Abstract
Background
Psoriasis is a common chronic inflammatory skin disease which lacks effective strategies for the treatment. Natural compounds with biological activities are good tools to identify new targets with therapeutic potentials. Acetyl-11-keto-β-boswellic acid (AKBA) is the most bioactive ingredient of boswellic acids, a group of compounds with anti-inflammatory and anti-cancer properties. Target identification of AKBA and metabolomics analysis of psoriasis helped to elucidate the molecular mechanism underlying its effect, and provide new target(s) to treat the disease.
Methods
To explore the targets and molecular mechanism of AKBA, we performed affinity purification, metabolomics analysis of HaCaT cells treated with AKBA, and epidermis of imiquimod (IMQ) induced mouse model of psoriasis and psoriasis patients.
Findings
AKBA directly interacts with methionine adenosyltransferase 2A (MAT2A), inhibited its enzyme activity, decreased level of S-adenosylmethionine (SAM) and SAM/SAH ratio, and reprogrammed one‑carbon metabolism in HaCaT cells. Untargeted metabolomics of epidermis showed one‑carbon metabolism was activated in psoriasis patients. Topical use of AKBA improved inflammatory phenotype of IMQ induced psoriasis-like mouse model. Molecular docking and site-directed mutagenesis revealed AKBA bound to an allosteric site at the interface of MAT2A dimer.
Interpretation
Our study extends the molecular mechanism of AKBA by revealing a new interacting protein MAT2A. And this leads us to find out the dysregulated one‑carbon metabolism in psoriasis, which indicates the therapeutic potential of AKBA in psoriasis.
Fund
The National Natural Science Foundation, the National Program on Key Basic Research Project, the Shanghai Municipal Commission, the Leading Academic Discipline Project of the Shanghai Municipal Education Commission.
Keywords: Psoriasis, AKBA, Metabolomics, MAT2A, Methionine cycle, One‑carbon metabolism
Graphical abstract
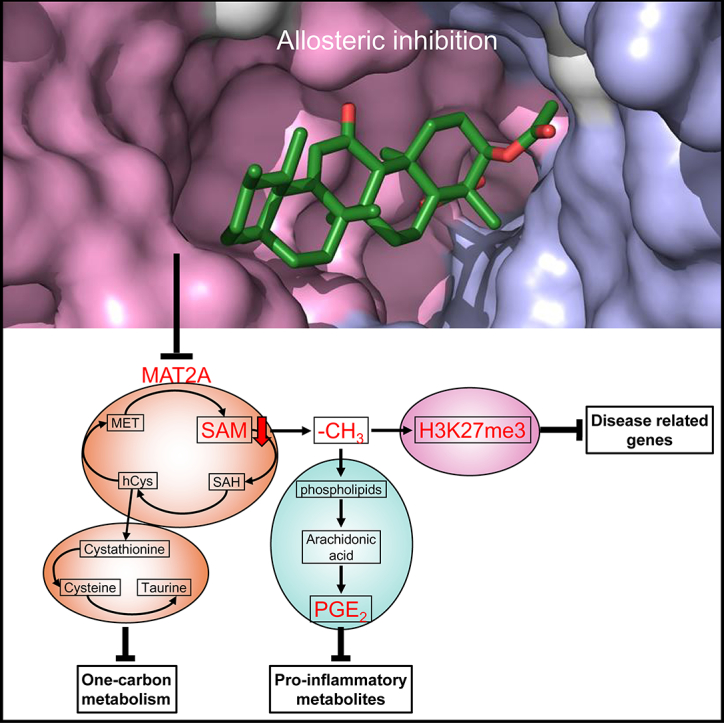
Research in context.
Evidence before this study
Psoriasis is a long-lasting autoimmune skin disease that urgently calls for effective treatment. AKBA, a natural compound extracted from gum resin of Boswellia species, has long been proved to be effective in inflammation and cancer models. Especially, we have confirmed its therapeutic effect in the CD18 hypomorphic mouse model of psoriasis. Further, we verified its effect in imiquimod (IMQ) induced mouse model of psoriasis, a universal in vivo model of psoriasis. However, the direct target(s) of AKBA still need to be fully explored. Besides, we hope to expand the targets for psoriasis therapy by elucidating the molecular mechanism of AKBA.
Added value of this study
Our study found MAT2A as one primary target of AKBA. We demonstrated AKBA directly interacted with MAT2A, inhibited its enzyme activity, and reprogrammed one‑carbon metabolism in HaCaT cells. Untargeted metabolomics of epidermis showed one‑carbon metabolism was activated in psoriasis patients and IMQ induced mouse model of psoriasis. The topical use of AKBA improved inflammatory phenotype of this mouse model probably through reregulating one‑carbon metabolism. Besides, the molecular docking and site-directed mutagenesis revealed one allosteric site of MAT2A which could be used for further compound screening.
Implications of all the available evidence
We provided evidence to indicate one‑carbon metabolism is dysregulated in the epidermis of psoriasis patients, which may induce the upregulation of proinflammatory metabolites as well as the construction of building blocks for rapid cell proliferation in psoriasis lesion. MAT2A and one‑carbon metabolism may be important targets for inhibiting the hyperproliferation of keratinocytes and improving the inflammatory phenotype of psoriasis.
Alt-text: Unlabelled Box
1. Introduction
The methionine adenosyltransferase (MAT) that catalyzes the synthesis of S-adenosylmethionine (SAM) from methionine and adenosine triphosphate (ATP) are conserved from bacteria to mammals [1]. There are three MAT genes in mammals: mat1A, mat2A encoding for the catalytic subunits MATα1 and MATα2, and mat2B encoding for the regulatory subunit MATβ [2]. Although MATα2 alone is able to produce SAM, an intermediate metabolite of methionine cycle, it forms MATαβ complex in vivo to synthesize SAM more efficiently [3]. SAM is the primary methyl donor for histone, DNA and RNA methylation to epigenetically regulate gene expression, and it is also involved in other metabolic pathways that require methyl moieties, including polyamine and lipid synthesis to exert control over cell growth and survival [4].
Besides up-regulation of methionine adenosyltransferase 2A (MAT2A) provides a growth advantage in colon cancer cells [5] and human hepatocellular carcinoma [3], serine, glycine and one‑carbon metabolism functionally act as drivers of cancer pathogenesis [4]. All these findings suggest that MAT2A and one‑carbon metabolism possibly have potentials as therapeutic targets. Indeed, two of the most widely used chemotherapies, methotrexate (MTX) and 5-fluorouracil, target one‑carbon metabolism to disrupt malignant hyperproliferation [6,7]. FIDAS agents, a group of stilbene derivatives, target MAT2A directly and exclusively, and repress CRC xenografts in nude mice [8,9].
Psoriasis is one of the most prevalent chronic skin diseases. Genetic predispositions and aberrant crosstalk between immune cells and keratinocytes contribute to the initiation and progression of the disease. Besides alternation in genes, epigenetics and proteomics, metabolomics was found greatly changed in patients with psoriasis [[10], [11], [12]]. Measurements of blood and skin samples of psoriasis patients showed significantly activated phospholipids and amino acids metabolism [13,14]. Among the most significantly altered metabolites, taurine was repeatedly reported to be increased either in blood or skin of patients with psoriasis [[15], [16], [17]]. Taurine is synthesized from cysteine through transsulfuration pathway. This pathway plus methionine cycle and folate cycle constitute one‑carbon metabolism which is responsible for distribution of carbon units into diverse cellular processes including cellular biosynthesis (lipids, nucleotides, proteins), maintenance of redox status, and epigenetic regulation through methylation of nucleic acids and proteins [4]. As methionine cycle provides methyl for generation of head groups of phospholipids, the aberrantly increased phospholipid and taurine metabolism seem to indicate an activated one‑carbon metabolism in psoriasis patients. Yet, whether this metabolism pathway is critical in the pathophysiology of psoriasis and whether the interference of this pathway harbors therapeutic potential are unexplored.
Acetyl-11-keto-β-boswellic acid (AKBA) is the most bioactive ingredient of boswellic acids [18]. These acids belong to a group of pentacyclic triterpenic acids that are purified from the gum resin of plants in the genus Boswellia which are usually found in dry and hot areas like India, Northern Africa and Middle East. Boswellic acids have long been used to treat inflammatory conditions and other ailments [19,20] since ancient time in China, India and Egypt [21]. Despite our previous study and others reported that some signaling pathways and/or proteins are involved in mechanisms of therapeutic effects of AKBA, such as NF-kappa B [22], 5-lipoxygense, topoisomerase, leukocyte elastase [23,24], LL-37 [25], HIF-1 [26], the direct target(s) of AKBA remain unclear.
Here we report that AKBA, a boswellic acid isolated from the gum resin of Boswellia species, binds directly and primarily to MAT2A. The site directed mutagenesis indicates that AKBA binds to an allosteric site of MAT2A in the dimer interface. AKBA inhibits MAT2A activity, decreases the production of SAM and SAM/SAH ratio, reprograms one‑carbon metabolism and reduces expression of H3K27me3 in HaCaT cells. Collectively, our data reveal that MAT2A is one primary target of AKBA, and AKBA ameliorates psoriasis-like phenotype of imiquimod (IMQ) induced mouse model probably through regulating one‑carbon metabolism and expression of disease related genes.
2. Materials and methods
2.1. Synthesis and characterization of biotinylated AKBA
A solution of AKBA (National Institutes for Food and Drug Control, 111,760) (1) (460.00 mg, 897.18 μmol, 1.00 eq) in SOCl2 (747.16 mg, 6.28 mmol, 7.00 eq) was stirred for 1 h at 90 °C. The reaction mixture was evaporated. The residue was diluted with toluene (15.00 mL) and then evaporated. This operation was repeated 2 times. To a solution of N-Boc-ethylenediamine (2) (172.48 mg, 1.08 mmol, 1.20 eq) and DMAP (49.32 mg, 403.73 μmol, 0.45 eq) in dry DCM (3 mL) was added drop-wise a solution of the above acyl chloride in dry DCM (3 mL) at 25 °C. The resulting mixture was stirred for 16 h at 25 °C. The mixture was evaporated to give N-(3-O-acetyl-11-keto-β-boswelloyl)-(2’-N-Boc)-ethylenediamine (3) (586.00 mg, crude). A solution of compound 3 (199.00 mg, 275.23 μmol, 1.00 eq) in HCl/dioxane (20.00 mL) was stirred for 0.5 h at 20 °C. The reaction mixture was evaporated to give the crude de-Boc product N-(3-O-acetyl-11-keto-β-boswelloyl)-ethylenediamine (4). To a mixture of the compound 4 (529.00 mg, 894.70 μmol, 1.00 eq), D-Biotin (5) (218.58 mg, 894.70 μmol, 1.00 eq) and DIEA (346.89 mg, 2.68 mmol, 3.00 eq) in dry DMF (2.00 mL) was added HATU (408.23 mg, 1.07 mmol, 1.20 eq). Then the resulting mixture was stirred for 16 h at 25 °C. The mixture was purified by prep-HPLC (neutral) to give biotinylated AKBA (6) (130.00 mg, 164.77 μmol, 18.42% yield, 99% purity) as a white solid. Characterization of biotinylated AKBA: 1H NMR (400 MHz, CDCl3) δ 7.35 (s, 1H), 6.78 (s, 1H), 6.58 (s, 1H), 5.84 (s, 1H), 5.54 (s, 1H), 5.28 (s, 1H), 4.52 (s, 1H), 4.33 (s, 1H), 3.50 (s, 2H), 3.28 (s, 2H), 3.14 (d, J = 4 Hz, 1H), 2,74 (d, J = 12.8 Hz, 1H), 2,53 (d, J = 12.8 Hz, 1H), 2.41 (d, J = 12 Hz, 1H), 2.09–2.22 (m, 4H), 1.96 (s, 4H), 1.47–1.74 (m, 10H), 1.18–1.42 (m, 19H), 1.09–1.11 (m, 7H), 0.95 (s, 4H), 0.82 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 13.29, 17.41, 18.37, 20.47, 21.12, 21.41, 24.39, 25.59, 27.49, 28.17, 28.88, 33.99, 37.35, 39.32, 43.80, 45.03, 46.53, 50.32, 55.69, 59.04, 60.19, 60.36, 61.74, 73.60, 130.46, 164.20, 165.00, 170.50, 174.73, 176.64, 199.25. LCMS (ESI): calculated for C44H68N4O6S [M + H]+, 781.49; found, 781.5.
