Abstract
Introduction
Therapeutic options for diffuse malignant peritoneal mesothelioma (DMPM) are limited to surgery and locoregional chemotherapy. Despite improvements in survival rates, patients eventually succumb to disease progression. We investigated splicing deregulation both as molecular prognostic factor and potential novel target in DMPM, while we tested modulators of SF3b complex for antitumor activity.
Methods
Tissue-microarrays of 64 DMPM specimens were subjected to immunohistochemical assessment of SF3B1 expression and correlation to clinical outcome. Two primary cell cultures were used for gene expression profiling and in vitro screening of SF3b modulators. Drug-induced splicing alterations affecting downstream cellular pathways were detected through RNA sequencing. Ultimately, we established bioluminescent orthotopic mouse models to test the efficacy of splicing modulation in vivo.
Results
Spliceosomal genes are differentially upregulated in DMPM cells compared to normal tissues and high expression of SF3B1 correlated with poor clinical outcome in univariate and multivariate analysis. SF3b modulators (Pladienolide-B, E7107, Meayamycin-B) showed potent cytotoxic activity in vitro with IC50 values in the low nanomolar range. Differential splicing analysis of Pladienolide-B-treated cells revealed abundant alterations of transcripts involved in cell cycle, apoptosis and other oncogenic pathways. This was validated by RT-PCR and functional assays. E7107 demonstrated remarkable in vivo antitumor efficacy, with significant improvement of survival rates compared to vehicle-treated controls.
Conclusions
SF3B1 emerged as a novel potential prognostic factor in DMPM. Splicing modulators markedly impair cancer cell viability, resulting also in potent antitumor activity in vivo. Our data designate splicing as a promising therapeutic target in DMPM.
Keywords: Mesothelioma, Therapy, SF3b modulation, Splicing, RNA-sequencing
1. Introduction
Malignant mesothelioma (MM) is a rare neoplasm of the lining of the pleural, peritoneal and pericardial cavity, as well as tunica vaginalis. Diffuse malignant peritoneal mesothelioma (DMPM) accounts for 10–30% of all cases of mesotheliomas, and its incidence is increasing worldwide [1]. This tumor is usually difficult to diagnose, both clinically and histologically, and is characterized by poor prognosis. Systemic chemotherapy is rarely effective [2], and cytoreductive surgery (CRS) combined with hyperthermic intraperitoneal chemotherapy (HIPEC) has become the new standard of care for selected patients. This treatment has improved survival (median OS of ~100 months), however, many patients still suffer of disease recurrence, especially when CRS fails to remove tumors from inaccessible abdominal loci [3]. Thus, novel therapeutic options to implement in the current surgical and HIPEC procedures are warranted.
Genomic studies were conducted on DMPM subsets by array-CGH, FISH and targeted DNA sequencing with the ultimate goal of finding new molecular targets. These studies reported that genetic alterations commonly found in pleural mesothelioma (BAP1, NF2, TP53 and CDKN2A mutations and/or copy number alterations) were also present in DMPM, although their frequencies vary according to the anatomical localization [[4], [5], [6]].
Recently, Bueno and collaborators carried out an extensive exome sequencing of a large collection of pleural mesothelioma specimens [7]. Besides the aforementioned MM-specific alterations, this study reported mutations affecting splicing factor 3b subunit 1 (SF3B1), which encodes an essential component of the spliceosome, as well as the histone methyltransferase SETD2 and the DEAD-box RNA helicases DDX51 and DDX3X, which are also involved in RNA processing and splicing. In addition, these analyses unraveled several MM-specific splice alterations, most of which were independent of splice site mutations. Collectively, these data highlight epigenetic and splicing deregulations as important features of MM pathogenesis and suggest their exploitation as novel antitumor strategy.
Alternative splicing (AS) is the essential process in eukaryotic gene expression by which non-coding intron sequences are removed from the pre-mRNA transcripts and specific exons are included or excluded from mature mRNAs. AS is controlled by a number of splicing factors (SFs) which recognize regulatory pre-mRNA sequences, assemble as a multi-protein complex called “spliceosome” and catalyze the reaction in a stepwise fashion. Multiple transcripts generated from a single gene can be translated into proteins, often executing distinct functions. Defects in splice factor genes, such as changes in expression levels or mutations, can produce aberrant mRNA splicing patterns on a genome-wide scale. This was demonstrated to be common in several cancers, mostly hematological malignancies [8].
Interestingly, these alterations have also prognostic significance [9] and contribute significantly to all hallmarks of tumorigenesis [10]. Therefore, splicing deregulation could represent a novel susceptibility to be explored for cancer therapy.
Pharmacological modulation of spliceosome activity via small molecules has shown potential in several pre-clinical studies [11]. FR901464, GEX1A and pladienolides are natural antitumor molecules targeting the SF3b complex. These compounds showed cytotoxic activity in the low nanomolar range in several tumor cell lines and reduction of tumor burden in vivo, which led to the synthesis of improved analogues such as meayamycins, spliceostatins and sudemycins [[12], [13], [14]].
The administration of E7107, a pladienolide derivative, in a Phase 1 trial stabilized the disease in 8 out of 40 cancer patients and reached partial response in one subject affected by metastatic pancreatic cancer [15]. Inhibition of splicing was observed during dose escalation, and the dose-limiting toxicity was primarily gastrointestinal related; however, visual impairment was reported in 2 patients [16]. A next generation splicing modulator, H3B-8800, is now in Phase 1 clinical trial (NCT02841540) [17].
Here we investigate for the first time the rationale of splicing as new target in DMPM by analyzing the expression of splice factors in patient tissues and correlating them with clinical outcome. Furthermore, we tested the antitumor effects of SF3b modulators in primary DMPM cell cultures and analyzed global changes in splicing patterns through RNA sequencing in vitro. Finally, we have established a unique orthotopic DMPM mouse model to monitor the efficacy of E7107 in vivo through innovative imaging techniques.
2. Materials and methods
2.1. Cell lines and culture conditions
Human epithelioid DMPM primary cultures (MesoII and STO) were derived from tumor samples of patients who underwent surgery [18] and maintained in DMEM/F12 in standard culturing conditions for <20 passages. Cells were routinely tested for mycoplasma at Microbiome (Amsterdam, the Netherlands) and resulted free of contamination. MesoII cells bear a SF3B1 heterozygous mutation (A1279S) which appears to be conserved and benign, according to several pathogenicity scores from the dbNSFP database (Supplementary Table 1). Co-transduction of cells with Fm and GC vectors and F-luc activity assessment were performed according to previously established methods [[19], [20], [21]].
2.2. Splicing modulators
Meayamycin-B (MAMB) was kindly provided by Prof. K. Koide (University of Pittsburgh). Pladienolide-B (PB) was purchased from Cayman Chemical (Ann Arbor, MI, USA). E7107 was provided by H3 Biomedicine (Cambridge, MA, USA).
2.3. RNAi and cell viability assessment
RNAi experiments were performed by using siRNA duplexes from the Dharmacon (Lafayette, CO, USA) SMARTpool siGENOME controls targeting UBB (M-013382-01) and NTp2 (D-001206-14) or two siGENOME siRNAs targeting SF3B1 (D-020061-07-0002 and D-020061-05-0002). Viability after transfection was measured with CellTiter-Blue reagent (Promega, Leiden, the Netherlands). Cell viability upon exposure to SF3b modulators was measured by sulforhodamine B assay [22].
Sequence D-020061-07-0002: CGAGUUUGCUUGGUCAGAA
Sequence D-020061-05-0002: AGGCGGACCAUGAUAAUUU
2.4. Cell cycle, apoptosis and proliferation assay
For the proliferation and apoptosis assay, cells were stained with 7-AAD (Via-Probe™, BD Bioscience, san Jose, CA, USA). To determine apoptosis, 7-AAD-stained cells were incubated with FITC-labeled AnnexinV antibody (Apoptest™, VPS Diagnostic, Hoeven, the Netherlands) in 5× diluted Annexin binding buffer (Invitrogen/ThermoFisher Scientific, Oregon, USA). Absolute cell count for proliferation assay was obtained by using Flow-Count™ Fluorospheres (Beckman Coulter, CA, USA).
