confirmed cases of A(H7N9) virus infection in a cluster. Definitions of a cluster index case, cluster secondary case, and sporadic case are available in the Supplementary Data.
We assumed that sporadic cases and cluster index cases acquired infection from exposure to either poultry or an environment contaminated by poultry (termed as “poultry” hereafter), while we classified the infection source for the cluster secondary cases as human, poultry, or unknown, based on the reported information of poultry exposure. If a cluster secondary case had contact with the cluster index case and did not share the same poultry exposure with the index case within 10 days before symptom onset, we classified the possible source of infection as human exposure; if the cluster secondary case had the same poultry exposure as the index case within 10 days before the onset of symptoms, or the interval between the illness onset dates of 2 successive cases was shorter than the lower bound of the estimated incubation period of 3 days [19], we classified the possible source of infection as poultry exposure. For cluster secondary cases where the source could not be identified, we classified the source of infection as unknown. The first wave was defined as from 1 February 2013 to 30 September 2013, and subsequent waves were defined as from 1 October to 30 September of the following year. We excluded data for wave 6 in the analysis, because only 3 sporadic human cases of A(H7N9) virus infection were reported when our analyses were completed as of 20 March 2018.
Statistical Analysis
The index case in each cluster was identified in the same way as sporadic cases. Hence, we compared secondary cluster cases with the combined group of sporadic cases and cluster index cases (sporadic/cluster index cases), using Wilcoxon test or Fisher’s exact test as appropriate, stratified by wave, to examine whether there were any systematic differences in epidemiological characteristics between the 2 types of cases. Clinical severity was assessed for cluster secondary cases and the sporadic/cluster index case, as well as for cases classified as having acquired a A(H7N9) virus infection from human exposure compared with poultry exposure, after excluding cases with an unknown infection source. A detailed clinical severity assessment is available in the Supplementary Data.
To assess epidemiological evidence for potential genetic susceptibility to A(H7N9) virus infection, family pedigrees were examined for the clusters with documented information on close contact with family members. Relative risks (RR) of infection of blood relatives vs non–blood relatives among close contacts of the index case were estimated by assuming that the probability of detecting infected symptomatic cases was the same for blood relatives and non–blood relatives. Definitions of blood relatives and non–blood relatives are available in the Supplementary Data. The RR of infection in related vs unrelated contacts was calculated as below. The Breslow-Day test was used to test for homogeneity of the RRs across waves.
Under the assumption of equal susceptibility of individuals and equal probability of case detection, we estimated the probability of infection given exposure, which led to the observed level of clustering. For a given probability of infection given exposure π, under the assumption of no genetic effect, the number of A(H7N9) cases in a household with size m follows a binomial distribution of Bin(m, π). Hence, we simulated the expected proportion of infection occurring in household clusters based on the household structure data of China, using previously published methods [20]. We used the previously published methods from Cauchemez et al to estimate the effective reproductive number Re [21], using 1 minus the proportion of all detected sporadic and index cases with any reported poultry exposure to provide an upper limit of Re and using 1 minus the proportion of index cases in a cluster with any reported poultry exposure as the lower limit of Re.
