Abstract
A recent meta-analysis revealed the contribution of the SIGLEC6 locus to the risk of developing systemic lupus erythematosus (SLE). However, no specific Siglec (sialic acid-binding immunoglobulin-like lectin) genes (Siglecs) have been implicated in the pathogenesis of SLE. Here, we performed in silico analysis of the function of three major protective alleles in the locus and found that these alleles were expression quantitative trait loci that enhanced expression of the adjacent SIGLEC12 gene. These data suggest that SIGLEC12 may protect against the development of SLE in Asian populations. Consistent with human genetic data, we identified two missense mutations in lupus-prone B6.NZM Sle1/Sle2/Sle3 (Sle1–3) mice in Siglece, which is the murine Siglec with the greatest homology to human SIGLEC12. Since the mutations resulted in reduced binding of Siglec E to splenic cells, we evaluated whether Siglece −/− mice had SLE phenotypes. We found that Siglece −/− mice showed increased autoantibody production, glomerular immune complex deposition and severe renal pathology reminiscent of human SLE nephropathy. Our data demonstrate that the Siglec genes confer resistance to SLE in mice and humans.
Keywords: Siglecs, CD24, HMGB1, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune inflammatory disease that predominately affects women of childbearing age. The prevalence of SLE ranges from 20 to 150 cases per 100 000 individuals, with a 10-year survival rate of ~70%.1 Production of autoantibodies against self-nucleic acids, such as double-stranded DNA (dsDNA), represents a serological hallmark of SLE.2 These autoantibodies contribute to the pathogenesis of SLE by forming immune complex deposits in different parts of the body, leading to inflammation and organ damage. Although the etiology of SLE remains elusive, genetic and environmental factors, as well as a failure to properly clear apoptotic cells leading to secondary necrosis and the release of nuclear autoantigens, challenges immunological tolerance, thereby exacerbating the risk of disease manifestation.3,4,5
Toll-like receptors (TLRs) recognize both pathogen-associated molecular patterns and danger-associated molecular patterns (DAMPs) and induce the production of inflammatory cytokines. TLRs play key roles in driving aberrant inflammation in response to DAMPs in SLE patients and SLE-prone mice.6,7,8 Sialic acid-binding immunoglobulin-like lectins (Siglecs) are sialic acid-recognizing cell surface receptors that are predominately expressed on immune and hematopoietic cells.9,10,11 They comprise a family of 14 receptors in humans and 9 receptors in mice,12 and have been shown to suppress TLR-mediated inflammatory responses to DAMPs.13,14,15 Siglecs have one or two extracellular N-terminal V-set Ig-like domains that bind to sialoside-containing structures with different specificities, as well as a C2-set Ig-like domain that contains a variable number of C2-type repeats.10,16 Many Siglecs have an intracellular immune receptor tyrosine-based inhibitory motifs. These motifs are phosphorylated by tyrosine kinases and are subsequently bound by SHP-1 and SHP-2 tyrosine phosphatases17 and the E3 ligase Cbl,7 thereby dampening TLR cell signaling in response to DAMPs. We have recently reported that Siglec E is directly associated with TLRs and regulates TLR-mediated induction of inflammatory responses, including endotoxemia.18
While the role for SIGLECS in SLE has not been systematically investigated, several lines of evidence suggest a potential role for Siglecs in the pathogenesis of SLE. First, we and others have reported that CD24, which encodes the first known natural ligand for a Siglec,13 affects the risk of developing SLE.19,20,21,22 Sialylated CD24 has been shown to interact with Siglec-G and human Siglec-10.23 This interaction attenuated proinflammatory TLR signaling in response to a variety of DAMPs released by damaged cells, such as nuclear protein high mobility group box 1 (HMGB1) and heat-shock proteins HSP70 and HSP90.14,15 Previous studies have demonstrated that, during cell damage and death, molecules such as HMGB1, HSPs and possibly HMGB1-containing nucleosomes induce the production of inflammatory cytokines in a TLR2-dependent manner, as well as the production of anti-dsDNA antibodies in BALB/c mice.6,24 Therefore, disruption of Siglecs and their sialylated ligands may promote autoimmunity. Second, a recent study showed that loss of Siglec-G expression in the SLE-prone MRL/lpr mouse strain moderately contributed to disease severity.25 Likewise, Siglecg single knockout mice show a massive increase in B1 B cells,26,27 and mice deficient for both Siglec-G and CD22 have an exacerbation of this phenotype and develop systemic autoimmunity with limited features of SLE.28 Third, mutations in sialic acid acetylesterase, the enzyme involved in modifying sialylated Siglec-G ligands, led to autoimmunity in mice and was associated with autoimmune diseases in humans.29 Nevertheless, no genetic polymorphisms of either Siglecg or SIGLEC10 genes have been reported to be associated with an increased risk of developing SLE.
Most recently, an association study including 4478 SLE cases and 12 656 controls from six East Asian cohorts identified SIGLEC6 as a major SLE risk locus among Asian populations.30 Our in silico analysis in this study suggests that both predisposing and protective alleles can be found within this region. Surprisingly, all protective alleles were found to be associated with enhanced expression of a SIGLEC12 gene. We also show that mice with significantly enhanced development of SLE due to expression of the alleles Sle1–3 have two mutations in the IgV-like domain of Siglec E, the closest known relative of human SIGLEC12 in the mouse. Targeted mutation of the Siglece gene in mice led to the development of lupus-like symptoms.
Materials and methods
Mice
Siglece −/− mice were generated by gene targeting from 129/Sv ES cells produced by the Mutant Mouse Regional Resource Center (MMRRC) at UC Davis (Davis, CA, USA), as described here (https://www.taconic.com/knockout-mouse/siglece-targeted). These mice were backcrossed to C57BL/6 mice for five generations. B6.NZM Sle1/Sle2/Sle3 (Sle1–3) mice were purchased from the Jackson Laboratory. All mice used were between 12 and 14 months of age. All mice were bred and maintained under specific pathogen-free conditions at the Children’s National Medical Center. All procedures were approved by the Animal Care and Use Committee of the Children’s National Medical Center.