2.2. Cell culture
HaCaT and sw1116 cells were cultured in DMEM growth medium, l-glutamine, high glucose, pyruvate (HyClone, SH30243.01) supplemented with 10% FBS and 1% Penicillian-Streptomycin. Mouse primary keratinocytes (mKCs) were isolated as previous reported [27]. All the cells were grown at 37 °C, 5% CO2.
2.3. Pulldown assay
HaCaT cells and mKCs were cultured in 6-well plates. AKBA in DMSO stock solution was added into culture media to pre-incubate cells for 1 h. Then biotin-AKBA or biotin were added into culture media to incubate cells for 1.5 h. Cells were washed twice with cold PBS before harvested and lysed in modified RIPA buffer (50 mM HEPES, 4 mM EDTA, 150 mM NaCl, 1% NP-40, pH 7.5) supplemented with protease and phosphatase inhibitors (Thermo, 78,440). After centrifugation at 12,000g for 15 min at 4 °C, the supernatant was collected and incubated with streptavidin-agarose beads in a tube revolver for 3 h at 4 °C. The beads were washed six times with modified RIPA buffer, and the bead-bound proteins were eluted, separated by SDS-PAGE, and visualized by silver staining. The protein-containing bands in the gel were excised, followed by in-gel digestion and analysis by LC-MS/MS.
2.4. MAT2A protein expression and purification
Human MAT2A expression plasmid in pET-28a (+) vector was transformed into Escherichia coli BL21 (DE3)-competent cells and expressed as a fusion protein containing an N-terminal TEV cleavable 6His-tag. Cells were grown at 37 °C in LB medium until OD600 = 4.0–6.0 and expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 20 °C. Cells were pelleted and resuspended in cold lysis buffer (25 mM HEPES pH 7.5, 0.5 M NaCl, 5% glycerol, 5 mM imidazole and 5 mM BME). The cell suspension was lysed with ultra-high-pressure cell disruptor and insoluble debris was removed by centrifugation at 14,000 rpm at 4 °C for 1 h. Proteins were eluted with 250 mM imidazole in lysis buffer. 6His-tag was removed by TEV cleavage overnight by dialysis against 25 mM HEPES pH 7.5, 200 mM NaCl, 5% glycerol, and 5 mM BME, which was followed by nickel reverse chromatography and dialysis to desalt. Untagged proteins were loaded onto a 5 mL HiTrap Q HP column (GE Healthcare). Proteins were eluted with a linear NaCl gradiat (25–500 mM). The eluted proteins were further purified by size-exclusion chromatography using a HiPrep 26/60 Superdex 200 (GE Healthcare). Fresh proteins were used for crystallization. Proteins for biochemical assays were stored at −80 °C in 25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM MgCl2, 5 mM KCl, 5% glycerol, and 2 mM DTT. Site-directed mutagenesis was performed with KOD-Plus-Mutagenesis Kit (TOYOBO) using pET28a-MAT2A as a template. These proteins were expressed in the Escherichia coli strain BL21 and purified.
2.5. Protein crystallization
The MAT2A-AKBA complex was prepared by mixing MAT2A at 5 mg/mL with 40 mM AKBA in DMSO at a final molar ratio of 4:1 AKBA:MAT2A. The mixture was incubated for 2 h on ice, then centrifuged at 14,000 rpm at 4 °C for 15 min to remove insoluble material. Using robot NT8, the complex was screened in sitting drop 350 nL + 350 nL drops. Good quality crystals grew in Jena Bioscience JBScreen Kinase 2 condition C8 (100 mM HEPES pH 7.5, 50 mM Lithium sulfate, 35% w/v polyethylene glycol 1000) at 20 °C.
2.6. Pocket detection and molecular docking
The potential binding sites on the MAT2A dimer were detected using the fpocket program. The predicted five top ranked binding sites were selected for docking studies. Molecular docking of AKBA to the five binding sites of MAT2A was performed using the AutoDock 4.2 program [28]. Polar hydrogen atoms were added to the MAT2A. Kollman united partial atomic charges were then assigned and the AutoDock atom types were defined for the protein using AutoDock Tools (ADT). Gasteiger charges were added to AKBA with the default root, rotatable bonds, and torsion setting using the TORSDOF module in ADT. The grid center was defined at the centroid of the five binding sites, and the numbers of grid points in the x, y, and z directions were set to 60, 60, and 60 with a spacing value of 0.375 Å. The Lamarckian genetic algorithm was used for the AKBA conformational search with identical docking parameters used in our previous protocols [29]. Fifty independent docking runs were conducted, and the binding energy was used to rank the docked AKBA in order of fitness.
2.7. MST assay
To test the binding affinity of AKBA and proteins, MST assay was conducted by using Monolish NT.115 (NanoTemper Technologies). Proteins were labeled with the Monolith NT™ Protein Labeling Kit RED (NanoTemper Technologies) according to the supplied labeling protocol. Labeled proteins were used at a concentration of ~ 165 nM. AKBA was titrated in 1:1 dilutions beginning at 25 μM, which contained 1.25% (v/v) DMSO. Samples were diluted in MAT assay buffer which contained 100 mM Tris-HCl pH 8.0, 20 mM MgCl2, and 200 mM KCl supplemented with DMSO at a final concentration of 1.25% to make sure that all samples contained the same DMSO concentration. For the measurement the samples were filled into premium coated capillaries.
2.8. BLI binding assays
BLI assays of MAT2A-AKBA interactions were performed using an Octet RED96 instrument (ForteBio). Biotin labeled AKBA was immobilized on Streptavidin coated biosensors in MAT assay buffer containing 100 mM Tris-HCl pH 8.0, 20 mM MgCl2 and 200 mM KCl. BSA was added to the assay buffer with a final concentration of 0.1 mg/mL to reduce non-specific interaction. The association of free MAT2A WT, K265AA, D258AA, K289BA, and K285BA with AKBA was measured at a range of protein concentrations (62.5, 125, 250, 500, and 1000 nM) for 800 s.
2.9. SAM synthesis activity assay by HPLC
SAM synthesis activity was assayed as previously reported with minor modifications [30,31]. Briefly, assays were performed with methionine, AKBA in DMSO, and 75 μg MAT2A in 150 μL end volume in MAT assay buffer at 37 °C. ATP was added into the system to initiate the reaction. The final concentration of MAT2A was 0.5 mg/ml, and for methionine and ATP were 5 mM. Then the reaction was stopped by addition of 150 μL 2% (v/v) cold HClO4 and subsequently neutralized with 24 μL 1 N NaOH. Separation of products was achieved by HPLC (Agilent 1260) using a Mercury SCX column (250 × 4.6 mm). The mobile phase consisted of 100 mM ammonium formate (pH was adjusted to 4.0 with formic acid). The formation of SAM was monitored at λ = 254 nm. The relative increase or decrease of SAM formation was analyzed by comparing peak areas of SAM auto integrated.
2.10. RNA interference
Custom and chemically modified small interfering RNA (siRNA) was designed to target human MAT2A (GenePharma). Non-specific siRNA duplex served as control (GenePharma). To knock down MAT2A in vitro, cultured HaCaT cells in 10 cm dish were transfected with 145 pmol siRNA (sense: GCUUGUGAAACUGU UGCUATT) by Lipofectamine RNAiMAX (Thermo, 13,778,030). The transfection duration was 48 h.
2.11. Human subjects
Psoriatic skin samples were obtained by punch biopsy from patients who were under local lidocaine anesthesia. Normal adult human skin specimens were taken from healthy donors who were undergoing plastic surgery. All of the individuals provided informed consent. The study was performed in accordance with the principles of the Declaration of Helsinki and approved by the Research Ethics Board of Xiangya Hospital of Central South University in Hunan, China.
2.12. Metabolite extraction
HaCaT cells were cultured in 10 cm dishes with 10 mL growth medium per dish. When cell confluency reached ~80%, 20 μL AKBA stock solution (10 mM, DMSO) was added into each dish to make the final concentration of AKBA 20 μM. The same volume of DMSO was added as control. After 12 h, cells were detached by 0.25% Trypsin/EDTA and collected by centrifugation. Then, 1 mL of extraction solvent (80% methanol/water) cooled to −80 °C was added to cell pellets. All metabolite extracts were centrifuged at 20,000 g at 4 °C for 10 min. Finally the solvent was evaporated in a SpeedVac Concentrator (Thermo, SPD1010–230). The cell extract was dissolved in methanol:acetonitrile:water (50:30:20, v/v/v, LC-MS grade, Fisher) solution. Samples were centrifuged at 20,000 g for 10 min at 4 °C and the supernatants were transferred to Liquid Chromatography (LC) vials. The injection volume was 1 μL. The metabolites of siRNA transfected HaCaT cells were extracted as described above.
Epidermis of human and mouse skin was separated from dermis as previously described [32]. Metabolites of human or mouse epidermis were extracted by adding 600 μL methanol:water (4:1, v/v) and 20 μL 2-Chloro-L-phenylalanine as internal standard substance and homogenizing. The mixtures were subjected to ultrasound exposure for 10 min in ice water bath and then placed at −20 °C for 30 min. After centrifugation at 13000 rpm for 15 min at 4 °C, 500 μL of supernatant was transferred to a new tube and evaporated in a Speed Vacuum. The tissue extract was dissolved in 200 μL 70% methanol/water. Samples were centrifuged at 13,000 rpm for 15 min at 4 °C and 150 μL of supernatants were transferred to LC vials, before 3 μL of the supernatant were injected into the UHPLC-LTQ Orbitrap instrument.
2.13. Chromatographic condition
The samples from HaCaT cells were run in a randomized order to prevent any run-dependent biases. Agilent Eclipse Plus-C18 RRHD (150 mm × 3.0 mm, 1.8 μm) was used for chromatographic separation and injection volume was 1 μL. Mobile phase A was aqueous formic acid (containing 0.1% (v/v) formic acid) and B was acetonitrile. The column was maintained at 30 °C and separation was achieved using the following gradient:0–1.0 min, 98% B; 1.0–5.0 min, 98%–60% B; 5.0–12.0 min, 60%–30% B; 12.0–15.0 min, 30%–5% B; 15.0–20.0 min, 5% B.