RESULTS
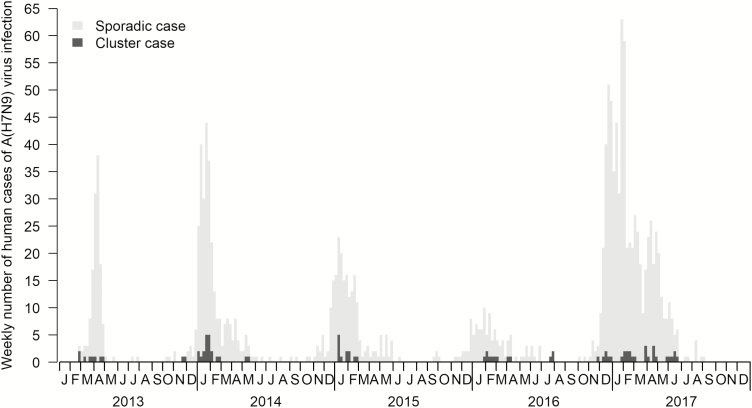
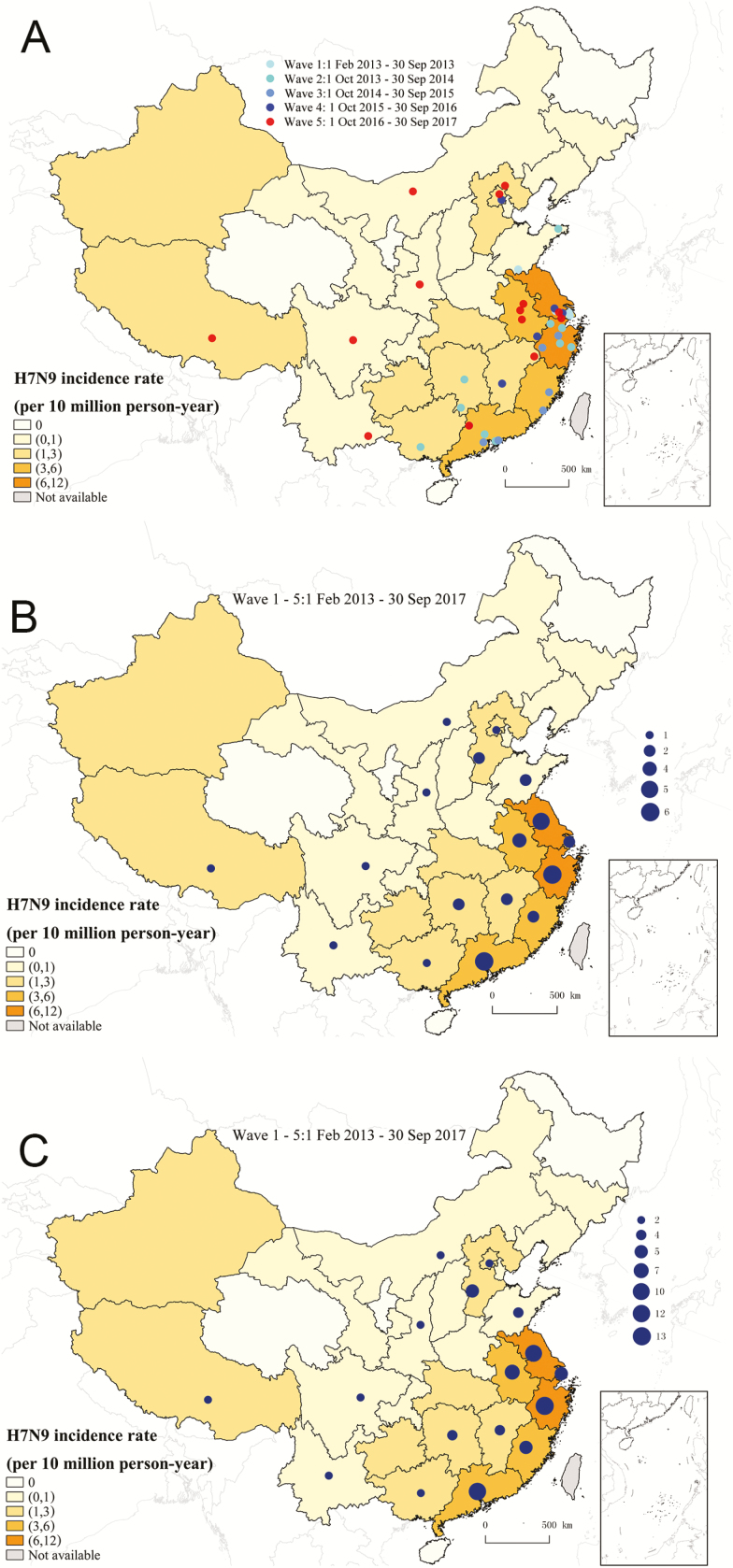
We identified 40 total clusters of cases of human infection with A(H7N9) virus, with 4, 11, 6, 5, and 14 clusters during waves 1 to 5, respectively (Table 1). The proportion of human cases occurring in clusters ranged from 3.9% to 10.5% across the 5 waves, with an overall proportion of 85/1523 (5.6%) (Table 1). The reported A(H7N9) cluster cases peaked during spring 2013 and during the winter-spring seasons of 2014–2017, and the peaks of sporadic A(H7N9) cases were at similar times (Figure 1). Human infections with the A(H7N9) virus were reported in all provinces/municipalities in mainland China, except for Qinghai, Ningxia, Heilongjiang, and Hainan provinces (Figure 2). The A(H7N9) cluster cases were primarily detected in eastern and southern China during the previous 4 waves, while new clusters were reported from western and northern provinces, including Tibet, Sichuan, Yunnan, Inner Mongolia, and Beijing, in the fifth wave (Figure 2).