Genetic analysis
Genomic DNA was extracted from three C57BL/6 and three B6.NZM Sle1/Sle2/Sle3 mice. Tail digestion was performed overnight at 55 °C in STE buffer (100 mm Tris, 5 mm EDTA, 0.2% sodium dodecyl sulfate, 200 mm NaCl, pH 8.5) and proteinase K. Genomic DNA was then purified using phenol–chloroform extraction. Amplification and sequencing primers were designed for each of the seven exons in Siglece (amplification primers—exon 1: 5′-, 5′-; exon 2: 5′-, 5′-; exon 3: 5′-, 5′-; exon 4: 5′-, 5′-; exon 5: 5′-, 5′-; exon 6: 5′-, 5′-; exon 7: 5′-, 5′-; sequencing primers—exon 1: 5′-, 5′-; exon 2: 5′-, 5′-; exon 3: 5′-, 5′-; exon 4: 5′-, 5′- ; exon 5: 5′-, 5′-; exon 6: 5′-, 5′-; exon 7: 5′-, 5′-. PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI, USA) for 40 cycles at 96 °C for 10 s, 57 °C for 30 s and 72 °C for 60 s.
Immunofluorescence analysis
For antinuclear antibody (ANA) level measurement, HeLa cells were seeded on coverslips and fixed with 4% paraformaldehyde (Sigma, St Louis, MO, USA) in phosphate-buffered saline (PBS) for 20 min at room temperature, followed by permeabilization with 0.1% Triton X-100 (Sigma) for an additional 10 min. After washing with PBS, cells were blocked with 10% fetal bovine serum in DMEM and stained with serum from 6-month-old mice diluted 1:1000. Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (IgG) (Invitrogen, Carlsbad, CA, USA) was used to detect ANA.
For evaluation of glomerular IgG, IgM and C3 deposition, kidneys were snap frozen in OCT medium directly after dissection. After sectioning using a cryostat, the 8-μm-thick frozen sections were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, followed by permeabilization with 0.3% Triton X-100 at room temperature for 15 min. After washing, the tissue sections were blocked with 3% normal goat serum (Sigma) and then stained with Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen), goat anti-mouse IgM (Santa Cruz Technology, Dallas, TX, USA) and rat anti-mouse C3 (Abcam, Cambridge, Cambridgeshire, UK). Deposits in the glomeruli were scored in a blinded manner on a scale of 0–4 (0=negative, 1=weak, 2=moderate, 3=strong, 4=maximal fluorescence) in five different fields for each kidney section. All images were acquired at the same exposure time to allow comparison among samples with an Olympus X51 microscope (Temple Hills, ML, USA).
Kidney histology
Kidneys from wild-type (WT), Siglece −/− and Sle1–3 mice were fixed in 10% formalin. After embedding, kidneys were sectioned at 5 μm thicknesses and stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS). PAS-stained kidneys were scored in a blinded manner for lupus nephritis. Sections were scored on a scale of 0–4 (0=normal, 1=mild, 2=moderate, 3=strong, 4=severe) for thickening of the glomerular basement membrane and mesangial matrix expansion.
dsDNA ELISA
Polystyrene plates where coated with poly-l-lysine overnight at 4 °C. After washing with PBS, plates were coated with 20 μg/ml calf thymus DNA diluted in ddH2O at 37 °C for 2 h. Plates were washed with PBS and blocked with 2% bovine serum albumin (BSA) in PBS at room temperature for 1 h. Serum samples were diluted 1:50 in 2% BSA in PBS and incubated overnight at 4 °C or at room temperature for 2 h. Plates were washed five times in PBS, and horse radish peroxidase-conjugated goat anti-mouse (GE Healthcare, Chicago, IL, USA) was added and incubated at room temperature for 1 h. After washing five times with PBS, 1-Step Ultra TMB-ELISA (Thermo Fisher Scientific, Waltham, MA, USA) substrate solution was added and incubated for 15 min. Reactions were stopped with 2 m HCl and measured to determine the optical density at 450 nm.
Binding assay
Splenocytes from C57BL/6 mice were isolated and homogenized into a single-cell suspension and incubated at 4 °C for 1 h with no protein, 0.25, 0.5, 1 or 2 μg/ml hIgG-Fc, Siglec-E-WT-Fc or Siglec-E-mutant-Fc diluted in 2% BSA in PBS. After incubation, the cell suspensions were washed three times with 2% BSA in PBS, followed by staining with phycoerythrin (PE) anti-human IgG Fc antibody (BioLegend, San Diego, CA, USA) at room temperature for 20 min. After washing, stained cells were analyzed on a BD Accuri C6 flow cytometer (Ann Arbor, MI, USA) and subsequently analyzed with the FlowJo Software (Ashland, OR, USA).
Production of inflammatory cytokines by macrophages
Peritoneal?macrophages from Siglece −/− and WT mice were isolated by lavage 3 days after intraperitoneal injection of 3% thioglycollate (Sigma). The cells were plated in 12-well plates at a density of 5 × 105 cells per well and cultured in RPMI medium containing 10% fetal bovine serum. The cells were treated with 5 μg/ml HMGB1 or PBS for 16 h before collection. The interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) cytokines in the medium were measured using mouse IL-6 and TNF-α ELISA Kits (R&D Systems, Minneapolis, MN, USA).
Bioinformatics
Statistical information for each of the quality-controlled evaluated single-nucleotide polymorphisms (SNPs) for discovery cohorts was derived from Supplementary Data in Sun et al. 30 and visualized using the Integrative Genomics Viewer (IGV) tool.31 The expression quantitative trait loci (eQTL) analysis was performed by the Portal for the Genotype-Tissue Expression (GTEx) project (http://www.gtexportal.org/home/). Isoform annotation and expression data of SIGLEC12 in different tissues were downloaded from the Ensembl project (http://www.ensembl.org) and the GTEx project (http://www.gtexportal.org/home/), respectively. The protein sequence of SIGLEC12 was compared against all mouse Siglec genes using Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with default parameters. Transcriptome RNA-seq data sets from multiple leukocyte subsets of humans and mice were obtained from the NCBI gene expression omnibus database (human immune cells: GSE64655; mouse B cells: GSE47703; mouse bone marrow dendritic cells: GSE83736; mouse bone marrow macrophages: GSE80160; mouse bone marrow monocytes: GSE86079; mouse natural killer cells: GSE52047; mouse CD4 T cells and CD8 T cells: GSE48138). The reference sequences used were genome and transcriptome sequences downloaded from the Ensembl website (http://www.ensembl.org/index.html, version GRCh38 for humans and version GRCm38 for mice). Corresponding gene expression levels (measured as fragments per kilobase per million mapped reads: FPKM) were calculated using HISAT2 and StringTie.
