Abstract
Recent studies suggest that, even within a single adipose depot, there may be distinct subpopulations of adipocytes. To investigate this cellular heterogeneity, we have developed multiple conditionally immortalized clonal preadipocyte lines from white adipose tissue of mice. Analysis of these clones reveals at least three white adipocyte subpopulations. These subpopulations have differences in metabolism and differentially respond to inflammatory cytokines, insulin, and growth hormones. These also have distinct gene expression profiles and can be tracked by differential expression of three marker genes: Wilms’ tumor 1, transgelin, and myxovirus 1. Lineage tracing analysis with dual‐fluorescent reporter mice indicates that these adipocyte subpopulations have differences in gene expression and metabolism that mirror those observed in the clonal cell lines. Furthermore, preadipocytes and adipocytes from these subpopulations differ in their abundance in different fat depots. Thus, white adipose tissue, even in a single depot, is comprised of distinct subpopulations of white adipocytes with different physiological phenotypes. These differences in adipocyte composition may contribute to the differences in metabolic behavior and physiology of different fat depots.
Keywords: adipose tissue, development, lineage tracing, metabolic syndrome, obesity
Subject Categories: Development & Differentiation, Metabolism
Introduction
The increasing prevalence of obesity is a major factor driving the worldwide epidemic of type 2 diabetes and metabolic syndrome. Adipose tissue not only stores energy, but also controls metabolism through secretion of hormones, cytokines, proteins, and microRNAs that affect the function of cells and tissues throughout the body (Klaus, 2004; Rosen & Spiegelman, 2006; Trujillo & Scherer, 2006; Stephens, 2012; Kusminski et al, 2016; Thomou et al, 2017). Accumulation of visceral white adipose tissue (WAT) leads to central obesity and is associated with insulin resistance and increased risk of metabolic disease, whereas accumulation of subcutaneous WAT leads to peripheral obesity and may even be protective of metabolic syndrome (Janssen et al, 2002; Fox et al, 2007). This is due, at least in part, to intrinsic functional differences between adipocytes in these depots. Indeed, transplantation of visceral versus subcutaneous adipose tissue to recipient mice can differentially modify host glucose tolerance and insulin sensitivity (Tran et al, 2008; Stanford et al, 2015). Adipocytes and preadipocytes from different WAT depots of mice and humans have different patterns of gene expression, many of which are in developmental and patterning genes. These differences have been shown to be conserved after multiple passages in vitro consistent with genetic programming (Gesta et al, 2006; Tchkonia et al, 2006; Macotela et al, 2012). In addition, different adipose depots secrete different combinations of hormones (adipokines), cytokines, and other molecules that can affect the function of tissues and metabolism throughout the body.
In addition to white adipose tissue, mammals have discrete collections of energy‐burning brown adipose tissue that differs both metabolically and in its gene expression profile, as well as intermediate cells referred to as beige or brite adipocytes, which are intermixed in some white fat depots and can be induced to become thermogenic by cold exposure or beta‐adrenergic stimulation (Cypess & Kahn, 2010; Schrauwen et al, 2015; Ramseyer & Granneman, 2016). While much attention has been paid to identifying differences between white, brown, and brite/beige adipocytes, there is growing evidence that there is functional heterogeneity among white adipocytes themselves. Thus, studies have shown that individual white adipocytes within a single depot may differ in insulin‐stimulated glucose uptake, maximal lipogenic rate, response to catecholamines, and uptake of free fatty acids (Salans & Dougherty, 1971; Gliemann & Vinten, 1974; Seydoux et al, 1996; Varlamov et al, 2015; Gao et al, 2017). Indeed, lineage tracing analyses have begun to uncover the differential developmental origins of adipocytes even within single fat depots (Chau et al, 2014; Sanchez‐Gurmaches & Guertin, 2014). Our laboratory recently showed that subpopulations of white adipocytes differ in their expression of the mesodermal homeobox gene Tbx15 and that this contributes to differences in oxidative phosphorylation and glycolysis in these cells (Lee et al, 2017a). This is distinct from the heterogeneity due to the presence of brown and brite adipocyte populations, although the latter may also have heterogeneity in thermogenic potential (Xue et al, 2015) and anabolic versus catabolic function (Lee et al, 2017b).