Table 1.
Characteristics of Avian Influenza A(H7N9) Clusters and Cluster Cases Identified in Mainland China Across 5 Epidemic Waves, 2013–2017
| Characteristic | Wave 1, Feb 2013–Sep 2013 (n = 135) | Wave 2, Oct 2013–Sep 2014 (n = 306) | Wave 3, Oct 2014–Sep 2015 (n = 219) | Wave 4, Oct 2015–Sep 2016 (n = 114) | Wave 5, Oct 2016–Sep 2017 (n = 749) | Wave 1–5, Feb 2013–Sep 2017 (n = 1523) |
|---|---|---|---|---|---|---|
| No. of clusters | 4 | 11 | 6 | 5 | 14 | 40 |
| n (%) of cases occurring in clusters | 9 (6.7) | 23 (7.5) | 12 (5.5) | 12 (10.5) | 29 (3.9) | 85 (5.6) |
| Cluster size (%) | ||||||
| Median (range) | 2 (2–3) | 2 (2–3) | 2 (2–2) | 2 (2–3) | 2 (2–3) | 2 (2–3) |
| Average (SD) | 2.2 (0.5) | 2.1 (0.3) | 2.0 (0.0) | 2.4 (0.6) | 2.1 (0.3) | 2.1 (0.3) |
| 2 cases | 3 (75.0) | 10 (90.9) | 6 (100.0) | 3 (60.0) | 13 (92.9) | 35 (87.5) |
| 3 cases | 1 (25.0) | 1 (9.1) | 0 (0.0) | 2 (40.0) | 1 (7.1) | 5 (12.5) |
| No. of cases by infection source (%) | ||||||
| Human | 5 (55.6) | 5 (21.7) | 3 (25.0) | 3 (25.0) | 6 (20.7) | 22 (25.9) |
| Poultry | 4 (44.4) | 12 (52.2) | 7 (58.3) | 5 (41.7) | 18 (62.1) | 46 (54.1) |
| Unknown | 0 (0.0) | 6 (26.1) | 2 (16.7) | 4 (33.3) | 5 (17.2) | 17 (20.0) |
| Secondary cases in blood relative and non–blood relative contacts of index casesa | ||||||
| Blood relative contact, infected | 4 | 8 | 4 | 5 | 8 | 29 |
| Blood relative contact, not infected | 3 | 22 | 15 | 9 | 10 | 59 |
| Non–blood relative, contact infected | 1 | 4 | 2 | 2 | 1 | 10 |
| Non–blood relative, contact not infected | 4 | 14 | 7 | 7 | 8 | 40 |
| Relative riskb (95% CI) | 2.86 (0.44, 18.48) | 1.2 (0.42, 3.42) | 0.95 (0.21, 4.25) | 1.61 (0.39, 6.58) | 4 (0.59, 27.25) | 1.65 (0.88, 3.09) |
Abbreviations: CI, confidence interval; SD, standard deviation.
aRefers to a blood relationship with the cluster index case. Index cases of the clusters are excluded. A blood relative relationship was defined as a parent/offspring, sibling, grandparent/grandchild, uncle/aunt, or niece/nephew. A non–blood relative contact was defined as a spouse, healthcare worker, son-/daughter-in-law, parent-in-law, or other unrelated family member.
bRelative risk of infection of blood relatives vs non–blood relatives.