Statistical analysis
Sample sizes were chosen based on past experience with the same models in the literature and our past experience with similar models. The specific tests used to analyze each set of experiments are indicated in the figure legends. For each statistical analysis, appropriate tests were selected on the basis of whether the data were normally distributed by using Shapiro–Wilk’s test. Data were analyzed using an unpaired two-tailed Mann–Whitney test or Student's t-test to compare data between two groups and two-way analysis of variance for two-way factorial design. Sample sizes were chosen with adequate statistical power on the basis of the literature and past experience. No samples were excluded from the analysis, and experiments were not randomized unless specified. In the graphs, y-axis error bars represent s.e.m., as indicated. Statistical calculations were performed using GraphPad Prism software (GraphPad Software, San Diego, CA, USA) or R software (https://www.r-project.org/).
Results
Hypermorphic Siglec alleles protect Asian individuals against SLE
Asian individuals are reportedly more susceptible to SLE, based on its increased incidence and severity in these populations.32 A high-density genotyping of immune-related genes involving 4478 SLE cases and 12 656 controls from six East Asian cohorts identified SIGLEC6 as a major SLE risk locus among Asian populations.30 As shown in Figure 1a, our reanalysis of that data revealed that most of the significant SNP clusters resided either in SIGLEC6 or in the intergenic regions between SIGLEC6 and SIGLEC12. The eight most prominent SNPs are shown in Table 1. Among these, five SNPs conferred susceptibility to SLE, while three SNPs conferred protection against SLE. Since none of the SNPs affected the coding sequence, we evaluated if any of them were eQTLs for surrounding genes. Surprisingly, while all protective SNPs resided in SIGLEC6, none of them affected the expression of SIGLEC6 (data not shown). All protective alleles were instead identified as eQTLs that enhanced the expression of SIGLEC12 (Figure 1b), but not any other genes within 1 Mb of the protective SNPs.33 These data raise the intriguing possibility that SIGLEC12 expression may suppress SLE.
Figure 1.
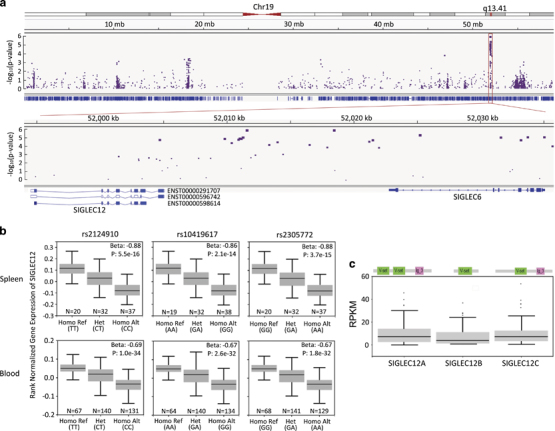
Regional association of Siglec gene family and SLE risk. (a) Regional association plots for chromosome 19 (top) and the hot region (bottom) of SLE. Plots of log10 (P-meta) versus genomic position. (b) Boxplots of SIGLEC12-normalized expression values for three significant eQTLs in a novel hot region in splenic and blood tissues. The β-value of the eQTLs is defined as the slope of the linear regression and is computed as the effect of the alternative allele (ALT) relative to the reference allele (REF). P-values were generated for each variant–gene pair by testing the alternative hypothesis that the slope of a linear regression model between genotype and expression deviated from 0. (c) Schematic diagram of Pfam domains of three isoforms of SIGLEC12 (top) and a boxplot of isoform expression values (RPKM) in splenic tissues using Genotype-Tissue Expression (GTEx) project data (bottom). eQTL, expression quantitative trait loci; RPKM, reads per kilobase million; Siglec, sialic acid-binding immunoglobulin-like lectin; SLE, systemic lupus erythematosus.
Table 1.
Meta-analysis results for top 8 loci in the SIGLEC region associated with SLE in Asian human cohorts
| Chr. | Position hg19 | SNP | Ref/Alt | MAF | Location | OR Korean | OR Han Chinese | OR Malaysian Chinese | P-value Meta-analysis | eQTL |
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 52011292 | Rs9676266 | C/A | 0.10(A) | Intergenic | 1.3 | 1.14 | 1.44 | 9.27E−06 | No |
| 19 | 52011329 | Rs9676272 | C/A | 0.13(A) | Intergenic | 1.3 | 1.15 | 1.39 | 8.96E−06 | No |
| 19 | 52011598 | Rs13343377 | C/A | 0.26(A) | Intergenic | 1.32 | 1.16 | 1.23 | 1.31E−06 | No |
| 19 | 52018474 | Rs6509546 | G/A | 0.25(A) | Intergenic | 1.32 | 1.15 | 1.17 | 1.26E−06 | No |
| 19 | 52025247 | Rs2124910 | T/C | 0.43(T) | Intron | 0.83 | 0.89 | 0.87 | 4.62E−06 | eQTL for SIGLEC12 |
| 19 | 52031648 | Rs10419617 | A/G | 0.40(A) | Synonymous | 0.83 | 0.89 | 0.91 | 9.09E−06 | eQTL for SIGLEC12 |
| 19 | 52033742 | Rs2305772 | G/A | 0.42(G) | Intron | 0.83 | 0.88 | 0.91 | 8.84E−06 | eQTL for SIGLEC12 |
| 19 | 52035059 | Rs12609761 | A/C | 0.15(C) | 5′-UTR variant | 1.29 | 1.18 | 1.35 | 8.44E−06 | No |
Abbreviations: Chr., chromosome; eQTL, expression quantitative trait loci; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism; UTR, untranslated region.