For cell cycle analysis, ethanol-permeabilized cells were incubated with RNAse A (100 μg/ml, QIAgen, Venlo, the Netherlands) and stained with propidium iodide (Sigma-Aldrich). In all assays fluorescence was measured using BD LSRFortessa and data analysis performed with FACS Diva software version 8.0.1.1 (BD Bioscience).
2.5. Wound healing assay
Cells were seeded in 96-well plates 24 h before the assay (MesoII: 3*104 cells/well; STO: 6*104 cells/well), wound tracks were made with a 96-pin scratcher and the cells were subsequently cultured for 24 h in complete medium with 15 nM, 30 nM or no PB (10 wells per condition). Images were taken immediately after the scratch and after 24 h of drug exposure with a Leica DMI300B migration station (Leica Microsystems, Eindhoven, the Netherlands) and analyzed with the Scratch Assay 6.2 software (Digital Cell Imaging Labs, Keerbergen, Belgium).
2.6. Spheroid aggregation assay
Spheroids with a diameter of approximately 300 μm were created in 96-well flat bottom plates coated with 1.5% agarose. DMEM/F12 medium was replaced with drug-free medium or medium containing 30 nM PB (6 wells per condition) every two or three days for 11 days. Images of spheroids were taken with an automated phase-contrast microscope (Universal Grab software, Digital Cell Imaging Labs).
To detect the amount of light passing through the spheroids, pixel intensities of 8-bit black/white-converted images were calculated using ImageJ Software (U.S. National Institutes of Health, Bethesda, Maryland, USA) and expressed as Mean Gray value (= sum of all Gray values of the spheroid selection divided by the pixels of that selection). Inhibition of cell aggregation for each drug-treated spheroid at Day 9 (“treated” in the formula below) was calculated by normalizing for the Mean Gray values of the sum control spheroids (“∑ control”, where “n” is the number of replicates in the formula below) as follows:
2.7. Gene expression analysis of DMPM cells
Transcriptomic data from MesoII and STO cell lines and 3 normal mesothelium tissues were generated using Illumina HumanHT-12 v4 arrays. We are aware of the limitation of comparing normal mesothelium tissues with primary cells. However our primary cells were analyzed at their first passages, and retained the main features of the originator tumors. Raw data were log2-transformed and normalized using the lumi package [23], then filtered and annotated. Raw and normalized data were deposited at Gene Expression Omnibus (accession number: GSE112154).
Differential gene expression was estimated using the limma Bioconductor package [24] in R environment. A t-value pre-ranked gene set enrichment analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp) was carried out using GO Biological Process ontology (c5.bp.v6.1) and KEGG common pathway (c2.cp.kegg.v6.1) gene sets of the Molecular Signature database (MSigDB). The Normalized Enrichment Score (NES) was used to measure the gene-sets enrichment.
2.8. RNA sequencing
Total RNA from cells and freshly frozen orthotopic tumor tissues was extracted with RNeasy mini Kit (QIAgen). RNA integrity was assessed with RNA 6000 Nano Kit on a 2100 Bioanalyzer system (Agilent Technologies Inc., Santa Clara, CA, USA). Sequencing libraries were prepared with the Illumina TruSeq Stranded mRNA Library Prep LT Kit (RS-122-2201, Illumina Inc., San Diego, CA, USA) and Agencount AMPure XP beads (Beckman Coulter). cDNA library size and concentration was measured by Bioanalyzer. Single-end, 100 bp-reads were obtained from Illumina RAPID Chip on HiSeq 2500 System (Illumina). The sequencing reaction yielded 40.1 ± 7.3 (average ± SD) million passing-filter raw reads/sample. Bioinformatic pipeline for data analysis is described here [22]. rMATS version 3.2.5 was used to detect unique AS events (ES, A5SS, A3SS and RI, with the exception of mutually exclusive exons) with FDR < 0.01, which were imported to Excel. The events were additionally filtered based on the sum of inclusion and skipping counts by using the median value of total counts (>350) as the cut-off value. Events below that value were discarded as supported by insufficient counts. Filtered AS events are collected in Supplementary Table 2 and raw AS events are collected in Supplementary Table 5. Z-score and hierarchical clustering of inclusion levels (Ψ) of AS events were calculated and visualized by using Perseus software [25].
Gene expression estimates from orthotopic tumor samples and parental cell line samples were calculated with EdgeR and visualized by ggplot, both R software from Bioconductor repository. Differential gene expression data of DMPM cells after treatment with PB are reported in Supplementary Table 3.
gProfiler version r1741_e90_eg37 (https://biit.cs.ut.ee/gprofiler/index.cgi) [26] was used for pathway analysis of genes affected by AS and selecting GO, KEGG and REACTOME databases (Supplementary Table 2). AS events were visualized as sashimi plots with IGV genome browser, http://software.broadinstitute.org/software/igv/home). cDNA synthesis and RT-PCR protocols are described here [22]. Primer sequences:
DNAJB1_Fw_ex2 (5′-GAACCAAAATCACTTTCCCCAAGGAAGG-3′);
DNAJB1_Rev_ex3 (5′-AATGAGGTCCCCACGTTTCTCGGGTGT-3′);
RON_Ex10_Fw (5′-CCTGAATATGTGGTCCGAGACCCCCAG-3′);
RON_Ex12_Rev (5′-CTAGCTGCTTCCTCCGCCACCAGTA-3′);
MCL-1_Fw (5′-GCCAAGGACACAAAGCCAAT-3′);
MCL-1_S/R (5′-GCTCCTACTCCAGCAACACC-3′).
RNApolymerase I D_ex1_Fw (5′-TGGAAGAGGATCAGGAGCTGG-3′).
RNApolymerase I D_ex2_Rev (5′-CATGAGCTCATTCAGGCCTCTC-3′).
BclX_ex2F (5′-GGGCATTCAGTGACCTGACA-3′).
BclX_ex3R (5′-GGGAGGGTAGAGTGGATGGT-3′).
2.9. Orthotopic mouse model and live imaging
Nu/nu female mice were purchased from Harlan Laboratories (Madison, WI, USA). Animal experiments were performed in accordance with the European Community Council Directive 2010/63/EU for laboratory animal care and the Dutch Law on animal experimentation. The working protocol was validated and approved by the local committee on animal experimentation of the VU University Medical Center (DEC HEMA14–01). At day 0, tumors were induced by injection of 2*106 fm/GC cells in the peritoneal cavity of the mice.
Bioluminescence imaging (BLI) was performed with a Bruker In-Vivo Xtreme Capture System and data was analyzed using Molecular Imaging Software (Bruker Corporation, Billerica, Massachusetts, USA). Mice were selected for PET-MRI and PET-CT on the basis of the intensity of the BLI signal. G-luc levels from plasma samples were measured with a luminometer immediately after adding 25 mg/ml coelenterazine (Alfa Aesar, Haverhill, MA, USA) as substrate. [18F]-FDG was produced routinely by BV Cyclotron VU (Amsterdam, The Netherlands) with a radiochemical purity of >97% and administered via intraocular injection (5 MBq/mouse). PET-CT and PET-MRI were performed with nanoPET-CT and nanoPET-MRI (Mediso, Budapest, Hungary). For PET-CT, a computed tomography (CT) scan was performed for 5 min, followed by tracer administration at the start of a dynamic PET scan of 60 min. For PET-MRI (field strength = 1 T), T1- and T2-weighed images were acquired for one hour after tracer administration.
PET data were normalized, and corrected for scatter, randomization, attenuation, decay and dead time. List-mode PET data were re-binned in 19 successive frames (4 × 5 s, 4 × 10s, 2 × 30s, 3 × 60s, 2 × 300 s, 3 × 600 s and 1 × 900 s), which were reconstructed using an iterative 3D Poisson ordered-subsets expectation maximization algorithm with four iterations and six subsets. Resulting images had a matrix size of 170 × 170 × 157 voxels, each with a dimension of 0.6 × 0.6 × 0.6 mm3. Data were normalized and images analyzed as previously described [27].
