Abstract
This paper describes the X-ray crystallographic structure of a derivative of the antibiotic teixobactin and shows that its supramolecular assembly through the formation of antiparallel β-sheets creates binding sites for oxyanions. An active derivative of teixobactin containing lysine in place of allo-enduracididine assembles to form amyloid-like fibrils, which are observed through a thioflavin T fluorescence assay and by transmission electron microscopy. A homologue, bearing an N-methyl substituent, to attenuate fibril formation, and an iodine atom, to facilitate X-ray crystallographic phase determination, crystallizes as double helices of β-sheets that bind sulfate anions. β-Sheet dimers are key subunits of these assemblies, with the N-terminal methylammonium group of one monomer and the C-terminal macrocycle of the other monomer binding each anion. These observations suggest a working model for the mechanism of action of teixobactin, in which the antibiotic assembles and the assemblies bind lipid II and related bacterial cell wall precursors on the surface of Gram-positive bacteria.
Graphical Abstract
The peptide antibiotic teixobactin has been the subject of intensive research efforts for its promise of addressing antibiotic-resistant Gram-positive pathogens such as MRSA and VRE (Figure 1).1,2,3,4,5,6,7,8,9 Teixobactin is thought to bind highly conserved prenyl-pyrophosphate-saccharide regions of lipid II and related membrane-bound cell wall precursors.1 Here we describe the first X-ray crystallographic structure of a full-length teixobactin analogue, which reveals an amphipathic amyloid-like assembly that acts as a multivalent receptor for sulfate anions. This crystallographic structure suggests a working model for the mechanism of action of teixobactin in which teixobactin forms fibrils or smaller assemblies that bind to the pyrophosphate groups of lipid II and related cell wall precursors on the bacterial cell membrane and thus disrupt cell wall biosynthesis. These findings should be of value both in understanding the mechanism of action of teixobactin and in rationally designing new antibiotics that target lipid II and related cell wall precursors.
Figure 1.
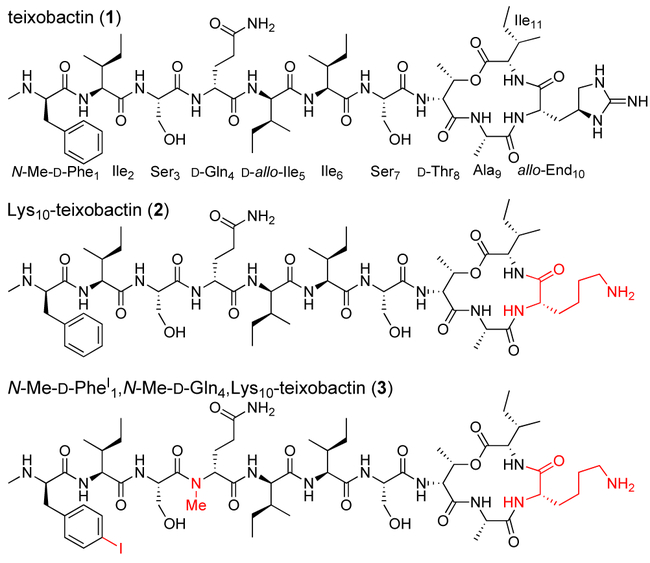
Teixobactin (1), Lys10-teixobactin (2), and N-Me-d-PheI1,N-Me-d-Gln4Lys10-teixobactin (3).
While studying structure-activity relationships among teixobactin analogues, we have observed that teixobactin and analogues with good antibiotic activity (low MIC values) form gels, while analogues with poor activity (high MIC values) do not.10 For example, Lys10-teixobactin (2), a homologue of teixobactin in which allo-enduracididine at position 10 is replaced with lysine (Figure 1), has an MIC of 0.5-1.0 μg/mL against S. aureus and forms a gel in PBS buffer, while d-Ala5,Lys10-teixobactin (MIC≥16 μg/mL) does not.10 This observation suggested that supramolecular assembly of teixobactin analogues could be involved in antibiotic activity.