Unlike most Siglecs, SIGLEC12 contains two IgV-like domains.34 As shown in Figure 1c, the SIGLEC12 gene encodes three major alternatively spliced forms in the spleen, each containing either one or both IgV-like domains. While isoform b lacks transmembrane and cytoplasmic domains, the more abundant isoforms, a and c, are predicted to encode transmembrane Siglec proteins, with two or one IgV-like domains, respectively.
We compared protein sequences encoded by the human SIGLEC12 gene and the mouse Siglec gene family by BLAST analysis. As shown in Figures 2a and b, the highest similarity was found between Siglec E and SIGLEC12, regardless of whether SIGLEC12A or SIGLEC12C amino-acid sequences were used for inquiries. In addition to amino-acid sequence homology, SIGLEC12C is similar to Siglec E in domain structures. Furthermore, SIGLEC12A and SIGLEC12C are expressed predominantly in human monocytes, which is analogous to mouse Siglece (Figure 2).
Figure 2.
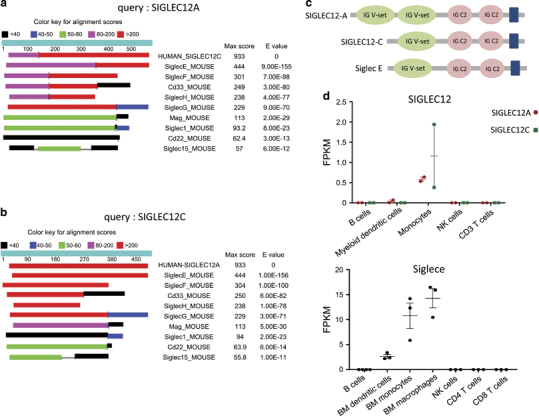
Homology analysis between human SIGLEC12 and mouse Siglec proteins. Comparison of the protein sequences encoded by human SIGLEC12A (a) and SIGLEC12C (b) and the mouse Siglec gene family using BLAST analysis. (c) Schematic diagram of Pfam domains of human SIGLEC12A, human SIGLEC12C and mouse Siglec E. (d) Dot plots of human SIGLEC12 and mouse Siglec E expression as measured by FPKM (fragments per kilobase per million mapped reads) in multiple leukocyte subsets. Siglec, sialic acid-binding immunoglobulin-like lectin.
B6.NZMSle1/Sle2/Sle3 (Sle1–3) mice carry hypomorphic mutations in the Siglece gene
Major mouse SLE susceptibility regions from NZM mice have been mapped to three regions. Specifically, genomic intervals on chromosomes 1 (Sle1), 4 (Sle2), and 7 (Sle3) have been strongly linked to the spontaneous development of lupus.35,36 Siglec E is present in a gene cluster within Sle3 on chromosome 7;37 therefore, we compared Siglece from C57BL/6 and Sle1–3 mice by Sanger sequencing of genomic DNA for all seven Siglec E exons. In Siglec E from Sle1–3 mice, we identified two single-residue mutations in exon 1, which encodes the N-terminal V-set Ig-like domain, and another mutation in exon 2, which encodes C2-set Ig-like domain-1 (Figures 3a and b). No mutations were found in other coding regions. As a result, three amino-acid replacements were found in the Siglec E protein (Figure 3c).
Figure 3.
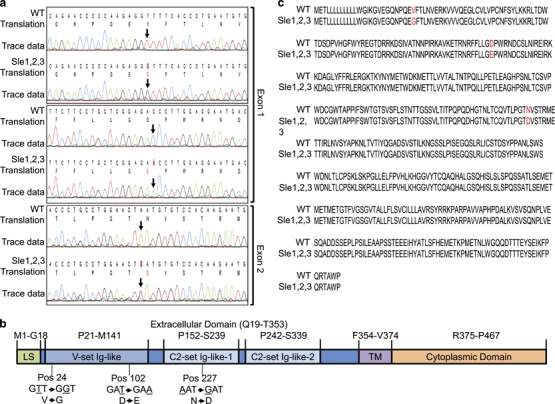
Missense mutations of Siglece coding region in B6.NZM Sle1/Sle2/Sle3 mice. (a) Sequencing reads were aligned using CLC Main Workbench. Arrows indicate SNP annotations. (b) Sequence alignment of Siglec E from C57BL/6 (WT) and B6.NZM Sle1/Sle2/Sle3 (Sle1–3) mice. Red indicates amino-acid substitutions. (c) Graphical representation of the Siglec E protein, which consists of a leader sequence (LS), a V-set Ig-like domain, C2-set Ig-like-1 and C2-set Ig-like-2 domains, a transmembrane domain (TM) and a cytoplasmic domain. Two amino-acid substitutions were in the V-set Ig-like domain, at positions 24 and 102, and one amino-acid substitution was in the C2-set Ig-like-1 domain at position 227. Siglec, sialic acid-binding immunoglobulin-like lectin; SNP, single-nucleotide polymorphism; WT, wild type.
Since the N-terminal V-set Ig-like domain mediates ligand binding, we expressed fusion proteins consisting of extracellular domains of WT, B6.NZM Sle1/Sle2/Sle3 mice and human IgG1-Fc (Figure 4a) and evaluated the impact of Siglece Sle mutations on Siglec E function. Siglec E from C57BL/6 (WT-SE-Fc) and Sle1–3 (Mutant-SE-Fc) was incubated with splenocytes, and binding was measured by flow cytometry. As shown in Figures 4b and c, Mutant-SE-Fc had reduced binding compared to WT-SE-Fc. Therefore, the B6.NZM Sle1/Sle2/Sle3 mice express a hypomorphic allele of Siglece. Since the mutations are predicted to be outside the ligand binding site, the reduced binding may be explained by subtle conformational changes caused by these mutations.
Figure 4.
