Dear Editor,
Most disease-associated genomic mutations are base substitutions and approximately half of pathogenic human single nucleotide polymorphisms (SNPs) are related to C-to-T substitutions in the ClinVar database.1 Base editors (BEs), which combine Cas9-D10A nickase and APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) or AID (activation-induced deaminase) cytidine deaminase family members,2 have been successfully applied to mediate C-to-T conversion in vitro and in vivo,3 providing a powerful tool to model or repair disease-related human SNPs. Yet, the editing scope of BE3 was limited by the low editing efficiency at GpC dinucleotides and/or in regions with high CpG methylation levels.2 We recently replaced rA1 with human APOBEC3A (hA3A) and then engineered hA3A isoform to develop a series of hA3A-BEs, including hA3A-BE3-Y130F, which has an editing window similar to BE3.4 As hA3A can deaminate both C and methylated C in various sequence contexts efficiently,5 hA3A-BE3-Y130F mediated efficient C-to-T base editing in GpC context and CpG context in vitro.4
According to ClinVar database,1 about half of pathogenic C-to-T substitutions in the human genome are located in GpC or CpG context. Furthermore, dramatic change of DNA methylation at CpG sites occurs during development.6 Thus, hA3A-BE3 may be an efficient tool for in vivo modeling or correcting disease-related human SNPs in G/C-rich regions, particularly in GpC context. Therefore, we chose hA3A-BE3-Y130F to generate pathogenic SNPs in mouse embryos to model genetic diseases.
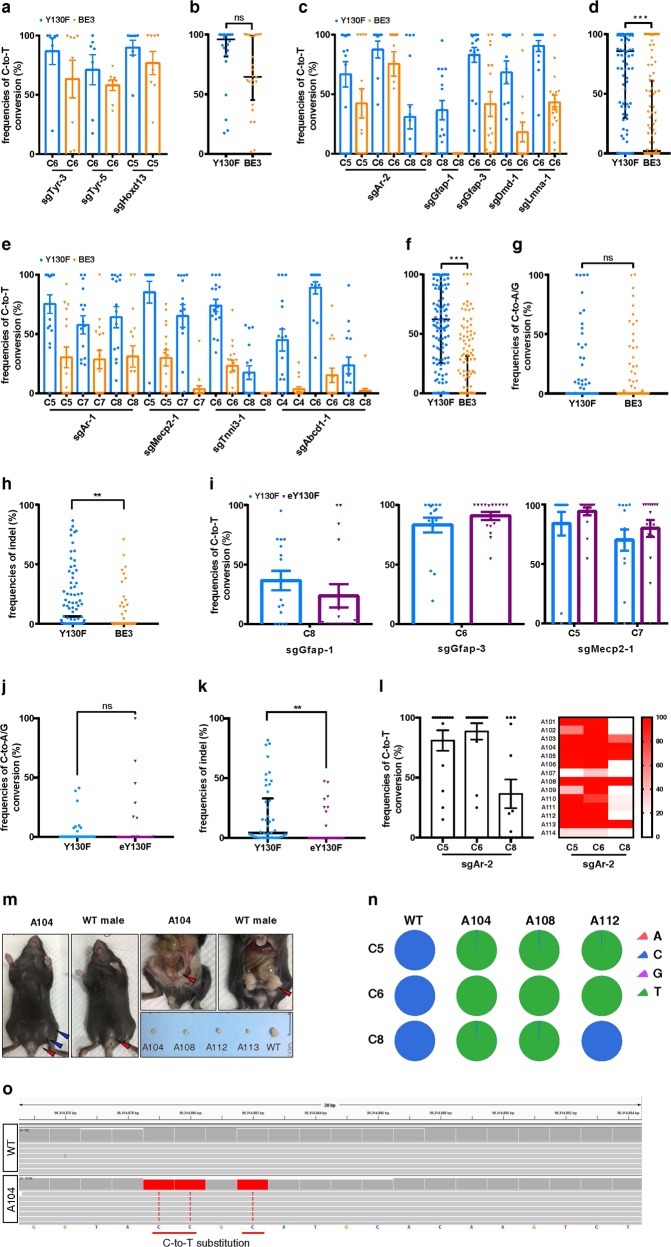
We first tested hA3A-BE3-Y130F in mouse embryos by targeting regular sites, where no GpC site is contained in editing window. Three sgRNAs, including two for Tyr and one for Hoxd13, were first tested in N2A cells (Supplementary information, Fig. S1a, b and Table S1). These sgRNAs may introduce mutations found in human patients (Supplementary information, Fig. S1c and Table S2). We then performed C-to-T editing in mouse embryos. The genomic DNA was edited at all tested pathogenic SNP sites and the editing efficiency of hA3A-BE3-Y130F was slightly but non-significantly higher than that of BE3 (median editing frequencies: ~88.89% vs. ~66.08%, Fig. 1a, b; Supplementary information, Fig. S2a, b).
