High-risk HPV infections lead to development of cervical cancer. This study identified the differential expression of 16 novel genes (LY6K, FAM83A, CELSR3, ASF1B, IQGAP3, SEMA3F, CLDN10, MSX1, CXCL5, ASRGL1, ELAVL2, GRB7, KHSRP, NOVA1, PTBP1, and RNASEH2A) in HPV-infected cervical tissue samples and keratinocytes. Eight of these genes (CDKN2A, ELAVL2, GRB7, HSPB1, KHSRP, NOVA1, PTBP1, and RNASEH2A) encode RNA-binding proteins. Further studies indicated that both HPV16 and HPV18 infections lead to the aberrant expression of selected RBP-encoding genes. We found that viral E6 and E7 decrease NOVA1 expression but that E7 increases RNASEH2A expression via E2F1. The altered expression of these genes may be utilized as biomarkers for high-risk (HR)-HPV carcinogenesis and progression.
KEYWORDS: CIN, cervical cancer, human papillomaviruses, RNA binding proteins, RNA-seq, RNASEH2A, viral oncoproteins
ABSTRACT
RNA-binding proteins (RBPs) control mRNA processing, stability, transport, editing, and translation. We recently conducted transcriptome analyses comparing normal (i.e., healthy) cervical tissue samples with human papillomavirus (HPV)-positive cervical cancer tissue samples and identified 614 differentially expressed protein-coding transcripts which are enriched in cancer-related pathways and consist of 95 known RBPs. We verified the altered expression of 26 genes with a cohort of 72 cervical samples, including 24 normal cervical samples, 25 cervical intraepithelial neoplasia grade 2 (CIN2) and CIN3 samples, and 23 cervical cancer tissue samples. LY6K (lymphocyte antigen 6 complex locus K), FAM83A (family member with sequence similarity 83), CELSR3, ASF1B, IQGAP3, SEMA3F, CLDN10, MSX1, CXCL5, ASRGL1, ELAVL2, GRB7, KHSRP, NOVA1, PTBP1, and RNASEH2A were identified as novel candidate genes associated with cervical lesion progression and carcinogenesis. HPV16 or HPV18 infection was found to alter the expression of 8 RBP genes (CDKN2A, ELAVL2, GRB7, HSPB1, KHSRP, NOVA1, PTBP1, and RNASEH2A) in human vaginal and foreskin keratinocytes. Both viral E6 and E7 decreased NOVA1 expression, but only E7 increased the expression of RNASEH2A in an E2F1-dependent manner. Proliferating cell nuclear antigen (PCNA) directs RNASEH2 activity with respect to DNA replication by removing the RNA primers to promote Okazaki fragment maturation, and two factors are closely associated with neoplasia progression. Therefore, we predict that the induction of expression of RNASEH2A via viral E7 and E2F1 may promote DNA replication and cancer cell proliferation.
INTRODUCTION
Cervical cancer is the second most common cancer among women worldwide (1). The implementation of tumor screening programs in developed countries over the past decades has dramatically reduced cervical cancer morbidity and mortality. Nevertheless, in resource-poor countries where cervical screening is not generally available or is of low quality and low coverage, cervical cancer remains a major malignancy among women (2). Approximately 527,600 incident cases of cervical cancer and 265,700 cervical cancer deaths are estimated to occur each year, with more than 80% of the cases arising in developing countries.
Persistent infection with oncogenic human papillomaviruses (HPVs) is known to be the causal factor for cervical cancer and its precursor lesions. HPVs also cause a subset of head and neck squamous cell carcinoma (HNSC) or oropharyngeal cancers (3). Fifteen mucosal HPV types are identified as oncogenic or high-risk (HR) HPVs, with HPV16 and HPV18 being the most prevalent HPVs in invasive cervical cancer (4). While HPV infection is very common, cervical cancer is comparatively rare and arises only in women in whom viral infection persists. Even among women with persistent, oncogenic HPV infection, only a fraction develop cervical cancer, and HPV detection by itself is not sufficient to identify those cases. Thus, there is a need for diagnostic markers that can be used for early diagnosis of persistent high-risk HPV infection and of HPV-associated precancer and cancer and for the development of intervention strategies for treatment of HPV-induced cancers.
Given the importance of transcriptional mechanisms during the development of cancer, it is generally assumed that the primary result of the presence of extracellular (extrinsic) signals, including virus infection, is alteration of gene transcription. However, an equally important step in the regulation of gene expression is posttranscriptional regulation of mRNAs. There are two major mechanisms. First, microRNAs (miRNAs), which predominantly bind to sequences in the 3′ untranslated regions (UTR) of mRNAs, influence mRNA stability and translation (5). We have previously characterized a number of miRNAs that were specifically dysregulated by oncogenic HPVs in cervical cancers and high-grade lesions as well in organotypic cultures derived from cervical or foreskin epithelial (E) cells harboring HPV16 and HPV18 genomes or HPV18 oncogenes (6–9). Second, RNA-binding proteins (RBPs) can control mRNA processing, stability, transport, editing, and translation (10, 11). The recent development of large-scale quantitative methods, especially next-generation sequencing and various proteomics technologies, facilitates genome-wide identification of RBPs (12–16). In humans, many diseases, including cancers, have been linked to defects or dysregulation in RBP expression or functions (17–19). However, despite the growing amount of data collected on RBPs (10, 11, 16), the expression atlas of RBPs in cervical cancer remains to be compiled. In this study, we investigated the differential gene expression levels in HPV-containing tissue samples relative to normal (i.e., healthy) tissue samples, with a particular emphasis on RBPs.
High-throughput RNA sequencing (RNA-seq) is an emerging technology that leverages the capabilities of next-generation sequencing to detect and quantify the entire transcriptome with a large number of “sequence reads” (20, 21). This approach presents unprecedented power for improving existing genome annotations; has significantly advanced our understanding of genomic organization, including novel transcripts, isoforms, and allele-specific expression (22); and could also be of profound value in terms of disease characterization. Transcriptomic profiling using RNA sequencing has previously been applied to cervical carcinogenesis (23, 24). However, the early studies were not comprehensive and were not necessarily validated for high-risk HPV infection. Here, we applied two different platforms of next-generation RNA-seq technologies to analyze the transcriptomes from normal cervical tissue samples and cervical cancer tissue samples. We have identified altered expression of a large set of host genes, including a number of RBP coding genes, attributable to HPV infections and cervical lesion progression. The altered expression of these genes could serve as potential biomarker for cervical cancer diagnosis, assessment, and therapy.
RESULTS
Transcriptomic signatures distinguish cancerous cervix cells from normal cervix cells.
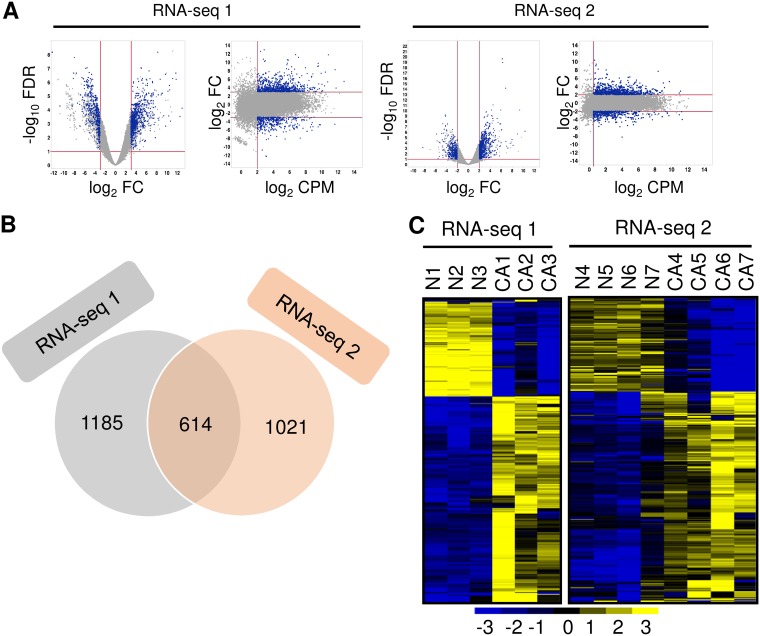
To investigate the transcriptomic signatures of normal and cancerous cervix cells and to avoid the variations from one technology to another, we conducted two RNA-seq analyses, one using poly(A)-positive [poly(A)+] RNA for library preparation by directional ligation sequencing (DeLi-Seq) (RNA-seq 1) and one using rRNA− total RNA for library preparation by TrueSeq stranded RNA sequencing (RNA-seq 2), for 14 different clinical cervical tissue samples, which consisted of 3 each of normal and cancerous cervical tissue samples for RNA-seq 1 and 4 each from a separate set of normal and cancerous cervical tissue samples for RNA-seq 2 (see Fig. S1A in the supplemental material). None of the normal cervical tissue samples had HPV infection, whereas all the cervical cancer tissue samples had HR-HPV infection, as determined by the use of Digene HPV genotyping kits (see Table S1 in the supplemental material). For RNA-seq 1, we obtained a total of 102 million high-quality raw reads from 6 samples. In the case of RNA-seq 2, we obtained a total of 633 million high-quality raw reads from 8 samples (Table S1). After processing RNA-seq raw data (NCBI GSE113942) and statistical analysis (see details in Materials and Methods), we obtained approximately ∼12 million and ∼34 million uniquely mapped reads per sample for RNA-seq 1 and RNA-seq 2 (Table S1), respectively. By applying stringent criteria (Fig. 1A; see also Fig. S1A), we demonstrated the presence of transcripts that were differentially expressed between normal control and cervical cancer tissue samples and identified 1,799 differentially expressed transcripts for RNA-seq 1 data (Fig. 1B; see also Table S2) and 1,635 for RNA-seq 2 data from cervical cancer tissue samples relative to normal cervix samples, respectively (Fig. 1B; see also Table S3). Among them, 614 transcripts were differentially expressed in both data sets (Fig. 1B; see also Table S4), with 416 being upregulated and 198 being downregulated in cervical cancer tissue samples (Fig. 1C).