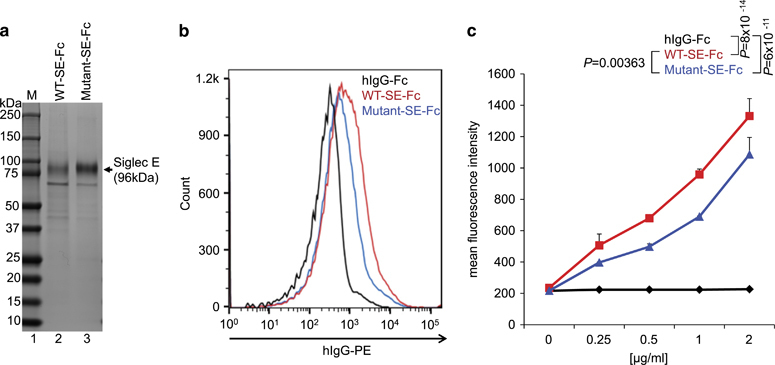
Siglec E from B6.NZM Sle1/Sle2/Sle3 mice has reduced ligand binding. (a) Coomassie blue staining of purified Siglec E from C57BL/6 (WT-SE-Fc, lane 2) and B6.NZM Sle1/Sle2/Sle3 (Mutant-SE-Fc, lane 3) mice. M=molecular weight marker (lane 1). (b) Splenocytes from C57BL/6 mice were incubated with 0.5 μg/ml of purified hIgG-Fc (control), WT-SE-Fc and Mutant-SE-Fc. Ligand binding was analyzed by flow cytometry. (c) Mean fluorescence intensity of splenocytes incubated with different concentrations of purified hIgG-Fc (control), WT-SE-Fc and Mutant-SE-Fc. Data in (b and c) are representative of three independent experiments involving different mice. Error bars in (c) represent s.e.m. of triplicate samples from the same mice (technical repeats), with statistical analysis by two-way ANOVA. ANOVA, analysis of variance; Siglec, sialic acid-binding immunoglobulin-like lectin; SNP, single-nucleotide polymorphism; WT, wild type.
Targeted mutation of the Siglece gene induces SLE-like phenotypes in mice
The production of autoantibodies, such as ANAs or anti-dsDNA, is a hallmark of SLE.2 Sle3 is the major regulator of the production of autoantibodies, including antinuclear and anti-dsDNA antibodies.36 To evaluate whether Siglec E deficiency leads to this phenotype, we used mice with targeted mutations of the Siglece gene.18 Mice were derived from Siglece +/− 129/Sv ES cells and then backcrossed to C57BL/6 for five generations (N5) before use. Moreover, we specifically removed the Caps11 null allele from 129/SV in our lines because of its critical role in non-canonical inflammasome activation.38 WT N5 mice were used as controls. Since the manifestations of SLE are more severe in female individuals, we monitored the production of autoantibodies in female mice. Since an indirect immunofluorescence assay is the most sensitive ANA detection method,39 we used this as the initial measure of autoantibody production. As shown in Figure 5a, ANA was easily detected in Siglece −/− mice but not in wild-type littermates. Interestingly, various staining patterns, such as nuclear membrane (21.4%, 3/14) and possible Golgi staining (7.1%, 1/14) were observed, suggesting a diverse production of autoantibodies in the Siglece −/− mice. The incidence of IgG ANA at 6 months of age was 57.1% (8/14) in sera from Siglece −/−, 12.5% (1/8) in wild type and 28.6% (2/7) in B6.NZM Sle1/Sle2/Sle3 mice (Figure 5b). The higher incidence in the Siglece −/− mice suggests a major role of the Siglece hypomorphic allele in the function of the Sle3 region that results in a predisposition to autoantibody production. Next, we assessed the levels of anti-dsDNA IgG autoantibodies using ELISA (enzyme-linked immunosorbent assay and found that there was a significant increase in anti-dsDNA production in Siglece −/− and B6.NZM Sle1/Sle2/Sle3 mice at 12 months of age compared with wild-type controls (Figure 5c).
Figure 5.
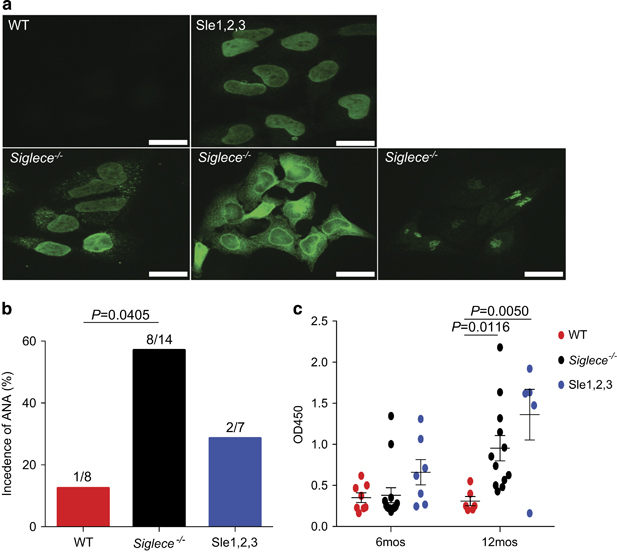
Increased production of autoantibodies in Siglec-E-deficient mice. (a) Autoantibody staining patterns using indirect fluorescence autoantibody assays in HeLa cells with sera derived from 6-month-old female WT, Siglece −/− and B6.NZM Sle1/Sle2/Sle3 (Sle1–3) mice at 1:1000 dilutions (scale bars, 20 μm; original magnification, x60). (b) Sera from 6-month-old female mice showed that ANA levels were markedly increased in Siglece −/− mice. WT, N=8; Siglece −/−, N=14; Sle1–3, N=7. (c) dsDNA autoantibody levels were assessed by ELISA with sera derived from 6-month-old and 12-month-old female mice. All sera were tested in the same assays. Each point represents a value from an individual mouse, and horizontal bars denote means with standard error of the mean (s.e.m.). Sample sizes were as follows: 6 months: WT, N=8; Siglece −/−, N=14; Sle1–3, N=7. 12 months: WT, N=6; Siglece −/−, N=12; Sle1–3, N=5. P-values at 6 months were analyzed using Mann–Whitney tests and were found to be nonsignificant. P-values at 12 months were calculated with a two-tailed unpaired Student’s t-test. Errors bar show deviations in biological repeats. ANA, antinuclear antibody; dsDNA, double-stranded DNA; ELISA, enzyme-linked immunosorbent assay; Siglec, sialic acid-binding immunoglobulin-like lectin; WT, wild type.
