Dear Editor,
Microbiome affects many aspects of human health and disease and elicits a wide range of host responses including remarkable epigenetic changes such as DNA methylation, histone modification and non-coding RNA expression.1 A still poorly explored area of microbiome-host interaction is the response of host RNA modification. N6-methyladenosine (m6A) is the most abundant mRNA modification in mammalian cells, occurring at ~3 modified adenosine residues per transcript. The m6A mapping and biology have been extensively studied recently.2 At the physiological level, m6A affects embryonic development, circadian clock, immuno-response, and others. At the cellular and molecular level, m6A affects all key aspects of mRNA processing, translation and decay. Importantly, m6A is a predominant, transcriptome-wide mark that is responsive to environmental changes; this dynamic m6A pattern is maintained by the writer enzyme complex containing the METTL3 and METTL14 proteins, and two eraser enzymes of FTO and ALKBH5.3,4
We investigated the host response marked by m6A in the transcriptome to the presence of microbiome in mice (Fig. 1a). We employed one group of germ-free (GF) mice to identify the host response to the absence, and the other group of specific pathogen-free (SPF) mice to identify the host response to the presence of microbiome. We validated the absence of gut microbiota in our GF mice by PCR of the representative 16S genes (Supplementary information, Fig. S1a). 16S rRNA gene amplicon sequencing of the SPF mice showed that all three mice in this group had similar bacterial compositions at the genus level, which were mainly blautia and roseburia (Supplementary information, Fig. S1b).
Fig. 1.
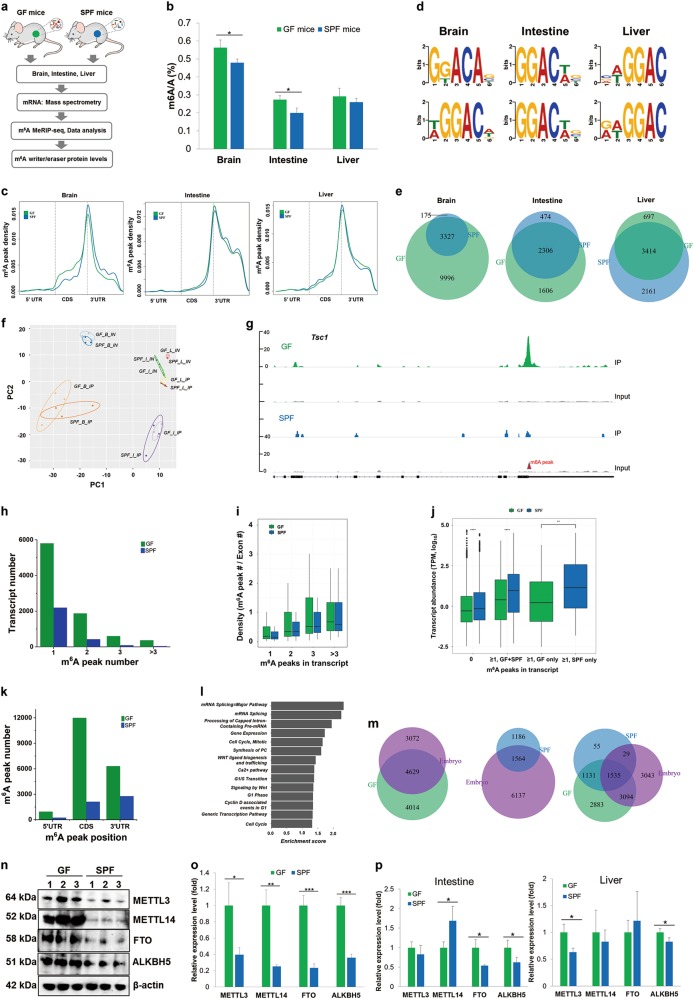
m6A methylome and writer/eraser expression in the germ-free (GF) and specific pathogen-free (SPF) mouse tissues. a Schematic representation of the study. b QQQ LC/MS measurement of total m6A/A ratio of polyA-selected and ribo-minus treated RNAs. Values are the means ± standard deviation (SD), n = 3, *P < 0.05, Student’s t-test. c m6A pattern distribution across the mRNA regions in brain, intestine and liver. m6A peaks were mapped back to the corresponding gene, and assigned as originated from 5′ UTR, coding region (CDS) or 3′ UTR. d Motif analysis of m6A peaks. Upper panel, GF tissues; lower panel, SPF tissues. e Venn diagram showing the differences of m6A peaks between GF and SPF samples. f Principal component analysis of input (IN) and IP samples. The label is for Sample_tissue_Seq, e.g., GF_B_IP stands for GF mouse, brain, m6A-IP. Tissue labels are: B, brain; I, intestine; L, liver. g Representative sequencing coverage of an mRNA in the brain showing a differential m6A peak in GF and SPF samples. h Transcript counts containing different m6A peak numbers in the brain. i m6A peak and exon density in the brain. j Abundance of m6A-containing transcripts in the brain. k mRNA m6A peak positions in the brain. l Reactome analysis of biological pathways of m6A-containing transcripts in the brain. m Venn diagram comparing the 4-week-old GF/SPF brain m6A peak-containing transcripts with those in the E13.5 embryonic brain. n Western blots of m6A writer proteins METTL3, METTL14, and eraser proteins FTO, ALKBH5 in the brain tissues. o Quantitation of m6A writer and eraser protein levels in the brain. Values are the means ± SD, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test. p Quantitation of m6A writer and eraser protein levels in the intestine and liver. Values are the means ± SD, n = 3, *P < 0.05, Student’s t-test
We harvested three tissues of GF and SPF mice of the same genetic background at 4 weeks of age, brain, intestine, and liver, and performed m6A analysis in polyA-selected RNA by liquid chromatography/mass spectrometry (LC/MS) to determine the total m6A/A ratios and by the m6A-MeRIP sequencing to determine the transcriptomic m6A pattern and distribution. These three tissues were selected based on their pervasive studies in the literature on the GF and SPF mouse physiology. The m6A/A ratios of the polyA-selected RNA are in the expected range of 0.2%–0.6%; brain showed the highest m6A content for both GF and SPF mice, and brain and intestine showed higher m6A content in the GF mice (Fig. 1b). The polyA-selected RNA in kidney also showed higher m6A content in the GF mice (Supplementary