Fig. 1.
hA3A-BE3-Y130F induces efficient C-to-T base editing in vivo. a, c, e Comparison of C-to-T editing efficiency induced by hA3A-BE3-Y130F and BE3 in the target regions having no GpC site (a), one GpC site (c), or two or three GpC sites (e) within the editing window in mouse embryos. Mean ± s.e.m. were from at least seven independent experiments. b, d, f Statistical analysis of the C-to-T editing frequency induced by hA3A-BE3-Y130F and BE3 in (a), (c), (e), respectively. n = 22 for hA3A-BE3-Y130F and n = 24 for BE3 (b), n = 79 for hA3A-BE3-Y130F and n = 80 for BE3 (d), and n = 62 for hA3A-BE3-Y130F and n = 64 for BE3 (f). g, h Statistical analysis of non-C-to-T base conversions (g) and indels (h) induced by hA3A-BE3-Y130F and BE3 in all embryos. n = 163 for hA3A-BE3-Y130F and n = 168 for BE3. i Comparison of C-to-T editing efficiency induced by hA3A-BE3-Y130F and hA3A-eBE-Y130F in the target regions having GpC dinucleotides in mouse embryos. Mean ± s.e.m. were from at least twelve independent experiments. j, k Statistical analysis of non-C-to-T base conversions (j) and indels (k) induced by hA3A-BE3-Y130F and hA3A-eBE-Y130F in mouse embryos. n = 46 for hA3A-BE3-Y130F, n = 44 for hA3A-eBE-Y130F. l C-to-T editing efficiency within the editing window induced by hA3A-BE3-Y130F and sgAr-2 in founder mice is displayed in column graph (mean ± s.e.m. were from fourteen founder mice) and heatmap. m Sexual reversal was observed in AIS-modeling founder mice. Left: A 4-week-old mouse (Founder A104) with female genitalia (red arrowhead) and nipples (blue arrowheads) and WT male with normal genitalia (red arrowhead); Right upper: Founder A104 with internal genitalia of male (red arrowhead) and smaller testis, and WT male with normal genitalia of male (red arrowhead); Right lower: Genitalia of Founders A104, A108, A112, A113, and WT male. n Base-editing product purities in Founder A104, A108, and A112 were determined by deep sequencing. o Confirmation of the on-target base editing by analyzing the whole-genome sequencing results of A104. b, d, f–h, j, k The median and interquartile range (IQR) are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ns not significant, Student’s two-tailed t-test
Next, we used hA3A-BE3-Y130F to introduce pathogenic SNPs in GpC context. Another 10 sgRNAs targeting eight genes were tested in N2A cells (Supplementary information, Figs. S3, S5). There were one GpC site within editing window for sgAr-2, sgGfap-1, sgGfap-3, sgDmd-1, and sgLmna-1; two for sgAr-1, sgMecp2-1, sgTnni3-1; three for sgRor2-1, sgAbcd1-1. Nine of these sgRNAs were then used in embryos and all of them are expected to introduce mutations seen in patients (Supplementary information, Figs. S3, S5 and Table S2). Notably, hA3A-BE3-Y130F induced significantly higher editing efficiencies than BE3 (median editing frequencies: ~65.65% vs. ~30.62% for the sgRNAs with one GpC site within the editing window, P < 0.001; ~59.25% vs. ~16.71% for the sgRNAs with two or three GpC sites within the editing window, P < 0.001) (Fig. 1c–f; Supplementary information, Figs. S4, S6).