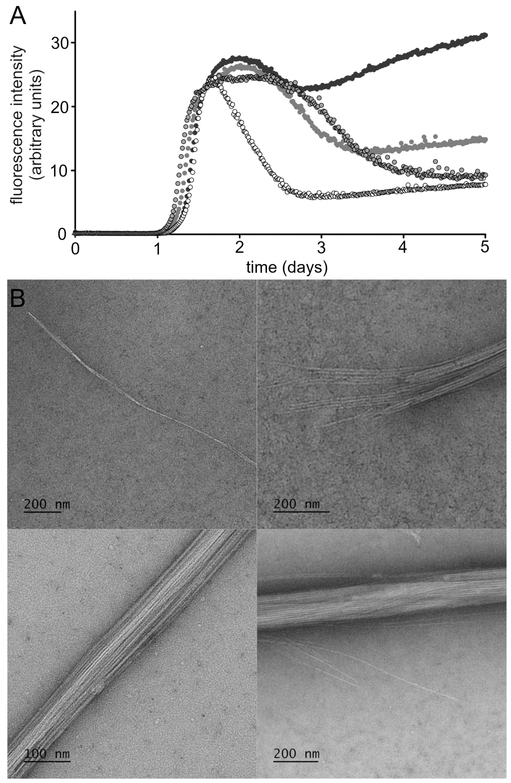
We began exploring the supramolecular assembly of teixobactin and its analogues by performing thioflavin T (ThT) fluorescence assays and transmission electron microscopy (TEM) studies upon Lys10-teixobactin. When we incubated Lys10-teixobactin with PBS buffer and ThT and monitored fluorescence, we observed a lag phase of ca. 1 day, followed by an increase in fluorescence (Figure 2A).11 This behavior is a hallmark of amyloidogenic peptides and proteins. To further explore the assemblies that formed, we performed TEM studies. TEM images of the aggregated Lys10-teixobactin revealed amyloid-like fibrils (Figure 2B). The fibrils range from individual or paired filaments, ca. 8 nm across, through bundles of filaments ca. 100-200 nm in diameter.
Figure 2.
(A) ThT fluorescence assay of Lys10-teixobactin (2, four replicate runs with 120 μM peptide in PBS buffer at pH 7.4). (B) TEM images of the fibrils formed by Lys10-teixobactin (2).
To further study teixobactin supramolecular assembly, we turned to X-ray crystallography. Although we had successfully crystallized a truncated teixobactin analogue containing only residues 6–11, all efforts to crystallize full-length teixobactin analogues failed, giving only amorphous aggregates.12 We postulated that N-methylation of the peptide backbone would attenuate the aggregation and permit the growth of crystals.13,14 We discovered that N-methylation of d-Gln4 indeed facilitated crystallization. We also incorporated an iodine atom in N-Me-d-Phe1 to give N-methyl-p-iodo-d-phenylalanine (N-Me-d-PheI1), to permit determination of the X-ray crystallographic phases.15,16 Figure 1 illustrates the structure of the resulting teixobactin analogue 3, a homologue of Lys10-teixobactin (2). Teixobactin analogue 3 does not form a gel and exhibits only modest activity against S. aureus (MIC=16 μg/mL).
We began our crystallization efforts by screening teixobactin analogue 3 in 864 conditions in a 96-well plate format using crystallization kits from Hampton Research (PEG/Ion, Index, and Crystal Screen). Rectangular rod-shaped crystals grew in conditions containing sulfate salts (Li2SO4, MgSO4, Na2SO4, K2SO4, (NH4)2SO4) and polyethylene glycol (PEG) 3,350. With further optimization in a 24-well plate format, 0.19 M Na2SO4 and 15% PEG 3,350 afforded crystals suitable for X-ray diffraction. Four X-ray diffraction datasets were acquired at the Stanford Synchrotron Radiation Lightsource (SSRL) at a wavelength of 2.07 Å. The datasets were processed using XDS17 and merged using BLEND18. The structure was solved by single-wavelength anomalous diffraction (SAD) phasing using the iodine anomalous signal from N-Me-d-PheI1. The structure was refined with REFMAC519 in the P212121 space group at 2.20 Å resolution. The asymmetric unit contains 32 crystallographically independent teixobactin analogue molecules, as well as 32 sulfate anions and 53 ordered water molecules.