For tissue samples, chromatography was carried out with an ACQUITY BEH C18 chromatography column (2.1 mm × 100 mm, 1.7 μm, Waters). The column temperature was maintained at 45 °C, and the gradient mobile phase conditions were composed of phase A (water with 0.1% formic acid) and phase B (acetonitrile containing 0.1% formic acid). The gradient for the separation of human samples was as follows: 0–1.5 min, 5–25% B; 1.5–10 min, 25–100% B; 10–13 min, 100–100% B; 13–13.5 min, 100–5% B; 13.5–14.5 min holding at 5% B. The gradient for the separation of mouse samples was as follows: 0–0.5, 1–1% B; 0.5–1 min, 1–30% B; 1–2.5 min, 30–60% B; 2.5–6.5 min, 60–90% B; 6.5–7 min, 90–93% B; 7–7.5 min, 93–97% B; 7.5–8.5 min, 97–100% B; 8.5–10.7 min, 100–100% B; 10.7–10.8 min, 100–1% B; 10.8–13 min holding at 1% B. The injection volume was 3 μL and the flow rate of the LC system was 0.4 mL min−1.
2.14. Mass spectrometry
The normalized extracts from HaCaT cells were analyzed using an Agilent 1290 HPLC that was coupled to an Agilent 6545 Quadrupole Time-of-flight (QToF) mass spectrometer (MS) with a Dual Jet Stream electrospray ionization (ESI) source. The samples were analyzed in positive-ion (ESI+) mode. Spectral peaks in the mass range from 50 to 1100 m/z were acquired. The run was performed at a flow rate of 300 μL min−1 and infused into the ion spray source held at 3.5 kV. Nitrogen was used as the drying gas at 325 °C at a flow rate of 8 L min−1. The nebulizer pressure and fragmentor voltage were maintained at 45 psig and 110 V, respectively.
The mass spectrometric system for epidermis of human samples was operated using a LTQ Orbitrap Mass Spectrometer equipped with an electrospray interface (Thermo Fisher Scientific). Both the positive-ion (ESI+) and negative-ion (ESI−) mode were used so as to monitor as many ions as possible. The capillary and source temperature were set at 350 °C, with a desolvation gas flow of 45 L h−1. Centroid data were collected from 50 to 1000 m/z with a 30,000 resolution. The optimal capillary voltage was set at 3000 V, and the cone voltage at 40 V. The mass spectrometric data of epidermis of mouse samples were collected using a Waters VION IMS Q-TOF Mass Spectrometer equipped with an electrospray ionization (ESI) source. Both the ESI+ and ESI− mode were used so as to monitor as many ions as possible. Capillary voltage, sampling cone and extraction cone were set at 1.0 kV, 35.0 V and 3.0 V, respectively. Low and High Collision Energy were set at 6.0 eV and 20–45 eV, respectively. The source temperature and desolvation temperature were set at 120 °C and 450 °C, respectively, with a desolvation gas flow of 900 L h−1. Centroid data were collected from 50 to 1000 m/z with a scan time of 0.1 s and interscan delay of 0.02 s over a 13 min analysis time.
2.15. Metabolomics and data analysis
Spectral data were extracted and aligned based on mass and retention time using MassHunter Profinder software B.08.00 (Agilent). The parameters for the molecular feature extraction included: a peak height of ≥1500 ion counts and possible ions with [M + H]+. The alignment also involved isotope grouping restrictions including: a peak spacing tolerance of 0.0020 m/z and 10.0 ppm and a maximum charge state of 1. It was also required that there were at least two or more ions for a single molecular feature. In addition, for binning and alignment purposes, a tolerance of 0.3 min for a retention time window was set along with a mass window of 20 ppm. Some of the post processing filters included: an absolute height filter of ≥3000 ion counts and the requirement for the molecular feature to be present in at least two out of three replicates in one experimental group.
The resulting aligned features were analyzed in MetaboAnalyst 4.0 (www.metaboanalyst.ca) and SIMCA-P + 14.0 (Umetrics, Umea, Sweden) for statistically significant differentiations between sampling conditions. The resulting m/z's were searched in METLIN database to determine possible candidate metabolites based on accurate mass and adduct.
2.16. 13C-methionine metabolite tracing
Before labeling study, the HaCaT cells were pre-incubated with 20 μM AKBA for 24 h. For labeling study, AKBA group cells were washed with PBS once and then the media was changed to DMEM (Thermo, 21,013,024) supplemented with 10% dialyzed FBS (Thermo, 26,400,044), 4 mM l-glutamine (Thermo, 25,030,081), 201 μM L-cystine (Sigma, C7352), 201 μM 13C5-methionine (CIL, CLM-893-H, standard DMEM concentration) and 20 μM AKBA. DMSO group cells were treated the same way but with 20 μL DMSO. After 1 h, to harvest intracellular metabolites, media was aspirated from the cultures and 2 mL pre-cold 80% methanol was added to each dishes directly. The dishes were stored at −80 °C for 2 h before the cells were collected by cell scraper. All metabolite extracts were centrifuged at 21000 g at 4 °C for 15 min. The supernatant was transferred to new tubes and evaporated in a SpeedVac Concentrator (Thermo, SPD1010–230). The cell extract was dissolved in acetonitrile:water (2:98, v/v, LC-MS grade, Fisher) solution. LC-MS analysis was performed on an Agilent 1290 Infinity LC system equipped with a Hypersil Gold C18 column (100 × 2.1 mm, 1.9 μm) connected to an Agilent 6545 Q-TOF Mass Spectrometer. Mass isotopomer distributions were obtained by integration of ion chromatograms.
2.17. Western blotting
Cells and epidermis of skin were lysed in RIPA lysis buffer (Cell Signaling Technology) supplemented with protease and phosphatase inhibitor cocktails (Thermo Scientific). Primary antibodies of MAT2A (ab177484, Abcam), MAT2B (ab109484, Abcam), H3K4me3 (ab8580, Abcam), H3K9me3 (ab8898, Abcam), H3K27me3 (ab6002, Abcam), H3K36me3 (ab9050, Abcam), H3 (ab1791, Abcam), Ki-67 (ab16667, Abcam), tubulin (2146, Cell Signaling Technology), and actin (3700S, Cell Signaling Technology) were used. HRP-labeled Goat Anti-Mouse IgG (H + L) (A0216, Beyotime), and HRP-labeled Goat Anti-rabbit IgG (H + L) (A0208, Beyotime) were used.
2.18. ChIP-Seq
ChIP-seq analysis of H3K27me3 was performed using chromatin prepared from HaCaT cells. Briefly, 2 × 107 HaCaT cells treated with DMSO (control) or 30 μM AKBA for 12 h were crosslinked and harvested for immunoprecipitation using the ChIP-IT Express Kit (53,008, Active Motif) according to the manufacturer's protocol. The following antibodies were used: H3K27me3 (07–449, Millipore), and rabbit IgG (12–370; Millipore). Raw sequence reads were initially processed by FastQC. For quality control, adapter sequences and poor quality reads were removed by using Cutadapt. Quality filtered reads were mapped to hg19 reference genome using BWA (0.7.10), and only uniquely mapped reads were kept. MACS2 (2.1.1) was used to call peaks with the sonicated input as a control and an initial threshold p-value of 0.01 as cutoff. Visualization of read count data was performed by converting raw BAM files to bigWig files using IGV tools. ChIP-seq data have been submitted to the Gene Expression Omnibus database under accession number GSE112142.
2.19. Mice
BALB/c mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. Mice were kept under specific-pathogen-free (SPF) conditions. Age- and sex- matched mice at 7–8 weeks of age were randomly used in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals with the approval (SYXK-2003-0026) of the Scientific Investigation Board of Shanghai Jiao Tong University School of Medicine, Shanghai, China. To ameliorate any suffering of mice observed throughout these experimental studies, mice were euthanized by CO2 inhalation. Mice received a daily topical dose of 62.5 mg imiquimod (IMQ) cream (5%) (MedShine, Chengdu, China) on the shaved back for seven consecutive days to induce psoriasiform lesions. From the eighth to seventeenth day, mice were treated with 100 mg 1% AKBA gel on shaved back for ten consecutive days. Control mice received the same dose of blank vehicle gel. All mice were administered 62.5 mg IMQ on shaved back every other day to maintain skin lesions for 10 days. The hair of mice were re-shaved after reappearing on the shaved area during assay. On the eighteenth day, mice were sacrificed, and skin samples on back were collected.
2.20. Gel preparation
For the preparation of AKBA topical gel, Carbomer U20, propylene glycol and glycerol were added to distilled water to swell. Triethanolamine was added to adjust pH and viscosity of the gel. AKBA dissolved in ethanol was dispersed in gel while stirring. The final concentration of carbomer, propylene glycol, glycerol, and AKBA were 1%, 2%, 2% and 1% (g/g), respectively.
2.21. RNA-seq
Total RNA of epidermis was extracted using TRIzol® Reagent (Invitrogen). The quantity and purity of RNA samples were evaluated using a NanoDrop 2000 Spectrophotometer (NanoDrop Technologies) and the integrity was verified using Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA libraries were prepared using the TruSeq RNA sample prep Kit (Illumina) and cluster generation was performed using the Illumina HiSeq 4000 PE Cluster Kit (Illumina). After quantification by TBS380 Picogreen (Invitrogen), the libraries were sequenced (300 cycles, paired-end sequencing) in the Illumina Hiseq X Ten instrument using a HiSeq 4000 SBS Kit (Illumina). Basecalls were performed using CASAVA version 1.8. Adapter sequences were removed by using SeqPrep, and then poor quality reads were filtered by sickle (version 1.200) for quality control. Quality filtered reads were then mapped to GRCm38 using Hiast2 (v2.1.0). The expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM). RSEM was used to quantify gene and isoform abundances. The statistical analyses of differential expression between samples were done using EdgeR. GO functional enrichment and KEGG pathway analysis were carried out by Goatools and KOBAS. RNA-seq data have been submitted to the Gene Expression Omnibus database under accession number GSE112248.
2.22. Histological analysis and immunohistochemistry
The mouse back skin was fixed in formalin and embedded in paraffin. Sections (6 μm) were stained with hematoxylin and eosin (H&E). Epidermal hyperplasia (acanthosis) was assessed as previously reported [33]. For immunohistochemistry, CD3 (T cells), Ki-67 (proliferation), Gr-1 (neutrophils) and F4/80 (macrophages) expression was evaluated in skin sections using anti-CD3 mAb (ab16669, Abcam), anti-mouse Ki-67 mAb (12,202, Cell Signaling Technology), anti-mouse Gr-1 mAb (ab2557, Abcam), or anti-mouse F4/80 mAb (14–4801-85, eBioscience), respectively, following the manufacturer's instructions. For counting dermal infiltrating cells or Ki-67+ cells, three areas in three sections of each sample were randomly taken, in which the number of infiltrating cells or Ki-67+ cells was calculated.
2.23. RT[2] Profiler PCR array and qPCR
Total RNA was extracted from epidermis of skin biopsies or cultured cells using the TRIzol reagent (Invitrogen). After generation of cDNA using SuperScript First-Strand Synthesis System (Invitrogen), the RT[2] Profiler PCR array for mouse chemokines and receptors was performed according to the manufacturer's instructions (Qiagen). A total of 84 key genes encoding chemokines and their receptors were included in the array. Data were analyzed by a web-based analysis program (Qiagen, www.qiagen.com/genes-and-pathways/data- analysis). qPCR was carried out with the FastStart Universal SYBR Green Master (Roche) in a ViiA 7 Real-Time PCR System (Applied Biosystems). The relative expression of target genes was confirmed using quantity of target gene/quantity of β-actin.
2.24. Statistical analysis
The data were analyzed with GraphPad Prism 5 and are presented as the mean ± SEM. Student's t-test was used when two conditions were compared. Probability values of <0.05 were considered significant; two-sided Student's t-tests was performed. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. Error bars depict SEM.