In the present study, we have used a combination of in vitro analysis of clonal preadipocyte cell lines from the Immortomouse™ and lineage tracing in vivo to investigate heterogeneity of white adipocyte subpopulations. Using this approach, we have identified at least three distinct subpopulations of white preadipocytes/adipocytes that are characterized by unique gene expression profiles and high expression of three different marker genes: Wilms’ tumor 1 (Wt1), transgelin (Tagln), and myxovirus 1 (Mx1). In vitro, these subpopulations have unique metabolic properties and differentially respond to exogenous stimuli. Lineage tracing analysis shows that these three preadipocyte markers define three independent white adipocyte subpopulations in vivo that retain the differential gene expression profiles and metabolic properties found in the clonal cell lines. These subpopulations are represented as different percentages of the preadipocyte and adipocyte population in different white adipose tissue depots. Thus, white adipose tissue is composed of molecularly and phenotypically distinct subpopulations of adipocytes which have cell autonomous differences in metabolic properties. This heterogeneity may modify the physiological impact of adipose tissue in different individuals and contribute to the variable response that increased adipose tissue mass has on the development of diabetes and metabolic syndrome.
Results
Cultured preadipocytes from immortomouse™ retain depot‐specific gene expression
We and others have previously observed that adipocytes and preadipocytes taken from different adipose tissue depots are characterized by differential expression of developmental and patterning genes (Gesta et al, 2006; Tchkonia et al, 2007; Dankel et al, 2010; Yamamoto et al, 2010; Macotela et al, 2012; Lee et al, 2013, 2017a; Ferrer‐Lorente et al, 2014). Consistent with these studies, in the white adipose tissue (WAT) of 8‐week‐old C57BL/6 mice, the expression of Short Stature Homeobox 2 (Shox2) and T‐box 15 (Tbx15) mRNA levels was about ~30‐fold higher in subcutaneous WAT depots (inguinal, scapular white) compared to visceral WAT (perigonadal, perirenal, mesenteric) depots, whereas HoxC8 and HoxA5 mRNA levels were ~2‐fold higher in perigonadal and perirenal WAT depots compared to subcutaneous WAT depots (Fig 1A).
Figure 1. Depot‐specific immortomouse cell lines retain the molecular signatures of the depot of origin.
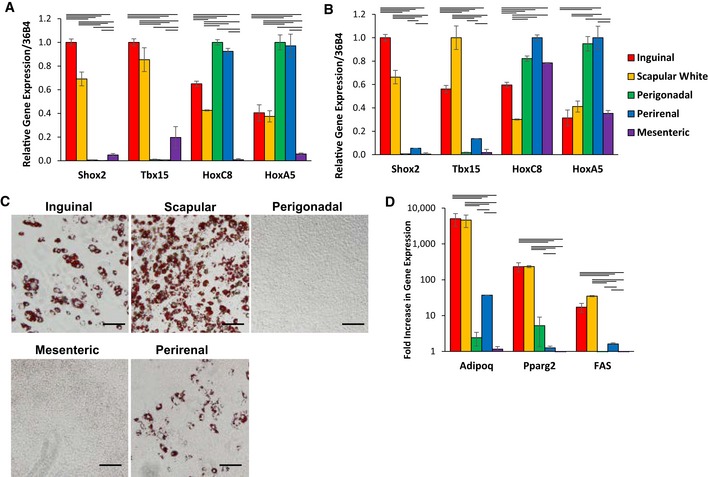
- Expression level of Shox2, Tbx15, HoxC8, and HoxC5, mRNA was compared using quantitative real‐time PCR (qPCR) of RNA isolated from different fat depots. Two subcutaneous depots (inguinal and scapular white), three intra‐abdominal fat depots (perigonadal, perirenal, and mesenteric), and interscapular brown adipose tissue of 8‐week‐old male C57BL/6 mice. Data are shown as mean ± SEM of six samples.
- Expression level of Shox2, Tbx15, HoxC8, and HoxC5, mRNA was compared using qPCR of RNA isolated from cultured stromovascular cells derived from adipose tissue of the Immortomouse™. The depot‐specific cell lines were made from inguinal, scapular white, perigonadal, perirenal, mesenteric, and interscapular brown adipose tissue of 8‐week‐old male mice. Data are shown as mean ± SEM of cell lines from three individual mice.
- Bright‐field image of depot‐derived cell lines after in vitro adipogenic differentiation and Oil Red O staining (Lee et al, 2017a). Original photographs were taken at 10× magnification (left panel). Scale bar = 100 μm.