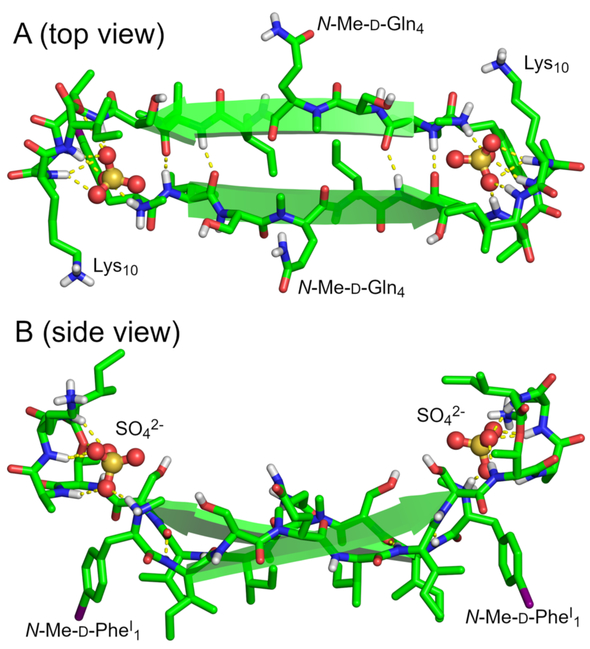
The 32 molecules of teixobactin analogue 3 form a double helix of β-sheet fibrils in which each fibril is composed of 16 peptide molecules. Each fibril may be thought of as comprising hydrogen-bonded dimers. Figure 3 illustrates the structure of a representative hydrogen-bonded dimer. In the dimer, two molecules of teixobactin analogue 3 come together to form an antiparallel β-sheet in which Ile2 hydrogen bonds with Ile6, N-Me-d-Gln4 pairs with N-Me-d-Gln4, and Ile6 hydrogen bonds with Ile2. The N-methyl groups of the two N-Me-d-Gln4 residues tilt upward, allowing the β-sheet to form in spite of the disruption of the hydrogen-bonding pattern. As a result, the β-sheet has four hydrogen bonds instead of six hydrogen bonds.
Figure 3.
X-ray crystallographic structure of a representative dimer of N-Me-d-PheI1,N-Me-d-Gln4,Lys10-teixobactin (3). (A) Top view. (B) Side view.
In the X-ray crystallographic structure, the dimer acts as a receptor for two sulfate anions. The amide NH groups of the macrocyclic ring of each monomer subunit act in conjunction with the N-terminus of the other monomer subunit to bind each sulfate anion. Each sulfate anion hydrogen bonds to the amide NH groups of d-Thr8, Ala9, Lys10, and Ile11 of one monomer subunit and the methylammonium group of the N-Me-d-PheI1 of the other subunit. The β-sheet dimer is amphipathic: the side chains of N-Me-d-PheI1, Ile2, d-allo-Ile5, and Ile6 create a hydrophobic surface, and the side chains of Ser3, N-Me-d-Gln4, and Ser7, as well as the N-terminal methylammonium group, create a hydrophilic surface. The macrocyclic rings and the sulfate anions lie above the hydrophilic surface.