Figure 1.
Epidemic curve of cluster and sporadic human cases of avian influenza A(H7N9) virus infection in mainland China, 2013–2017.
Figure 2.
Geographic distribution of clusters and cluster cases of human infection with avian influenza A(H7N9) virus in mainland China, 2013–2017. A, Clusters by epidemic waves. B, clusters in all epidemic waves. C, cluster cases in all epidemic waves.
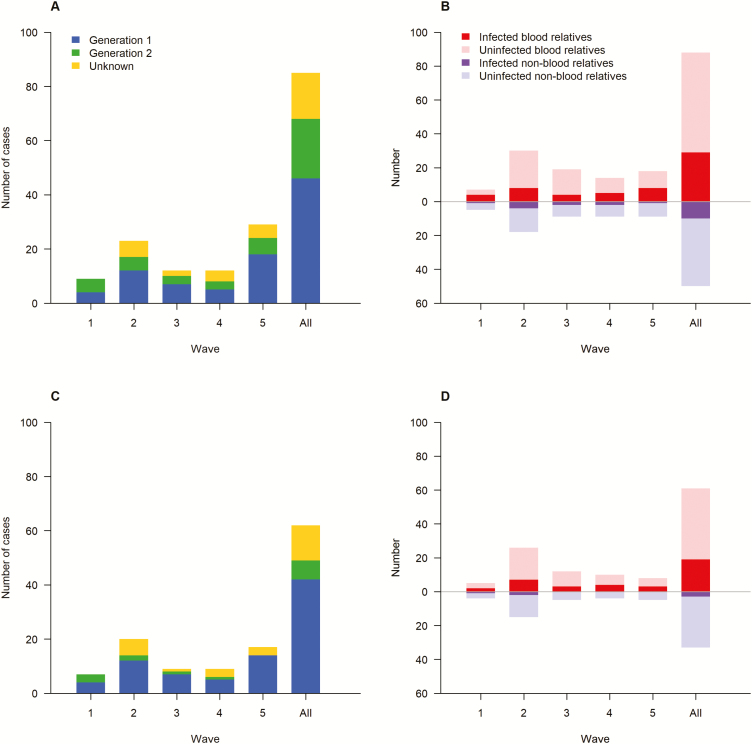
Cluster sizes remained small across waves, with a maximum cluster size of 3 cases and a median cluster size of 2 (Table 1). None of the clusters involved potential transmission beyond 2 generations (Figure 3A and 3C). Half of the cluster cases were classified as having acquired infection from poultry-related exposures and 26% of cluster cases were classified as having acquired infection from exposure to an infected human case, while the infection source for the remaining cluster cases was classified as unknown (Table 1). The proportions of identified cluster cases that were among blood relatives were generally higher than those among non–blood relatives (Figure 3B and 3D), although none of the RRs were statistically significant (Table 1). RRs of infection in blood relatives versus non–blood relatives were consistent across waves (χ2 = 2.66, P = .617).
Figure 3.
Generations of cluster cases of avian influenza A(H7N9) virus infection and infection status in blood and non–blood relatives across 5 epidemic waves in mainland China, 2013–2017. A, generations of cluster cases among family relatives. B, infection status in blood and non–blood relatives among family relatives. C, generations of cluster cases among household members. D, infection status in blood and non–blood relatives among household members.
The individuals in sporadic/index cases tended to be older than those in secondary cases (median, 58 years vs 40 years; P < .001; Supplementary Table S1). However, the age difference was reduced in the fifth wave, with individuals in sporadic/index cases only 4 years older, on average, than those in secondary cases (Supplementary Table S1). Overall, males accounted for a higher proportion of those infected in sporadic/index cases than secondary cases (71% vs 51%, relatively; P = .007). Individuals from sporadic/index cases were more likely to have chronic medical conditions than those from secondary cases (36% vs 13%, relatively; P = .002; Supplementary Table S1). Individuals from sporadic/index cases had higher proportions of any poultry exposure, occupational exposure and visiting a live poultry market, than secondary cases, but the 2 groups were comparable for exposure to backyard poultry (Supplementary Table S1). Exposure to sick/dead poultry was more common for individuals from the sporadic/index cases than for those from the cluster secondary cases in waves 1–4 (P < .001), while the trend was the opposite in wave 5, but without statistical significance (P = .373).