- qPCR analysis for adiponectin (Adipoq), peroxisome proliferator‐activated receptor gamma (Pparγ2), and fatty acid synthase (FAS) in RNA isolated from depot‐derived cell lines after in vitro differentiation. Data are shown as mean ± SEM of three cell lines/group.
To investigate how adipocyte heterogeneity and differences in the expression of developmental genes might impact the biology of adipocytes and preadipocytes, we created preadipocyte cell lines from the stromovascular fraction (SVF) isolated from the scapular white, inguinal, perigonadal, perirenal, and mesenteric fat pads of 6‐week‐old male Immortomouse™ (Jat et al, 1991). The Immortomouse™ harbors a transgene that expresses the thermolabile simian virus 40 (Sv40) large T antigen tsA58 under the control of the interferon‐inducible murine H‐2K b promoter. This allows the isolation of conditionally immortalized cell lines directly from the transgenic mouse. These cells undergo reversible immortalization when grown under permissive conditions (33°C, with interferon‐gamma), but stop dividing and are capable of differentiation when grown under standard conditions (37°C, in the absence of interferon‐gamma). qPCR of these cell lines under standard conditions demonstrated that the differential gene expression of developmental genes between white adipose tissue depots was retained in the cell lines derived from these depots. Thus, Tbx15 and Shox2 mRNA expression were high in cell lines derived from subcutaneous fat, while HoxC9 and HoxA5 mRNA expression were higher in cells derived from visceral adipose tissue depots (Fig 1B). Oil Red O staining of these cell lines after in vitro differentiation demonstrated that those derived from subcutaneous WAT depots (inguinal, scapular white) were able to differentiate into adipocytes, whereas the visceral immortalized preadipocytes showed little to no differentiation (Fig 1C). These results were confirmed by qPCR of markers of differentiation, including adiponectin (Adipoq), peroxisome proliferator‐activated receptor gamma 2 (Pparγ2), and fatty acid synthase (Fas), which were induced only during adipogenic differentiation of cell lines from subcutaneous fat and not mesenteric or perigonadal fat, with a slight induction in the perirenal fat cell line (Fig 1D).
Clonal analysis of adipocytes derived from immortomouse™ adipose tissue
To investigate the heterogeneity of white preadipocytes in more detail, after the second passage, subcutaneous and perigonadal depot‐specific cell lines from the Immortomouse were subjected to cloning by ungated flow sorting of single cells into wells of a cell culture plate. This yielded 94 clonal cell lines—45 from subcutaneous fat and 49 from perigonadal fat. After induction of differentiation with dexamethasone, IBMX, insulin, rosiglitazone, T3, and indomethacin (Lee et al, 2011), 31 of the subcutaneous and 18 of the perigonadal clones were highly and reproducibly adipogenic, as evidenced by the accumulation of neutral lipids visualized by Oil Red O staining (Fig 2A). Since 37% of the isolated perigonadal depot‐derived clones could differentiate, but no differentiated cells were observed in the original pool of cells from which the clones were selected, this suggests the presence of cells releasing some anti‐adipogenic factor which inhibits differentiation in the mixed cell culture.
Figure 2. Clonal cell lines represent three distinct subtypes of adipocytes based on differential gene expression.
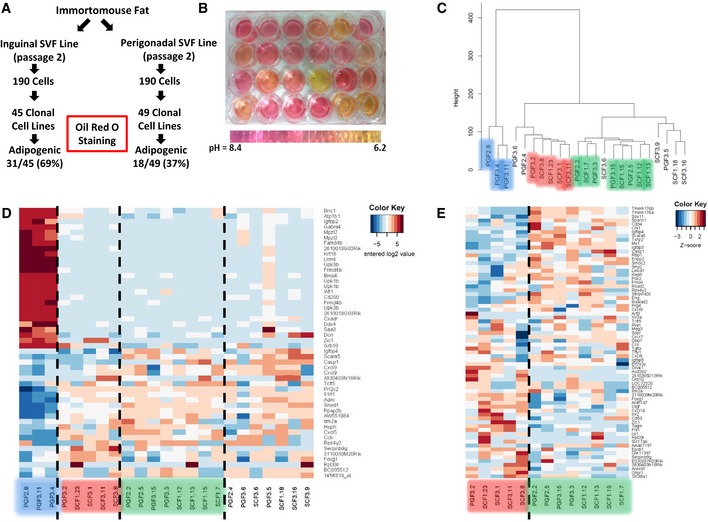
- Schematic depicting the experimental design of establishing clonal cell lines from the SVF isolated from inguinal and perigonadal clonal cell lines.