FIG 1.
The transcriptomic signatures of normal cervix versus cervical cancer. (A) Smear plot visualization of differential expression analyses of cancer versus normal transcriptomes in RNA-seq 1 and RNA-seq 2. Transcripts used for the Venn diagram presented in panel B are highlighted in blue. FC, fold change; FDR, false-discovery rate; CPM, counts per million. (B) Venn diagram depicting the overlapped gene transcripts differentially expressed between RNA-seq 1 and RNA-seq 2. (C) Heat maps showing 614 overlapped, differentially expressed gene transcripts with normalized expression values crossing all normal cervical tissue samples and cervical cancer tissue samples. Left, RNA-seq 1; right, RNA-seq 2.
Comparison of transcriptomic signatures of normal cervix and cervical cancer. (A) Schematic illustration of the strategies used to discover and define bona fide differentially expressed gene transcripts in cervical cancer. (B) Identification of the altered expression of RNA-binding protein genes in cervical cancer. A schematic illustration of the strategies used to find differentially expressed RBP genes in cervical cancer is shown. Download FIG S1, PDF file, 0.2 MB (164.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Total RNA reads and mapped reads in each normal cervix and cervical cancer sample. Download Table S1, XLS file, 0.02 MB (25.5KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 1,799 gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 1. Download Table S2, XLS file, 0.5 MB (552KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 1,635 gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 2. Download Table S3, XLS file, 0.5 MB (524KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 614 overlapped gene transcripts differentially expressed in both RNA-seq 1 and RNA-seq 2. Download Table S4, XLSX file, 0.2 MB (163.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Core pathways influenced by the deregulated transcripts in cervical cancer.
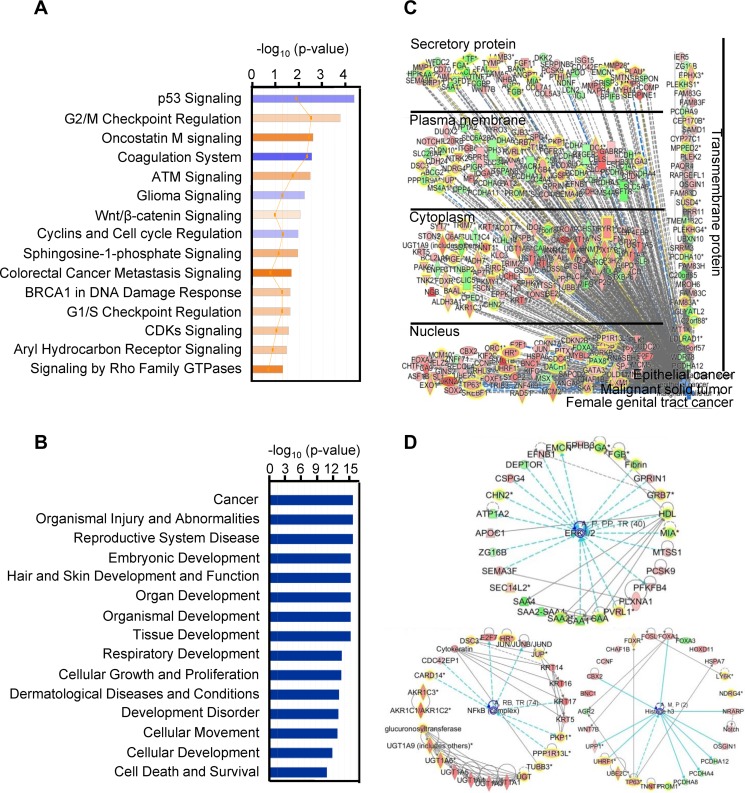
To determine the biological relevance of the 614 differentially expressed gene transcripts obtained from the cancer versus normal tissue samples from the two RNA-seq data sets described above, we performed Ingenuity pathway analysis (IPA) and identified several pathways enriched in cancer tissue samples over normal tissue samples (Fig. 2A), including p53 signaling and cell cycle pathways, targeted by viral oncoproteins E6 and E7, the basis of HPV oncogenesis. Disease and function analysis demonstrated that the cancer-related pathway was the pathway that was mainly affected (Fig. 2B). As a consequence of focusing on pathways related to malignant solid tumor, epithelial cancer, and female genital tract cancer, the combined results showed that the cancer signaling-associated pathway transcripts were also found in cervical cancers. The altered gene transcripts in cervical cancer can also be grouped by their protein distributions in the compartment of the infected cells (Fig. 2C). The network analysis showed that the top three dysregulated networks in cervical cancer are mainly associated with the extracellular signal-regulated kinase 1 (ERK1) and 2 (ERK1/2), histone H3, and NFкB networks (Fig. 2D).
FIG 2.
IPA enrichment analysis of the 614 gene transcripts with differential changes in cervical cancer. (A) Canonical pathway analysis, showing the 15 most significant activated (orange) and inhibited (blue) pathways in cervical cancer. (B) Disease and function analysis of the identified 614 gene transcripts in cervical cancer, showing the 15 main influenced diseases and functions. (C) Combined analysis performed with the terms “malignant solid tumor,” “epithelial cancer,” and “female genital tract cancer,” showing the influenced gene transcripts in cervical cancer as grouped by their protein distributions in the infected cells. Red, upregulated; green, downregulated; orange, predicted activation; blue, predicted inhibition; gray, effect not predicted. (D) Top three networks of interest (ERK1/2, histone H3, and NFкB) were generated. Red, higher expression in cervical cancer; green, lower expression in cancer tissue samples. The solid and dashed lines in panels C and D represent direct and indirect relationships, respectively. An asterisk after a gene symbol indicates multiple transcript IDs for the same gene.
Validation of the differentially expressed genes in cervical tissue samples.
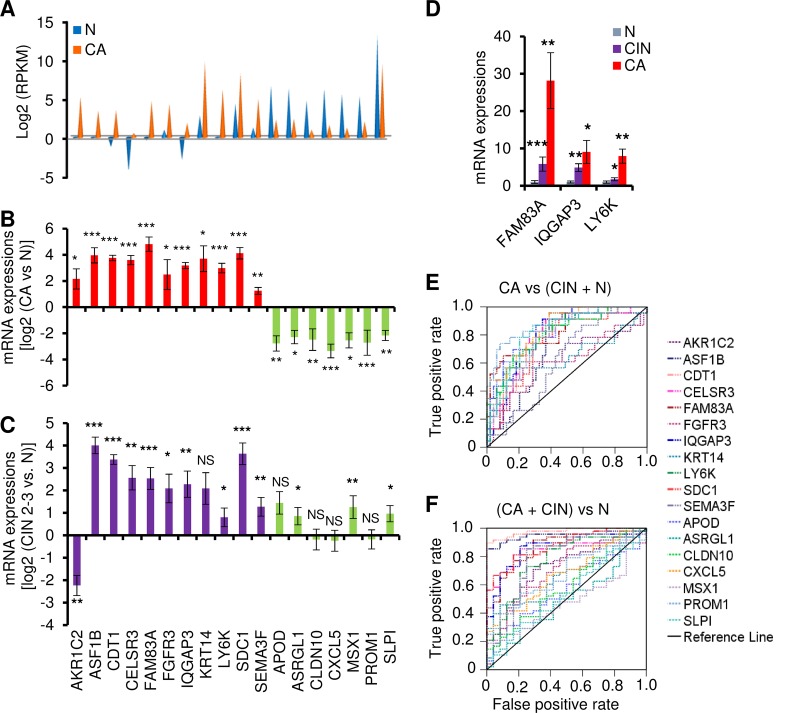
To verify the differentially expressed genes identified by RNA-seq, we examined 72 additional human cervical tissue samples, including 24 normal cervical tissue samples, 23 cervical cancer tissue samples, and 25 precancer lesions (cervical intraepithelial neoplasia grade 2 [CIN2] to CIN3) for 18 genes selected from the genes corresponding to the 614 transcripts that were expressed differentially from cervical cancer tissue samples relative to normal cervical tissue samples (Fig. 3A). We chose these 18 genes on the basis of the main pathways and top network analyses, as well as of the function of these genes. Among these 18 genes, 10 (LY6K, FAM83A, CELSR3, ASF1B, IQGAP3, SEMA3F, CLDN10, MSX1, CXCL5, and ASRGL1) have not been reported in association with cervical cancer. The remaining eight genes (KRT14, AKR1C2, CDT1, SDC1, FGFR3, APOD, SLPI, and PROM1) have been implicated in the development of cervical cancer or HPV infection and were therefore chosen as positive controls. TaqMan reverse transcription-quantitative PCR (RT-qPCR) validated all 18 genes for differential expression (P < 0.05) in normal cervix cells relative to cervical cancer cells as determined by RNA-seq (Fig. 3B). In addition, 13 of 18 genes in 25 precancer lesions (CIN2 to CIN3) also showed an expression pattern similar to that seen with cancer tissue samples (Fig. 3C), except AKR1C2. Interestingly, we observed that the increased expression of FAM83A, IQGAP3, and LY6K was closely associated with cervical lesion progression (Fig. 3D).