2.10. Patient material and tissue microarray (TMA) construction and immunohistochemistry (IHC)
Tumor tissues of 64 DMPM patients treated with CRS and HIPEC in the National Cancer Institute of Milan (from August 1995 to October 2013) were selected for pathological examination. Normal mesothelium tissues were obtained from surgeries of patients affected by benign familiar adenomatous polyposis. TMAs were constructed as described in [28] by using a tissue-arraying instrument (Beecher Instruments, Silver Springs, MD, USA). Monoclonal SF3B1 antibody (Cat. D221–3) was purchased from MBL, Medical & Biological Laboratories CO. LTD. (Japan).
2.11. Statistical analysis and graphs
Clinical outcome was correlated with N-stage, histological subtype, performance status, cytoreduction and SF3B1 expression by means of a univariate analysis using a Mantel-Cox (Log rank, Chi-Square) test. Survival rates were calculated using the Kaplan-Meier method.
A multivariate model to assess the prognostic relevance of SF3B1 expression was obtained by selecting independent prognostic factors among all clinical and surgical parameters (selected as covariates) and using the backward stepwise elimination (Wald) method.
Statistical analysis for clinical data was performed with SPSS version 22 (IBM Corporation, NY, USA). Graphs and statistical analysis for in-vitro experiments were produced with GraphPad Prism version 6.
3. Results
3.1. Splicing factors are upregulated in DMPM and SF3B1 expression is an independent prognostic factor
In search for potential therapeutic targets, we performed gene expression profiling between two primary epithelioid DMPM cell cultures, namely MesoII and STO [29], and three normal mesothelium tissues, which resulted in a total of 4042 genes differentially expressed, of which 2217 up-regulated and 1825 down-regulated in the DMPM cells.
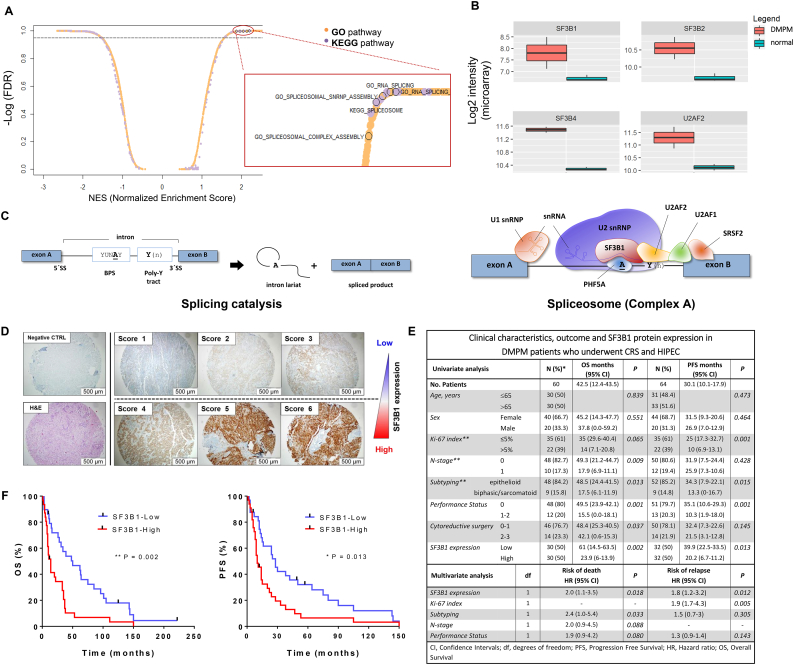
Pathways involved in RNA splicing and spliceosome complex assembly were overrepresented among genes up-regulated in DMPM cells, as revealed by gene set enrichment analysis (GSEA, Fig. 1A). In particular, MesoII and STO cells express higher mRNA levels of members of the SF3b family and U2AF2 compared to normal tissues (Fig. 1B). These genes encode for splicing factors which recognize pre-mRNA transcripts and assist the recruitment of the spliceosome machinery to regulatory sequences (Fig. 1C).
Fig. 1.
Core spliceosomal genes are upregulated in DMPM cells and SF3B1 is an independent predictor of the treatment outcome.
(A) Scatter plot showing normalized enrichment score (NES) and relative significance (FDR) of both GO and KEGG pathways in DMPM cell lines compared to normal specimens, as from GSEA. In the magnification, dots referred to splicing-related pathways are bordered in black. (B) Box plots reporting gene expression values of 4 selected splicing factors in DMPM cells and normal mesothelium samples.
(C) Schematic representation of cis-acting elements orchestrating the pre-mRNA splicing reaction and early spliceosome assembly (Complex A). A typical pre-mRNA is composed of exonic sequences (blue boxes) interspaced by intronic regions (thin black line) which are defined by 5′ and 3′ splice sites (5`SS and 3`SS) and two additional regulatory consensus sequences (BPS: branchpoint sequence; Poly-Y: polypyrimidine tract). The process of splicing results in the junction of adjacent exonic sequences by two subsequent trans-esterification reactions and the removal of the intron in the form of a lariat. The splicing reaction is catalyzed by splice factor proteins (SF and SRSF), small nuclear ribonucleoproteins (snRNP) and auxiliary factor proteins (AF), which interact by forming dynamic complexes with the pre-mRNA. For simplicity, only Complex A is shown here.
U1 snRNP binds to the 5`SS via base-pairing interactions between small nuclear RNA (snRNA) and pre-mRNA; auxiliary U2AF2 and U2AF1 factors bind to the Poly-Y tract and to the 3`SS respectively; U2 snRNP binds to the BPS and base-pairing interactions are stabilized by SF3b complex. SF3B1 and PHFA5 (SF3B14), in particular, make contact with the “invariant” adenine of the BPS (“A”), which initiates the first transesterification reaction [51] [52].
(D) Representative pictures of TMA cores (original magnification 20×) obtained from paraffin-embedded DMPM specimens. The figure illustrates the scoring system used to assess SF3B1 protein expression upon incubation with an anti-SF3B1 specific antibody, as described in the methods.
(E) Clinicopathologic characteristics and SF3B1 protein expression in relation to the treatment outcome (overall survival – OS; progression-free survival – PFS) in DMPM patients. The results of univariate and multivariate Cox regression analysis are shown.
* There were 56 deaths (event rate of 93.3%), while four patients were lost at follow-up after disease progression at last contact. The percentages for each clinicopathological characteristic are calculated according to these data.
** Data on Ki-67 index, subtyping and N-stage were not available for 7, 3 and 4 cases, respectively.
(F) Kaplan-Meier analysis of SF3B1 protein expression in relation to OS and PFS in DMPM patients. P-values were determined with the Log-rank (Mantel-Cox) test.
We then investigated whether the expression levels of SF3B1 protein correlate with the treatment outcome of DMPM patients. To this end, we determined SF3B1 expression by tissue microarrays (TMAs) performed on histological specimens of 64 DMPM patients (Fig. 1E and Supplementary S1A).
The expression levels of SF3B1 protein varied among specimens and patients were categorized according to their SF3B1 levels using a 6-grade scoring system described in Fig. 1D and Supplementary Fig. S1B. The univariate Cox regression analysis revealed a significant correlation between high SF3B1 level and shorter overall and progression-free survival (Fig. 1F). Importantly, multivariate analysis, including well-established predictors of the treatment outcome in DMPM, confirmed the independent prognostic value of SF3B1 (Fig. 1E).
Altogether, these data demonstrate that DMPM is characterized by elevated expression of splicing factors and that high level of SF3B1 protein is associated with inferior treatment outcome. Therefore, targeting SF3b complex in these patients could constitute an attractive therapeutic option.