By analyzing the deep-sequencing data of all 331 edited embryos (163 for hA3A-BE3-Y130F and 168 for BE3), we also determined the non-C-to-T conversions (i.e., C-to-A/G) and insertions/deletions (indels). We found that both hA3A-BE3-Y130F and BE3 induced non-C-to-T base conversions (4.62% vs. 4.15%), and indels (9.72% vs. 4.83%) (Fig. 1g, h; Supplementary information, Tables S3, S4). Interestingly, no indel or non-C-to-T conversion was detected in a large part of embryos (Supplementary information, Tables S3, S4), showing that the yield of side-products varied among embryos. Further, although both BE3 and hA3A-BE3-Y130F induced some side-products, they can be reduced by additional uracil-DNA glycosylase inhibitor (UGI).7,8 We have fused three copies of the 2 A (self-cleaving peptide)-UGI sequence to the C-terminus of hA3A-BE3-Y130F to develop hA3A-eBE-Y130F in a recent study.4 To test this construct in embryos, the mRNA encoding hA3A-eBE-Y130F together with three selected sgRNAs, i.e., sgGfap-1, sgGfap-3, and sgMecp2-1, were microinjected into embryos. A total of 44 blastocysts (16 for sgGfap-1, 16 for sgGfap-3, and 12 for sgMecp2-1) were collected and tested. The results showed that the additional free UGI did not affect the editing efficiency (median editing frequencies: ~48.03% vs. ~46.52% for hA3A-eBE-Y130F and hA3A-BE3-Y130F, respectively) (Fig. 1i; Supplementary information, Fig. S7). Further analysis showed that hA3A-eBE-Y130F induced similar non-C-to-T conversions to hA3A-BE3-Y130F (~4.89% vs. ~3.75%; Fig. 1j), but much less indels as expected (~5.38% vs. ~18.54%; Fig. 1k). Thus, additional free UGI produced by hA3A-eBE-Y130F ensures much purer editing products in individual embryos.
To generate a mouse model of androgen insensitivity syndrome (AIS), the mRNA encoding hA3A-BE3-Y130F together with sgAr-2 were injected into zygotes, making pathogenic mutations in GpC context. Forty embryos were transplanted into two surrogate mothers and fourteen offsprings were obtained (Supplementary information, Fig. S8). Notably, single C-to-T substitution at position 6 or simultaneous C-to-T substitutions at positions of both 5 and 8 in the editing window can create a R754C mutation. The Sanger sequencing results showed that all pups were edited at C6 position at an average editing frequency of 89%, whereas at two synonymous mutation sites C5 and C8, the average editing frequencies were 81% and 36%, respectively (Fig. 1l; Supplementary information, Fig. S8c). Eleven founders (A101, A102, A103, A104, A105, A106, A108, A109, A111, A112, and A113) harbored 100% editing at C6 (Supplementary information, Fig. S8a). Among these founders, A104, A108, and A113 contained homogenous mutations at positions 5, 6, and 8, and A101 and A112 at positions 5 and 6 (Supplementary information, Fig. S8a). Also, non-C-to-T mutations at positions 5 and 8 were observed at frequencies of 15%, 5% and 20% in only three of all fourteen founders (A102, A106, and A107, Supplementary information, Fig. S8a, g), leading to unwanted amino acid change at Y753. In addition, indels were observed in only two of fourteen founders (A107 and A114, Supplementary information, Fig. S8a, g).