- Representative picture of preadipocyte clonal cell lines showing media color 48 h after last media change. Cells in all wells were 100% confluent when media was changed.
- Hierarchical clustering of preadipocyte clonal cell line gene expression using Ward's agglomeration.
- Heatmap depicting the 50 most variable genes among all preadipocyte clonal cell lines. Red represents high expression of genes, while blue represents low expression of genes.
- Heatmap depicting the 50 most variable genes among Type 2 and 3 preadipocyte clonal cell lines. Red represents high expression of genes, while blue represents low expression of genes.
An interesting and unexpected finding was that during routine culture of the subcutaneous and visceral/perigonadal clonal cell lines, we observed extreme variation in media acidification rates that was unrelated to the fat pad of origin, the differentiation capacity of the cells, or the rate of their proliferation, suggesting metabolic heterogeneity (Figs 2B and EV1A). To further investigate this possibility, 24 clonal cell lines (12 each from subcutaneous and perigonadal fat) were selected based on variable media acidification rates, and their mRNA expression pattern determined by microarray analysis (Fig EV1B). The expression data were subjected to clustering analysis utilizing three different approaches: hierarchical clustering with Ward's minimum variance, the average‐based agglomeration method, and affinity propagation (Frey & Dueck, 2007). Based on a consensus of the three algorithms, we identified three subgroups termed Types 1, 2, and 3 and highlighted in blue, red, and green in Figs 2C and EV1C and D. While Type 1 preadipocytes were solely perigonadal in origin, clones from both subcutaneous and perigonadal depots were found in the Type 2 and 3 clusters. The most dramatic differences in gene expression were between Type 1 preadipocytes and the other two groups, as can be visualized by a heatmap of the 50 most differential genes and by principal component analysis (Figs 2D and EV1E), however, significant differences were also present between Types 2 and 3, as visualized by a heatmap of the 50 most differential genes between these two groups (Fig 2E). Despite these differences in gene expression, the preadipocyte lines from each of the three clusters were reproducibly adipogenic, as visualized by Oil Red O staining of neutral lipids, when subjected to adipocyte differentiation conditions. In addition, all three clusters were also able to differentiate into osteoblasts and chondrocytes in vitro using conditions that induce bone and cartilage development (Boquest et al, 2005) as visualized by Alizarin Red S staining of calcium deposition and Alcian Blue of acidic mucins, respectively (Fig EV1F). Thus, although the cells were derived from the SVF of adipose tissue, under optimized conditions of differentiation, all three subtypes of cells behaved as multipotent mesenchymal stem cells and were not limited to adipocyte differentiation.
Figure EV1. Microarray and clustering analysis of Immortomouse™ clonal preadipocyte cell lines.
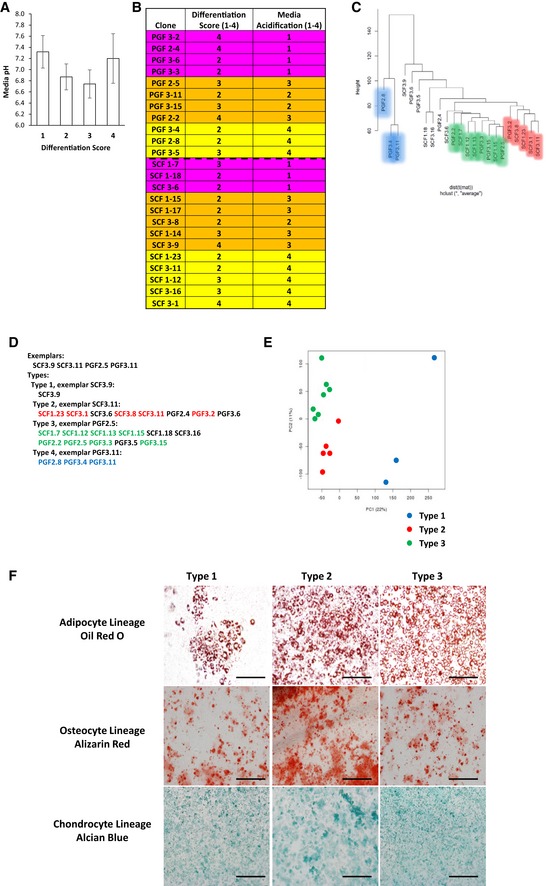
- pH measurement of confluent preadipocyte cell lines with different adipogenic differentiation potential. The cells were scored on a scale from 1 to 4 for adipogenic differentiation after Oil Red O staining (1: < 25%; 2: 25–50%; 3: 50–75%, and 4: 75–100% of cells differentiated). Data are shown as mean ± SEM of 4–10 cell lines/group.