3.2. Splicing modulators targeting SF3b complex induce DMPM cell death
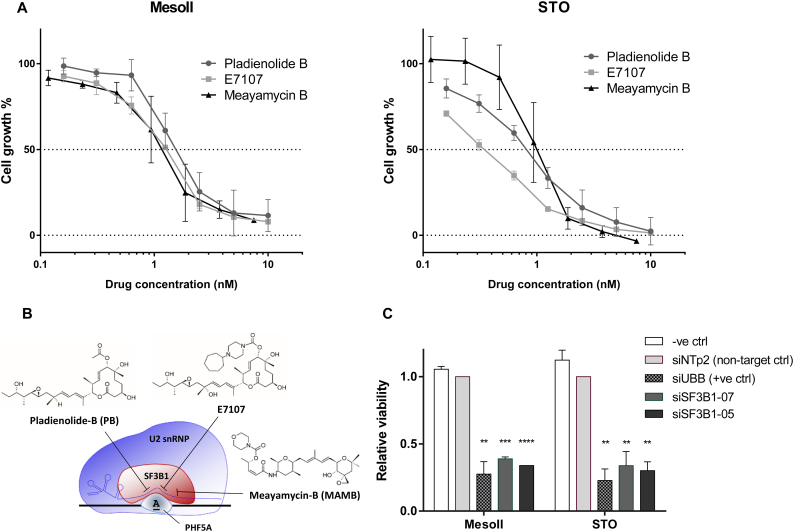
To address this hypothesis, we tested the effects of three modulators of SF3b complex activity (Pladienolide-B, E7107 and Meayamycin-B, depicted in Fig. 2B) on the viability of MesoII and STO cells. Remarkably, both primary cell cultures were extremely sensitive to all spliceosome modulators with IC50 values within low nanomolar ranges (Fig. 2A). Of note, siRNA-mediated knockdown of SF3B1 drastically decreased cell viability compared to non-targeting control, thus underlining the key role of SF3B1 in cell viability (Fig. 2C).
Fig. 2.
Splicing modulators induce DMPM cell death through SF3b-specific modulation.
(A) Dose-dependent inhibition of cell growth by Pladienolide-B (PB), E7107 and Meayamycin-B (MAMB) in two DMPM primary cell lines (MesoII and STO). Cell viability was determined using SRB assay upon 72 h treatment with increasing concentrations of splicing modulators. Data points represent mean percentage of cell growth relative to untreated cells ± SD of three independent experiments. IC50 values ± SD (drug concentration that inhibits 50% of cell growth) were determined by graphical interpolation of dose-response curves and reported as follows: 1.57 ± 0.29 nM and 0.82 ± 0.1 nM for PB, 1.27 ± 0.04 nM and 0.34 ± 0.08 nM for E7107, 1.19 ± 0.43 nM and 1.00 ± 0.3 nM for MAMB, in MesoII and STO cells respectively.
(B) Chemical structures of PB, E7107 and MAMB and schematic representation of their targets SF3B1 and PHF5A. The molecules destabilize U2 snRNP assembly at the interface between BPS, U2 snRNA and SF3b complex [13].
(C) Cell viability 5 days after transfection with two siRNAs targeting SF3B1 (siSF3B1–07 and siSF3B1–05) compared to non-targeting control siRNA (siNTp2, set to 1) and a positive control (siUBB). Data are derived from two independent experiments. Asterisks indicate statistical significance compared with siNTp2 (****P < 0.0001, ***P < 0.001, **P < 0.01 Student's t-test).
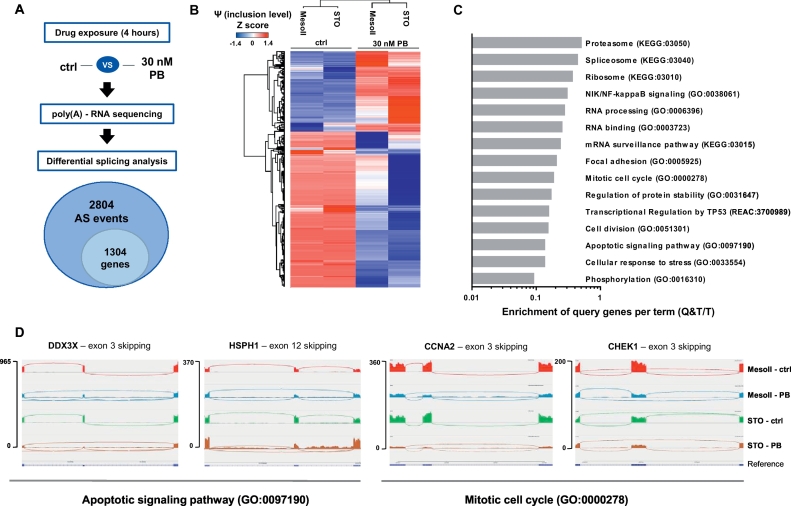
3.3. Pladienolide-B induces splicing alterations in relevant cellular pathways
To investigate the downstream effects of spliceosome modulation, we performed high-throughput sequencing of total mRNA of MesoII and STO cells treated with 30 nM Pladienolide-B (PB, Fig. 3A). A 4-h exposure time was sufficient to induce intron retention of the DNAJB1 transcript, known to be affected by PB [14](Supplementary Fig. S2). Differential splicing analysis between untreated and PB-treated samples detected 2804 significant AS events, predominantly skipped exons (ES) and retained introns (RI, Supplementary Fig. S3), which affected 1304 genes. The magnitude of each AS event is expressed as inclusion level (or Percentage Spliced In, Ψ, [30]), measured by comparing untreated and PB-treated samples and visualized as a heat-map in Fig. 3B.
Fig. 3.
Transcriptome-wide (global) splicing changes induced by PB in DMPM. MesoII and STO cells were treated with 30 nM PB for 4 h, followed by RNA sequencing-based analysis of global splicing.
(A) Schematic workflow, including differential splicing analysis of RNA sequencing data (using rMATS) and its output. Total number of significant AS events and the affected genes are shown. AS events were considered significant when FDR < 0.01 and total supporting read counts (sum of counts in all the analyzed samples supporting both inclusion and skipping) per splicing event were above 350 (median value).
(B) Hierarchical clustering based on inclusion levels (or Percentage Spliced In, Ψ) of significant splicing events calculated by rMATS and Z-score-normalized.
(C) Major biological processes affected by PB-induced AS events as determined by gProfiler [26]. Gray bars represent the enrichment score per each term (P < 0.05) expressed as Enrichment of Query genes per Term (Q&T/T).
(D) Sashimi plots depicting PB-induced splicing alterations in selected genes involved in cell cycle (CCNA2, CHEK1) and apoptosis regulation (HSPH1, DDX3X). IGV plots were set to same scale for all samples. For PB-treated samples, the number of junction reads connecting the leftmost directly to the rightmost exons are higher than the ones connecting to the central exon, indicating increase in ES.
Pathway analysis performed on AS events revealed several processes involved in regulation of gene expression (e.g. splicing, ribosome biogenesis/translation and protein degradation by proteasome), cell proliferation and survival, cell cycle control, response to stress and apoptosis (Fig. 3C-D).
Interestingly, among PB-induced AS events we found transcripts encoding kinases with tumorigenic functions in MM [31] such as AKT1, RAF1, MAP2K1 and ABL1 (Supplementary Table 4). Collectively, our results demonstrate that PB treatment perturbs the global splicing landscape of DMPM cells and affects genes essential for disease pathogenesis and survival of malignant cells.
3.4. Splicing modulation affects essential cellular functions in DMPM
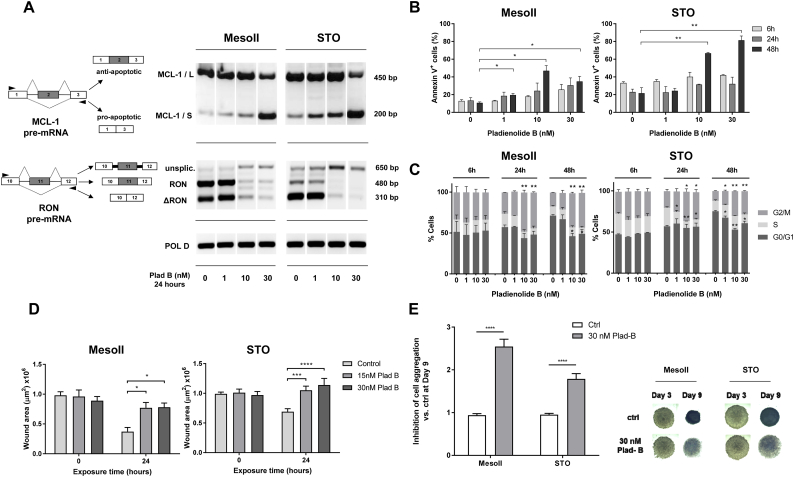
One of the best documented effects of spliceosome modulation is the skipping of exon 2 of the apoptotic gene MCL-1 which is translated into a truncated protein isoform exhibiting pro-apoptotic functions [32]. Accordingly, MCL-1/S (short) variant was induced in DMPM cells after 24-h PB exposure in both MesoII and STO cells, reaching its highest peak at 30 nM (Fig. 4A). Similarly, the transcript of the apoptotic gene BCL-X was converted into its pro-apoptotic splice variant (BCL-X/S) upon PB exposure, although this effect seems to be less prominent at high drug concentrations (Supplementary Fig. S4). This suggests some functional redundancy between apoptosis regulators.