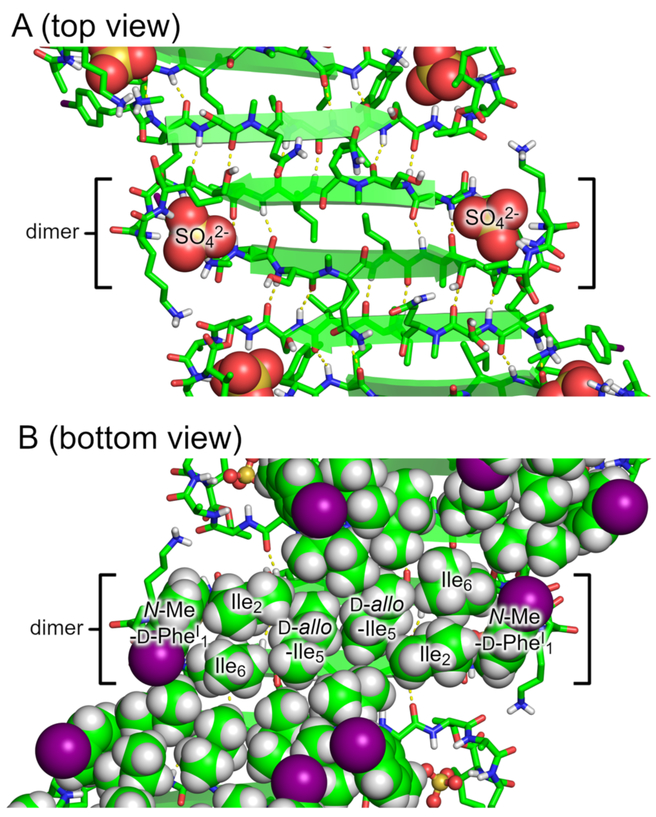
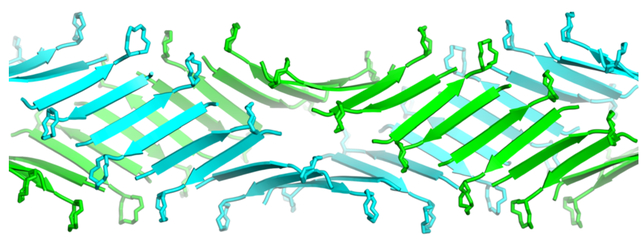
Sixteen molecules of teixobactin analogue 3 assemble to form each β-sheet fibril (Figure 4). The molecules assemble in an antiparallel fashion to form an extended amphiphilic β-sheet, with the hydrophobic residues on one face and hydrophilic residues on the other face. At each β-sheet interface between the dimers, Ser3 hydrogen bonds with Ser7, d-allo-Ile5 hydrogen bonds with d-allo-Ile5, and Ser7 hydrogen bonds with Ser3. Each dimer interface is thus shifted by two residues, which results in an offset fibril structure.20 (In an aligned fibril structure, N-Me-d-PheI1 would hydrogen bond with Ser7, Ser3 would hydrogen bond with d-allo-Ile5, d-allo-Ile5 would hydrogen bond with Ser3, and Ser7 would hydrogen bond with N-Me-d-PheI1.)
Figure 4.
β-Sheet fibril formed by N-Me-d-PheI1,N-Me-d-Gln4Lys10-teixobactin (3). (A) Top view. (B) Bottom view with hydrophobic side chains shown as spheres.
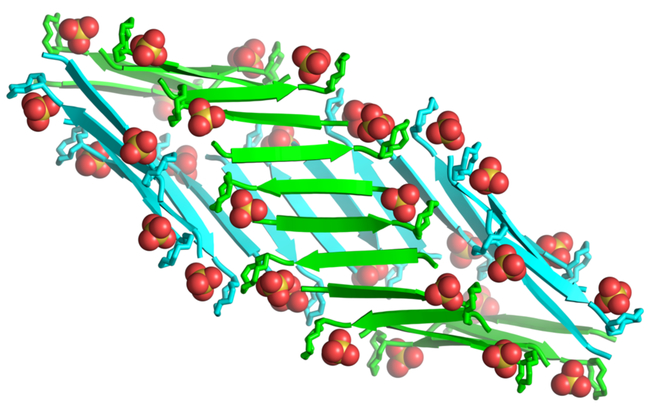
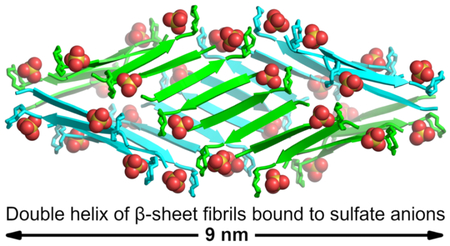
Two β-sheet fibrils wrap around each other to form a right-handed double helix of β-sheets, with the hydrophobic surfaces in the interior and the hydrophilic surfaces on the exterior (Figure 5). Each double helix contains 32 molecules of teixobactin analogue 3 and corresponds to the asymmetric unit. The double helices are discrete structures in the crystal lattice and are not part of extended superstructures. The double helix is ca. 9 nm in length and ca. 4 nm in diameter in the middle, tapering to ca. 2 nm at the two ends. The ends of the double helix are closed, but the middle has a central cavity of ca. 1 nm in diameter and ca. 5 nm in length that is surrounded by the hydrophobic side chains of N-Me-d-PheI1, Ile2, d-allo-Ile5, and Ile6 (Figure S5). The ordered water molecules surround the hydrophilic exterior of the double helix.