FIG 3.
Experimental validation of selected 18 genes in cervical tissue samples. (A) Distribution of expression levels of 18 selected genes from the overlapped 614 gene transcripts, being measured by RNA-seq with RPKM. (B and C) TaqMan RT-qPCR validation of the 18 genes in 72 clinical cervical tissue samples, including 24 normal (N) cervix, 23 cervical cancer (CA), and 25 precancerous (CIN2 to CIN3) tissue samples. Experimental validation data of selective gene expression levels in human cervical samples are shown as mean ± SD log2 levels of cancer group versus normal group (B) or CIN2 to CIN3 group versus normal group (C). NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) The expression of FAM83A, IQGAP3, and LY6K mRNAs was significantly increased along with the cervical lesion progression from CIN2 to CIN3 to cervical cancer compared to the normal cervical tissue samples. (E and F) Receiver operating characteristic (ROC) curves of the 18 gene transcripts, showing good discrimination in identifying cancer group versus CIN2-CIN3 plus normal group (E) or cancer group plus CIN2-CIN3 versus normal group (F). AKR1C2, aldo-keto reductase family 1 member C2; APOD, apolipoprotein D; ASF1B, anti-silencing function 1B histone chaperone; ASRGL1, asparaginase like 1; CDKN2A, cyclin-dependent kinase inhibitor 2A; CDT1, chromatin licensing and DNA replication factor 1; CELSR3, cadherin epidermal growth factor LAG seven-pass G-type receptor 3; CLDN10, claudin 10; CXCL5, C-X-C motif chemokine ligand 5; ELAVL2, ELAV-like RNA binding protein 2; FAM83A, family member with sequence similarity 83; FGFR3, fibroblast growth factor receptor 3; GRB7, growth factor receptor-bound protein 7; HSPB1, heat shock protein family B (small) member 1; IQGAP3, IQ motif containing GTPase-activating protein 3; KHSRP, KH-type splicing regulatory protein; KRT14, keratin 14; LY6K, lymphocyte antigen 6 family member K; MSX1, Msh homeobox 1; NOVA1, NOVA alternative splicing regulator 1; PROM1, prominin 1; PTBP1, polypyrimidine tract binding protein 1; RNASEH2A, RNase H2 subunit A; SDC1, syndecan 1; SEMA3F, semaphorin 3F; SLPI, secretory leukocyte peptidase inhibitor syndecan 1.
Among the 18 genes, we compared the levels of sensitivity and specificity using receiver operating characteristic (ROC) analysis in order to determine their diagnostic value in identifying precursor lesions or cervical cancer. The TaqMan RT-qPCR results were plotted and analyzed by ROC analysis according to comparisons of normal plus CIN2 to CIN3 versus cervical cancer samples or normal versus CIN2 to CIN3 plus cervical cancer samples. As shown in Fig. 3 and Table S5, by using an AUC (area under the ROC curve) value of >0.70 as a cutoff, we identified 14 genes (LY6K, FAM83A, CELSR3, ASF1B, IQGAP3, CLDN10, MSX1, CDT1, APOD, SLPI, PROM1, SDC1, ASRGL1, and CXCL5) as being specific for the cervical cancer group over the CIN2 to CIN3 plus normal group (Fig. 3E) and 9 genes (LY6K, FAM83A, CELSR3, ASF1B, IQGAP3, CDT1, SEMA3F, SDC1, and FGFR3) as being specific for the cervical cancer plus CIN2 to CIN3 group over the normal group (Fig. 3F). Thus, data in this grouping analysis suggest that LY6K, FAM83A, CELSR3, ASF1B, IQGAP3, CDT1, and SDC1 are more likely specific biomarkers for HPV infection-induced CIN2 to CIN3 and beyond, whereas the differential expression levels of CLDN10, MSX1, SEMA3F, APOD, SLPI, PROM1, ASRGL1, FGFR3, and CXCL5 are presumably associated with cervical lesion progression.
Predicted host genes with validated differential expression might be useful biomarkers for diagnosis of cervical lesion progression. Download Table S5, XLSX file, 0.01 MB (12.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of altered expression of RNA-binding protein (RBP) genes in cervical cancer.
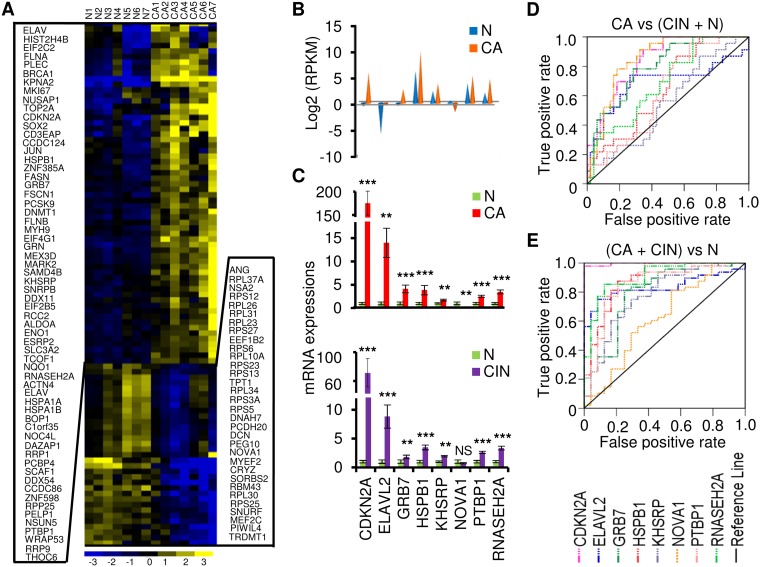
We next sought to characterize RBP genes that showed significant expression differences between normal cervix and cervical cancer samples. To apply more-stringent criteria, seven normal tissue samples and seven cancer tissue samples were combined. In this analysis, the significance of the data from the differentially expressed genes is likely independent of the methods used for library preparation and of the RNA-seq platforms. By following the strategies shown in Fig. S1B and using 2-fold change (FC) as a threshold, we identified 3,610 overlapped differentially expressed transcripts from the two RNA-seq data sets (Table S6). By annotating them against 1,502 known RBPs (11, 14, 15, 25), we found 205 transcripts from 95 RBP genes (Fig. 4A; see also Table S7). Eight RBP genes (CDKN2A, ELAVL2, GRB7, HSPB1, KHSRP, NOVA1, PTBP1, and RNASEH2A) not validated in Fig. 3B were chosen based on their function and expression abundance for further validation with the same 72 clinical cervical samples (24 normal cervical tissue samples, 25 CIN2 to CIN3 tissue samples, and 23 cervical cancer tissue samples) as described above. ELAVL2, GRB7, KHSRP, NOVA1, PTBP1, and RNASEH2A are well-studied RBPs, while CDKN2A and HSPB1 were recently identified as RBPs by the Markus Landthaler group and Matthias Hentze group, respectively (14, 15, 25). Among them, ELAVL2, KHSRP, NOVA1, PTBP1, and RNASEH2A have not been reported in association with cervical cancer. Dysregulation of the remaining RBPs (CDKN2A, GRB7, and HSPB1) has been extensively implicated in the development of cervical cancer. The 8 RBPs are summarized in Fig. 4B, with data showing differential expression levels in cervical cancer tissue samples relative to normal tissue samples as determined by RNA-Seq. TaqMan RT-qPCR confirmed that the expression levels of 7 RBP genes were significantly increased and that NOVA1 was the only gene whose expression level was significantly decreased in 23 cervical cancer tissue samples over the 24 normal cervical tissue samples (Fig. 4C). The seven RBP genes with increased expression in cervical cancer also exhibited elevated expression in 25 CIN2 to CIN3 tissue samples compared to normal tissue samples (Fig. 4C), indicating that altered RBP expression appears at the early stages of viral carcinogenesis in the cervix. Analyses of RBP pathways and networks showed that these RBPs are mainly involved in top 7 cellular functions, including KAN response to arsenic trioxide, mitogen-activated protein (MAP) signaling, Whiteford pediatric cancer markers, Burton adipogenesis 3, cervical cancer proliferation, cytoskeleton organization and biogenesis, and cytoskeletal protein binding, among which more than 50% of the gene counts in each pathway are the RBP genes identified in this report (Table S8). To confirm the contribution of the eight RBP genes to cervical carcinogenesis, we applied ROC analysis with our TaqMan RT-qPCR data to assess the potential utility of the gene expression of each validated RBP in early diagnosis of cervical lesions. As shown in Fig. 4 and Table S5, by using an AUC value of >0.70 as a cutoff, we demonstrated that the altered expression levels of CDKN2A, GRB7, and NOVA1 were specific for the cervical cancer group over the normal plus CIN2 to CIN3 group (Fig. 4D). However, compared with the normal cervical cell group, the CIN2 to CIN3 plus cervical cancer group displayed altered expression levels of all 7 of the 8 RBPs, except NOVA1 (Fig. 4E; see also Table S5), indicating that all 7 RBP genes with altered expression could be specific biomarkers for CIN2 to CIN3 and beyond.