Fig. 4.
PB induces apoptosis and cell cycle arrest and impairs DMPM cell migration and aggregation which is associated with alternative splicing of several genes involved in these processes.
(A) Splicing profiles of MCL-1 and RON for MesoII and STO cells assessed by RT-PCR after 24 h incubation with 1, 10 and 30 nM PB. Schemes of pre-mRNA structures with primer annealing sites (black triangles) and predicted PCR products are shown on the left.
(B) PB-induced apoptosis in MesoII and STO cells treated at 1, 10 and 30 nM for 24 and 48 h. Cells were stained with 7AAD and AnnexinV-FITC and analyzed by flow cytometry. Bars represent mean percentage of apoptotic (AnnexinV-positive) cells in each condition ± SEM of three independent experiments. Asterisks indicate statistical significance compared to untreated cells at 48 h (**P < 0.01, *P < 0.05, Student's t-test).
(C) Cell cycle distribution of cells stained with propidium iodide as measured by flow cytometry at several time points (6, 24 and 48 h) after treatment with 1, 10 and 30 nM PB. Bars represent mean percentages of cells in G0/G1, S and G2/M phases of at least two independent experiments (**P < 0.01, *P < 0.05, Student's t-test).
(D) Wound healing assay performed for MesoII and STO cells incubated with 15 and 30 nM PB for 24 h after creating wound tracks in 96 well plates. Bars represent mean wound area ± SEM of three independent experiments (at least 10 wells per condition in each experiment). Asterisks indicate statistical significance compared to untreated cells at 24 h (****P < .0001, ***P = 0.001, *P = 0.05, Student's t-test).
(E) Inhibition of cell aggregation measured in spheroids after 9 days incubation with 30 nM PB. Bars on the left represent normalized mean gray values of digital pictures ± SD (N = 6, ****P < 0.0001, Student's t-test). Representative images of spheroids at Day 3 (start treatment) and Day 9 for both MesoII and STO cells are shown on the right.
Concomitant with the altered splicing of these apoptotic regulators, the percentage of apoptotic cells (AnnexinV-positive) increased significantly upon 48-h PB treatment. Apoptosis induction in MesoII and STO cells was dose-dependent and reached 35% and 81% respectively (Fig. 4B). In addition, we observed a block in G1 and G2 phase and a concomitant reduction of cells in the S phase (Fig. 4C). These data confirmed that cell cycle progression is heavily affected by spliceosome modulation, as reported in previous studies [13] and provokes a drastic reduction of cell proliferation (Supplementary Fig. S5).
Migration processes are also influenced by alternative splicing [33]. In particular, we found that MesoII and STO cells express the truncated variant of the proto-oncogene RON (Fig. 4A). ∆RON plays a pivotal role in cancer cell motility due to its constantly activated kinase function [34] and might underlie the typical expansive growth pattern of DMPM [1]. Interestingly, a 24-h exposure to 10 and 30 nM PB caused intron retention in RON transcript and decrease in transcript abundance, which is probably due to nonsense-mediated decay (NMD) of the aberrantly spliced transcript (Supplementary Fig. S6 and Supplementary Table 5). To assess the functional effects of RON mis-splicing, we performed a 24-h wound-healing assay and we showed that incubation with PB resulted in a statistically significant inhibition of cell migration compared to untreated controls (Fig. 4D), consistent with the observed splicing modulation of RON. This effect was more pronounced in STO cells, which express higher levels of the ∆RON variant compared to its wild type transcript.
Ultimately, spliceosome modulation affects pathways controlling focal adhesions (Fig. 3D and Supplementary Fig. S7). To investigate the effects of the drug on cells organized in tridimensional structures, we imaged DMPM-derived spheroids exposed to 30 nM PB (Fig. 4E). Untreated spheres appear compact and dense, while treated ones appear more loose, especially at the outer layers, indicating progressive deterioration of cell-cell interactions. Inhibition of cell-cell aggregation was measured by detecting the amount of light passing through the spheroids (see Methods). PB-treated spheres show 2- to 3-fold higher inhibition levels compared to untreated controls (Fig. 4E).
Altogether, these data indicate that splicing modulation in vitro affects molecular processes involved in cell migration, cell-cell interaction and cell cycle progression which ultimately lead to apoptosis. Consequently, we evaluated the antitumor efficacy of SF3b-modulating agents in vivo.
3.5. Establishment of in vivo orthotopic DMPM mouse models
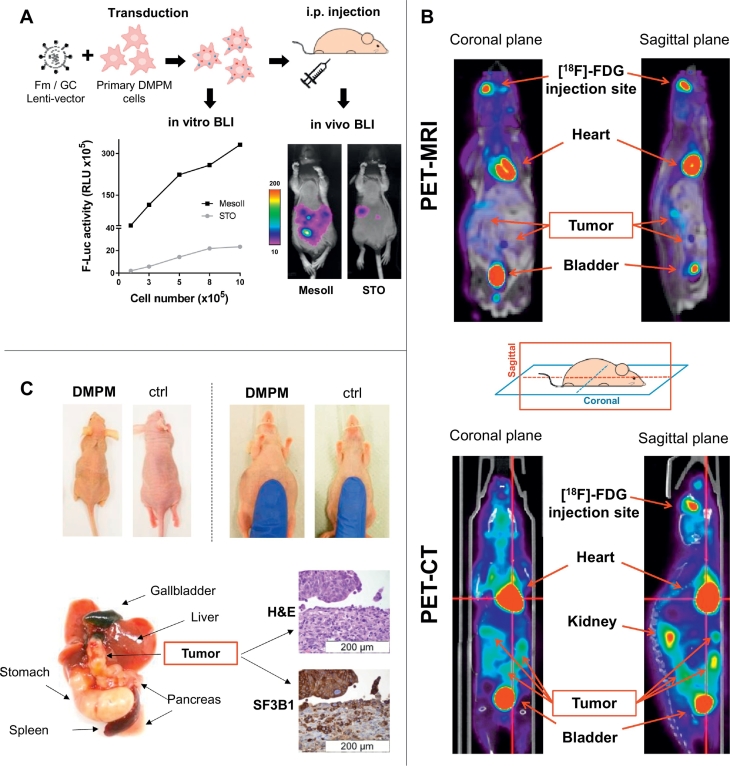
We first developed two novel bioluminescent (BLI) orthotopic mouse models in order to monitor the tumor growth over time. MesoII and STO cells were co-transduced with two lentiviral vectors expressing Firefly luciferase-mCherry (Fm) and Gaussia luciferase-CFP (GC), as previously described [[19], [20], [21]]. Intensity of the BLI signal in vitro was directly proportional to the number of cells (Fig. 5A) and they successfully engrafted in the peritoneal cavity of immune deficient mice (100% take rate). MesoII models showed superior transduction efficiency compared to STO and were used in the following experiments.
Fig. 5.
Characterization of bioluminescent orthotopic DMPM in vivo model.
(A) Primary DMPM cells were transduced with Fm/GC vectors and tested for in vitro F-Luc activity before implantation after two consecutive passages. The BLI signal was proportional to the number of cells. STO cells showed lower transduction efficiency compared to MesoII, which was also confirmed by in vivo F-Luc activity 7 days after cell implantation.
(B) PET-CT and PET-MRI of MesoII model after [18F]-FDG intraocular administration. Tumors were localized throughout the entire peritoneal and retro-peritoneal cavity (site of injection).
(C) Clinicopathological features of the in vivo DMPM model. Macroscopic and histopathological (hematoxylin/eosin and SF3B1 staining) examination of orthotopic tumor specimens. DMPM mice at Day 50 after cell implantation showed jaundice, cachexia and accumulation of ascitic fluids in the peritoneal cavity. Some animals presented high tumor load in proximity of the gastrointestinal tract which interferes with the normal digestive functions. SF3B1 staining showed high protein expression of the orthotopic tumor.