Figure 5.
Double helix of β-sheet fibrils formed by N-Me-d-PheI1,N-Me-d-Gln4,Lys10-teixobactin (3). Sulfate anions are shown as spheres.
The X-ray crystallographic structure of the discrete double helix of β-sheets formed by teixobactin analogue 3 suggests a molecular model for the assembly of teixobactin analogue 2 into the filaments and fibrils observed by TEM (Figure 2). In this model, teixobactin analogue 2 assembles to form extended networks of β-sheet fibrils, which wrap around each other to form extended double helices of β-sheets. Figure 6 illustrates this model. Unlike the discrete structures formed by N-methylated analogue 3, these double helices persist for many hundreds of nanometers and contain thousands of molecules. These fibrils further wrap or bundle together to form the fibrils and bundles observed by TEM. Although the N-methyl group in teixobactin analogue 3 does not prevent β-sheet formation, it impedes the formation of extended fibrils by reducing the stability of the β-sheets that form.
Figure 6.
Crystallographically based molecular model of an extended double helix of β-sheet fibrils formed by teixobactin analogue 2 and observed by TEM (Figure 2).
The amphipathic assembly formed by teixobactin analogue 3 explains many of the previously reported structure-activity relationships in teixobactin analogues.10,21,22 Our laboratory has previously reported that substituting residues 1, 2, 5, 6, and 7 with l- or d-alanine dramatically reduces or eliminates the antibiotic activity of Lys10-teixobactin, while substituting residues 3 and 4 with l- or d-alanine has much smaller effects upon activity.10 Similar effects have been observed upon replacement of residues 2–7 with l- or d-lysine.21 The densely packed hydrophobic surface formed by residues 1, 2, 5, and 6 on the interior of the double helix of β-sheet fibrils (Figure 4B) explains why mutating any of these bulky hydrophobic residues to l- or d- alanine or lysine disrupts supramolecular assembly and causes loss of activity. The hydrophilic side chains of residues 3 and 4 are on the hydrophilic exterior of the double helix of β-sheet fibrils (Figure 4A) and are substantially more tolerant of substitution. The hydrophilic side chain of residue 7 is also on the hydrophilic exterior of the double helix of β-sheet fibrils, however the X-ray crystallographic structure does not appear to explain the loss of activity upon mutating this residue to Ala or Lys. Additional studies have reported that substituting l-amino acids for d-amino acids at residues 1, 4, and 5 in Arg10-teixobactin also dramatically reduces or eliminates antibiotic activity.22 Each of these stereochemical mutations disrupts the amphipathic β-sheet formed by residues 1–7 and causes loss of antibiotic activity.
The X-ray crystallographic structure of teixobactin analogue 3, in conjunction with the observation that Lys10-teixobactin (2) forms amyloid-like fibrils, suggest that supramolecular assembly may be involved in the antibiotic activity of teixobactin. We thus propose a working model for the antibiotic activity of teixobactin in which teixobactin forms dimers, higher-order assemblies, or fibrils through antiparallel β-sheet interactions.23 The dimers or dimer subunits create binding sites for the pyrophosphate groups of lipid II and related membrane-bound cell wall precursors, perhaps adhering strongly to the surface through contacts with multiple lipid molecules.24 In the binding site, the amide NH groups of residues 8–11 of one teixobactin molecule in the dimer and the N-terminus of the other teixobactin molecule interact with each bound pyrophosphate group. In teixobactin (1), the guanidinium group of allo-End10 may make additional contacts to the pyrophosphate group.