information, Fig. S2a). The higher m6A content in the brain tissue was also observed in GF and SPF mice that were 10 weeks old (Supplementary information, Fig. S2b) and even 2 years old (Supplementary information, Fig. S2c). Our m6A-MeRIP results of all three tissues (Supplementary information, Table S1) showed the well-known m6A pattern across the mRNA transcripts such as the strong enrichment of m6A peaks at the junction of coding region (CDS) and 3′ UTR (Fig. 1c). We identified the m6A-containing transcripts that were present in all three GF or SPF mouse groups as “high confidence” data and used only these for further analysis (Supplementary information, Fig. S3). We recovered the known m6A installation consensus sequence, RRACH (R = A/G, H = A/C/U) among the m6A peaks with a preference of guanosine 5′ to the m6A site (Fig. 1d). We validated our sequencing results by quantitative RT-PCR of specific transcripts (Supplementary information, Fig. S4a). Our sequencing result was also consistent with the expected mRNA expression difference of GF versus SPF mouse reported in the literature5,6 (Supplementary information, Fig. S4b). These results validated the high-quality nature of our m6A-MeRIP data.
We identified several differences in m6A patterns between the GF and SPF tissues. First, the m6A peak distributions had distinct shapes among these tissues (Fig. 1c). When benchmarked against the m6A cluster near the stop codon, the GF brain m6A occurrence was higher in the CDS region compared to the SPF brain. Second, the m6A installation consensus sequences in GF and SPF tissues were deviated in brain, but identical in intestine and liver (Fig. 1d). Third, only 25% of the m6A peaks in the GF brain overlapped with those in SPF brain, whereas > 59% of the m6A peaks overlapped in GF and SPF intestine and liver (Fig. 1e). The large brain m6A peak differences was also shown by principal component analysis and their statistical significance in gene ontology analysis across all tissues (Fig. 1f and Supplementary information, Fig. S5), and was not derived from global transcript expression differences (Supplementary information, Fig. S6). On the other hand, SPF brain may have higher m6A modification fraction for some common m6A peaks (Supplementary information, Fig. S7). All together, these results indicate that the presence of microbiome has a profound influence on the cellular mRNA m6A patterns in a tissue-dependent manner. Alteration of the m6A pattern is most pronounced in the brain, where the m6A methylome is substantially reduced in the presence of microbiome.
We performed in-depth analysis for the GF and SPF brain tissues to further elucidate the m6A alteration in the mRNA (a representative read coverage plot shown in Fig. 1g). Among the 8643 and 2750 m6A-containing transcripts, 67 and 80% had only one m6A peak in the GF and SPF brains, respectively, and the GF/SPF transcript ratio was 2.6 (Fig. 1h). However, this GF/SPF ratio for the transcripts containing two or more m6A peaks steadily increased to 4.4 for two, 6.7 for three, and 9.2 for more than three m6A peaks. GF brain transcripts also had a broader distribution of m6A peak/exon ratios (Fig. 1i). These results indicate that more m6A clusters are present in individual GF than SPF brain transcripts. The abundance of the m6A-containing transcripts was lower in GF than SPF brain (Fig. 1j), which might be associated with a major known role of m6A in accelerating mRNA decay.7 More m6A peaks were present in all three mRNA regions in GF than SPF brain (Fig. 1k). The m6A location in different mRNA regions has been associated with different functions. For example, m6A in the 5′ UTR enhances translation through eIF3-dependent recruitment of the ribosome;8 m6A in the 3′ UTR regulates mRNA stability and translation efficiency that depend on the m6A reader proteins YTHDF1, YTHDF2 and YTHDF3; m6A in the CDS regulates splicing that involves the m6A reader proteins YTHDC1, hnRNPC and hnRNPG,9,10 and codon-dependent translational efficiency.11 The large increase of the m6A peaks in the GF brain therefore could affect the m6A-dependent mRNA function in several different ways. This multifaceted m6A effects in the brain were consistent with the reactome analysis of biological pathways that showed the top enriched categories for m6A-containing transcripts, including mRNA splicing, cell cycle and signaling (Fig. 1l).