To determine the spatial localization of the tumor masses, we performed a rotational BLI analysis (MARS system - Supplementary Video 1). Moreover, BLI was complemented with PET-CT and MRI analyses (Supplementary Video 2 and 3, respectively), performed 10 days after cell implantation. DMPM-derived tumors were detectable in the whole abdominal cavity as multiple nodules, thus reproducing the diffusion pattern of the clinical disease (Fig. 5B).
After about 40 days, untreated mice manifested the typical symptoms of DMPM, such as jaundice and accumulation of ascitic fluid in the peritoneal cavity. Macroscopic analysis of animal tissues revealed nodules in close proximity of the gastrointestinal tract, as previously reported [6,18](Fig. 5C). These lesions interfered with digestive processes, thus causing gallbladder enlargement and cachexia. To investigate whether our mouse models recapitulate the histopathology and genomic profiles of human DMPM, we performed both IHC and RNA-sequencing analyses. DMPM xenografts were comparable to surgical material resected from the human originator tumors and maintained high SF3B1 protein expression (Fig. 5C).
3.6. Anti-tumor activity of E7107 in DMPM xenografts
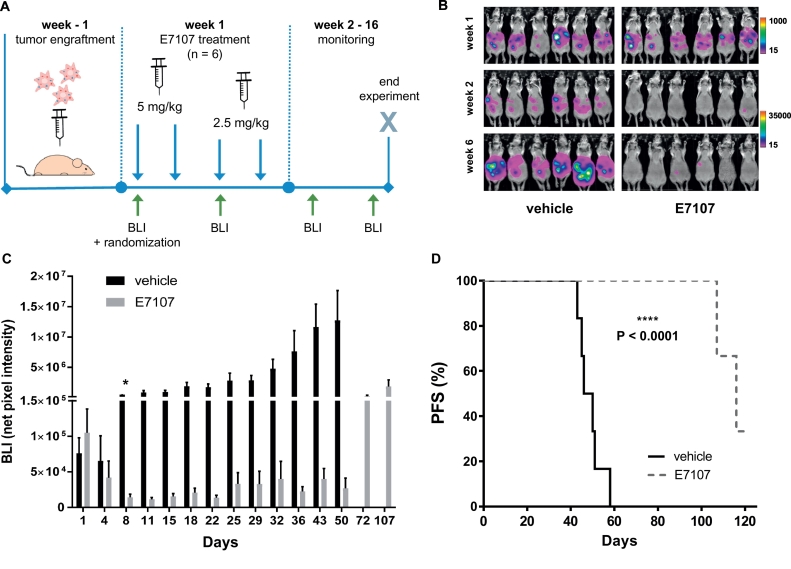
After stratification of the mice into groups with comparable BLI signal, E7107 was administered intraperitoneally according to treatment regime (Fig. 6A).
Fig. 6.
E7107 induced tumor regression and extended survival in an orthotopic DMPM mouse model.
(A) Experimental setup: bioluminescent DMPM cells were injected into the peritoneal cavity of immune deficient mice and randomized after one week (6 animals per arm). E7107 was administered four times (two doses of 5 mg/kg and two doses of 2.5 mg/kg) and tumor growth monitored twice a week until day 50, then every 3 weeks.
(B) Representative BLI pictures at week 1, 2 and 6. BLI intensity scales were adjusted in respect to signal.
(C) Quantification of BLI signal. Bars represent mean net pixel intensities ± SEM of each group. Statistical significance between the two groups was achieved at Day 8 (*P = 0.001) and persisted until termination of the experiment.
(D) Kaplan-Meier analysis progression-free survival (PFS) of E7107 and vehicle-treated groups. P-value (****P < 0.0001) was determined by the Log-rank (Mantel-Cox) test.
To avoid body weight loss potentially leading to discontinuation of the study, the initial dose was halved after two consecutive injections (Supplementary Fig. S8). BLI was monitored twice per week until day 50, then every 3 weeks. Starting already from Day 8, mean F-luc intensity was significantly decreased (P = 0.0011) in mice receiving E7107 as compared to untreated control mice (Fig. 6B and C). Importantly, the measurement of G-luc intensities in blood paralleled F-luc read out (i.e. G-luc intensity in the treatment group was 180-fold decreased compared to the control group at day 43, Supplementary Fig. S9).
In addition, RT-PCR of tumor samples from mice showed that E7107 treatment altered the splicing profiles of the same genes affected by PB and E7107 exposure in vitro (Supplementary Fig. S10 and S11).
Tumor growth inhibition was reflected in a significantly longer survival of treated mice. As illustrated by the Kaplan-Meier analysis in Fig. 6D, animals in the E7107-treated group showed an increase in survival rate compared to vehicle-treated group (P < 0.0001). Animals from the control group were sacrificed according to humane end-point (accumulation of ascitic fluid, cachexia, high tumor load and obstruction of gastrointestinal tract) after approximately 50 days. Remarkably, at the end of the experiment, three mice from the treatment group were clear of tumor cells, while two mice died from disease progression and one died from infection (no tumor masses were detected during post-mortem examination).
4. Discussion
In this study for the first time we demonstrate the high potential of targeting the spliceosome in DMPM. Initially, we found that primary DMPM cells show elevated expression of splice factor genes when compared with normal mesothelium tissues. These findings extend previous studies, showing up-regulation of SFs with pro-tumorigenic roles in several neoplasms including lung, colon and breast cancer [9]. However, different components of the splicing machinery may also act as a tumor suppressor and controversial data on their prognostic role have been reported in some tumor types. In particular, SF3B1 mutations are associated with a favorable prognosis in patients affected by myelodysplastic syndromes, in contrast to chronic lymphocytic leukemia where mutated SF3B1 dictated a poor prognosis and was associated with fludarabine resistance [10].
Interestingly, in our analysis performed on a large cohort of DMPM patients we found that the overexpression of SF3B1 protein is associated with significantly worse prognosis and is an independent risk factor of both death and disease progression.
SF3B1 is the most commonly mutated spliceosomal gene, both in hematological and solid cancers; mutations occur in 20% of myelodysplastic syndromes [35] and above 22% of uveal melanoma cases [36]. Darman and collaborators [37] showed that SF3B1 hotspot mutations induce aberrant 3` splice site selection. This in turn results in non-sense mediated decay for approximately 50% of the transcripts as well as production of aberrant proteins in cancer cells. Based on current data, we now speculate that not only mutations, but also the overexpression of SF3B1 perturbs splicing catalysis and results in the synthesis of pro-tumorigenic splice variants, which may constitute a vulnerability of cancer cells. Splice-switching oligonucleotides and RNA interference are suitable tools to target such variants in vitro [38], however it remains difficult to use these agents in the clinical setting due to limited stability in plasma and intracellular uptake [11]. Therefore we focused on small antitumor molecules targeting SF3b complex based on the assumption that their activity would be more clinically relevant in tumors where the expression of these factors is altered.
Two recent studies interrogating the molecular structure of U2 snRNP bound with PB and E7107 revealed that the conformation of the SF3B1 catalytic pocket, in association with PHF5A, is critical for BPS recognition and these compounds compete for the pre-mRNA substrate in a dose-dependent fashion [39,40].
Next to the potent in vitro antitumor activity of PB, E7107 and MAMB in two primary DMPM cultures, we also provide the first AS landscape of DMPM, as result of the perturbation by SF3b modulation. The PB-induced AS events affecting essential cellular pathways are considered as critical for MM pathogenesis [31] and are associated with in vitro anti-proliferative activity.
In particular, dysregulation of apoptosis has been demonstrated to play a crucial role in DMPM chemoresistance [29]. For instance, we found that DDX3X was one of the genes affected upon splicing modulation within the apoptotic pathway. This gene encodes a RNA helicase involved in pre-mRNA splicing and apoptotic signaling and is mutated in a subset of DMPM patients [4,7]. Interestingly, we identified a PB-induced transcript variant of this gene lacking an in frame exon (NM_001193417.2) which is most likely translated.
In addition, heat-shock protein (HSP) family members exert important anti-apoptotic roles [41] and seem to be up-regulated in MM [42]. Our analysis showed exon 3 skipping of the HSPH1 member, thus adding additional evidence to the pro-apoptotic effect of SF3b modulation.