Interestingly, male genital was absent in all founder mice (Supplementary information, Fig. S8e). However, five pups (Founder A104, A108, A110, A112, and A113) were Sry positive (Supplementary information, Fig. S8d), suggesting that these five mutant mice recapitulated the AIS-like sex reversal phenotype. Indeed, upon autopsy, these mice displayed smaller testis compared to the wild type (WT; Fig. 1m). Genotyping using the genomic DNA of four organs (heart, kidney, intestine, and testis) from the four autopsied founders (A104, A108, A112, and A113) and the genomic DNA of the tails from WT mice confirmed that the four autopsied founders all contained AIS mutations (Supplementary information, Fig. S8b).
Since hA3A-BE3-Y130F induced non-C-to-T base conversions and indels in some embryos, we wondered whether these side-products were propagated to adults. Thus, we deep-sequenced the target sites in Founders A104, A108, and A112, and found that the on-target C-to-T editing was almost all 100% (Fig. 1n). Importantly, no non-C-to-T base conversion or indel was detected (Fig. 1n; Supplementary information, Fig. S8f and Tables S4, S5), indicating that the high editing efficiency of hA3A-BE3-Y130F can lead to pure editing products in a part of mice.
To comprehensively analyze base-editing specificity, we performed whole-genome sequencing (WGS) for Ar mutant mice A104. A total of 4,830,040 and 4,014,959 SNPs were detected in the genomes of WT and A104, respectively (Supplementary information, Fig. S9a). After filtering out dbSNPs (naturally occurring variants in the SNP database) and unintended base substitutions (C-to-G/A and G-to-T/C), we examined whether the remaining SNPs located at the on- or potential off-target sites. Among 3040 predicted off-target sites, no C-to-T base substitution was uniquely found in the A104 genome (Supplementary information, Fig. S9b). In contrast, the on-target C-to-T base editing was detected in A104 but not in WT genome (Supplementary information, Fig. S9b; Fig. 1o). These results indicated that hA3A-BE3-Y130F can mediate base editing with high specificity in vivo.
In summary, we demonstrated that both hA3A-BE3-Y130F and hA3A-eBE-Y130F can induce efficient and precise in vivo base editing, particularly at the pathogenic SNPs in GpC context, which can be employed to generate disease models. Thus, these two base editors expand the scope of current in vivo base editing system.
Supplementary information
Acknowledgements
We thank members of Huang lab, Sun lab, and Chen lab for helpful discussions. We are grateful to Dr. Tian Chi for excellent manuscript editing. This work is supported by the National Key R&D Program (2016YFA0500903 and 2016YFC0905901), National Postdoctoral Program for Innovative Talents (BX201700266), Local Grants (16JC1420200 and 17JC1420103) and the Leading Talents of Guangdong Province (2016LJ06S386).
Author contributions
X.H., Q.S., and J.C. conceived, designed, and supervised the project. J.L., Z.L., and S.H. performed most experiments with the help of X.W., G.L., Y.X., W.Y., and Y.Z.; S.H. performed the deep sequencing data analyses and bioinformatics analysis. Z.K. and H.M. provided expert technical assistance. J.L., S.H., J.C., and X.H. wrote the paper with inputs from all authors. X.H., Q.S., and J.C. managed the project.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jianan Li, Zhen Liu, Shisheng Huang
Contributor Information
Jia Chen, Email: chenjia@shanghaitech.edu.cn.
Qiang Sun, Email: qsun@ion.ac.cn.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0133-4.
References
- 1.Landrum MJ, et al. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, et al. Nat. Commun. 2018;9:2338. doi: 10.1038/s41467-018-04768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. Nat. Biotechnol. 2018;36:946–949. doi: 10.1038/nbt.4198. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter MA, et al. J. Biol. Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, et al. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, et al. Cell Res. 2017;27:1289–1292. doi: 10.1038/cr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komor AC, et al. Sci. Adv. 2017;3:1–10. doi: 10.1126/sciadv.aao4774. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.