FIG 4.
Identification and validation of the altered expression of RNA-binding protein genes in cervical cancer. (A) Heat map of the 95 differentially expressed RBP genes with normalized expression values across 7 normal cervical tissue samples and 7 cervical cancer tissue samples. (B) RNA-seq expression values (in RPKM) for selected 8 RBP genes. (C) TaqMan RT-qPCR validation of 8 RBP genes in 24 normal cervix samples, 25 cervical precursor lesion samples (CIN2 to CIN3), and 23 cervical cancer tissue samples. Data are shown as means (2−ΔΔCt) ± SD. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D and E) ROC curves of 8 RBP gene expression detection, showing good discrimination of cancer group from CIN2-CIN3 plus normal group (D) and cancer plus CIN2-CIN3 group from normal group (E).
Identification of 3,610 overlapped gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 1 and RNA-seq 2. Download Table S6, XLS file, 1.4 MB (1.5MB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 205 gene transcripts from 95 RBP genes differentially expressed in RNA-seq 1 and RNA-seq 2. Download Table S7, XLSX file, 0.1 MB (58.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicted RBP pathways involved in cervical carcinogenesis. Download Table S8, XLSX file, 0.03 MB (34.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNASEH2A and NOVA1 are HR-HPV-regulated RBP genes in raft culture tissue samples.
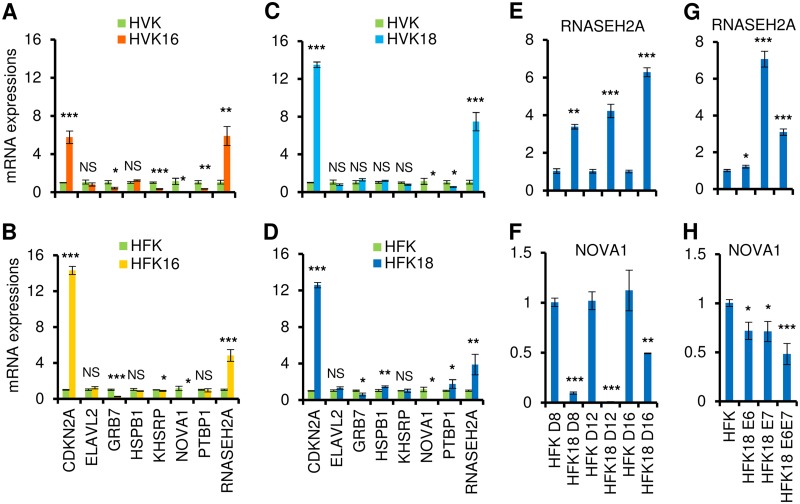
To examine whether HR-HPV infection influences the altered expression of these 8 RBP genes, raft cultures of human vaginal keratinocytes (HVK) with or without HPV16 infection were studied. As shown in Fig. 5A, HPV16 infection led to increased expression of CDKN2A and RNASEH2A and to decreased expression of GRB7, KHSRP, NOVA1, and PTBP1. To minimize the potential experimental variances due to cell types, we expanded the procedure to include raft cultures of human foreskin keratinocytes (HFK) with or without HPV16 infection. Although the expression of PTBP1 in HPV16-infected HFK was not consistent with its expression in HPV16-infected HVK raft cultures, all other genes in both types of the cells showed similar levels of upregulation or downregulation in response to HPV16 infection (Fig. 5B).
FIG 5.
HPV16 or HPV18 infection and viral E6 or E7 affect the expression of RBPs in raft cultures. Total tissue RNA from each raft culture was extracted and used for the study. (A to D) Expression of 8 RBP genes in 10-day-old raft tissue samples derived from human vaginal keratinocytes (HVK) (A [HVK and HVK16] and C [HVK and HVK18]) or human foreskin keratinocytes (HFK) (B [HFK and HFK16] and D [HFK and HFK18]) was validated by TaqMan RT-qPCR and compared to expression of raft cultures of HVK or HFK cell lines containing HPV16 (A and B) or HPV18 (C and D). (E to H) Validation by TaqMan RT-qPCR of RNASEH2A and NOVA1 expression in primary HFK-derived raft cultures with or without productive HPV18 infection or HPV18 oncogene expression. HPV18 genomes were excised from recombinant plasmid in primary HFK by Cre-LoxP recombination. (E and F) Raft cultures with or without productive HPV18 infection were prepared within a week of DNA transfection and followed by TaqMan RT-qPCR detection of the expression of RNASEH2A (E) and NOVA1 (F) in primary HFK-derived raft tissue samples on day 8 (D8), day 12 (D12), and day 16 (D16). (G and H) TaqMan RT-qPCR detection of the expression of RNASEH2A (G) and NOVA1 (H) in day 11 raft cultures derived from primary HFK acutely transduced with a retrovirus expressing HPV18 E6, E7, or E6E7 or with an empty control retrovirus. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. The data in all panels represent means ± SD determined in duplicate from three independent raft tissue samples.
Similar studies were conducted with raft cultures of HVKs with or without HPV18 infection. We found increased expression of CDKN2A and RNASEH2A and reduced expression of NOVA1 and PTBP1 in HPV18-infected HVK raft tissue samples (Fig. 5C). However, in raft cultures of the HFK with or without HPV18, the following variations in the selected RBP genes were found: the expression levels of CDKN2A, ELAV2, HSPB1, PTBP1, and RNASEH2A were increased but those of GRB7 and NOVA1 were reduced (Fig. 5D). We concluded that the expression levels of CDKN2A, RNASEH2A, and NOVA1 were commonly altered by HPV16 or HPV18 infection in raft cultures of HVKs or HFKs.
We further confirmed the dynamic expression change of RNASEH2A and NOVA1 in raft cultures of HFKs in the course of productive HPV18 infection. Cultures were harvested on day 8 (D8), D12, and D16. In this system, viral DNA amplifies to high copy numbers on or after day 10, when viral oncogene activity starts to wane (26). Abundant capsid protein is detected in the desquamated strata on or after day 16, signifying a robust production of progeny virus (26). RNASEH2A showed sustained, increased expression (Fig. 5E) over the time course of the infection. In contrast, the level of expression of NOVA1 showed a dramatic decrease on day 8 and day 12, when viral oncogenes are highly expressed, but was restored to half of that of uninfected cells on day 16, possibly due to the significant downregulation of the viral oncogenes (Fig. 5F).
Expression of viral oncogenes E6 and E7 is a key event in the malignant progression of HR-HPV-infected cervical cells and is associated with multiple alterations in cellular pathways (27). To investigate whether the increased or decreased expression of RNASEH2A and NOVA1 genes was regulated by viral E6 or E7, raft cultures derived from HFKs that were acutely transduced with retrovirus and stably expressed HPV18 E6, E7, or E6E7 or an empty vector were examined by TaqMan RT-qPCR. We found that RNASEH2A expression was elevated primarily in the presence of viral oncoprotein E7 (Fig. 5G), whereas NOVA1 expression was decreased in the presence of either oncoprotein E6 or oncoprotein E7 (Fig. 5H). These results indicate that both RNASEH2A and NOVA1 respond to HPV infection and that their altered expression levels in cervical cancer cells could be attributed to viral E6 and E7.