This model shares a number of features in common with those observed for other antibiotics that target lipid II and related cell wall precursors, including ramoplanin and nisin.25,26 Ramoplanin forms fibrils with lipid II analogues, and supramolecular assembly through the formation of antiparallel β-sheet dimers is thought to be important in its mechanism of action. 27,28,29,30,31 Nisin binds the pyrophosphate group of lipid II by means of a pyrophosphate cage formed by amide NH groups in and adjacent to the 16-membered lanthionine A ring.32,33
The unique pattern of hydrophobicity and stereochemistry of residues 1–7 of teixobactin makes fibril formation possible. By having evolved a d-l-l-d-d-l-l pattern of stereochemistry with a hydrophobic-hydrophobic-hydrophillic-hydrophilic-hydrophobic-hydrophobic-hydrophilic pattern of side chains, Eleftheria terrae has achieved an amyloidogenic non-ribosomal peptide that can assemble to form amphiphilic β-sheets and amyloid-like fibrils that can bind oxyanions. On the basis of our crystal structure, we have proposed a working model for the mechanism of action of teixobactin involving the formation of β-sheet dimers or higher-order supramolecular assemblies. We further recognize that the crystallographic observation of supramolecular assembly 34,35 and its potential involvement in antibiotic activity 36,37 does not assure its biological relevance.38,39 We envision the model put forth here to be worthy of further study and anticipate reporting these studies in due course.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Nicholas Chim and Huiying Li for helpful advice with X-ray crystallography and Dr. Li Xing for helpful assistance with TEM imaging. This work was supported by the National Institutes of Health (grants 1R21AI121548 and 1R56AI137258). H. Y. acknowledges the Allegan for fellowship support. M. W. acknowledges the support from the Ministry of Science and Higher Education, Republic of Poland (Mobility Plus grant no. 1647/MOB/V/2017/0). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, NIGMS (including P41GM103393).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b07709.
Procedures for the synthesis of N-Me-d-PheI1,N-Me-d-Gln4,Lys10-teixobactin (3), MIC assays, solubility assays, ThT fluorescence assays, and TEM imaging; characterization data (HPLC, ESI-MS) for analogue 3; details of X-ray crystallographic data collection, processing, and refinement.
Crystallographic coordinates of analogue 3 were deposited into the Protein Data Bank with PDB code 6E00 (data collected on a synchrotron source at 2.07 Å wavelength).
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schäberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Homma T; Nuxoll A; Gandt AB; Ebner P; Engels I; Schneider T; Götz F; Lewis K; Conlon BP Dual Targeting of Cell Wall Precursors by Teixobactin Leads to Cell Lysis. Antimicrob. Agents Chemother. 2016, 60, 6510–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong Y; Sun X; Gao H; Meyer KJ; Lewis K; Rao Y Developing Equipotent Teixobactin Analogues against Drug-Resistant Bacteria and Discovering a Hydrophobic Interaction between Lipid II and Teixobactin. J. Med. Chem. 2018, 61, 3409–3421. [DOI] [PubMed] [Google Scholar]
- 4.Jin K; Sam IH; Po KHL; Lin D; Ghazvini Zadeh EH; Chen S; Yuan Y; Li X Total synthesis of teixobactin. Nat. Commun. 2016, 7, 12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parmar A; Lakshminarayanan R; Iyer A; Mayandi V; Leng Goh ET; Lloyd DG; Chalasani MLS; Verma NK; Prior SH; Beuerman RW; Madder A; Taylor EJ; Singh I Design and Syntheses of Highly Potent Teixobactin Analogues against Staphylococcus aureus, Methicillin-Resistant Staphylococcus aureus (MRSA), and Vancomycin-Resistant Enterococci (VRE) in Vitro and in Vivo. J. Med. Chem. 2018, 61, 2009–2017. [DOI] [PubMed] [Google Scholar]
- 6.Abdel Monaim SAH; Jad YE; El-Faham A; de la Torre BG; Albericio F Teixobactin as a scaffold for unlimited new antimicrobial peptides: SAR study. Bioorganic Med. Chem 2018, 26, 2788–2796. [DOI] [PubMed] [Google Scholar]
- 7.Yang H; Chen KH; Nowick JS Elucidation of the Teixobactin Pharmacophore. ACS Chem. Biol 2016, 11, 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kåhrström CT Antimicrobials: A new drug for resistant bugs. Nat. Rev. Microbiol 2015, 13, 126–127. [DOI] [PubMed] [Google Scholar]
- 9.Wen P; Vanegas JM; Rempe SB; Tajkhorshid E Probing Key Elements of Teixobactin-Lipid II Interactions in Membrane. Chem. Sci 2018, 9, 6997–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen KH; Le SP; Han X; Frais JM; Nowick JS Alanine scan reveals modifiable residues in teixobactin. Chem. Commun 2017, 53, 11357–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upon further incubation, the fluorescence declines variably. This subsequent change in fluorescence may reflect further reorganization of the amyloid-like fibrils that form, such as assembly into the bundles of filaments that are observed by TEM.