- Clonal preadipocyte cell lines used for microarray analysis. Cells were scored on a scale from 1 to 4 for adipogenic differentiation after Oil Red O staining (1: < 25%; 2: 25–50%; 3: 50–75%, and 4: 75–100% of cells differentiated) and 1–4 for media acidification by media color based on scale in Fig 2B (1: pH < 6.5; 2: pH from 6.5 to 6.9; 3: pH from 7.0 to 7.6; 4: pH > 7.7). Cells were selected for differentiation scores of 2 or greater and variable media acidification rates.
- Affinity propagation clustering of preadipocyte clonal cell line gene expression. Exemplars are representatives of their cluster.
- Hierarchical clustering utilizing the average method of agglomeration of preadipocyte clonal cell line gene expression.
- Principal component analysis of preadipocyte clonal cell line gene expression.
- Bright‐field image of clonal preadipocyte lines after in vitro adipogenic, osteogenic, and chondrogenic differentiation. Adipocytes, osteocytes, and chondrocytes were stained with Oil Red O, Alizarin Red, and Alcian Blue, respectively. Original photographs were at 10× magnification (left panel). Scale bar = 100 μm.
Adipocyte subtypes display distinct gene expression profiles
To confirm differences in gene expression between the different adipocyte subtypes, we performed qPCR analysis of the most differential genes highly expressed in one type of preadipocyte. These showed that compared to other types, the Type 1 preadipocytes have ~100‐fold higher expression of three marker genes—Wilms’ tumor 1 (Wt1), leucine‐rich repeat neuronal 4 (Lrrn4), and uroplakin 3b (Upk3b). During adipogenic differentiation of these clones, the levels of Wt1 and Lrrn4 levels decreased by ~50% but remained significantly higher in Type 1 clones than in the other clusters (Fig 3A–C). Wt1, Lrrn4, and uroplakin 3b are known markers of primary mesothelial cells, which have been shown to be able to give rise to adipocytes both in vitro and in vivo (Kanamori‐Katayama et al, 2011; Lansley et al, 2011; Chau et al, 2014). On bright‐field microscopy, undifferentiated Type 1 cells displayed the cobblestone morphology of mesothelial cells, whereas Type 2 and 3 cells were fibroblast‐like in morphology (Fig 3D).
Figure 3. Clonal cell line clusters exhibit distinct gene expression profiles and morphology.
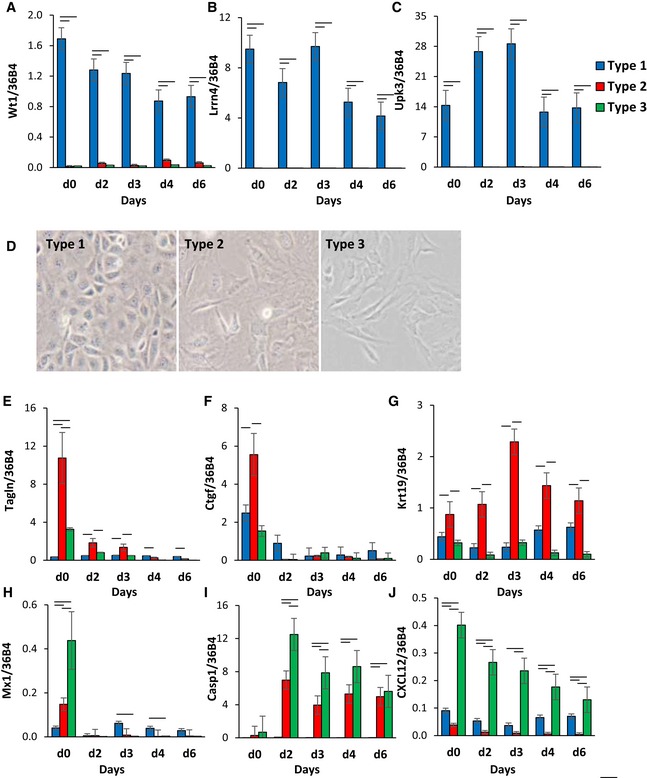
-
A–CExpression of Wt1, leucine‐rich repeat neuronal 4 (Lrrn4), uroplakin 3b (Upk3b) in Immortomouse clonal cell line clusters as preadipocytes (d0) and during 6 days of adipogenic differentiation. Data are shown as mean ± SEM of 3–7 cell lines/group. Bars indicate significant differences between groups (P < 0.05; paired two‐way ANOVA).