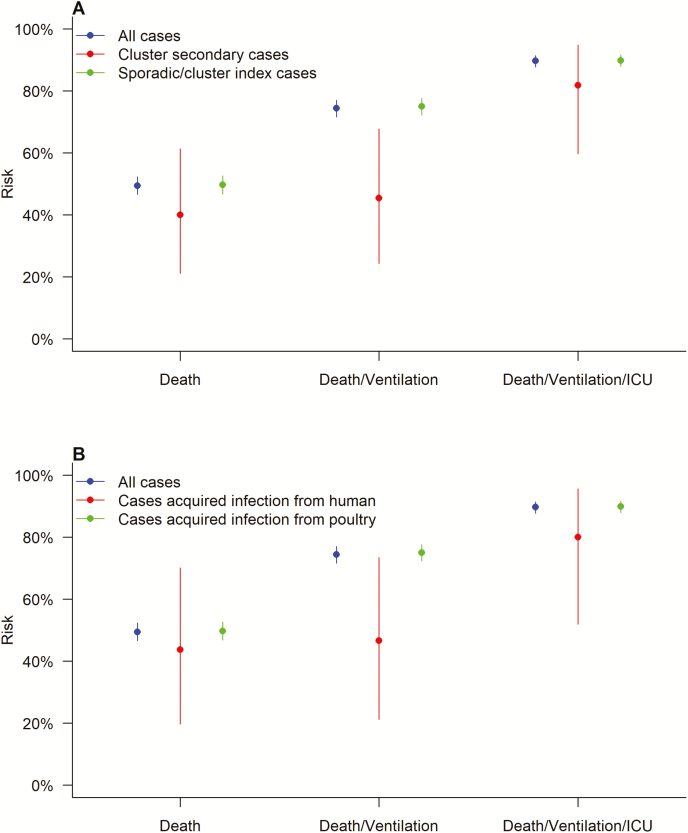
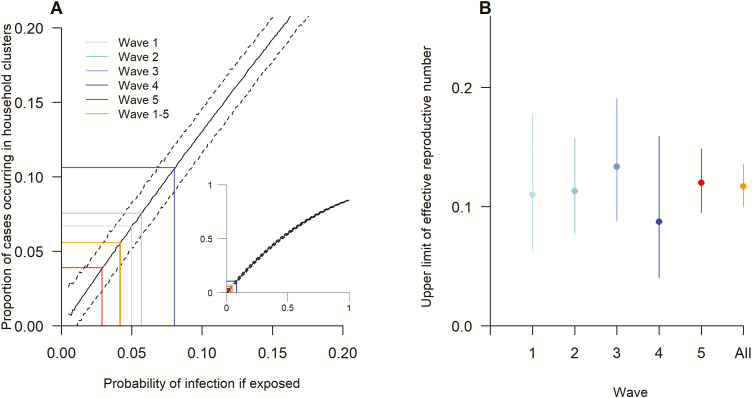
The point estimates of risks of death, mechanical ventilation, or intensive care unit admission in sporadic/index cases were all higher than the risks in cluster secondary cases, although the difference was statistically significant only for death/mechanical ventilation (χ2 = 8.40, P = .004; Figure 4). Cases classified as having acquired infection from poultry exposure had higher point estimates of risks (although non-significant) for deaths, mechanical ventilation, or intensive care unit admission than cases classified as having acquired infection from exposure to other human cases (Figure 4). Similar results were found in the sensitivity analyses, in which cases with unknown sources of infection were either classified as resulting from poultry or from human cases (Supplementary Figure S1). If equal susceptibility of every individual is assumed, the probability of infection of 4.2% (95% CI: 3.0%, 5.3%) is compatible with the observed percentage of cases that occurred in clusters (5.6%). The estimated probability of infection, given exposure, was lower in the fifth wave compared with the previous 4 waves (Figure 5A). The overall upper limit of Re for A(H7N9) virus among humans was 0.12 (95% CI: 0.10, 0.14), without a significant difference across waves (χ2 = 1.52, P = .822; Figure 5B). The overall lower limit of Re was 0.03 (95% CI: 0, 0.14), and the 95% CIs for wave-stratified Re estimates were wide due to the limited number of cluster index cases identified in each wave (Supplementary Table S2). The wave-specific lower limit of Re was not significantly different (χ2 = 6.58, P = .160).