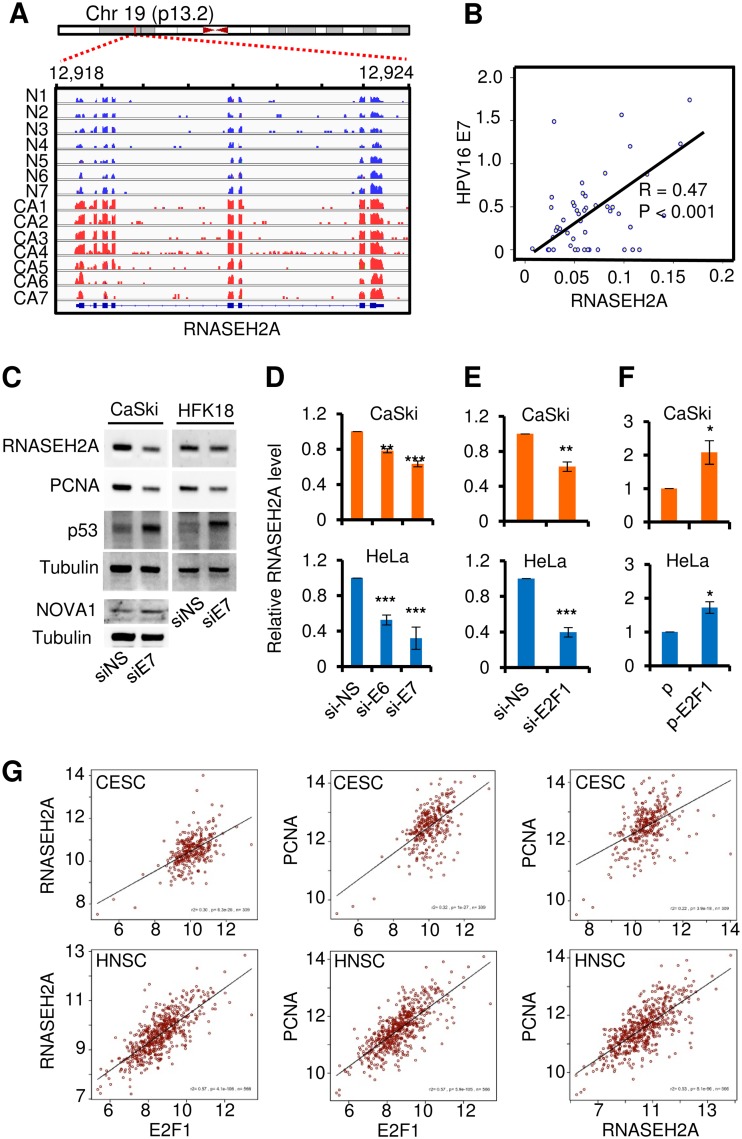
HPV oncoproteins, RNASEH2A, and E2F1.
RNASEH2A is abundantly expressed in cervical cancer tissue samples (Fig. 6A). To confirm whether this abundant expression of RNASEH2A is also highly correlated with HPV E7 in clinical samples, we detected HPV16 E7 mRNA levels in the same 25 CIN2 to CIN3 samples and 23 cervical cancer tissue samples. As expected, we found a positive correlation between HPV16 E7 and RNASEH2A mRNA (Fig. 6B). This positive correlation was confirmed by small interfering RNA (siRNA) knockdown of E7 expression in both HPV16-positive CaSki cells and HPV18-immortalized human foreskin keratinocytes by Western blot analyses (Fig. 6C). Because HPV E7 activates proliferating cell nuclear antigen (PCNA) (28) and decreases NOVA1 expression (Fig. 5H), knockdown of E7 expression in these cells also reduced PCNA levels and increased NOVA1 expression (Fig. 6C). The role of E6 and E7 in the increase of RNASEH2A expression was further validated at the RNA level by RT-qPCR after siRNA knocking down of the expression of HPV16/18 E6 and E7 in CaSki and HeLa (HPV18-positive) cells. We discovered that both E6 and E7 knockdowns reduced RNASEH2A RNA expression (Fig. 6D) but that viral E7 of both HPV16 and HPV18 was more potent than E6 in upregulation of RNASEH2A expression. To provide additional evidence for a causal relationship between HPV E7 and RNASEH2A, RNAi was used to knock down the expression of E2F1, a well-known direct target of E7 (29), in CaSki and HeLa cells. The results revealed that knockdown of E2F1 expression led to repression of RNASEH2A expression (Fig. 6E). In contrast, ectopic expression of E2F1 using an expression vector significantly increased the expression of RNASEH2A (Fig. 6F). Together, the results of these studies suggest that the increased expression of RNASEH2A in HR-HPV-infected cervical tissue samples/cells is primarily from E2F1 via viral oncoprotein E7.
FIG 6.
RNASEH2A is regulated by viral E7 via E2F1 and its expression in association with E2F1 and PCNA in cervical and endocervical cancer and head and neck squamous cell carcinoma. (A) Read coverage maps showing the RNA expression levels of all 7 cervical cancer samples and 7 normal samples for RNASEH2A. Distributions of RNASEH2A reads (blue for normal, red for cancer) along with RNASEH2A transcript structure were visualized using the Integrative Genomics Viewer (IGV). (B) Correlation between RNASEH2A and HPV16 E7 mRNAs in 48 clinical cervical lesion samples, including 25 CIN2-CIN3 and 23 cervical cancer tissue samples. (C) siRNA-specific knockdown of HPV16 or HPV18 E7 expression in CaSki (HPV16+) cells or HPV18-immortalized human foreskin keratinocytes (HFK18) with respect to expression of RNASEH2A, PCNA, and NOVA1 was evaluated by Western blotting. si-NS, nonspecific siRNA; si-E7, HPV16 or HPV18 E7-specific siRNA. Knockdown efficiency of E7 expression was evaluated by analysis of increased expression of p53 due to the E7 siRNA also targeting the overlapped bicistronic E6 RNA (53); tubulin served as a sample loading control. (D) siRNA-specific knockdown of HPV16 or HPV18 E6 or E7 in CaSki (HPV16+) or HeLa (HPV18+) cells with respect to RNASEH2A expression was evaluated by TaqMan RT-qPCR. si-NS, nonspecific siRNA; si-E6, HPV16 or HPV18 E6-specific siRNA; si-E7, HPV16 or HPV18 E7-specific siRNA. (E and F) Specific-siRNA knockdown (E) or ectopic expression of E2F1 (F) in CaSki or HeLa cells with respect to RNASEH2A expression was evaluated by TaqMan RT-qPCR. si-NS, nonspecific siRNA; si-E2F1, E2F1-specific siRNA; p, control vector; p-E2F1, E2F1-expression vector. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The relative expression levels of RNASEH2A in panels D, E, and F are shown as means ± SD for each treatment group determined in duplicate from three independent experiments, with the level in the si-NS (D and E) or vector control (F) group being set to 1. (G) Increased coexpression of RNASEH2A, PCNA, and E2F1 in cervical squamous cell carcinoma, endocervical adenocarcinoma (CESC), and head and neck squamous cell carcinoma (HNSC). Data were extracted from the TCGA data sets using TCGA2STAT R package version 1.2 (https://CRAN.R-project.org/package=TCGA2STAT).
Given that HPV E7 activates PCNA expression via E2F (28) and that the cancer cells display increased DNA replication and cell proliferation, we hypothesized that the increased expression levels of both RNASEH2A and PCNA might be a common phenotype for both HPV-induced cervical cancer cells and head and neck cancer cells. Subsequently, we analyzed The Cancer Genome Atlas (TCGA) data sets for expression correlations among RNASEH2A, PCNA, and E2F1 genes in cervical squamous cell carcinoma/endocervical adenocarcinoma and head/neck squamous cell carcinoma. As expected, we found that their gene expression levels were highly correlated (Fig. 6G) and therefore displayed a strong association.
DISCUSSION
The era of next-generation sequencing has immensely advanced our understanding of the development of cancer. Here, we applied high-throughput RNA deep sequencing methods to examine seven normal cervical tissue samples and seven cervical cancer tissue samples and to illustrate the expression of approximately 19,000 coding genes. We identified at least 614 coding genes that were differentially expressed in cervical cancer, with 95 genes encoding RBPs from the 1,502 known human RBP genes (11, 14, 15, 25).
A unique feature of our study was the parallel analysis of the full spectra of normal and cervical cancer cells using two RNA-seq methods. Though there were some levels of variability in the two data sets due to different sets of clinical cervical samples and different methods being used for library preparations, we were able to identify 614 host genes with altered expression from two RNA-seq analyses and validated the altered expression of 26 genes, including 8 RBP genes, in an expanded panel of 72 human cervical tissue samples. Consistent with the RNA-seq data, all of the validated genes showed similar expression patterns across the multiple stages of cervical carcinogenesis, indicating that the combined two RNA-seq data were reliable for further transcriptomic analysis. By plotting our validated RT-qPCR data using the ROC program, we found that the altered expression of 16 genes, including 7 RBP genes, corresponded to high AUC values and, statistically, high sensitivity and specificity in the identification of high-grade CIN and cancer tissue samples compared to normal cervical tissue samples. We also found that 17 genes, including 3 RBP genes (CDKN2A, GRB7, and NOVA1), possessed a high AUC value and thus high sensitivity and specificity to discriminate cervical cancer tissue samples from CIN2 to CIN3 and normal cervical tissue samples.
To our knowledge, many of these genes reported in this study were not previously reported in association with cervical cancer. For instance, expression of FAM83A (family member with sequence similarity 83), a novel human tumor-specific gene in lung and breast cancer (30, 31), was found to be significantly increased along with the cervical lesion progression from CIN to cervical cancer compared with normal cervix levels. Similarly, LY6K (lymphocyte antigen 6 complex locus K), a testicular cancer antigen and a novel target and diagnostic biomarker for breast cancer, lung cancer, and esophageal squamous cell carcinomas (32, 33), was also found to be overexpressed during cervical lesion progression. More importantly, of the 3,610 differentially expressed coding gene transcripts identified in cervical cancer from this study, we determined that 5.7% represented RBP gene transcripts. RBPs control nearly all aspects of RNA fate, including RNA processing, stability, and/or translation. Dysregulation of their expression or function causes a broad spectrum of human pathologies and syndromes (19, 34). Among the validated 8 RBP genes, some are better known for their conventional functions. For instance, CDKN2A encodes p16INK4A, an inhibitor for the assembly of cyclin D and cyclin-dependent kinase 4 (CDK4) or CDK6 and a well-established biomarker for high-grade CIN and cervical cancer. HSPB1 encodes heat shock protein p27, which protects cell survival under stress conditions. GRB7 encodes a growth factor receptor-binding protein that interacts with epidermal growth factor receptor and ephrin receptor. However, novel roles of CDKN2A, HSPB1, and GRB7 in posttranscriptional regulation have been suggested previously (14, 15, 25). Finding of the altered expression of ELAVL2, KHSRP, NOVA1, PTBP1, and RNASEH2A also suggests their possible roles in cervical carcinogenesis. Collectively, the concomitant analyses of these differentially expressed genes, including regulatory RBP genes, during cervical cancer transformation may open new windows for diagnostic and/or therapeutic interventions.