-
DBright‐field images of preadipocyte cell lines under standard culture conditions. The photographs were taken at 10× magnification.
-
E–JExpression of Tagln, connective tissue growth factor (Ctgf), keratin 19 (Krt19), Mx1, caspase 1 (Casp1), and C‐X‐C motif chemokine ligand 12 (Cxcl12) in Immortomouse clonal cell line clusters as preadipocytes (d0) and during 6 days of adipogenic differentiation. Data are shown as mean ± SEM of 3–7 cell lines/group. Bars indicate significant differences between groups (P < 0.05; paired two‐way ANOVA).
Type 2 clones were marked by ~2‐ to 3‐fold high expression of transgelin (Tagln) and connective tissue growth factor (Ctgf) and keratin 19 (Krt19) compared to Type 1 and 3 clones. The expression of both Tagln and Ctgf was decreased by 90–95% during adipogenic differentiation, indicating that the expression of these markers is primarily in the adipocyte precursors and lost in the mature adipocytes, while Krt19 expression remained high in Type 2 clones even after differentiation (Fig 3E–G).
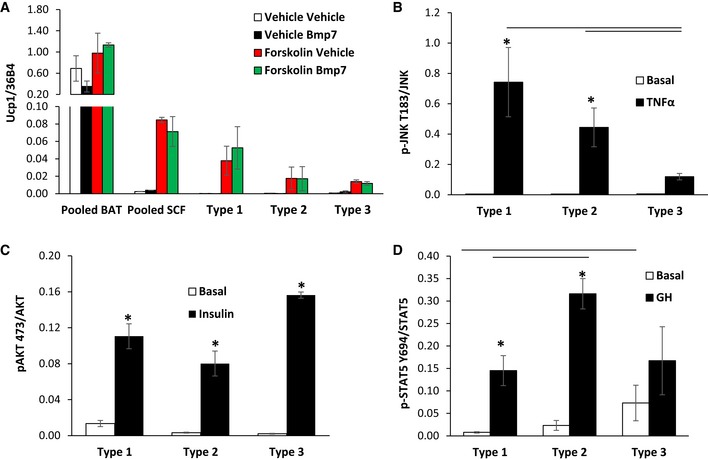
Type 3 clones were marked by high expression of myxovirus 1 (Mx1), caspase 1 (Casp1), and chemokine (C‐X‐C motif) ligand 12 (Cxcl12). During adipocyte differentiation, the expression of both Mx1 and Cxcl12 decreased, while mRNA levels of Casp1 increased over 20‐fold (Fig 3H–J). These differences were not related to browning potential. Thus, treatment with inducers of brown/brite adipocytes, including BMP7 (Tseng et al, 2008) or forskolin (Wu et al, 2012), resulted in no significant differences in uncoupling protein 1 (Ucp1) expression between the three types of adipocytes and was low compared to the levels in brown adipocytes (Fig EV2A). Taken together, these clonal cell lines represent three subpopulations of white preadipocytes with distinct gene expression profiles in vitro, which are independent of browning potential.
Figure EV2. Measurement of Ucp1 expression and quantitation of signaling in preadipocyte cell cultures in response to exogenous stimuli.
-
AqPCR analysis for Ucp1 in RNA isolated from Immortomouse adipocyte cell lines: pooled brown (BAT), pooled subcutaneous (SCF), Type 1, Type 2, and Type 3, after in vitro differentiation. Cells were treated ± 3.3 nM BMP7 for 3 days prior to differentiation or treated ± 10 μM forskolin for 4 h. Data are shown as mean ± SEM of 2–7 cell lines per group.
-
B–DQuantitation of Western blots depicted in Fig 4I. Data are shown as mean ± SEM of three independent cell lines per group; each experiment was performed at least twice. Bars indicate significant differences between groups (P < 0.05; two‐way ANOVA) in all panels. Asterisks indicate significant differences between basal and treated samples (P < 0.05; Student's t‐test).