Figure 4.
Clinical severities of laboratory-confirmed cases of avian influenza A(H7N9) virus infection in mainland China, stratified by (A) cluster secondary cases or sporadic/cluster index cases or (B) cases with an infection acquired from a human or poultry source. Abbreviation: ICU, intensive care unit.
Figure 5.
Infection risk and the estimated effective reproductive numbers for avian influenza A(H7N9) virus across 5 epidemic waves. A, proportion of cases that occurred in household clusters by the probability of infection given exposure. B, estimated upper limit of the effective reproductive number Re.
DISCUSSION
Our study provides a comprehensive description of human clusters of A(H7N9) virus infection and a quantitative assessment of the risk of human-to-human transmissibility of A(H7N9) virus across 5 epidemic waves in mainland China. The relatively small cluster size and Re suggest that human-to-human transmissibility of the A(H7N9) virus remained limited during the 2016–2017 wave. The surge of reported human cases of A(H7N9) virus infection in the fifth wave is, therefore, most likely explained by sporadic A(H7N9) virus transmission from poultry to humans in a wider geographic area, rather than by enhanced transmissibility between humans. Our finding of unchanged human-to-human A(H7N9) transmission risk is consistent with a previous descriptive analysis [17]. However, our study further advances the understanding of A(H7N9) virus transmission by providing quantitative, wave-specific estimates of the lower and upper limits of Re and the probability of infection given exposure among the population at risk, using modeling and simulation techniques.
We observed that the peak of A(H7N9) cluster cases occurred at about the same time with the peak of sporadic A(H7N9) cases across waves, yet the proportion of human cases occurring in clusters in wave 5 was significantly lower than the proportions in waves 2 and 4. This phenomenon does not mean that human-to-human transmissibility of A(H7N9) virus was reduced in the fifth wave, since estimates of Re are comparable across waves. Under-ascertainment and under-reporting of human cluster cases of A(H7N9) virus infection, particularly those resulting in mild illnesses not requiring hospitalization, might be a more plausible explanation, although a well-designed population-based sero-epidemiology study would be necessary to test this hypothesis. Individuals from sporadic/index cases were, on average, older than those from secondary cases, and the age difference declined in the fifth wave, suggesting that the detection and reporting bias towards the older population was reduced in the fifth wave.
The finding of lower point estimates of risks of death or mechanical ventilation for cluster secondary cases compared to sporadic/index cases is likely due to different case detection approaches for sporadic/index and secondary cases of A(H7N9) virus infection. Sporadic/index mild or moderately ill cases were not fully captured, because case finding was focused on hospitalized patients with severe illnesses. A case-series analysis of mild to moderate illnesses from A(H7N9) virus infection, detected by China’s national sentinel surveillance system for influenza-like illnesses, supports the existence of a “clinical iceberg” phenomenon for A(H7N9) virus infections, with most mild illness cases likely going undetected [22]. Persons with a non-severe illness might not seek medical care or not be suspected, and not tested, for the A(H7N9) virus infection. In contrast, clinically mild or moderately ill secondary cases were more likely to be detected through the active 10-day monitoring conducted among close contacts for each severely ill index case of laboratory-confirmed A(H7N9) virus infection. Any illness in a close contact during the 10 days prompted A(H7N9) virus testing. This enhanced case detection among close contacts of a severely ill index case resulted in a higher probability of detecting secondary cases with mild to moderate illnesses. Another possible explanation for the lower severities for secondary cases could be through attenuated virulence as a result of A(H7N9) virus adaption to human hosts [23], although there is currently no evidence that the A(H7N9) viruses infecting secondary cases differ from the viruses infecting sporadic/index cases.