H&E and PAS staining revealed sclerotic nephritis, heavy proteinaceous deposits in the mesangium, tubular cast formation and diffuse proliferation of glomerular cells in both Siglece −/− and B6.NZM Sle1/Sle2/Sle3 mice (Figures 6a and b). Semiquantitative pathological scoring of PAS-stained kidneys revealed that both Siglece −/− and B6.NZM Sle1/Sle2/Sle3 mice had more severe glomerular damage than WT controls (Figure 6c). Immunofluorescence staining showed increased IgG, IgM and C3 glomerular deposition in Siglece −/− and B6.NZM Sle1/Sle2/Sle3 mice (Figures 6d and e).
Figure 6.
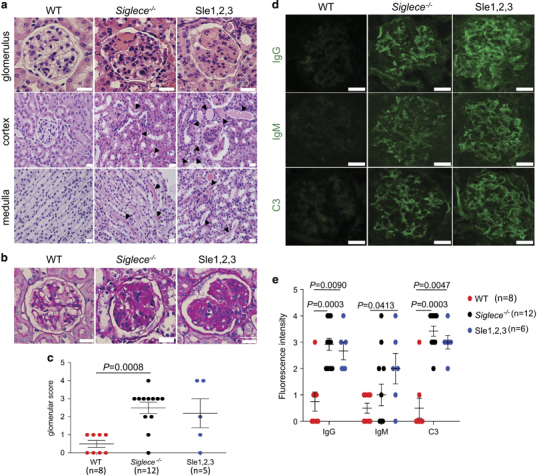
Increased renal pathology in Siglece −/− mice. (a) Kidney sections from 12-month-old female WT, Siglece −/− and B6.NZM Sle1/Sle2/Sle3 (Sle1–3) were stained with H&E. Arrows in Siglece −/− and B6.NZM Sle1/Sle2/Sle3 (Sle1–3) cortex and medulla indicate amyloid deposition. (b) PAS-positive material in the glomerular capillary lumen is shown in Siglece −/− and B6.NZM Sle1/Sle2/Sle3 (Sle1–3) mice. (c) Glomerular disease was scored from 0 to 4 for each mouse. Mice aged between 10–12 months were scored based on PAS staining. WT, N=8; Siglece −/−, N=12; Sle1–3, N=5. (d) Glomerular immune deposits were detected by immunofluorescence staining for IgG, IgM and complement C3. Representative images are shown (scale bars, 20 μm; original magnification, x60). Immunofluorescence staining was analyzed in triplicate. (e) Glomerular fluorescence intensity was scored from 0 to 4, in a double-blind manner. Each point represents a value from an individual mouse, and horizontal bars denote means with standard error of the mean (s.e.m.). WT, N=8; Siglece −/−, N=12; Sle1–3, N=6. P-values in (c and e) were calculated with a two-tailed unpaired Student’s t-test. Error bars show deviations in biological repeats. H&E, hematoxylin and eosin; Ig, immunogobulin; PAS, periodic acid-Schiff; WT, wild type.
Targeted mutation of Siglece increased the production of inflammatory cytokines by macrophages in response to HMGB1
HMGB1 has been implicated in the pathogenesis of SLE in mice and humans.6 Given the critical role of TLR2 and TLR4 in SLE40 and host responses to HMGB1,6 and the function of Siglec E in regulating the response to TLR2 and TLR4 ligands from pathogens,18 we determined whether targeted mutation of Siglece exacerbated the production of inflammatory cytokine by murine macrophages in response to HMGB1. As shown in Figure 7, Siglece −/− macrophages produced approximately twice as much TNFα (Figure 7a) and IL-6 (Figure 7b) in response to HMGB1.
Figure 7.
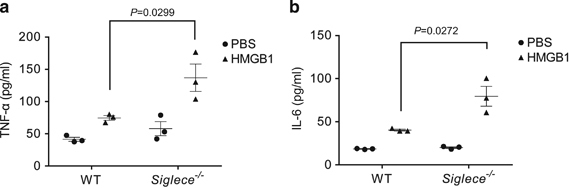
Targeted mutation of Siglece increases macrophage responses to HMGB1, as indicated by increased production of TNF-α (a) and IL-6 (b). Data shown indicate means and s.e.m. from a representative experiment with three mice per group and were reproduced twice. P-values were determined by two-tailed Student’s t-test. Errors bar show deviations in biological repeats. HMGB1, high mobility group box 1; IL, interleukin; PBS, phosphate-buffered saline; TNF-α, tumor necrosis factor-α; WT, wild type.
Discussion
Collectively, our data and in silico analyses demonstrate that the murine Siglece and human SIGLEC12 genes protect against the development of SLE. These data provide the first genetic evidence linking these two genes to SLE pathogenesis.
Since ~50% of humans have frameshift mutations in the first IgV-like domain, it has been suggested that SIGLEC12 may be a pseudogene in a large proportion of humans.41 Since this SNP has not been shown to be associated with an increased risk of SLE, the protective role of SIGLEC12 may not be immediately clear. However, apart from the SIGLEC12 isoform SIGLEC12A, which contains two IgV domains, we found equally abundant SIGLEC12A and SIGLRC12C isoforms devoid of the first IgV-encoding exon. Since SIGLEC12B does not contain the mutated exon, this splicing effectively neutralizes the impact of the frameshift mutation while simultaneously preserving the second IgV-like domain. Therefore, there is no known null allele of SIGLEC12 in humans.
Since the first IgV domain of full-length SIGLEC12 lacks a critical arginine at residue 122 (R122>C122), which is important in mediating Siglec binding to sialic acid,37 it is unclear whether it is a sialoside-recognizing lectin like the other Siglecs. However, although Angata et al. 37 showed that back-mutation of C122 to Y122 enhanced SIGLEC12 binding to sialic acid probes, Yu et al. 34 reported that SIGLEC12 can recognize sialic acid on red blood cells, perhaps through a glutamine in an analogous position in the second IgV-like domain.