The role of HR-HPV in the etiology of invasive cervical cancer has been well established. The continuous expression of HR-HPV oncoproteins E6 and E7 is critical in the malignant progression of cervical cancer. Using HVK- or HFK-derived raft culture tissue samples with or without HPV16 and HPV18 infection, we further corroborated that the altered expression of RBP genes, including CDKN2A, RNASEH2A, and NOVA1, is attributable to HPV infection. The difference in the progression from precancer and cancer tissue samples to raft cultures in RBP expression was noticed, and this difference could have been a consequence of the fact that the raft culture was originally developed to study keratinocyte differentiation for HPV productive infection, and not for HPV oncogenesis study, despite the fact that HPV-infected rafts display cell proliferation and dysplastic cell morphologies similar to those seen in early CIN.
Of particular importance was the finding that HPV18 E6 or E7 or both alter the expression of RNASEH2A and NOVA1, which may affect the ability of important posttranscriptional networks to govern cell fate. We further proved the connection between E7 and RNASEH2A and demonstrated that silencing or overexpression of transcription factor E2F1, a critical effector of the HPV E7/RBP pathway, led to significant downregulation or upregulation of RNASEH2A expression, respectively. RNase H2 is a trimer of subunits A, B, and C, with the A subunit being the catalytically active subunit and the B subunit interacting with the proliferating cell nuclear antigen (PCNA) (35). PCNA directs RNase H2 activity with respect to DNA replication by removing the RNA primers to promote Okazaki fragment maturation (36) and has been widely used as a hallmark for virus E7 expression (28). We indeed found correlated increases in expression of both RNASEH2A and PCNA in cervical cancer and HNSC as well as in most of other cancer types (see Fig. S2 in the supplemental material). As all cancer cells lack a functional G1 checkpoint, the induced expression of RNASEH2A and PCNA via viral E7 and E2F1 may speed DNA replication and shorten the cell cycling time, thereby promoting cancer cell proliferation.
Expression correlations among genes of RNASH2A, E2F1, and PCNA within each cancer type. Data were extracted from the TCGA datasets using TCGA2STAT R package version 1.2 (https://CRAN.R-project.org/package=TCGA2STAT). Pearson correlation analysis was run using the lm function from the R statistical package (https://www.R-project.org/) on log-normalized gene expression values. LAML, acute myeloid leukemia; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; LGG, brain lower-grade glioma; BRCA, breast invasive carcinoma; CESE, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; LCML, chronic myelogenous leukemia; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiform; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; MESO, mesothelioma; MISC, miscellaneous; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THYM, thymoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. Download FIG S2, PDF file, 4.2 MB (4.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NOVA1 (Neuro-oncological ventral antigen 1) is a neuron-specific RNA-binding splicing regulator, which binds to the YCAY elements of the target pre-mRNAs to selectively enhance or suppress exon exclusion (37, 38). Apart from neuronal functions, increasing evidence has shown that NOVA1 is also involved in carcinogenesis (39–41). Here, we demonstrated that NOVA1 was downregulated in CIN2-CIN3 and cervical cancer tissue samples compared with normal tissue samples. Of importance, a dramatic decrease of NOVA1 expression was found in raft culture tissue samples with HPV16 or HPV18 infection and was partially related to viral E6 and E7 expression. Data suggest that high-risk HPV infection might also affect host gene expression at the posttranscription level by affecting expression of host splicing factors NOVA1, PTBP1, KHSRP, and ELAVL2. However, further studies are needed to address the functional roles of these RBPs in HPV-induced cervical pathogenesis.
In summary, this report describes comprehensive genome-wide transcriptomic changes from normal cervix to cervical cancer. By validation of the selected 26 genes, including eight RBP genes, with respect to their expression levels in clinical tissue samples, we discovered their differentiated expression in association with cervical lesion progression and HPV infections. Our data offer a new insight into the multifaceted roles of HPV in cervical cancer biology and the mechanisms by which these RBPs might act in cervical malignant transformation. We believe that this information will lay the groundwork for a broad range of future studies.
MATERIALS AND METHODS
Human patient samples.
Cervical samples, including 7 normal and 7 cancer tissue samples for RNA sequencing and another panel of cervical tissue samples containing 24 normal, 23 cancer, and 25 CIN tissue samples for validation by TaqMan qPCR analysis, were all collected from the Women’s Hospital, Zhejiang University School of Medicine. For collection of CIN2 to CIN3 samples, the surface of cervix was treated with 5% acetic acid solution and 2% iodine solution to improve visualization of the abnormal areas. Colposcopy was then performed, and biopsy samples were taken directly from the abnormal area for further pathological examination. We chose a tissue sample from the large lesion area where CIN was pathologically graded. All tissue samples were used in accordance with the Institutional Review Board procedures of the hospital. Informed consent was obtained from each participant prior to the study. All tissue samples were snap-frozen and stored at −80°C until use.
RNA isolation and sequencing.
RNA was isolated from each human tissue sample by the use of TRIzol (Invitrogen, CA, USA) according to the instructions provided by the manufacturer. Total RNA quality and quantity were verified by the use of a model 2100 Bioanalyzer (Agilent Technologies, CA, USA) and a NanoDrop ND-1000 spectrometer (Thermo Scientific, DE, USA), respectively.
RNA-seq 1 was performed as described previously (42, 43). Briefly, polyadenylated [poly(A)+] RNA was obtained with two rounds of poly(A) selection using Dynabeads Oligo(dT)25 (Invitrogen). DeLi-Seq was used to prepare the sequencing library and then purified by the use of a Zymo DNA Clean and Concentrator-5 kit (Irvine, CA). The products (∼200 to ∼300 bp) were sequenced using Illumina/Solexa Genome Analyzer II.
For RNA-seq 2, libraries were prepared using a TruSeq stranded total RNA sample preparation kit with Ribo-Zero depletion (Illumina, CA, USA). In brief, high quality total RNA (1 µg) was subjected to Ribo-Zero depletion, fragmented, and then subjected to reverse transcription (RT). Double-stranded cDNAs were A-tailed and ligated with Illumina sequencing adapters. Subsequently, the ligated products were enriched by PCR and size-selected by agarose gel electrophoresis. The products (∼200 to ∼400 bp) were sequenced by the use of an Illumina Hi-seq 2500 platform with a paired 50-mer sequencing modality.
Read mapping and transcriptome analyses.
The raw sequence reads in fastq format (NCBI GSE113942) were mapped to a human reference genome (hg19; GRCh37) by the use of Tophat (44) v2.0.11(-g 1), using the Bowtie aligner (v2.2.1.0) with the following parameter settings: -N 0, -L 20, -i S,1,1.25, -n-ceil L,0,0.15, and -gbar 4. Only uniquely mapped reads were assigned to the UCSC (University of California, Santa Cruz) Refseq (hg19) transcripts by Bedtools (v2.22.0). The transcript raw reads were inputted into the Bioconductor edgeR package (45) followed by filtering out low-abundance transcripts with low counts per million (CPM) values. CPM is defined as the read count of a given transcript per million mapped reads of the corresponding RNA-seq library. After normalization on the basis of trimmed means of the M-values (TMM) crossing all samples of the data set, we employed the Exact Test for detection of the transcripts differentially expressed between the normal control and cancer groups. Differentially expressed gene transcripts were obtained at the log2 fold change (FC) cutoff value (FC = 8 or 4) with a 10% false-discovery rate (FDR) (for RNA-seq 1, |log2 FC| ≥ 8 and log2 CPM ≥ 2 [10% FDR]; for RNA-seq 2, |log2FC| ≥ 4 and log2 CPM ≥ 0.5 [10% FDR]) (Fig. 1A). The FDR was controlled by the Benjamini-Hochberg (BH) procedure. Each normalized transcript expression level was defined as log2 reads per kilobase per million (RPKM) and was compared with the quantitative PCR expression value for the validation assay.
We selected RNA-binding protein genes from the literature (14, 15, 25). The normalized reads from multiple transcripts of each gene were averaged to represent composite gene expression. The unsupervised hierarchical clustering analysis used the Euclidean distance as the similarity measure.
Ingenuity pathway analysis.