3. Results
3.1. Identification of AKBA binding proteins
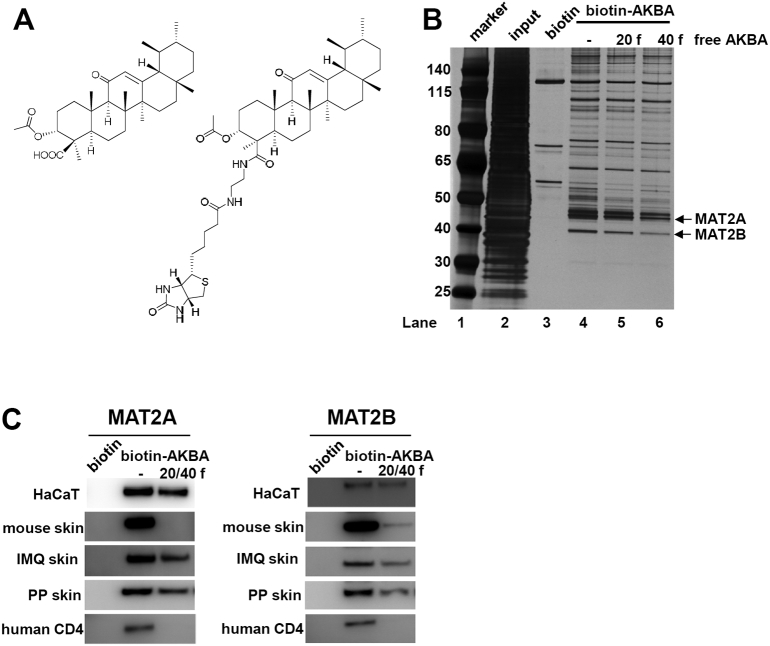
Boswellic acids have been used in traditional medicine to treat various inflammatory diseases for centuries [21]. Experimental data from animal models confirmed not only anti-inflammatory effects, but also pro-apoptotic and anti-proliferative properties of these small molecule compounds [34,35]. We previously reported that AKBA, the principal bioactive ingredient in boswellic acids, alleviates the skin inflammation in a CD18 hypomorphic mouse model of psoriasis [22]. Despite diverse pharmacological properties, the direct target(s) of AKBA was still undefined. To identify proteins interacting with AKBA, we synthesized biotinylated AKBA to facilitate affinity purification using streptavidin (SA)-agarose beads (Fig. 1A; Fig. S1). Biotinylation did not compromise the cellular activity of AKBA evaluated by CCK-8 tests (Fig. S2). We incubated HaCaT cells with Biotin-AKBA in the absence or presence of free AKBA, and subsequently SA beads were added into cell lysates. We found that the free AKBA compound competed away the binding sites on target proteins, and the yields of “pull-down” proteins were clearly reduced, suggesting that these proteins specifically interact with AKBA (Fig. 1B lane 4–6). Tandem mass spectrometry (MS) analysis identified MAT2A and MAT2B as two target proteins with the highest scores (Table S1). Identities of these proteins were further confirmed by immunoblotting in various cell types and tissues (Fig. 1C).
Fig. 1.
AKBA Binds to MAT2A and MAT2B. (A) Chemical structure of AKBA and biotinylated AKBA. (B) Biotinylated AKBA-binding proteins are purified from HaCaT cells by using streptavidin-agarose beads. Biotin is added into HaCaT cells as controls. Twenty and forty fold excess of free AKBA is also added to the cells to bind proteins competitively. (C) Species and cell type independent interaction of MAT2A and MAT2B with biotin-AKBA. Twenty or forty fold free AKBA is added to extracts during incubation to compete binding sites of protein.
3.2. AKBA binds to MAT2A directly at an allosteric site
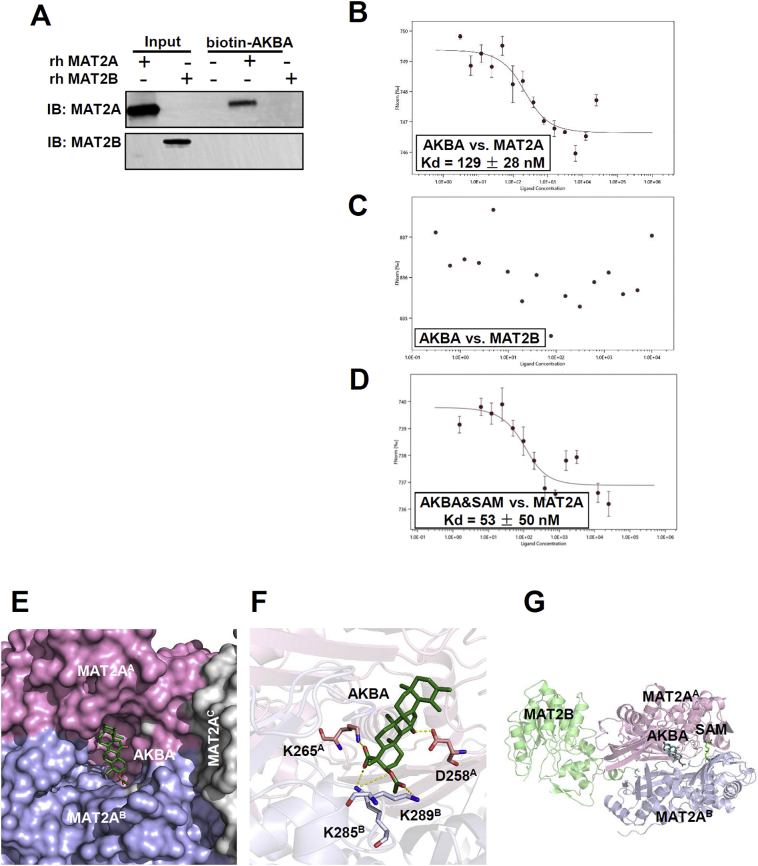
In principle, MAT2A and MAT2B form a complex to function in vivo [3,36]. To determine which protein directly binds to AKBA, we incubated biotin-AKBA with purified recombinant protein of MAT2A or MAT2B. We showed that AKBA interacts directly with MAT2A and indirectly with MAT2B through its association with MAT2A (Fig. 2A). The equilibrium dissociation constant (Kd) of MAT2A-AKBA interaction was measured to be 129 ± 28 nM by microscale thermophoresis (MST) assay (Fig. 2B). When testing Kd of MAT2B and AKBA, the data were random and cannot be fitted (Fig. 2C), confirming that AKBA did not interact with MAT2B directly, but indirectly through its interaction with MAT2A.
Fig. 2.
AKBA Binds at An Allosteric Site on MAT2A. (A) Purified recombinant human MAT2A and MAT2B are incubated with biotinylated AKBA and streptavidin beads respectively. The bound proteins are eluted and immunoblotted. (B) MST analysis of the binding affinity between AKBA and MAT2A. The fitted Kd value is shown. Error bars are SEM of three independent experiments. (C) MST analysis of the binding affinity between AKBA and MAT2B. (D) Binding affinity of AKBA and MAT2A while SAM is in the system. The fitted Kd value is shown. Error bars are SEM of three independent experiments. (E) Binding model of AKBA and MAT2A shows that AKBA bind at the interface of MAT2A dimer. One subunit of the dimer is shown as MAT2AA in purple and the other is shown as MAT2AB in cyan. MAT2AC shown in white is another subunit of the MAT2A tetramer. The A, B or C in superscript mean different subunit of MAT2A complex. (F) Four residues around the predicted binding site are mutated in MAT2A and assessed for AKBA binding. The corresponding Kd values are listed in Table S3. (G) The binding site of AKBA does not overlap with the active site (SAM) or the binding site of MAT2B.
To further determine whether AKBA was a competitive inhibitor of MAT2A, we added SAM to the binding assay system. Because SAM is synthesized from methionine and ATP in a reaction catalyzed by MAT2A, it occupies the locations of both the substrates [2]. The results of the MST assay showed that the binding affinity of AKBA to MAT2A was not impaired by SAM, suggesting an allosteric mechanism (Fig. 2D).
To identify the binding site of AKBA in MAT2A, we tried to solve the structure of MAT2A-AKBA complex. Unfortunately, we could not obtain the co-crystallization of the complex. The structure we got showed only MAT2A to 1.74-Å resolution (PDB 6FWB) (Table S2). In consistence with prior structures, MAT2A crystallized in a dimer-of-dimers tetramer configuration [3]. We next used the fpocket program to detect the potential binding sites on the MAT2A [37]. The predicted five top ranked binding sites were then selected for molecular docking studies. The binding site with the lowest binding energy for the MAT2A-AKBA interaction was at the interface between the dimer subunits (Fig. 2E). To further validate the binding model, four MAT2A residues were mutated (Fig. 2F). The mutagenesis studies and subsequent affinity measurement using biolayer interferometry showed that the replacement of Lys265 by Ala reduced its binding affinity towards AKBA by 1.66-fold for a loss of salt bridge. Replacement of Asp258 and Lys289 with Ala decreased the binding affinity by 3.71- and 4.05-fold, for disruption of one hydrogen bond respectively (Table S3). Besides, replacement of Lys285 with Ala increased the binding affinity by 1.56-fold (Table S3). The variation of the binding affinity with every mutation supported the binding model which showed that AKBA bound in the middle of the symmetric MAT2A homodimer interface, approximately 21 Å from the enzyme active site (location of SAM) (Fig. 2G). Together, these data indicated that AKBA bound to MAT2A directly at a novel allosteric site on the interface between the dimer subunits.
3.3. AKBA inhibits MAT2A function
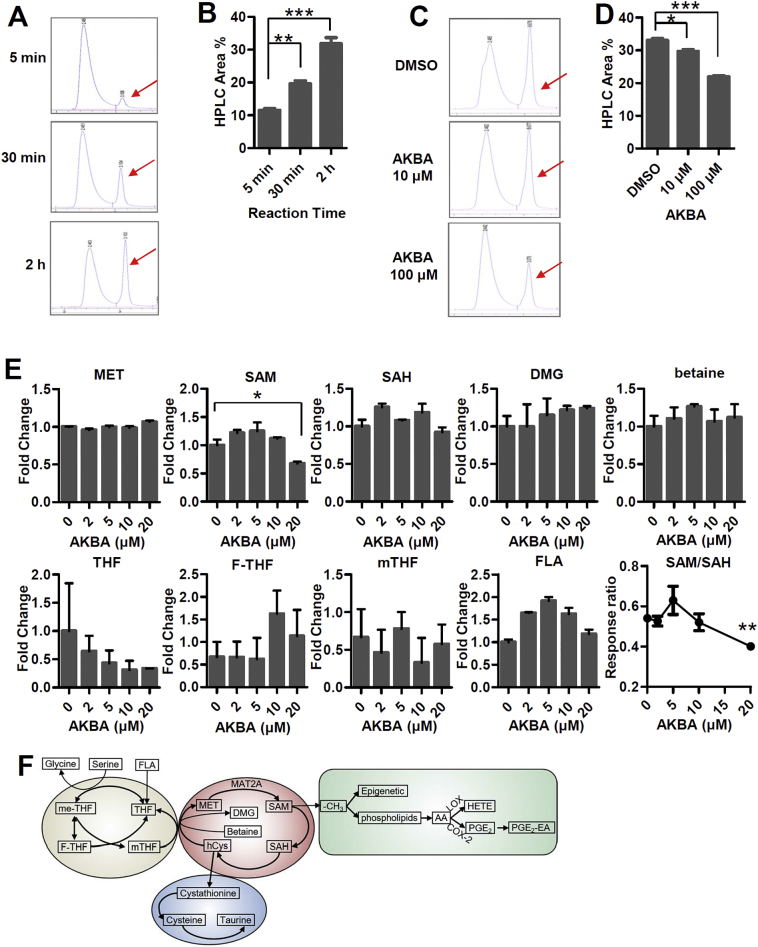
We next explored whether AKBA binding enhances or impairs the enzymatic activity of MAT2A. An HPLC-based protocol was used to directly detect SAM formation [30,31]. MAT2A was incubated with its substrates which were methionine and ATP, and SAM was formed and its concentration was increased over the reaction time (Fig. 3A and B). After 2 h, it almost reached maximum production. Thus we used 2 h as the reaction time in the following assays. When the concentration of AKBA in the system was increased, the formation of SAM was decreased significantly (Fig. 3C and D), indicating that AKBA inhibits MAT2A to synthesize SAM. We next investigated the capability of AKBA to modulate SAM synthesis in HaCaT cells. HaCaT cells were treated with 20 μM AKBA for 12 h, which was followed by methanol/water extraction and analyzed by LC-MS. The relative levels of metabolites in methionine cycle including one‑carbon metabolism, such as SAM, S-adenosylhomocysteine (SAH), dimethylglycine (DMG), betaine, methionine (MET), folic acid (FLA), tetrahydrofolate (THF), 10-formyltetrahydrofolate (F-THF), and 5-methyltetrahydrofolate (mTHF) were measured (Fig. 3E and F). SAM levels exhibited a significant decay at concentration of AKBA as 20 μM. While the levels of MET, SAH, DMG and betaine remained almost unchanged. The SAM/SAH ratio also decreased at AKBA concentration as 20 μM (Fig. 3E). The levels of metabolites of folate cycle, including THF, F-THF, mTHF and FLA, fluctuated somehow with the increasing concentration of AKBA, but without significant differences, indicating that the folate cycle was largely uncoupled to the methionine cycle in HaCaT cells (Fig. 3E). These data suggested that AKBA inhibited MAT2A function and consequently decreased the level of SAM and SAM/SAH ratio.