Furthermore, previous literature [18] showed that targeting key apoptotic pathways and genes such as survivin might be an effective approach in DMPM therapy. We therefore encourage investigating AS modulation in combination with standard HIPEC procedures.
In agreement with previous data on the impact of PB on cell cycle regulation, we found PB-induced AS of CCNA2, which mediates cell cycle progression through G2/M phase. However, we report the skipping of exon 3 instead of exon 5, as found earlier by Effenberger et al. [12].
Another relevant gene emerging from this analysis was CHEK1, affected at the level of exon 3. This gene controls checkpoint-mediated cell cycle arrest and activation of DNA repair and is a potential new target in MM treatment to overcome resistance to chemotherapy or radiation [43].
Interestingly, epithelioid DMPM is characterized by epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial reverse transition (MErT) features such as migration and cell-cell adhesion [44]. Migration processes are controlled by transcription factors, tyrosine kinases and, as recently reviewed, also by alternatively spliced transcripts such as ∆RON [33,34,45,46]. Our data clearly show that SF3b modulation impairs the splicing pattern of this variant and this effect is reflected by reduction in cell migratory capacities in vitro.
Collectively, our in vitro assays show that the extent of SF3b modulation depends on the concentration of the compound and exposure times. It is therefore important to design preclinical studies to identify optimal therapeutic concentrations and clarify the role of pro-apoptotic splice variants which mediate cytotoxic effects. In addition, it is pivotal to characterize AS events in patient specimens compared to matched healthy tissues. This may result in the identification of novel DMPM-specific splice variants and the development of more targeted antitumor approaches. Furthermore, several studies suggest that specific subtypes of cancers could be particularly sensitive to splicing modulation, most prominently those characterized by high MYC expression. Hsu and collaborators discovered that MYC-driven tumors could be potentially vulnerable to spliceosome modulators because of the increased transcription rates and the consequent engulfment of the spliceosome machinery [47]. FISH analyses showed that mesotheliomas carry an amplification of the 8q24 chromosomal region, which includes the c-MYC locus (frequencies vary between 15% [48] and 48% [5], of which 64% occur in DMPM cases). Hence, future studies should determine whether these patients are hypersensitive to splicing modulation.
In line with the observation by Deraco et al. [28] that DMPM has low proliferating activity and that a Ki-67 index above 5% is associated with dismal OS and PFS, our data show a significant correlation between Ki-67 values above 5% and significantly shorted PFS. This suggests that SF3b modulators may be effective especially in highly aggressive cases due to their antiproliferative activity. Moreover, PB was effective in vitro in cisplatin-resistant SCLC cells, irrespective of their MYC status [49], and Lambert et al. [50] reported that cisplatin and PB synergize in eradicating breast cancer cells. Therefore, splicing modulation could constitute a promising option for chemotherapy-resistant cases of DMPM and it may synergize with current standard treatments. Ultimately, our data support the implementation of splicing modulators into the clinic, since we showed that in vivo administration of E7107 with four consecutive intraperitoneal injections leads to complete eradication of the tumor masses in 50% of tested animals throughout the study. In the light of this finding, we anticipate that locoregional administration of spliceosome modulators in solid tumors may be more effective than intravenous delivery. This might mitigate adverse events, especially those related to central nervous system, although this has not been investigated in the present study.
In conclusion, novel therapeutic approaches are urgently needed for DMPM, a rare disease characterized by poor survival rates and refractoriness to current therapeutic regimes. Our clinical data enrich the current understanding of DMPM pathobiology and assign new prognostic roles to SF3B1, which prompt validation studies and may be extended to other spliceosome components. Finally, our in vitro and in vivo efficacy data support the rationale that drugs modulating the spliceosomal activity and its downstream pathways may be applicable to future clinical practice in DMPM.
The following are the supplementary data related to this article.
Rotational BLI scan (MARS system) of an orthotopic DMPM model.
PET-CT scan of two orthotopic DMPM models.
MRI scan of an orthotopic DMPM model.
Single nucleotide variants detected in MesoII and STO cells by RNA-seq (ANNOVAR annotation).
Differential splicing analysis output (rMATS) in DMPM cells after 4h treatment with 30 nM Pladienolide-B.
Differential gene expression analysis output (edgeR) in DMPM cells after 4h treatment with 30 nM Pladienolide-B.
PB induces aberrant splicing of protein kinases involved in the pathogenesis of malignant mesothelioma.
rMATS raw output.
Supplementary figures.
Financial support
This work was supported by the Cancer Center Amsterdam (CCA) Foundation grants 2013 and 2015 (JC, EG), ‘the Law Offices of Peter G. Angelos Grant’ from the Mesothelioma Applied Research Foundation (MARF), United States (EG), Italian Association for Cancer Research, AIRC/Start-Up grant, Italy (EG), and Tuscany Region grant FAS-DIAMANTE, Italy (NF and EG).
Declaration of interests
Silvia Buonamici is employee of H3 Biomedicine.
Author contributions
EG and RS designed and coordinated the study. RS, AW and EG wrote the manuscript. RS, BE, MB and EAZvdL performed in vitro experiments. RS and TL performed in vivo experiments and imaging analyses within the animal facilities coordinated by CFM. NZ, PG, MD and SK provided patient specimens, clinical data and tissue microarrays (TMA). NF performed TMA staining and SF3B1 scoring. EG performed survival analysis and statistics. PG performed microarray experiments and data analysis. RS performed RNA-seq experiments and analysis of splicing data.
JC, GJLK, GJP, GJ and TW critically reviewed the manuscript.
Acknowledgments
Dr. Luca E. Pollina (AOUP, Pisa) for IHC analysis.
Stefano Percio and Leticia G. Leon for assistance in bioinformatic analyses.
Naomi Petersen for assistance with animal experiments and Marry Lin for assistance with in vitro experiments.
References
- 1.García-Fadrique A., Mehta A., Mohamed F., Dayal S., Cecil T., Moran B.J. Clinical presentation, diagnosis, classification and management of peritoneal mesothelioma: a review. J Gastrointest Oncol. 2017 Oct;8(5):915–924. doi: 10.21037/jgo.2017.08.01. http://jgo.amegroups.com/article/view/15305/13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander H.R., Li C.Y., Kennedy T.J. Current management and future opportunities for peritoneal metastases: peritoneal mesothelioma. Ann Surg Oncol. 2018;25(8):2159–2164. doi: 10.1245/s10434-018-6337-5. [DOI] [PubMed] [Google Scholar]
- 3.Gilani S.N.S., Mehta A., Garcia-Fadrique A., Rowaiye B., Jenei V., Dayal S. Outcomes of cytoreductive surgery with hyperthermic intraperitoneal chemotherapy for peritoneal mesothelioma and predictors of survival. Int. J. Hyperthermia. 2018;34(5):578–584. doi: 10.1080/02656736.2018.1434902. [DOI] [PubMed] [Google Scholar]
- 4.Joseph N.M., Chen Y.-Y., Nasr A., Yeh I., Talevich E., Onodera C. Genomic profiling of malignant peritoneal mesothelioma reveals recurrent alterations in epigenetic regulatory genes BAP1, SETD2, and DDX3X. Mod Pathol. 2017;30(2):246–254. doi: 10.1038/modpathol.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeda M., Kasai T., Enomoto Y., Takano M., Morita K., Nakai T. Comparison of genomic abnormality in malignant mesothelioma by the site of origin. J Clin Pathol. 2014;67(12):1038–1043. doi: 10.1136/jclinpath-2014-202465. [DOI] [PubMed] [Google Scholar]
- 6.Chirac P., Maillet D., Leprêtre F., Isaac S., Glehen O., Figeac M. Genomic copy number alterations in 33 malignant peritoneal mesothelioma analyzed by comparative genomic hybridization array. Hum Pathol. 2016;55:72–82. doi: 10.1016/j.humpath.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Bueno R., Stawiski E.W., Goldstein L.D., Durinck S., De Rienzo A., Modrusan Z. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 8.Sveen A., Kilpinen S., Ruusulehto A., Lothe R.A., Skotheim R.I. Aberrant RNA splicing in cancer; Expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35(19):2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 9.Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojtuszkiewicz A., Assaraf Y.G., Maas M.J., Kaspers G.J., Jansen G., Cloos J. Pre-mRNA splicing in cancer: the relevance in oncogenesis, treatment and drug resistance. Expert Opin Drug Metab Toxicol. 2015;11(5):673–689. doi: 10.1517/17425255.2015.993316. [DOI] [PubMed] [Google Scholar]