Whether susceptibility to avian influenza A virus infections in humans has a heritable component remains undetermined. Clustering of human cases of the A(H5N1) virus infection among blood relatives suggests that genetic susceptibility may play a role, while a simulation study reported that the high proportion of clusters among blood relatives is expected without increased genetic susceptibility [24]. Our study observed a higher proportion of secondary A(H7N9) virus infections among blood relatives, but the RR of infection in blood relatives versus non–blood relatives was not statistically significant, suggesting limited genetic susceptibility to A(H7N9) virus infections. Further in-depth studies on genetic susceptibility to avian influenza A(H7N9) virus infections are needed.
Our study has several limitations. First, our classification of the infection source for A(H7N9) cases was based on reported epidemiological characteristics, and thus some misclassification errors were inevitable. Second, data on poultry exposure were missing for 2–23% of A(H7N9) cases across waves, which may have affected the estimates of Re using the method developed by Cauchemez et al [21]. Third, potential under-ascertainment of A(H7N9) cluster cases during the fifth wave may have impacted our estimates of the probability of infection after exposure. Fourth, a total of 28 human cases of HPAI A(H7N9) virus infection had been reported as of 14 July 2017, with 27 cases reported in mainland China [4]. Stratified analyses of Re estimates by LPAI and HPAI A(H7N9) cases would allow comparison of any transmissibility differences between LPAI and HPAI A(H7N9) viruses, but we lacked the data to exactly distinguish the HPAI A(H7N9) cases in our line list. Nevertheless, we did search the literature for reports of human cases of HPAI A(H7N9) virus infection to match the cases in our line list. Under 1000 scenarios of matching, the upper limits of Re were not statistically different for HPAI and LPAI viruses.
We concluded that the risk of human-to-human A(H7N9) virus transmission remained low throughout the 5 epidemic waves in China. However, more human infections with the A(H7N9) virus are expected, and ongoing monitoring and assessment of the evolving virology and transmission dynamics of the A(H7N9) virus, both to and between humans, is of public health importance for pandemic influenza preparedness and response.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
Funding. This work was supported by the National Science Fund for Distinguished Young Scholars [grant number 81525023 to H. Y.]; Program of Shanghai Academy/Technology Research Leader [grant number 18XD1400300 to H. Y.]; U.S. National Institutes of Health [grant number U19 AI51915 to H. Y. ]; Harvard Center for Communicable Disease Dynamics from the National Institute of General Medical Sciences [grant number U54 GM088558 to B. J. C.]; Health and Medical Research Fund of the Health, Welfare, and Food Bureau of the Hong Kong Special Administrative Region Government [grant number 14131432 to B. J. C.]; Research Grants Council of the Hong Kong Special Administrative Region, China [project number T11-705/14N to B. J. C.]; a commissioned grant from the Health and Medical Research Fund of the Health, Welfare, and Food Bureau of the Hong Kong SAR Government; and the National Natural Science Foundation of China [grant numbers 81402731 to H. Y. and 81602936 to X. W.].