Fig. 3.
AKBA Represses Enzymatic Activity of MAT2A. (A) Representative HPLC result of kinetic SAM formation 5 min, 30 min and 2 h after reaction began. The red arrows show the peak of SAM. (B) The peak areas (%) of SAM with reaction time by HPLC. Data represent mean ± SEM from three independent experiments. Significant differences are indicated: **p < 0.01, ***p < 0.001, two-tailed Student's t-test. (C) Representative HPLC result of SAM formation 2 h after reaction began in presence of different concentrations of AKBA. The red arrows show the peak of SAM. (D) The peak areas (%) of SAM in presence of 0, 10 and 100 μM AKBA. Data represent mean ± SEM from three independent experiments. Significant differences are indicated: *p < 0.05, ***p < 0.001, two-tailed Student's t-test. (E) Relative concentration of one‑carbon cycle metabolites in HaCaT cells treated with different concentrations of AKBA. All error bars are computed from SEM (n = 3). (F) Metabolites of one‑carbon metabolism and downstream metabolites affected by methyl donation.
3.4. AKBA suppresses methionine metabolism
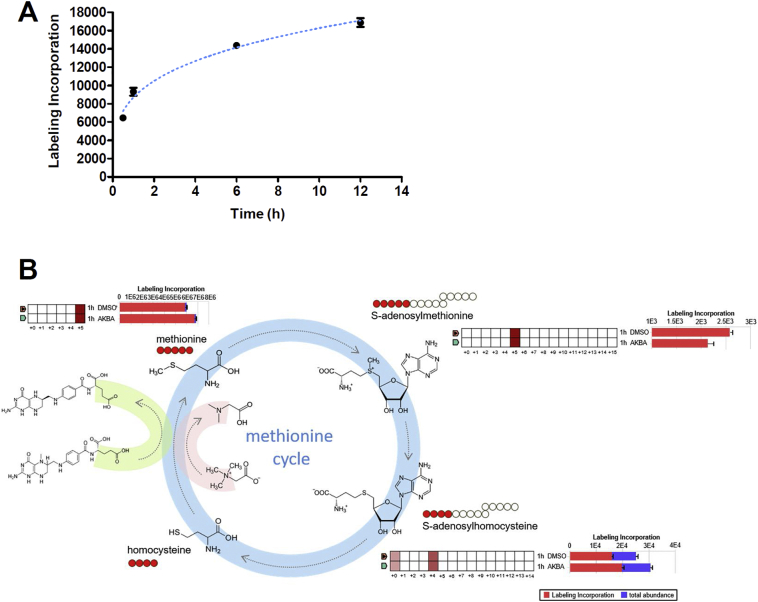
To assess whether AKBA could regulate methionine cycle, we performed stable isotope tracing analysis by adding 13C-labeled methionine to culture medium with dialyzed FBS and tracing the labeled 13C in metabolites using LC-MS. First of all, we assessed the incorporation of methionine tracer into SAM with time. To our surprise, this process was very quick. Only 0.5 h after adding 13C-labeled methionine to culture medium, substantial amount of label incorporation of M + 5 SAM was detected (Fig. 4A). After 12 h, the process of incorporation of methionine tracer almost reached the plateau. So we chose 1 h as the time point to evaluate the effect of AKBA on 13C-labeled methionine incorporation into SAM and other downstream metabolites of methionine cycle. Methionine and ATP being converted to SAM by MAT2A is the only reaction to form SAM in organism, so all the five carbons of methionine are transferred to SAM. 13C5-methionine tracing in HaCaT cells revealed that M + 5 enrichment of SAM was decreased, and M + 4 SAH was increased by AKBA at 1 h (Fig. 4B). Meanwhile, the consumption of M + 5 methionine was abolished by AKBA treatment (Fig. 4B), suggesting AKBA directly regulated the levels of metabolites of methionine cycle by interfering with the reaction of SAM formation.
Fig. 4.
AKBA Regulates Methionine Cycle. (A) Kinetic of 13C5-methionine labeling into SAM. (B) Tracing analysis using 13C-labeled methionine by LC-MS. The quilt plots and horizontal bar charts to the side of each structural formulas summarize isotopologue abundance and tracer incorporation. The two samples (1 h DMSO and 1 h AKBA) are displayed on the y-axis of the quilt plot, and the isotopologues are shown on the x-axis. The fill colour intensity indicates relative abundance of tracer incorporation. The bar chart shows the total tracer incorporation as a proportion of the total metabolite abundance (sum of all isotopologues detected). The schematic diagrams show the corresponding isotopomer transition from 13C-labeled methionine, and the red circles represent the number of detected 13C-labeled carbon.
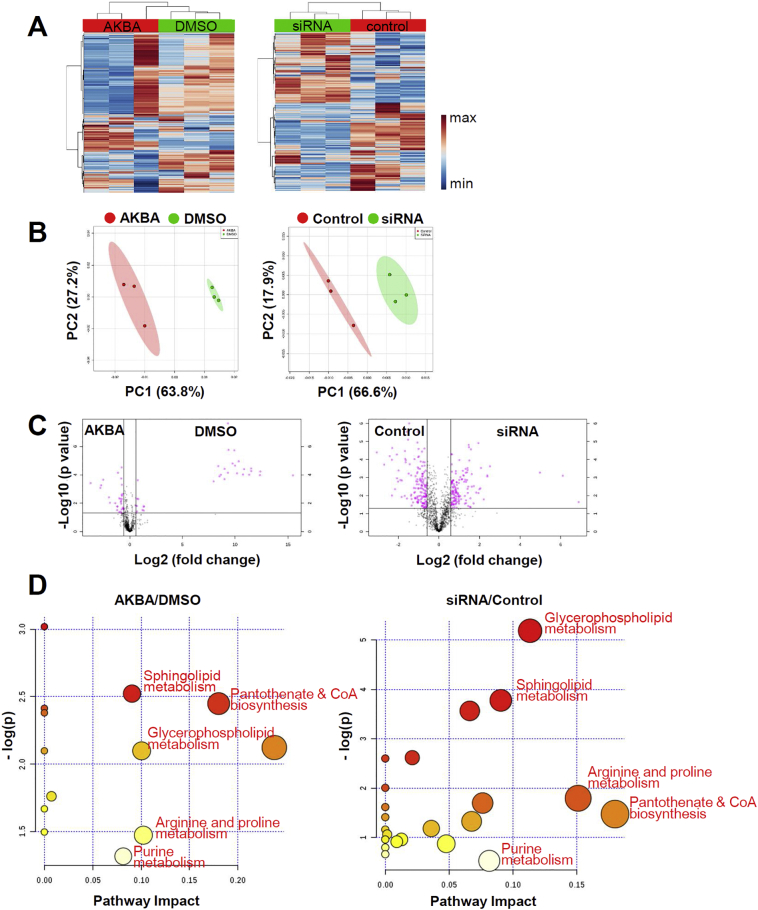
3.5. Comparative metabolomics of AKBA and siRNA of MAT2A treated HaCaT cells
Thus far we have shown that AKBA binds to MAT2A in an allosteric site which has not been reported before and that the binding impedes the formation of SAM. Deregulated SAM causes diverse biological alternations in metabolism, genome integrity and redox status [4,38]. We therefore hypothesized if AKBA primarily targeted MAT2A, the effects it conferred on reprogramming the metabolism of cells would resemble the effects of silencing MAT2A. We incubated HaCaT cells with AKBA at 20 μM or the same volume of DMSO as control for 12 h. The duration of RNA interference was 48 h using specific siRNA that targeted MAT2A or scramble siRNA as control. Metabolites of these four groups of cells were extracted by the same procedure and analyzed by a liquid chromatography, high-resolution mass spectrometry (LC-HRMS) metabolomics technology as one batch. We detected 1522 features in positive mode (Fig. 5A). Principal component analysis (PCA) was performed to evaluate the separation between the AKBA and DMSO treated cells, as well as the MAT2A siRNA and scramble siRNA transfected cells. The unsupervised multivariate analysis revealed significant differences between every two groups (Fig. 5B). Differential analysis comparing AKBA and DMSO treated HaCaT cells identified a set of 78 differentially expressed metabolites (Fig. 5C, left panel). When comparing MAT2A siRNA and scramble siRNA transfected HaCaT cells, there were 284 differentially expressed metabolites (Fig. 5C, right panel). Pathway analysis was performed using MetaboAnalyst. Notably, the top metabolic pathways with impact factor ≥ 0.08 of AKBA/DMSO group overlapped perfectly with those of siRNA/control group with only one exception. These five pathways of importance including sphingolipid metabolism, glycerophospholipid metabolism, pantothenate and CoA biosynthesis, arginine and proline metabolism and purine metabolism were listed in red in Fig. 5D. Together these findings demonstrated that the changes of metabolic pattern induced by AKBA and siRNA of MAT2A were similar in HaCaT cells.
Fig. 5.
Comparative Analysis of Metabolomic Alternations of AKBA and siRNA of MAT2A Treated HaCaT Cells. (A) Heatmap of all metabolites. The colour indicates metabolite expression value, blue: lowest, red: highest. (B) PCA score plots derived from LC-QTOFMS spectra concerning AKBA (red), DMSO (green) and siRNA (green), and control (red) samples in positive mode. (C) Volcano plots of differential metabolites between AKBA and DMSO, siRNA of MAT2A and scrambled control. Each circle represents a metabolite. Pink colour refers to significantly regulated metabolites (p value <.05 and fold change >1.5). (D) The pathway impact plot based on 78 (AKBA/DMSO) and 284 (siRNA/control) differential metabolites using MetaboAnalyst 4.0. Redder colors represent lower p-values, and larger circles represent higher impact factors. Low p-values and large pathway impact factors indicate that the pathway is greatly influenced.