Since SIGLEC12 is closely related to Siglece, it is tempting to speculate that our mouse genetic data regarding Siglece explains the protective SLE alleles identified in chromosome 19.30 Consistent with this hypothesis, our in silico analysis of cellular distributions suggested that SIGLEC12 and Siglece have similar expression patterns. Furthermore, as discussed above, frameshift mutations and alternative splicing independently generate a SIGLEC12C protein that is similar to Siglec E in domain structure in the mouse. Nevertheless, given the rapid evolution among CD33 families of Siglecs in humans and mice, it is premature to definitively designate Siglec E as the ortholog of the Siglec12C isoform.
It is of note that early studies using Siglece-deficient mice did not reveal phenotypes suggestive of SLE.42,43 It is unclear if other laboratories have investigated the potential involvement of Siglec E in the pathogenesis of SLE. However, it is worth noting that the 129/Sv background contains a null allele for caspase-11, which is a critical regulator of inflammation.38 To avoid confounding factors, we specifically screened out caspase-11-null alleles in early generations of mice during backcrossing. It is worth investigating whether the elimination of caspase-11 mutations allowed us to reveal the critical role of Siglec E in the pathogenesis of SLE.
Another potential caveat concerns whether other 129/Sv genes may confound the phenotype of the Siglec E-deficient mice. We consider this unlikely, as the controls used for this study were WT mice from the same generations of backcrossing, which is the standard approach to minimize this confounding factor. It is important to note that the putative alleles in the 129/Sv background are revealed only if the C1q gene is deleted,44 which will not be able to cause SLE-like symptoms and pathology as described herein. Furthermore, the 129/Sv allele affects glomerulonephritis but not the production of autoantibodies,44 while Siglece deletion caused both glomerulonephritis and autoantibody production.
The significant protection conferred by Siglec E is best understood in the context of TLR signaling in the pathogenesis of SLE as we have previously shown that Siglec E binds and negatively regulates the function of multiple TLRs, including TLR2 and TLR4.18 Experimental evidence in lupus-prone animal models suggests a role for TLR2 and TLR4, which bind components of bacterial cell walls.6 TLR4-deficient mice and, to a lesser extent, TLR2-deficient mice have shown much less severe disease phenotypes with significantly reduced production of ANAs, decreased renal lesions and decreased MZ B cells compared with wild-type mice.40,45 TLR4-deficient mice also had reduced inflammatory cytokine production, decreased anti-dsDNA antibody levels and attenuated nephritis in pristane-induced lupus.46 In addition, studies have shown that upregulation of TLR4 results in a strong induction of lupus-like disease.47 Furthermore, TLR2 and TLR4 may contribute to the production of anti-dsDNA autoantibodies by binding to HMGB1-containing nucleosomes.6,24 By showing the strong impact of the Siglece mutation on macrophage responses to HMGB1, our work provides an immunological basis that may explain how SIGLEC genes control the SLE risk.
Acknowledgements
This study was supported by Grants (AI064350, AG036690) from the National Institutes of Health.
Author contributions
RF and P Zhang generated all of the data with assistance from WW and PY. YL and P Zheng supervised the study and wrote the manuscript along with RF and P Zhang.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Pan Zheng, Email: pzheng@oncoimmune.com.
Yang Liu, Email: yangl@oncoimmune.com.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Holman HR, Kunkel HG. Affinity between the lupus erythematosus serum factor and cell nuclei and nucleoprotein. Science. 1957;126:162–163. doi: 10.1126/science.126.3265.162. [DOI] [PubMed] [Google Scholar]
- 3.Munoz LE, Janko C, Schulze C, Schorn C, Sarter K, Schett G, et al. Autoimmunity and chronic inflammation—two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev. 2010;10:38–42. doi: 10.1016/j.autrev.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Silva MT. Secondary necrosis: the natural outcome of the complete apoptotic program. FEBS Lett. 2010;584:4491–4499. doi: 10.1016/j.febslet.2010.10.046. [DOI] [PubMed] [Google Scholar]
- 5.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 6.Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, et al. Induction of inflammatory and immune responses by HMGB1–nucleosome complexes: implications for the pathogenesis of SLE. J Exp Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152:467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Boyd CR, Orr SJ, Spence S, Burrows JF, Elliott J, Carroll HP, et al. Siglec-E is up-regulated and phosphorylated following lipopolysaccharide stimulation in order to limit TLR-driven cytokine production. J Immunol. 2009;183:7703–7709. doi: 10.4049/jimmunol.0902780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocker PR, Clark EA, Filbin M, Gordon S, Jones Y, Kehrl JH, et al. Siglecs: a family of sialic-acid binding lectins. Glycobiology. 1998;8:v. doi: 10.1093/oxfordjournals.glycob.a018832. [DOI] [PubMed] [Google Scholar]
- 10.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 11.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 12.Jandus C, Simon HU, von Gunten S. Targeting siglecs—a novel pharmacological strategy for immuno- and glycotherapy. Biochem Pharmacol. 2011;82:323–332. doi: 10.1016/j.bcp.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science (New York, NY) 2009;323:1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Chen GY, Zheng P. CD24-Siglec G/10 discriminates danger- from pathogen-associated molecular patterns. Trends Immunol. 2009;30:557–561. doi: 10.1016/j.it.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Chen GY, Zheng P. Sialoside-based pattern recognitions discriminating infections from tissue injuries. Curr Opin Immunol. 2011;23:41–45. doi: 10.1016/j.coi.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaji T, Teranishi T, Alphey MS, Crocker PR, Hashimoto Y. A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues. A comparison with Siglec-9. J Biol Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 17.Ulyanova T, Shah DD, Thomas ML. Molecular cloning of MIS, a myeloid inhibitory siglec, that binds protein-tyrosine phosphatases SHP-1 and SHP-2. J Biol Chem. 2001;276:14451–14458. doi: 10.1074/jbc.M011650200. [DOI] [PubMed] [Google Scholar]