“Canonical pathway” analysis and “Disease and Function enrichment” analysis were performed using Ingenuity pathway analysis (IPA; Ingenuity Systems Inc.) version 2.0 software (46). To perform IPA, all 614 overlapping gene transcripts (see Table S5 in the supplemental material) that were differentially expressed between normal and cervical cancer groups were uploaded to the IPA database with the corresponding RefSeq identifier (ID) and log2 FC (fold change) value. Canonical pathway analysis was used to identify the most significantly modulated networks across the subject groups. Functional analysis identified the biological roles and/or diseases that are most significant with respect to the data set.
Human primary keratinocytes and organotypic (raft) epithelial cultures.
Primary human vaginal keratinocytes (HVKs) and primary human foreskin keratinocytes (HFKs) were isolated from adult vaginectomy and newborn circumcision tissue specimens, respectively, as previously described (29). Keratinocytes were grown in monolayer culture by using epithelial (E) medium plus epidermal growth factor (5 ng/ml) in the presence of J2 3T3 feeder cells treated with mitomycin C (4 µg/ml). Keratinocyte lines stably maintaining HPV16 and HPV18 DNA following electroporation were made as previously described (47). Organotypic (raft) epithelial culture tissue samples derived from HPV16- and HPV18-immortalized HVK or HFK were prepared as described previously (47). The stratified and differentiated cultures were collected free from collagen (no fibroblasts) on day 10 and frozen on dry ice for total cell RNA preparation and for histology and differentiation analyses as described in a previous report (48).
Additional raft cultures of primary HFKs with productive HPV18 infection were also analyzed. Genomic HPV18 plasmid was generated by Cre-loxP-mediated recombination in transfected HFKs followed by 3 days of selection as described previously (26). Within a week of transfection, the selected HFKs were used to prepare raft cultures. The epithelial tissue samples were collected on day 8 (D8), D12, and D16. Uninfected HFK raft cultures were developed in parallel. The tissue samples in one set were formalin fixed and paraffin embedded. Thin sections were cut to evaluate histology and cell differentiation (26). Only the infected cultures were hyperproliferative as expected and positive for the E7-induced PCNA and cyclin B1 in the differentiated strata and basal cells. The uninfected cultures had signals only in the basal stratum. Abundant viral capsid protein L1 was detected in the desquamated strata in the day 16 infected cultures, validating the identification of a productive infection.
Plasmids pLJd-HPV-18URR-E6, pLC-HPV-18URR-E7, and pLJd-HPV-18URR-E6E7 have been described previously (49). Retroviruses derived from the vectors mentioned above were prepared as described previously (50). Primary HFKs were acutely infected with the retroviruses and selected with G-418 (300 μg/ml) for 2 days. The selected primary HFKs were used to establish epithelial raft cultures, which were then harvested on day 11.
TaqMan real-time quantitative PCR assays.
Real-time PCR TaqMan gene expression assays (Applied Biosystems) were used for quantitative validation of host gene transcripts in clinical samples and raft cultures. In brief, 2 µg of total RNA from each sample was subjected to reverse transcription using a SuperScript first-stand synthesis kit (Invitrogen) according to the manufacturer’s instructions. After reverse transcription, PCR products were amplified from the cDNA samples using TaqMan gene expression Master Mix together with TaqMan gene expression assays on a StepOne Plus real-time PCR system. TaqMan gene expression assays for 26 genes, including 8 RBP genes, were obtained from Applied Biosystems. The probes span exon-exon junctions and amplify only spliced RNA products to avoid amplifying any contaminated residual genomic DNA in our RNA samples. Gene enrichment was calculated using the threshold cycle (2−ΔΔCT) method in relation to the housekeeping GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene transcripts. The mean threshold CT value corresponding to the values determined for a given gene from 24 normal cervical tissue samples after normalization served as a basal level to calculate a relative level of the gene transcripts detected in each clinical sample. Data are presented as a bar graph with means ± standard deviations (SD) for each group. The significance of the relative mRNA levels among clinical tissue groups was analyzed using the nonparametric Mann-Whitney U test, while the significance of the relative mRNA levels in raft culture tissue groups and cell line groups was analyzed using Student’s t test.
ROC curve analysis.
Receiver operating characteristic (ROC) analysis (51) is widely used for evaluation of diagnostic performance. For ROC curve analysis, the TaqMan gene expression validation results (2−ΔΔCT) determined for all 26 genes, including 8 RBP genes, in 24 normal cervix, 25 CIN2 to CIN3, and 23 cervical cancer tissue samples were uploaded to IBM SPSS20.0 software. To compare normal subjects and CIN2 to CIN3 subjects with cancer subjects, we grouped normal tissue samples plus CIN2 to CIN3 tissue samples as group 1 and cervical cancer tissue samples as group 2. The data were then applied for ROC curve analysis. Similarly, for comparisons of normal tissue samples and CIN2 to CIN3 plus cancer tissue samples, we grouped the normal tissue samples as group 1 and the CIN2 to CIN3 plus cancer tissue samples as group 2. The area under the ROC curve (AUC), one of the most commonly used summary indices of the ROC curve, is interpreted as the average value of sensitivity (true-positive fraction [TPF]) for all possible values of specificity (false-positive fraction [FPF]) as follows: AUC < 0.5, not significant; AUC > 0.5, significant (0.5 to 0.7, low significance; 0.7 to 1.0, high significance).
Cell transfections, siRNAs, and Western blotting.
CaSki cells (ATCC CRL-1550), representing a cervical cancer cell line with an integrated HPV16 genome (500 to 600 copies/cell), HeLa cells (ATCC CCL-2), representing a cervical cancer cell line with an integrated HPV18 genome (10 to 50 copies/cell), and HPV18-immortalized HFKs were chosen to knock down the expression of endogenous proteins. In general, cells at the active-proliferation stage were transfected twice in a 24-h interval with 80 nM siGenome SMARTpool siRNAs (Dharmacon) targeting E6, E7, or E2F1. A nontargeting, scrambled siRNA (Dharmacon) was used as a negative control. The siRNA transfection was performed using LipoJet in vitro transfection reagent (SignaGen, MD, USA). Cell lysates and total RNA were prepared after 24 h of the second siRNA knockdown. The cell lysates were analyzed by Western blotting with the following corresponding antibodies: rabbit polyclonal anti-RNASEH2A (catalog no. A104-149A; Bethyl Laboratories, Montgomery, TX), anti-NOVA1 (catalog no. ab97368; Abcam, Cambridge, MA), and anti-P53 (catalog no. 10442-1-AP; ProteinTech, Rosemont, IL) and mouse monoclonal anti-PCNA (clone PC10) (catalog no. sc-56; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-β-tubulin (clone Tub 2.1) (catalog no. T5201; Sigma-Aldrich, St. Louis, MO). Total RNA was analyzed for RNASEH2A expression by real-time RT-qPCR.
CaSki and HeLa cells were also transfected with 2 μg of an E2F1 expression vector (pRcCMV-HA-E2F1; Addgene, Watertown, MA) with X-tremeGENE HP DNA transfection reagent (Roche), and total RNA from the cells with E2F1 overexpression was analyzed for RNASEH2A expression by real-time RT-qPCR.
All experiments were performed in duplicate and repeated at least three times.
TCGA data analysis.
TCGA RNA-seq (level 3) gene expression data in the form of normalization of RNA-seq by expectation-maximization (RSEM) values from all cancer data sets were downloaded from the Firehose Broad GDAC database (http://gdac.broadinstitute.org/; DOI for data release, 10.7908/C11G0KM9) using the TCGA2STAT package in Bioconductor (52) for R (R package version 1.2; https://CRAN.R-project.org/package=TCGA2STAT). The data were then used to find correlations among genes RNASEH2A, E2F1, and PCNA within each cancer type, and the Pearson correlation analysis was run using the lm function from the R statistical package (https://www.R-project.org/) on log-normalized gene expression values.
ACKNOWLEDGMENTS
This study was supported by the Intramural Research Program of NCI/NIH (1ZIASC010357 to Z.M.Z.), by the National Key Research and Development Program of China (2016YFC1302900 to W.L.), the NSF of China (81702552 to J.X.), and the Medical and Health Science and Technology Project of Zhejiang Province (2017KY099 to J.X.). H.-K.W., N.S.B., and L.T.C. were supported by NIH grant CA83679 and funds from the Anderson Family Endowed Chair to the University of Alabama at Birmingham (UAB).
We have no potential conflicts of interest to disclose.
Footnotes
Citation Xu J, Liu H, Yang Y, Wang X, Liu P, Li Y, Meyers C, Banerjee NS, Wang H-K, Cam M, Lu W, Chow LT, Xie X, Zhu J, Zheng ZM. 2019. Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. mBio 10:e02687-18. https://doi.org/10.1128/mBio.02687-18.
Contributor Information
Xiang-Jin Meng, Virginia Polytechnic Institute and State University.
James Pipas, University of Pittsburgh.
Xuefeng Liu, Georgetown University.