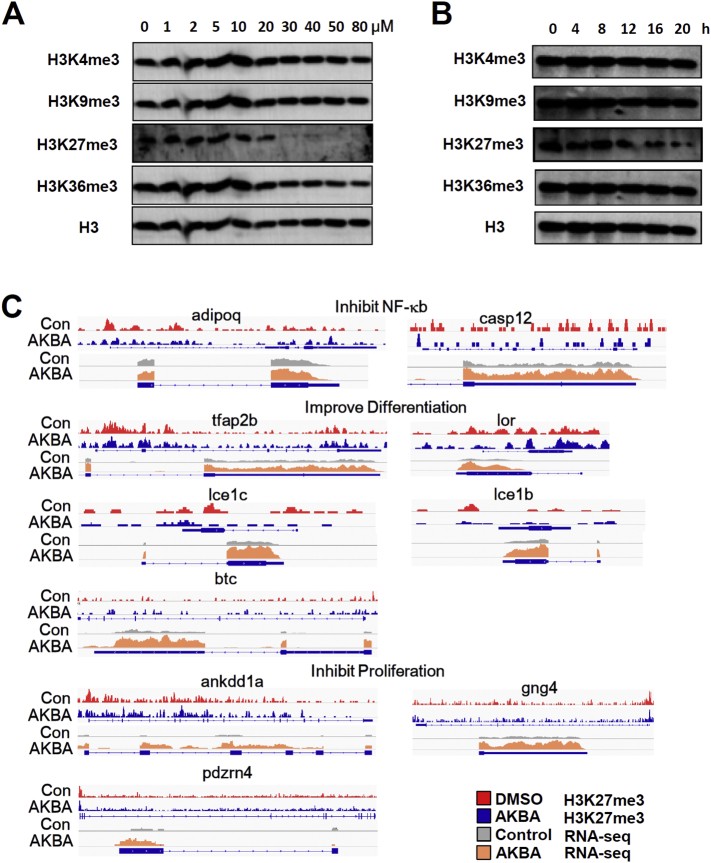
3.6. Altered methionine cycle affects histone methylation
One‑carbon metabolism, especially the concentration of SAM and SAM/SAH ratio regulate histone methylation, especially tri-methylation at lysine-4 on histone H3 (H3K4me3) and tri-methylation at lysine-27 on histone H3 (H3K27me3) [[39], [40], [41]]. So, we next investigated whether the decreased SAM and SAM/SAH ratio induced by AKBA might cause alternations in histone methylation. We profiled dose-dependent expression levels of histone methylation markers, tri-methylation at H3K4/9/27/36, in response to increased concentrations of AKBA in HaCaT cells. We found that H3K27me3 and H3K36me3 exhibited decreased methylation, with H3K27me3 exhibiting the greater change (Fig. 6A). H3K27me3 began to decrease at concentration of AKBA between 10 and 20 μM, consistent with the LC-MS data of SAM formation (Figs. 3E and 6A). We next monitored the dynamics of these markers. H3K27me3 began to decrease after 12 h of AKBA treatment (Fig. 6B). To further test the generality of this observation, we considered the response of H3K27me3 to AKBA treatment on mouse primary keratinocytes. Similar to HaCaT cells, AKBA also induced dose response and dynamic decrease of H3K27me3 (Fig. S3). It was shown that AKBA inhibited proliferation of HaCaT cells with IC50 of 27.3 μM (Fig. S2). We therefore elucidated whether changes of methylation metabolism were due to non-specific effects on proliferation. The previous CCK-8 assays were carried out at 24 h. When the duration was 12 h as in H3K27me3 assay, high concentration of AKBA (> 50 μM) only moderately inhibited proliferation of HaCaT cells (< 30% inhibition ratio) (Fig. S4A). Likewise, AKBA at the concentration of 20 μM only slightly inhibited HaCaT cell proliferation (~30% inhibition ratio) at different time points (Fig. S4B). Western blot and flow cytometry assays using Ki-67 antibody showed similar results (Fig. S4C and S4D). These data indicated that the decrease of H3K27me3 was not induced by the limited impact on cell proliferation by AKBA.
Fig. 6.
AKBA Alters Gene Expression by Decreasing H3K27me3. (A) Concentration dependent effects of AKBA on histone methylation. (B) Dynamics of histone methylation in presence of 20 μM AKBA. Representative of three independent experiments is shown in (A) and (B). (C) Changes in H3K27me3 ChIP-Seq signals with corresponding changes in gene expression from RNA-seq for psoriasis related genes.
H3K27me3 is a repressive marker for transcription. We next sought to determine the precise location of H3K27me3 in the genome using chromatin immunoprecipitation with sequencing (ChIP-Seq) and the consequence of gene expression by RNA-sequencing (RNA-Seq). We found that some psoriasis associated genes were enriched for loss of H3K27me3 at promoters, with resulting enhanced expression. Notably, adipoq and casp12 that were reported to inhibit NF-κB, tfap2b, lor, lce1c, lce1b and btc that were reported to improve differentiation, ankdd1a, gng4 and pdzrn4 that were reported to inhibit proliferation were responsive to the decrease in promoter H3K27me3 (Fig. 6C). Overall, these results indicated that AKBA-induced downregulation of SAM and SAM/SAH ratio might be predictive of tri-methylation level at H3K27, and consequently affected chromatin state and gene expression which was related to pathological process of psoriasis.
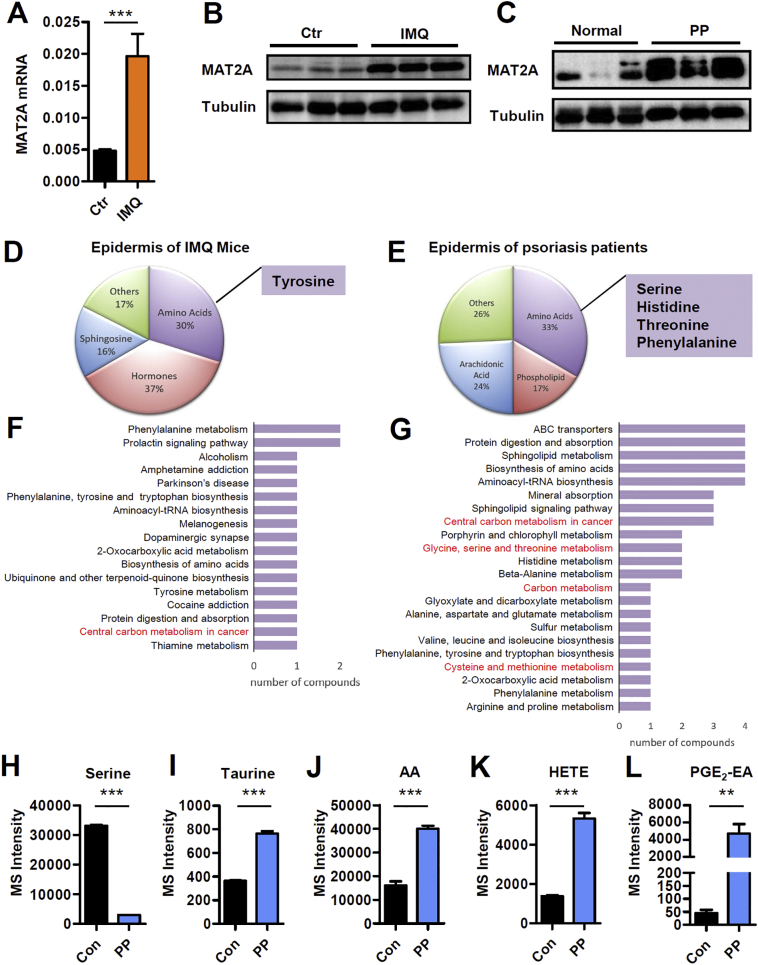
3.7. One-carbon metabolism of IMQ induced mouse model of psoriasis and psoriasis patients
We next questioned whether one‑carbon metabolism was altered in lesional skin of IMQ mice or psoriasis patients. MAT2A was the key enzyme of methionine cycle of one‑carbon metabolism and also the target protein of AKBA. We therefore sought to determine the expression level of MAT2A in inflamed skin of psoriasis. It was found that both the mRNA and protein level of MAT2A were increased in the epidermis of IMQ induced mouse model of psoriasis as well as psoriasis patients compared with their respective controls (Fig. 7A-C). We then used an UPLC-QTOF/MS based untargeted metabolomic technology to generate a profile of >140 differential metabolites of inflamed skin. It was found that principal-components analysis (PCA) of all samples showed a complete separation between metabolites from inflamed and control skin (Fig. S5), indicating IMQ could induce profound changes to metabolic profile of epidermis just like psoriasis. The KEGG pathway enrichment analysis of mouse metabolomics revealed that the 57 changed pathways of IMQ mice were mainly related to amino acid, hormones and sphingosine (Fig. 7D). And the 66 changed pathways of psoriasis patients mainly involved amino acids, arachidonic acid and phospholipids (Fig. 7E). As in both cases differentially regulated amino acids contributed greatly to metabolic shifting in the epidermis, we further assessed the amino acids related KEGG pathways. In the epidermis of IMQ mice, one of these pathways was central carbon metabolism which was related to one‑carbon units (Fig. 7F). In the epidermis of psoriasis patients, four pathways that were relative to one‑carbon units were shown: central carbon metabolism in cancer, glycine, serine and threonine metabolism, carbon metabolism and cysteine and methionine metabolism (Fig. 7G). Furthermore, we found serine, an essential one‑carbon donor and input of one‑carbon metabolism was significantly decreased in the epidermis of lesional skin of psoriasis patients (Fig. 7H), and taurine, an output of trans-sulphuration pathway of one‑carbon metabolism, was increased (Fig. 7I), indicating an activated one‑carbon metabolic pathway. Consistently, higher concentrations of taurine were reported by independent metabolomic studies of psoriasis patients compared to control skin or serum [14]. In addition, phospholipid metabolism was also aberrantly regulated in the epidermis of psoriasis (Fig. 7E). Interestingly, the generation of phospholipids depends on one‑carbon metabolism to provide the head group for lipids [4]. Membrane phospholipids are one of the two origins of arachidonic acid (AA). Besides, AA and its downstream metabolites including prostaglandins, thromboxane, hydroxyeicosatetraenoic acids (HETE) and leukotrienes are a family of potent mediators of inflammation. Particularly, they are indicated to play an important role in the pathogenesis of psoriasis [42]. So, the levels of AA and its derivatives could be potentially regarded as symbols of status of one‑carbon metabolism in psoriasis. We observed significant increase in AA, HETE and prostaglandin E2-ethanolamide (PGE2-EA) in the epidermis of psoriasis patients (Fig. 7J-L). Taken together, these findings demonstrated that one‑carbon metabolism was activated in the epidermis of psoriasis, making this pathway a potential target for psoriasis therapy.
Fig. 7.
MAT2A Expression and Metabolism Profile of Epidermis Derived from IMQ Mice and Psoriasis Patients. (A) qPCR analysis of MAT2A mRNA expression in epidermis of mice treated with blank vehicle control or imiquimod. Significant differences are indicated: two-tailed Student's t-test, n = 9–11 per group (mean ± SEM). (B) Western blotting analysis of MAT2A in epidermis of imiquimod induced mouse model of psoriasis. (C) Western blotting analysis of MAT2A in epidermis of psoriasis patients. (D) The dominant deregulated metabolites in epidermis of lesional skin from imiquimod induced mouse model of psoriasis compared with untreated mice (n = 6). (E) The dominant deregulated metabolites in epidermis of lesional skin from psoriasis patients compared with healthy controls (n = 6). (F) The most changed KEGG pathways involved with amino acids in epidermis of lesional skin from imiquimod mouse model. Those are related to one‑carbon metabolism are marked in red. (G) The most changed KEGG pathways involved with amino acids in epidermis of lesional skin from psoriasis patients. Those are related to one‑carbon metabolism are marked in red. (H-L) One‑carbon metabolism related metabolites in psoriasis patients compared with healthy controls. Significant differences are indicated: **p < 0.01, ***p < 0.001, two-tailed Student's t-test, n = 6 per group, (mean ± SEM).