The GF brain may represent an under-developed state due to the lack of its microbiome exposure.12–14 To obtain insight into whether this idea applies to m6A in the transcriptome, we compared our results with the published m6A patterns from the mouse embryonic brain15 using only the high-confidence m6A-containing transcripts from both studies (Fig. 1m). The E13.5 embryonic brain (7701) had a comparable amount of m6A-containing transcripts to the 4-week-old GF brain (8,643); the overlap between them was 60% for embryonic brain and 54% for GF brain. The embryonic brain (7701) had a much higher amount of m6A-containing transcripts than the 4-week-old SPF brain (2750); the overlap between them was 20% for embryonic brain and 57% for SPF brain. Ninety-seven percent of all GF and SPF brain m6A-containing transcripts overlapped, which explained the similar overlapping percentage of GF and SPF with the embryonic transcripts. These results suggest that in regards to m6A modification, the GF brain more closely resembles the embryonic brain than the SPF brain of the same age.
To obtain mechanistic understanding of m6A changes in GF and SPF tissues, we measured the levels of the mRNA m6A writer proteins METTL3 and METTL14, and the m6A eraser proteins FTO and ALKBH5 by western blot. We found that both m6A writer proteins and both m6A eraser proteins were highly overexpressed in the GF brain compared to the SPF brain (Figs. 1n, o, and Supplementary information, Fig. S8). In contrast to brain, the differential expression of these proteins in the intestine and liver was much less noticeable without a uniform trend (Fig. 1p). These results correlate well with the finding that the brain has the largest difference in the m6A pattern among the three tissues examined here. Furthermore, the simultaneous overexpression of the m6A writer complex and the erasers in the same tissue should increase the ability to rapidly tune the m6A pattern upon environmental changes.
In summary, here we show that the microbiome has a strong effect on host m6A mRNA modification. Among the brain, intestine and liver tissues, the largest effect is present in the brain, which is associated with overexpression of both m6A writer and eraser proteins; this result suggests that the brain tissue may be more sensitive to adjust the m6A methylome in response to the microbiome than other tissues. Future studies will reveal the specific microbial species and the molecular mechanisms that regulate the host m6A methylome.
Supplementary information
Acknowledgements
We thank Dr. Hilary Morrison for assistance in 16S rRNA gene sequencing. We are grateful to Drs. Katya Frazier and Vanessa Leone from Dr. Eugene B. Chang’s laboratory for providing mouse tissues and insightful discussion. This work was supported by National Institutes of Health (K01 DK111764 to XYW, RM1 HG008935 to TP), DoD/CDMRP (BC160450 to TP), and the National Natural Science Foundation of China (31741033 and 91753129 to GZL).
Author contributions
X.Y.W. and T.P. designed and initiated the study. Y.L. and G.Z.L. analyzed sequencing data. X.Y.W. and W.J.C. performed experiments. H.L.S. helped prepare m6A sequencing libraries. A.M. performed 16S rRNA gene sequencing. A.M.E. analyzed 16S rRNA gene sequencing data. C.H., helped design the study. C.H. and A.M.E. helped interpret the data. X.Y.W., G.Z.L. and T.P. wrote the paper.
Competing interests
The authors declare no competing interests.
Contributor Information
Xiaoyun Wang, Email: xwang13@uchicago.edu.
Guan-Zheng Luo, Email: luogzh5@mail.sysu.edu.cn.
Tao Pan, Email: taopan@uchicago.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-018-0127-2.
References
- 1.Romano KA, et al. Cell. Host. Microbe. 2017;22:279–290. doi: 10.1016/j.chom.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roundtree IA, Evans ME, Pan T, He C. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Hsu PJ, Chen YS, Yang YG. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng X, et al. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selwyn FP, Cui JY, Klaassen CD. Drug Metab. Dispos. 2015;43:1572–1580. doi: 10.1124/dmd.115.063545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan WH, et al. Genome Med. 2018;10:27. doi: 10.1186/s13073-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, et al. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer KD, et al. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao W, et al. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Liu N, et al. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J, et al. Nat. Struct. Mol. Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz Heijtz R, et al. Proc. Natl. Acad. Sci. USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braniste V, et al. Sci. Transl. Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luczynski P, et al. Int. J. Neuropsychopharmacol. 2016;19:pyw020. doi: 10.1093/ijnp/pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon KJ, et al. Cell. 2017;171:877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.