- 12.Yang H; Du Bois DR; Ziller JW; Nowick JS X-ray crystallographic structure of a teixobactin analogue reveals key interactions of the teixobactin pharmacophore. Chem. Commun. 2017, 53, 2772–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer R; Li H; Nowick JS X-ray Crystallographic Structures of Trimers and Higher-Order Oligomeric Assemblies of a Peptide Derived from Aβ17-36. J. Am. Chem. Soc 2014. 136, 5595–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer RK; Kreutzer AG; Salveson PJ; Li H; Nowick JS X-ray Crystallographic Structures of Oligomers of Peptides Derived from β2-Microglobulin. J. Am. Chem. Soc 2015. 137, 6304–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson MB; Brown DB; Vasquez CA; Ziller JW; Johnston KM; Weiss GA Synthesis and Explosion Hazards of 4-Azido-l-phenylalanine. J. Org. Chem. 2018, 83, 4525–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malkov AV; Stončius S; MacDougall K, N.; Mariani, A.; McGeoch, G. D.; Kočovský, P. Formamides derived from N-methyl amino acids serve as new chiral organocatalysts in the enantioselective reduction of aromatic ketimines with trichlorosilane. Tetrahedron 2006, 62, 264–284. [Google Scholar]
- 17.Kabsch W XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foadi J; Aller P; Alguel Y; Cameron A; Axford D; Owen RL; Armour W; Waterman DG; Iwata S; Evans G Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov GN; Vagin AA; Dodson EJ Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 20.Sangwan S; Zhao A; Adams KL; Jayson CK; Sawaya MR; Guenther EL; Pan AC; Ngo J; Moore DM; Soriaga AB; Do TD; Goldschmidt L; Nelson R; Bowers MT; Koehler CM; Shaw DE; Novitch BG; Eisenberg DS Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 8770–8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel Monaim SAH; Jad YE; Ramchuran EJ; El-Faham A; Govender T; Kruger HG; de la Torre BG; Albericio F Lysine Scanning of Arg10–Teixobactin: Deciphering the Role of Hydrophobic and Hydrophilic Residues. ACS Omega 2016, 1, 1262–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmar A; Prior SH; Iyer A; Vincent CS; Van Lysebetten D; Breukink E; Madder A; Taylor EJ; Singh I Defining the molecular structure of teixobactin analogues and understanding their role in antibacterial activities. Chem. Commun 2017, 53, 2016–2019. [DOI] [PubMed] [Google Scholar]
- 23.Lewandowski et al. have concurrently reported NMR-based structural studies of teixobactin in aqueous and membranelike environments, both with and without lipid II and lipid II analogues. [Öster, C.; Walkowiak, G. P.; Hughes, D. E.; Spoering, A. L.; Peoples, A. J.; Catherwood, A. C.; Tod, J. A.; Lloyd, A. J.; Herrmann, T.; Lewis, K.; Dowson, C.; Lewandowski, J. R. Structural studies suggest aggregation as one of the modes of action for teixobactin. Chem. Sci. 2018, Accepted Manuscript (DOI: 10.1039/C8SC03655A)] These studies indicate that teixobactin, in the presence of lipid II, rapidly aggregates and the residues 2–6 rearrange from random coil like conformation to a more extended β-strand like conformation. [DOI] [PMC free article] [PubMed]
- 24.A 2:1 teixobactin:lipid II binding stoichiometry was reported in the original 2015 Nature paper on teixobactin (reference 1). The crystallographic observation of putative pyrophosphate binding sites created by dimers could potentially support either a 2:1 or a 2:2 teixobactin:lipid II stoichiometry, depending on how the binding of one molecule of lipid II to the dimer affects the accessibility of the other site of the dimer.