Potential conflicts of interest. H. Y. has received investigator-initiated research funding from Sanofi Pasteur, GlaxoSmithKline, bioMérieux Diagnostic Product (Shanghai), and Yichang HEC Changjiang Pharmaceutical Company. B. J. C. has received research funding from Sanofi Pasteur. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gao GF. From “A”IV to “Z”IKV: attacks from emerging and re-emerging pathogens. Cell 2018; 172:1157–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Q, Zhou L, Zhou M, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 2014; 370:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention. Influenza risk assessment tool (IRAT). Available at: https://www.cdc.gov/flu/pandemic-resources/national-strategy/risk-assessment.htm. Accessed 12 November 2017. [Google Scholar]
- 4. Yang L, Zhu W, Li X, et al. Genesis and spread of newly emerged highly pathogenic H7N9 Avian viruses in Mainland China. J Virol 2017; 91:e01277–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Human infection with avian influenza A(H7N9) virus—China. Available at: http://www.who.int/csr/don/27-february-2017-ah7n9-china/en/#. Accessed 27 February 2017. [Google Scholar]
- 6. Shi J, Deng G, Kong H, et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res 2017; 27:1409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Subbarao K. Avian influenza H7N9 viruses: a rare second warning. Cell Res 2018; 28:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kile JC, Ren R, Liu L, et al. Update: increase in human infections with Novel Asian lineage Avian influenza A(H7N9) viruses during the fifth epidemic—China, October 1, 2016–August 7, 2017. MMWR Morb Mortal Wkly Rep 2017; 66:928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X, Jiang H, Wu P, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 2017; 17:822–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou L, Ren R, Yang L, et al. Sudden increase in human infection with avian influenza A(H7N9) virus in China, September–December 2016. Western Pac Surveill Response J 2017; 8:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iuliano AD, Jang Y, Jones J, et al. Increase in human infections with Avian influenza A(H7N9) virus during the fifth epidemic - China, October 2016-February 2017. MMWR Morb Mortal Wkly Rep 2017; 66:254–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou L, Tan Y, Kang M, et al. Preliminary epidemiology of human infections with highly pathogenic Avian influenza A(H7N9) virus, China, 2017. Emerg Infect Dis 2017; 23:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang M, Lau EHY, Guan W, et al. Epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus in Guangdong, 2016 to 2017. Euro Surveill 2017; 22:30568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu J, Zhu Y, Zhao B, et al. Limited human-to-human transmission of avian influenza A(H7N9) virus, Shanghai, China, March to April 2013. Euro Surveill 2014; 19:20838. [DOI] [PubMed] [Google Scholar]
- 15. Qin Y, Horby PW, Tsang TK, et al. Differences in the epidemiology of human cases of avian influenza A(H7N9) and A(H5N1) viruses infection. Clin Infect Dis 2015; 61:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu B, Havers FP, Zhou L, et al. Clusters of human infections with Avian influenza A(H7N9) virus in China, March 2013 to June 2015. J Infect Dis 2017; 216(Suppl 4):S548–54. [DOI] [PubMed] [Google Scholar]
- 17. Zhou L, Chen E, Bao C, et al. Clusters of human infection and human-to-human transmission of Avian influenza A(H7N9) virus, 2013–2017. Emerg Infect Dis 2018; 24:397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu H, Cowling BJ, Feng L, et al. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 2013; 382:138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Virlogeux V, Li M, Tsang TK, et al. Estimating the distribution of the incubation periods of human avian influenza A (H7N9) virus infections. Am J Epidemiol 2015; 182:723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horby P, Sudoyo H, Viprakasit V, et al. What is the evidence of a role for host genetics in susceptibility to influenza A/H5N1?Epidemiol Infect 2010; 138:1550–8. [DOI] [PubMed] [Google Scholar]
- 21. Cauchemez S, Epperson S, Biggerstaff M, Swerdlow D, Finelli L, Ferguson NM. Using routine surveillance data to estimate the epidemic potential of emerging zoonoses: application to the emergence of US swine origin influenza A H3N2v virus. PLoS Med 2013; 10:e1001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ip DK, Liao Q, Wu P, et al. Detection of mild to moderate influenza A/H7N9 infection by China’s national sentinel surveillance system for influenza-like illness: case series. BMJ 2013; 346:f3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown NF, Wickham ME, Coombes BK, Finlay BB. Crossing the line: selection and evolution of virulence traits. PLoS Pathog 2006; 2:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pitzer VE, Olsen SJ, Bergstrom CT, Dowell SF, Lipsitch M. Little evidence for genetic susceptibility to influenza A (H5N1) from family clustering data. Emerg Infect Dis 2007; 13:1074–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.