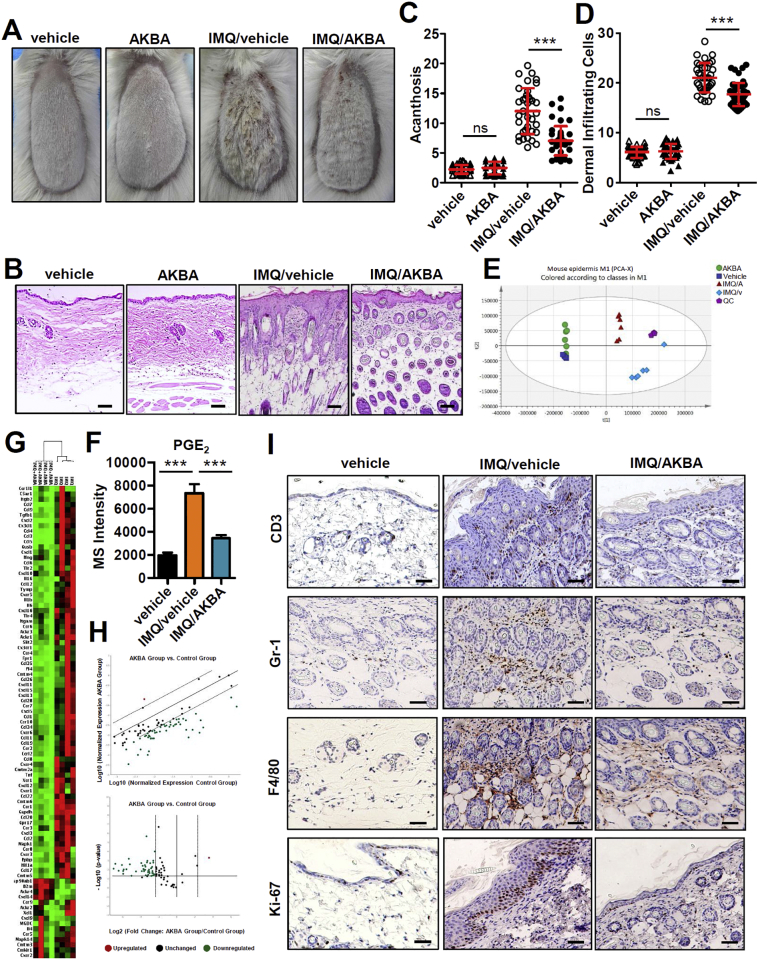
3.8. The therapeutic effect of AKBA in IMQ mice
Since MAT2A was increased in both mRNA and protein level in the epidermis of IMQ induced mouse model of psoriasis (Fig. 7A and B), we hypothesized that the MAT2A inhibitor, AKBA, harbored the therapeutic potential on this model. We developed a gel formulation for topical use of AKBA on the back skin of mice pretreated with IMQ. The AKBA gel substantially improved the psoriasis-like skin phenotype in the IMQ mice (Fig. 8A). The epidermal thickness (acanthosis) and dermal cellular infiltrates were reduced significantly in mice treated with AKBA gel compared with vehicle treated controls (Fig. 8B-D). Meanwhile, the topical use of AKBA did not changed the spleen weight or body weight of either control mice or IMQ mice compared with blank vehicle, indicating minimal toxicity during the assay (Fig. S6). PCA analysis of metabolites in epidermis showed that AKBA barely changed metabolomics in normal mice skin, but profoundly changed the metabolomic pattern of IMQ mice, indicating a regulatory effect of this compound (Fig. 8E). The metabolites of methionine cycle could not be detected by this method. But we found the level of PGE2 in the epidermis was enhanced by IMQ, and then decreased by treatment of AKBA (Fig. 8F). AKBA barely inhibit COX-2 [43], the enzyme converted AA to PGE2. So, the decreased level of PGE2 was likely due to inactivated synthesis of phospholipids caused by inadequate source of SAM. Additionally, we validated 84 genes that encode chemokines and their receptors using Mouse Chemokines & Receptors PCR arrays (RT [2] Profiler PCR Array Mouse Chemokines & Receptors). It was found that the expression of most of the 84 genes in epidermis were decreased significantly compared to blank vehicle treated control (Fig. 8G and H). Accordingly, immunohistochemistry staining revealed that AKBA gel remarkably decreased the infiltration of F4/80+ macrophages, CD3+ T cells and Gr-1+ neutrophils in inflamed skin lesions (Fig. 8I; Fig. S7A-C). Notably, the expression of Ki-67, a marker strictly associated with cell proliferation, was dramatically decreased in the AKBA treated mice, indicating that the excessive proliferation of basal keratinocytes induced by IMQ was reduced (Fig. 8I; Fig. S7D). Together, these findings indicated that topical use of AKBA was effective in ameliorating psoriasis-like inflammation induce by IMQ. In particular, AKBA reprogrammed the deregulated metabolic profile of IMQ induced mouse model of psoriasis. Considering MAT2A was the primary target of AKBA, it was likely that AKBA regulated levels of SAM and SAM/SAH ratio to affect various biological processes which needed methyl transfer, including epigenetic regulation, phospholipid generation and so on, whose consequences involved alternation in psoriasis-related genes and pro-inflammatory metabolites.
Fig. 8.
AKBA represses imiquimod induced skin inflammation in mice. (A) Representative phenotype of blank vehicle and AKBA treated control mice, and blank vehicle and AKBA treated IMQ-induced mice. Experiment is repeated independently for three time, n = 4–5 per group. (B) Representative H&E staining of every group. (C, D) Acanthosis (C) and dermal cellular infiltrates (D) of skin samples of every group.(E) Overview of PCA score plot obtained from vehicle treated normal mice (blue square), AKBA treated normal mice (green circle), vehicle treated IMQ mice (light blue square) and AKBA treated IMQ mice (red triangle). (F) PGE2 in epidermis of vehicle treated normal mice, vehicle treated IMQ mice, and AKBA treated IMQ mice. Significant differences are indicated: ***p < 0.001, two-tailed Student's t-test, n = 6 per group, (mean ± SEM). (G) Heat map of RT [2] profiler PCR array gene expression of chemokines in epidermis of mouse skin, n = 4. (H) The scatter plot (upper panel) and volcano plot (lower panel) of normalized expression of 84 chemokines and receptors genes of mice epidermis by RT [2] profiler PCR array gene expression. Blank vehicle treated IMQ mice are control group. IMQ mice treated with AKBA are experimental group. Fold regulation threshold is set to 5 and p-value cut off is 0.05. The central line indicates unchanged gene expression. The dotted line indicates the selected fold regulation threshold. Data points beyond the dotted lines meet the selected fold regulation threshold. The red, black, and green dots represent the upregulated, unchanged and downregulated genes, respectively. (I) Immunohistochemical staining analysis of CD3 (T cells), Gr-1 (neutrophils), F4/80 (macrophages) and Ki-67 (proliferation) in skin of vehicle treated normal mice and lesional skin of IMQ-induced mice treated with blank vehicle and AKBA. Scale bar: 50 μm.
4. Discussion
In human hepatocellular carcinoma MAT2A and MAT2B are dramatically upregulated, and silencing MAT2A results in cancer cell death [44]. Besides, there are increasing studies reporting that MAT2A confers a cell growth advantage in various human cancers and are important for differentiation and apoptosis [3,5]. These data suggest that MAT2A has great potential as a therapeutic target. So far, PF-9366 screened by Pfizer Inc. is the most potent and specific inhibitor of MAT2A [45]. Although PF-9366 efficiently inhibits cellular SAM production, it drivers an adaptive and compensatory transcriptional upregulation of MAT2A, which consequently fails to inhibit cell proliferation. Interestingly, we have found that AKBA does not provoke upregulation of MAT2A in response to its inhibition (Fig. S8). This could be due to the different binding sites of PF-9366 and AKBA on MAT2A. PF-9366 binds an allosteric site on MAT2A that overlaps with the binding site for MAT2B. So, PF-9366 binding on MAT2A prevented formation of MATαβ complex, which could possibly upregulate MAT2A in response to compensation mechanism. While the binding site of AKBA on MAT2A was away from the C-terminal tail of MAT2B (Fig. S9), AKBA did not impede the formation of MATαβ complex. The structure model was verified by the fact that the biotinylated AKBA could pull down the complex of MAT2A and MAT2B from cells. The binding model of AKBA may explain why it does not upregulate MAT2A in response to the inhibition.
Different from screening the entire chemical library to identify candidates directly against the drug target, we used the strategy of fishing target proteins by natural compound derived from traditional medicine. Traditional medicines have been used for thousands of years and proved to be effective with low toxicity. These natural compounds helped us find out not only target proteins with great therapeutic potentials but also novel druggable pockets. Like the one at which AKBA binds to MAT2A, it could be used as a target pocket to guide the optimization of the structure of small molecular compounds in the future.
One‑carbon metabolism status is able to influence histone methylation [39,41]. Methionine restriction decreased SAM production as well as H3K4me3 and altered gene expression in HCT116 cells [39]. RNAi of SAM synthetases provoked severe reduction in methylation of H3K9, K27, and K36 in C. elegans40. Threonine provided glycine and acetyl-coenzyme A needed for SAM synthesis [41]. Depletion of threonine from the culture medium of mouse embryonic stem cells (mESCs) decreased accumulation of SAM and decreased H3K4me3 [41]. Interestingly, in the same experiment, depletion of methionine did not influence any of the histone methylation markers including H3K4me3, H3K4me2, H3K4me1, H3K9me3, H3K27me3, H3K36me3, or H3K79me3 [41], indicating that the effect of one‑carbon metabolism on histone methylation was cell-dependent. The underlying mechanism needs to be further explored. The dysregulated one‑carbon metabolism and SAM metabolism were also reported to influence DNA methylation [46]. The effect of AKBA on DNA methylation will be subjected to our next study.
Unlike AKBA, one‑carbon metabolism inhibitors MTX and 5-fluorouracil inhibit folate cycle or the downstream of folate cycle to disrupt the synthesis of DNA [47,48]. These cytotoxic drugs are very commonly associated with adverse effects such as mucositis and ulceration. While, the acute oral, acute dermal and dose-dependent 90-day subchronic toxicity studies of 30% AKBA containing Boswellia serrata extract did not show any adverse effects in all organs tested, indicating the broad spectrum safety of AKBA [49].
In summary, through binding an allosteric pocket which does not overlaps the MAT2B binding site, inhibition of MAT2A by small molecular compound reprograms methionine cycle of one‑carbon metabolism without further triggering an adaptive upregulation of MAT2A. By this way, AKBA decreases the metabolic outputs with pro-inflammatory properties by regulating one‑carbon metabolism in IMQ induced mouse model of psoriasis. Our study identifies a direct target of AKBA and explains the mechanism of action in psoriasis, which result in new translational opportunities for drug development.