- 11.Salton M., Misteli T. Small Molecule Modulators of pre-mRNA Splicing in Cancer Therapy. Trends Mol Med. 2016;22(1):28–37. doi: 10.1016/j.molmed.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Effenberger K., Anderson D.D., Bray W.M., Prichard B.E., Ma N., Adams M.S. Coherence between cellular responses and in vitro splicing inhibition for the anti-tumor drug pladienolide b and its analogs. J Biol Chem. 2014;289(4):1938–1947. doi: 10.1074/jbc.M113.515536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnal S., Vigevani L., Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11(11):847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 14.Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3(9):570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 15.Eskens F.A.L.M., Ramos F.J., Burger H., O'Brien J.P., Piera A., De Jonge M.J.A. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res. 2013;19(22):6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 16.Hong D.S., Kurzrock R., Naing A., Wheler J.J., Falchook G.S., Schiffman J.S. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32(3):436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 17.Seiler M., Yoshimi A., Darman R., Chan B., Keaney G., Thomas M. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med. 2018;24(4):497–504. doi: 10.1038/nm.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Cesare M., Cominetti D., Doldi V., Lopergolo A., Deraco M., Gandellini P. Anti-tumor activity of selective inhibitors of XPO1/CRM1-mediated nuclear export in diffuse malignant peritoneal mesothelioma: the role of survivin. Oncotarget. 2015;6(15):13119–13132. doi: 10.18632/oncotarget.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wurdinger T., Badr C., Pike L., de Kleine R., Weissleder R., Breakefield X.O. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rijn S., Nilsson J., Noske D.P., Vandertop W.P., Tannous B.A., Würdinger T. Functional multiplex reporter assay using tagged Gaussia luciferase. Sci Rep. 2013;3(1):1046. doi: 10.1038/srep01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avan A., Caretti V., Funel N., Galvani E., Maftouh M., Honeywell R.J. Crizotinib inhibits metabolic inactivation of gemcitabine in c-Met-driven pancreatic carcinoma. Cancer Res. 2013;73(22):6745–6756. doi: 10.1158/0008-5472.CAN-13-0837. [DOI] [PubMed] [Google Scholar]
- 22.Sciarrillo R., Wojtuszkiewicz A., Kooi I.E., Gómez V.E., Boggi U., Jansen G. Using RNA-sequencing to detect novel splice variants related to drug resistance in in vitro cancer models. J Vis Exp. 2016;9(118):e54714. doi: 10.3791/54714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 24.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13(Suppl. 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H. G:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gent Y.Y.J., Weijers K., Molthoff C.F.M., Windhorst A.D., Huisman M.C., Smith D.E.C. Evaluation of the novel folate receptor ligand [18 F] fluoro-PEG-folate for macrophage targeting in a rat model of arthritis. Arthr Res Ther. 2013;15:R37. doi: 10.1186/ar4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deraco M., Cabras A., Baratti D., Kusamura S. Immunohistochemical Evaluation of Minichromosome Maintenance Protein 7 (MCM7), Topoisomerase II, and Ki-67 in Diffuse Malignant Peritoneal Mesothelioma patients using Tissue Microarray. Ann Surg Oncol. 2015;22(13):4344–4351. doi: 10.1245/s10434-015-4498-z. [DOI] [PubMed] [Google Scholar]
- 29.Zaffaroni N., Costa A., Pennati M., De Marco C., Affini E., Madeo M. Survivin is highly expressed and promotes cell survival in malignant peritoneal mesothelioma. Cell Oncol. 2007;29(6):453–466. doi: 10.1155/2007/456839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen S., Park J.W., Lu Z., Lin L., Henry M.D., Wu Y.N. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci. 2014;111(51):E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Assis L.V.M., Locatelli J., Isoldi M.C. The role of key genes and pathways involved in the tumorigenesis of Malignant Mesothelioma. Biochim Biophys Acta - Rev Cancer. 2014;1845(2):232–247. doi: 10.1016/j.bbcan.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y., Trivedi S., Ferris R.L., Koide K. Regulation of HPV16 E6 and MCL1 by SF3B1 inhibitor in head and neck cancer cells. Sci Rep. 2014;4:6098. doi: 10.1038/srep06098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chabot B., Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212(1):13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P.M. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20(6):881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D. Somatic SF3B1 Mutation in Myelodysplasia with Ring Sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32(2):204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D. Cancer-associated SF3B1 hotspot mutations induce cryptic 3’ splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 38.Jyotsana N.H.M. Exploiting differential RNA splicing patterns: a potential new group of therapeutic targets in cancer. Expert Opin Ther Target. 2018;22(2):107–121. doi: 10.1080/14728222.2018.1417390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finci L.I., Zhang X., Huang X., Zhou Q., Tsai J., Teng T. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 2018;32(3–4):309–320. doi: 10.1101/gad.311043.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cretu C., Agrawal A.A., Cook A., Will C.L., Fekkes P., Smith P.G. Structural basis of splicing modulation by antitumor macrolide compounds. Mol Cell. 2018;70(2):265–273. doi: 10.1016/j.molcel.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Takayama S., Reed J.C., Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;22:9041–9047. doi: 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]
- 42.Kaiser L.R., Albelda S.M., Singhal S., Wiewrodt R., Malden L.D., Amin K.M. Gene expression profiling of malignant mesothelioma. Clin Cancer Res. 2003;9(8):3080–3097. [PubMed] [Google Scholar]
- 43.Røe O.D., Anderssen E., Sandeck H., Christensen T., Larsson E., Lundgren S. Malignant pleural mesothelioma: Genome-wide expression patterns reflecting general resistance mechanisms and a proposal of novel targets. Lung Cancer. 2010;67(1):57–68. doi: 10.1016/j.lungcan.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 44.Bozzi F., Brich S., Dagrada G.P., Negri T., Conca E., Cortelazzi B. Epithelioid peritoneal mesothelioma: a hybrid phenotype within a mesenchymal-epithelial/epithelial-mesenchymal transition framework. Oncotarget. 2016;7(46):75503–75517. doi: 10.18632/oncotarget.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collesi C., Santoro M.M., Gaudino G., Comoglio P.M. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16(10):5518–5526. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biamonti G., Bonomi S., Gallo S., Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT) Cell Mol Life Sci. 2012;69(15):2515–2526. doi: 10.1007/s00018-012-0931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu T.Y.T., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riquelme E., Suraokar M.B., Rodriguez J., Mino B., Lin H.Y., Rice D.C. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9(7):998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suda K., Rozeboom L., Yu H., Ellison K., Rivard C.J., Mitsudomi T. Potential effect of spliceosome inhibition in small cell lung cancer irrespective of the MYC status. PLoS One. 2017;12(2):e0172209. doi: 10.1371/journal.pone.0172209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert C.A., Garbacki N., Colige A.C. Chemotherapy induces alternative transcription and splicing: Facts and hopes for cancer treatment. Int J Biochem Cell Biol. 2017;91:84–97. doi: 10.1016/j.biocel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimi A., Abdel-Wahab O. Molecular pathways: Understanding and targeting mutant spliceosomal proteins. Clin Cancer Res. 2017;23(2):336–341. doi: 10.1158/1078-0432.CCR-16-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agrawal A.A., Yu L., Smith P.G., Buonamici S. Targeting splicing abnormalities in cancer. Curr Opin Genet Dev. 2018;48:67–74. doi: 10.1016/j.gde.2017.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rotational BLI scan (MARS system) of an orthotopic DMPM model.
PET-CT scan of two orthotopic DMPM models.
MRI scan of an orthotopic DMPM model.
Single nucleotide variants detected in MesoII and STO cells by RNA-seq (ANNOVAR annotation).
Differential splicing analysis output (rMATS) in DMPM cells after 4h treatment with 30 nM Pladienolide-B.
Differential gene expression analysis output (edgeR) in DMPM cells after 4h treatment with 30 nM Pladienolide-B.
PB induces aberrant splicing of protein kinases involved in the pathogenesis of malignant mesothelioma.
rMATS raw output.
Supplementary figures.