- 18.Chen GY, Brown NK, Wu W, Khedri Z, Yu H, Chen X, et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. eLife. 2014;3:e04066. doi: 10.7554/eLife.04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YH, Bae SC. Association between functional CD24 polymorphisms and susceptibility to autoimmune diseases: a meta-analysis. Cell Mol Biol (Noisy-le-grand) 2015;61:97–104. [PubMed] [Google Scholar]
- 20.Piotrowski P, Lianeri M, Wudarski M, Lacki JK, Jagodzinski PP. CD24 Ala57Val gene polymorphism and the risk of systemic lupus erythematosus. Tissue Antigens. 2010;75:696–700. doi: 10.1111/j.1399-0039.2010.01447.x. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez E, Abelson AK, Sabio JM, González-Gay MA, Ortego-Centeno N, Jiménez-Alonso J, et al. Association of a CD24 gene polymorphism with susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2007;56:3080–3086. doi: 10.1002/art.22871. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Lin S, Rammohan K, Liu Z, Liu JQ, Liu RH, et al. A di-nucleotide deletion in CD24 confers protection against autoimmune diseases. PLoS Genet. 2007;3:e49. doi: 10.1371/journal.pgen.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, et al. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29:428–435. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urbonaviciute V, Voll RE. High-mobility group box 1 represents a potential marker of disease activity and novel therapeutic target in systemic lupus erythematosus. J Intern Med. 2011;270:309–318. doi: 10.1111/j.1365-2796.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- 25.Bokers S, Urbat A, Daniel C, Amann K, Smith KG, Espéli M, et al. Siglec-G deficiency leads to more severe collagen-induced arthritis and earlier onset of lupus-like symptoms in MRL/lpr mice. J Immunol. 2014;192:2994–3002. doi: 10.4049/jimmunol.1303367. [DOI] [PubMed] [Google Scholar]
- 26.Ding C, Liu Y, Wang Y, Park BK, Wang CY, Zheng P, et al. Siglecg limits the size of B1a B cell lineage by down-regulating NFkappaB activation. PLoS One. 2007;2:e997. doi: 10.1371/journal.pone.0000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, et al. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol. 2007;8:695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- 28.Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. CD22 x Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol (Baltimore, Md: 1950) 2010;184:3618–3637. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- 29.Surolia I, Pirnie SP, Chellappa V, Taylor KN, Cariappa A, Moya J, et al. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 2010;466:243–247. doi: 10.1038/nature09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet. 2016;48:323–330. doi: 10.1038/ng.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakes RW, Bae SC, Louthrenoo W, Mok CC, Navarra SV, Kwon N. Systematic review of the epidemiology of systemic lupus erythematosus in the Asia-Pacific region: prevalence, incidence, clinical features, and mortality. Arthritis Care Res (Hoboken) 2012;64:159–168. doi: 10.1002/acr.20683. [DOI] [PubMed] [Google Scholar]
- 33.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Z, Lai CM, Maoui M, Banville D, Shen SH. Identification and characterization of S2V, a novel putative siglec that contains two V set Ig-like domains and recruits protein-tyrosine phosphatases SHPs. J Biol Chem. 2001;276:23816–23824. doi: 10.1074/jbc.M102394200. [DOI] [PubMed] [Google Scholar]
- 35.Mohan C, Yu Y, Morel L, Yang P, Wakeland EK. Genetic dissection of Sle pathogenesis: Sle3 on murine chromosome 7 impacts T cell activation, differentiation, and cell death. J Immunol. 1999;162:6492–6502. [PubMed] [Google Scholar]
- 36.Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, et al. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- 37.Angata T, Hingorani R, Varki NM, Varki A. Cloning and characterization of a novel mouse Siglec, mSiglec-F: differential evolution of the mouse and human (CD33) Siglec-3-related gene clusters. J Biol Chem. 2001;276:45128–45136. doi: 10.1074/jbc.M108573200. [DOI] [PubMed] [Google Scholar]
- 38.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 39.Emlen W, O'Neill L. Clinical significance of antinuclear antibodies: comparison of detection with immunofluorescence and enzyme-linked immunosorbent assays. Arthritis Rheum. 1997;40:1612–1618. doi: 10.1002/art.1780400910. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Yin H, Zhao M, Lu Q. TLR2 and TLR4 in autoimmune diseases: a comprehensive review. Clin Rev Allergy Immunol. 2014;47:136–147. doi: 10.1007/s12016-013-8402-y. [DOI] [PubMed] [Google Scholar]
- 41.Mitra N, Banda K, Altheide TK, Schaffer L, Johnson-Pais TL, Beuten J, et al. SIGLEC12, a human-specific segregating (pseudo)gene, encodes a signaling molecule expressed in prostate carcinomas. J Biol Chem. 2011;286:23003–23011. doi: 10.1074/jbc.M111.244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz F, Pearce OM, Wang X, Samraj AN, Läubli H, Garcia JO et al. Siglec receptors impact mammalian lifespan by modulating oxidative stress. Elife 2015; 4. [DOI] [PMC free article] [PubMed]
- 43.McMillan SJ, Sharma RS, McKenzie EJ, Richards HE, Zhang J, Prescott A, et al. Siglec-E is a negative regulator of acute pulmonary neutrophil inflammation and suppresses CD11b beta2-integrin-dependent signaling. Blood. 2013;121:2084–2094. doi: 10.1182/blood-2012-08-449983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidari Y, Bygrave AE, Rigby RJ, Rose KL, Walport MJ, Cook HT, et al. Identification of chromosome intervals from 129 and C57BL/6 mouse strains linked to the development of systemic lupus erythematosus. Genes Immun. 2006;7:592–599. doi: 10.1038/sj.gene.6364335. [DOI] [PubMed] [Google Scholar]
- 45.Lartigue A, Colliou N, Calbo S, François A, Jacquot S, Arnoult C, et al. Critical role of TLR2 and TLR4 in autoantibody production and glomerulonephritis in lpr mutation-induced mouse lupus. J Immunol. 2009;183:6207–6216. doi: 10.4049/jimmunol.0803219. [DOI] [PubMed] [Google Scholar]
- 46.Summers SA, Hoi A, Steinmetz OM, O'Sullivan KM, Ooi JD, Odobasic D, et al. TLR9 and TLR4 are required for the development of autoimmunity and lupus nephritis in pristane nephropathy. J Autoimmun. 2010;35:291–298. doi: 10.1016/j.jaut.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Liu B, Yang Y, Dai J, Medzhitov R, Freudenberg MA, Zhang PL, et al. TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J Immunol. 2006;177:6880–6888. doi: 10.4049/jimmunol.177.10.6880. [DOI] [PubMed] [Google Scholar]