Lucia Pirisi-creek, University of South Carolina.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. 2015. Global cancer statistics, 2012. CA Cancer J Clin 65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. 2013. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. Eur J Cancer 49:3262–3273. doi: 10.1016/j.ejca.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Hennessey PT, Westra WH, Califano JA. 2009. Human papillomavirus and head and neck squamous cell carcinoma: recent evidence and clinical implications. J Dent Res 88:300–306. doi: 10.1177/0022034509333371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 5.Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. 2008. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Wang H-K, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. 2009. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA 15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Meyers C, Guo M, Zheng ZM. 2011. Up-regulation of p18Ink4c expression by oncogenic HPV E6 via p53-miR-34a pathway. Int J Cancer 129:1362–1372. doi: 10.1002/ijc.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, Tuschl T, Zheng ZM. 2014. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci U S A 111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentze MW, Castello A, Schwarzl T, Preiss T. 2018. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 11.Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerstberger S, Hafner M, Tuschl T. 2013. Learning the language of post-transcriptional gene regulation. Genome Biol 14:130. doi: 10.1186/gb-2013-14-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. 2012. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 16.Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. 2016. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kechavarzi B, Janga SC. 2014. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol 15:R14. doi: 10.1186/gb-2014-15-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira B, Billaud M, Almeida R. 2017. RNA-binding proteins in cancer: old players and new actors. Trends Cancer 3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Roberts A, Pimentel H, Trapnell C, Pachter L. 2011. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 21.Oshlack A, Robinson MD, Young MD. 2010. From RNA-seq reads to differential expression results. Genome Biol 11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denoeud F, Aury JM, Da SC, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P, Scarpelli C, Jaillon O, Artiguenave F. 2008. Annotating genomes with massive-scale RNA sequencing. Genome Biol 9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Royse KE, Zhi D, Conner MG, Clodfelder-Miller B, Srinivasasainagendra V, Vaughan LK, Skibola CF, Crossman DK, Levy S, Shrestha S. 2014. Differential gene expression landscape of co-existing cervical pre-cancer lesions using RNA-seq. Front Oncol 4:339. doi: 10.3389/fonc.2014.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin W, Feng M, Li X, Zhong P, Guo A, Chen G, Xu Q, Ye Y. 2017. Transcriptome profiling of cancer and normal tissues from cervical squamous cancer patients by deep sequencing. Mol Med Rep 16:2075–2088. doi: 10.3892/mmr.2017.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castello A, Fischer B, Hentze MW, Preiss T. 2013. RNA-binding proteins in Mendelian disease. Trends Genet 29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Wang HK, Duffy AA, Broker TR, Chow LT. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev 23:181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 28.Noya F, Chien WM, Wu X, Banerjee NS, Kappes JC, Broker TR, Chow LT. 2002. The promoter of the human proliferating cell nuclear antigen gene is not sufficient for cell cycle-dependent regulation in organotypic cultures of keratinocytes. J Biol Chem 277:17271–17280. doi: 10.1074/jbc.M112441200. [DOI] [PubMed] [Google Scholar]
- 29.Hwang SG, Lee D, Kim J, Seo T, Choe J. 2002. Human papillomavirus type 16 E7 binds to E2F1 and activates E2F1-driven transcription in a retinoblastoma protein-independent manner. J Biol Chem 277:2923–2930. doi: 10.1074/jbc.M109113200. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y, Pang X, Zhang Y, Chen W. 2005. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia 7:1073–1080. doi: 10.1593/neo.05406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, Bissell MJ. 2012. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest 122:3211–3220. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda R, Enokida H, Chiyomaru T, Kikkawa N, Sugimoto T, Kawakami K, Tatarano S, Yoshino H, Toki K, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M. 2011. LY6K is a novel molecular target in bladder cancer on basis of integrate genome-wide profiling. Br J Cancer 104:376–386. doi: 10.1038/sj.bjc.6605990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Ito H, Miyagi Y, Nakayama H, Fujita M, Hosokawa M, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. 2007. Cancer-testis antigen lymphocyte antigen 6 complex locus K is a serologic biomarker and a therapeutic target for lung and esophageal carcinomas. Cancer Res 67:11601–11611. doi: 10.1158/0008-5472.CAN-07-3243. [DOI] [PubMed] [Google Scholar]
- 34.Gerstberger S, Hafner M, Ascano M, Tuschl T. 2014. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol 825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. 2009. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res 37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Hu J, Wang J, Kong D. 2017. Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J Biol Chem 292:4777–4788. doi: 10.1074/jbc.M116.758599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 38.Buckanovich RJ, Yang YY, Darnell RB. 1996. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci 16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EK, Yoon SO, Kim SH, Yang WI, Cho YA, Kim SJ. 2016. Upregulated neuro-oncological ventral antigen 1 (NOVA1) expression is specific to mature and immature T- and NK-cell lymphomas. J Pathol Transl Med 50:104–112. doi: 10.4132/jptm.2016.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim EK, Yoon SO, Jung WY, Lee H, Kang Y, Jang YJ, Hong SW, Choi SH, Yang WI. 2017. Implications of NOVA1 suppression within the microenvironment of gastric cancer: association with immune cell dysregulation. Gastric Cancer 20:438–447. doi: 10.1007/s10120-016-0623-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YA, Zhu JM, Yin J, Tang WQ, Guo YM, Shen XZ, Liu TT. 2014. High expression of neuro-oncological ventral antigen 1 correlates with poor prognosis in hepatocellular carcinoma. PLoS One 9:e90955. doi: 10.1371/journal.pone.0090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ni T, Tu K, Wang Z, Song S, Wu H, Xie B, Scott KC, Grewal SI, Gao Y, Zhu J. 2010. The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One 5:e15271. doi: 10.1371/journal.pone.0015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Wang X, Ni T, Wang F, Lu W, Zhu J, Xie X, Zheng ZM. 2013. Human papillomavirus type 58 genome variations and RNA expression in cervical lesions. J Virol 87:9313–9322. doi: 10.1128/JVI.01154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas S, Bonchev D. 2010. A survey of current software for network analysis in molecular biology. Hum Genomics 4:353–360. doi: 10.1186/1479-7364-4-5-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLaughlin-Drubin ME, Meyers C. 2005. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med 119:171–186. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- 48.Alam S, Bowser BS, Conway MJ, Israr M, Ryndock EJ, Xi LF, Meyers C. 2010. Downregulation of Cdc2/CDK1 kinase activity induces the synthesis of noninfectious human papillomavirus type 31b virions in organotypic tissues exposed to benzo[a]pyrene. J Virol 84:4630–4645. doi: 10.1128/JVI.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev 9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 50.Banerjee NS, Chow LT, Broker TR. 2005. Retrovirus-mediated gene transfer to analyze HPV gene regulation and protein functions in organotypic “raft” cultures. Methods Mol Med 119:187–202. doi: 10.1385/1-59259-982-6:187. [DOI] [PubMed] [Google Scholar]
- 51.Zweig MH, Campbell G. 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39:561–577. [PubMed] [Google Scholar]
- 52.Wan YW, Allen GI, Liu Z. 2016. TCGA2STAT: simple TCGA data access for integrated statistical analysis in R. Bioinformatics 32:952–954. doi: 10.1093/bioinformatics/btv677. [DOI] [PubMed] [Google Scholar]
- 53.Tang S, Tao M, McCoy JP Jr, Zheng ZM. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol 80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of transcriptomic signatures of normal cervix and cervical cancer. (A) Schematic illustration of the strategies used to discover and define bona fide differentially expressed gene transcripts in cervical cancer. (B) Identification of the altered expression of RNA-binding protein genes in cervical cancer. A schematic illustration of the strategies used to find differentially expressed RBP genes in cervical cancer is shown. Download FIG S1, PDF file, 0.2 MB (164.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Total RNA reads and mapped reads in each normal cervix and cervical cancer sample. Download Table S1, XLS file, 0.02 MB (25.5KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 1,799 gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 1. Download Table S2, XLS file, 0.5 MB (552KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 1,635 gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 2. Download Table S3, XLS file, 0.5 MB (524KB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 614 overlapped gene transcripts differentially expressed in both RNA-seq 1 and RNA-seq 2. Download Table S4, XLSX file, 0.2 MB (163.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicted host genes with validated differential expression might be useful biomarkers for diagnosis of cervical lesion progression. Download Table S5, XLSX file, 0.01 MB (12.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 3,610 overlapped gene transcripts differentially expressed between cervical cancer and normal cervix in RNA-seq 1 and RNA-seq 2. Download Table S6, XLS file, 1.4 MB (1.5MB, xls) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of 205 gene transcripts from 95 RBP genes differentially expressed in RNA-seq 1 and RNA-seq 2. Download Table S7, XLSX file, 0.1 MB (58.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicted RBP pathways involved in cervical carcinogenesis. Download Table S8, XLSX file, 0.03 MB (34.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression correlations among genes of RNASH2A, E2F1, and PCNA within each cancer type. Data were extracted from the TCGA datasets using TCGA2STAT R package version 1.2 (https://CRAN.R-project.org/package=TCGA2STAT). Pearson correlation analysis was run using the lm function from the R statistical package (https://www.R-project.org/) on log-normalized gene expression values. LAML, acute myeloid leukemia; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; LGG, brain lower-grade glioma; BRCA, breast invasive carcinoma; CESE, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; LCML, chronic myelogenous leukemia; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiform; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; MESO, mesothelioma; MISC, miscellaneous; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THYM, thymoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. Download FIG S2, PDF file, 4.2 MB (4.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.