Funding sources
This work is supported by grants from the National Natural Science Foundation of China (No: 81502723, No: 31330026, No: 31570922, No: 81725018), the National Program on Key Basic Research Project (973 program, No: 2014CB541905), the Shanghai Municipal Commission (201740063), the Leading Academic Discipline Project of the Shanghai Municipal Education Commission (No: J50208 and No: J50207), the Open Research Fund of State Key Laboratory of Translational Medicine And Innovative Drug Development (No: 2015(329)). The funders did not play a role in manuscript design, data collection, data analysis, interpretation or writing of the manuscript.
Author contributions
J.B., Y.G. and HL.W. designed the research, analyzed the data and wrote the paper. J.B. conducted most of the experiments; F.L. performed human skin metabolomics assay; Z.X. performed animal study; Z.W. and A.Z. helped with protein crystallization; S.L. and J.Z. performed the molecular docking; L.C., Q.Y., H.Z., Z.W., Q.L., W.C., Y.S., L.N. and H.W. helped with the experimental details. HL.W. supervised the study.
Declaration of interests
The authors declare no competing financial interests.
Acknowledgements
We gratefully acknowledge members of the Structure Biology Platform and Metabolomics Platform of Core Facility of Basic Medical Science of SJTU for their support. We thank National Center for Protein Science Shanghai, Molecular Biology Platform of Shanghai Institute of Biochemistry and Cell Biology of CAS, LC-MS Platform of Instrumental Analysis Center of SJTU for their help.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.036.
Appendix A. Supplementary data
Supplementary material
References
- 1.Landgraf B.J., Booker S.J. Biochemistry: the ylide has landed. Nature. 2013;498(7452):45–47. doi: 10.1038/nature12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray B., Antonyuk S.V., Marina A. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc Natl Acad Sci U S A. 2016;113(8):2104–2109. doi: 10.1073/pnas.1510959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray B., Antonyuk S.V., Marina A. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ. 2014;1(Pt 4):240–249. doi: 10.1107/S2052252514012585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H., Xia M., Lin M. Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells. Gastroenterology. 2007;133(1):207–218. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 6.Chabner B.A., Roberts T.G., Jr. Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5(1):65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 7.Pinedo H.M., Peters G.F. Fluorouracil: biochemistry and pharmacology. J Clin Oncol. 1988;6(10):1653–1664. doi: 10.1200/JCO.1988.6.10.1653. [DOI] [PubMed] [Google Scholar]
- 8.Sviripa V.M., Zhang W., Balia A.G. 2′,6′-Dihalostyrylanilines, pyridines, and pyrimidines for the inhibition of the catalytic subunit of methionine S-adenosyltransferase-2. J Med Chem. 2014;57(14):6083–6091. doi: 10.1021/jm5004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Sviripa V., Chen X. Fluorinated N,N-dialkylaminostilbenes repress colon cancer by targeting methionine S-adenosyltransferase 2A. ACS Chem Biol. 2013;8(4):796–803. doi: 10.1021/cb3005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollock R.A., Abji F., Gladman D.D. Epigenetics of psoriatic disease: a systematic review and critical appraisal. J Autoimmun. 2017;78:29–38. doi: 10.1016/j.jaut.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Lundberg K.C., Fritz Y., Johnston A. Proteomics of skin proteins in psoriasis: from discovery and verification in a mouse model to confirmation in humans. Mol. Cell. Proteom. 2015;14(1):109–119. doi: 10.1074/mcp.M114.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong A.W., Wu J., Johnson M.A. Metabolomics in psoriatic disease: pilot study reveals metabolite differences in psoriasis and psoriatic arthritis. F1000Research. 2014;3:248. doi: 10.12688/f1000research.4709.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S., Hinchliffe T.E., Wu T. Biomarkers of an Autoimmune Skin Disease--Psoriasis. Genomics Proteomics Bioinformatics. 2015;13(4):224–233. doi: 10.1016/j.gpb.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottas A., Fishman D., Okas T.L., Kingo K., Soomets U. The metabolic analysis of psoriasis identifies the associated metabolites while providing computational models for the monitoring of the disease. Arch. Dermatol. Res. 2017 Sep;309(7):519–528. doi: 10.1007/s00403-017-1760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sitter B., Johnsson M.K., Halgunset J., Bathen T.F. Metabolic changes in psoriatic skin under topical corticosteroid treatment. BMC Dermatol. 2013;13:8. doi: 10.1186/1471-5945-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamleh M.A., Snowden S.G., Grapov D. LC-MS metabolomics of psoriasis patients reveals disease severity-dependent increases in circulating amino acids that are ameliorated by anti-TNFalpha treatment. J Proteome Res. 2015;14(1):557–566. doi: 10.1021/pr500782g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.H., Orenberg E.K., Faull K.F., Wade-Jardetzky N.G., Jardetzky O. 1H NMR spectroscopy: an approach to evaluation of diseased skin in vivo. J Invest Dermatol. 1989;92(2):210–216. doi: 10.1111/1523-1747.ep12276738. [DOI] [PubMed] [Google Scholar]
- 18.Du Z., Liu Z., Ning Z. Prospects of boswellic acids as potential pharmaceutics. Planta Med. 2015;81(4):259–271. doi: 10.1055/s-0034-1396313. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Tawab M., Werz O., Schubert-Zsilavecz M. Boswellia serrata: an overall assessment of in vitro, preclinical, pharmacokinetic and clinical data. Clin Pharmacokinet. 2011;50(6):349–369. doi: 10.2165/11586800-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Ammon H.P. Modulation of the immune system by Boswellia serrata extracts and boswellic acids. Phytomedicine. 2010;17(11):862–867. doi: 10.1016/j.phymed.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Roy N.K., Deka A., Bordoloi D. The potential role of boswellic acids in cancer prevention and treatment. Cancer Lett. 2016;377(1):74–86. doi: 10.1016/j.canlet.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Syrovets T., Kess D. Targeting NF-kappa B with a natural triterpenoid alleviates skin inflammation in a mouse model of psoriasis. J Immunol. 2009;183(7):4755–4763. doi: 10.4049/jimmunol.0900521. [DOI] [PubMed] [Google Scholar]
- 23.Safayhi H., Rall B., Sailer E.R., Ammon H.P. Inhibition by boswellic acids of human leukocyte elastase. J Pharmacol Exp Ther. 1997;281(1):460–463. [PubMed] [Google Scholar]
- 24.Wildfeuer A., Neu I.S., Safayhi H. Effects of boswellic acids extracted from a herbal medicine on the biosynthesis of leukotrienes and the course of experimental autoimmune encephalomyelitis. Arzneimittelforschung. 1998;48(6):668–674. [PubMed] [Google Scholar]
- 25.Henkel A., Tausch L., Pillong M. Boswellic acids target the human immune system-modulating antimicrobial peptide LL-37. Pharmacol Res. 2015;102:53–60. doi: 10.1016/j.phrs.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Lakka A., Mylonis I., Bonanou S., Simos G., Tsakalof A. Isolation of hypoxia-inducible factor 1 (HIF-1) inhibitors from frankincense using a molecularly imprinted polymer. Invest New Drugs. 2011;29(5):1081–1089. doi: 10.1007/s10637-010-9440-4. [DOI] [PubMed] [Google Scholar]
- 27.Kunnumakkara A.B., Nair A.S., Sung B., Pandey M.K., Aggarwal B.B. Boswellic acid blocks signal transducers and activators of transcription 3 signaling, proliferation, and survival of multiple myeloma via the protein tyrosine phosphatase SHP-1. Mol. Cancer Res. 2009;7(1):118–128. doi: 10.1158/1541-7786.MCR-08-0154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Khan M.A., Ali R., Parveen R., Najmi A.K., Ahmad S. Pharmacological evidences for cytotoxic and antitumor properties of Boswellic acids from Boswellia serrata. J Ethnopharmacol. 2016;191:315–323. doi: 10.1016/j.jep.2016.06.053. [DOI] [PubMed] [Google Scholar]
- 29.Legros H.L., Jr., Halim A.B., Geller A.M., Kotb M. Cloning, expression, and functional characterization of the beta regulatory subunit of human methionine adenosyltransferase (MAT II) J Biol Chem. 2000;275(4):2359–2366. doi: 10.1074/jbc.275.4.2359. [DOI] [PubMed] [Google Scholar]
- 30.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamarthapu V., Rao K.V., Srinivas P.N., Reddy G.B., Reddy V.D. Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli. Biochim Biophys Acta. 2008;1784(12):1949–1958. doi: 10.1016/j.bbapap.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Schlesier J., Siegrist J., Gerhardt S. Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis. BMC Struct Biol. 2013;13:22. doi: 10.1186/1472-6807-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ducker G.S., Rabinowitz J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mentch S.J., Mehrmohamadi M., Huang L. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22(5):861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towbin B.D., Gonzalez-Aguilera C., Sack R. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150(5):934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 36.Shyh-Chang N., Locasale J.W., Lyssiotis C.A. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikai K. Psoriasis and the arachidonic acid cascade. J Dermatol Sci. 1999;21(3):135–146. doi: 10.1016/s0923-1811(99)00042-0. [DOI] [PubMed] [Google Scholar]
- 38.Siemoneit U., Hofmann B., Kather N. Identification and functional analysis of cyclooxygenase-1 as a molecular target of boswellic acids. Biochem Pharmacol. 2008;75(2):503–513. doi: 10.1016/j.bcp.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q., Wu K., Zhu Y., He Y., Wu J., Liu Z. Silencing MAT2A gene by RNA interference inhibited cell growth and induced apoptosis in human hepatoma cells. Hepatol Res. 2007;37(5):376–388. doi: 10.1111/j.1872-034X.2007.00041.x. [DOI] [PubMed] [Google Scholar]
- 40.Quinlan C.L., Kaiser S.E., Bolanos B. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13(7):785–792. doi: 10.1038/nchembio.2384. [DOI] [PubMed] [Google Scholar]
- 41.Cuyas E., Fernandez-Arroyo S., Verdura S. Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene. 2018;37(7):963–970. doi: 10.1038/onc.2017.367. [DOI] [PubMed] [Google Scholar]
- 42.Rajagopalan P.T., Zhang Z., McCourt L., Dwyer M., Benkovic S.J., Hammes G.G. Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics. Proc Natl Acad Sci U S A. 2002;99(21):13481–13486. doi: 10.1073/pnas.172501499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 44.Lalithakumari K., Krishnaraju A.V., Sengupta K., Subbaraju G.V., Chatterjee A. Safety and Toxicological Evaluation of a Novel, standardized 3-O-Acetyl-11-keto-beta-Boswellic Acid (AKBA)-Enriched Boswellia serrata Extract (5-Loxin(R)) Toxicol Mech Methods. 2006;16(4):199–226. doi: 10.1080/15376520600620232. [DOI] [PubMed] [Google Scholar]
- 45.Yan S., Xu Z., Lou F. NF-kappaB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat Commun. 2015;6:7652. doi: 10.1038/ncomms8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris G.M., Huey R., Lindstrom W. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu S., Huang W., Wang Q. The structural basis of ATP as an allosteric modulator. PLoS Comput Biol. 2014;10(9) doi: 10.1371/journal.pcbi.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai J., Liu Z., Xu Z. Epigenetic downregulation of SFRP4 contributes to epidermal hyperplasia in psoriasis. J Immunol. 2015;194(9):4185–4198. doi: 10.4049/jimmunol.1403196. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H., Lou F., Yin Q. RIG-I antiviral signaling drives interleukin-23 production and psoriasis-like skin disease. EMBO Mol Med. 2017;9(5):589–604. doi: 10.15252/emmm.201607027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material