- 25.Breukink E; de Kruijff B Lipid II as a target for antibiotics. Nat. Rev. Drug Discov 2006, 5, 321–332. [DOI] [PubMed] [Google Scholar]
- 26.de Kruijff B; van Dam V; Breukink E Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids 2008, 79, 117–121. [DOI] [PubMed] [Google Scholar]
- 27.Lo MC; Men H; Branstrom A; Helm J; Yao N; Goldman R; Walker S A new mechanism of action proposed for ramoplanin. J. Am. Chem. Soc 2000, 122, 3540–3541. [Google Scholar]
- 28.Lo MC; Helm JS; Sarngadharan G; Pelczer I; Walker S A new structure for the substrate-binding antibiotic ramoplanin. J. Am. Chem. Soc 2001, 123, 8640–8641. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y; Helm JS; Chen L; Ye X-Y; Walker S Ramoplanin inhibits bacterial transglycosylases by binding as a dimer to lipid II. J. Am. Chem. Soc 2003, 125, 8736–8737. [DOI] [PubMed] [Google Scholar]
- 30.Walker S; Chen L; Hu Y; Rew Y; Shin D; Boger DL Chemistry and biology of ramoplanin: a lipoglycodepsipeptide with potent antibiotic activity. Chem. Rev 2005, 105, 449–476. [DOI] [PubMed] [Google Scholar]
- 31.Hamburger JB; Hoertz AJ; Lee A; Senturia RJ; McCafferty DG; Loll PJ; A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential Lipid II docking interface. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 13759–13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu ST; Breukink E; Tischenko E; Lutters MA; de Kruijff B; Kaptein R; Bonvin AM; Nuland NA The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol 2004, 11, 963–967. [DOI] [PubMed] [Google Scholar]
- 33.Watson JD; Milner-White EJ A novel main-chain anion-binding site in proteins: The nest. A particular combination of phi, psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2002, 315, 171–182. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GM; Jones PG; Kennard O; Williams DH; Smith GA Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 1978, 271, 223–225. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer M; Schneider TR; Sheldrick GM Crystal structure of vancomycin. Structure 1996, 4, 1509–1515. [DOI] [PubMed] [Google Scholar]
- 36.Sharman GJ; Try AC; Dancer RJ; Cho YR; Staroske T; Bardsley B; Maguire AJ; Cooper MA; O'Brie DP; Williams. D. H. The Roles of Dimerization and Membrane Anchoring in Activity of Glycopeptide Antibiotics against Vancomycin-Resistant Bacteria. J. Am. Chem. Soc 1997, 119, 12041–12047. [Google Scholar]
- 37.Cooper MA; Williams DH Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem. Biol. 1999, 6, 891–899. [DOI] [PubMed] [Google Scholar]
- 38.Ge M; Chen Z; Onishi HR; Kohler J; Silver LL; Kerns R; Fukuzawa S; Thompson C; Kahne D Vancomycin Derivatives That Inhibit Peptidoglycan Biosynthesis Without Binding d-Ala-d-Ala. Science 1999, 284, 507–511. [DOI] [PubMed] [Google Scholar]
- 39.Kahne D; Leimkuhler C; Lu W; Walsh C Glycopeptide and Lipoglycopeptide Antibiotics. Chem. Rev 2005, 105, 425–448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.