White adipocyte subtypes have differential metabolic properties and response to exogenous stimuli
While all of three types of white adipocyte precursor cells had the ability to differentiate, quantitation of triglyceride content after differentiation revealed that Type 1 adipocytes accumulated ~50% less triglyceride per well than Type 1 and 3 preadipocytes (Fig 4A). This occurred despite the fact that there were no significant differences in uptake of [H3] 2‐deoxyglucose in either the basal or insulin‐stimulated states in the Type 1–3 preadipocytes (Fig 4B). After differentiation, however, both Type 2 and 3 adipocytes showed a significant increase in glucose uptake following insulin stimulation, which was not observed in Type 1 cells (Fig 4C). Similarly, insulin‐stimulated de novo lipogenesis, as measured by incorporation of [C14]‐labeled glucose in lipid, was greater in Type 2 and 3 adipocytes, although this did not quite reach statistical significance due to variability between cell lines (Fig 4D). On the other hand, all three cell types responded to isoproterenol stimulated lipolysis, with no differences between the adipocyte subtypes (Fig 4D and E).
Figure 4. Adipocyte subtypes exhibit differential phenotypic characteristics and signaling in response to exogenous stimuli.
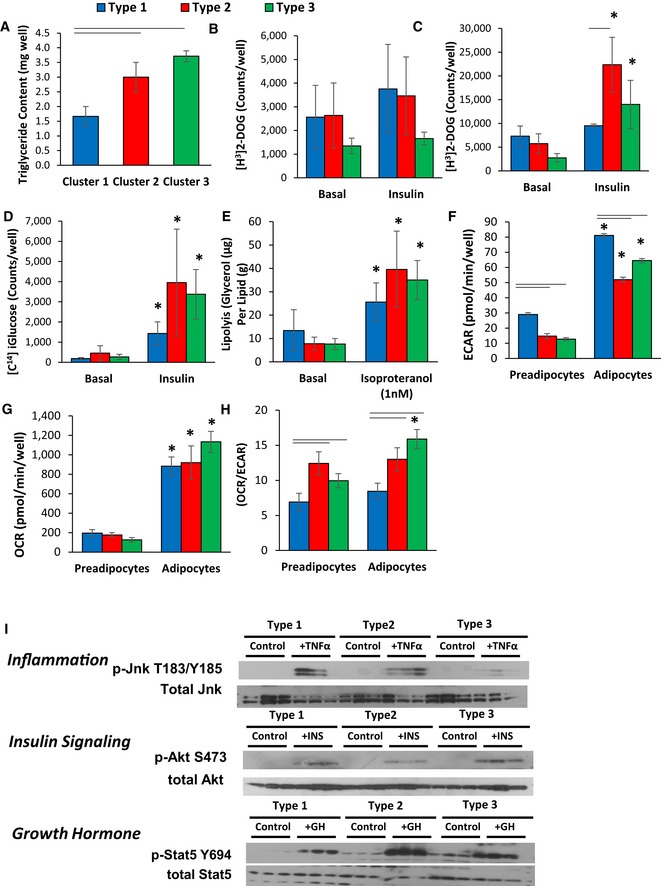
- Triglyceride content of cell cultures after 6 days of differentiation using the protocol described in Materials and Methods. Data are shown as mean ± SEM of 3–7 samples, and the entire experiment was repeated twice.
- [3H]‐labeled 2‐deoxyglucose uptake in preadipocyte cell lines in the basal state and after pretreatment for 20 min with 100 nM insulin. Uptake was measured for 1 h, and data are shown as mean ± SEM of 3–7 samples and repeated twice.
- [3H]‐labeled 2‐deoxyglucose uptake in adipocytes after 6 days of adipogenic differentiation in the basal state and after stimulation for 20 min with 100 nM insulin. Uptake was measured for 1 h, and data are shown as mean ± SEM of 3–7 samples. The experiment was repeated twice.
- [14C] D‐glucose incorporation into lipids in adipocytes after 6 days of adipogenic differentiation in the basal state and after stimulation for 20 min with 100 nM insulin. [14C] D‐glucose incorporation was measured after 3 h, and data are shown as mean ± SEM of 3–7 samples. The experiment was repeated twice.
- Lipolysis rates after stimulation with 1 nM isoproterenol as measured by glycerol release from adipocytes after 6 days of adipogenic differentiation. Data are shown as mean ± SEM of 3–7 replicates and normalized by lipid content of the cells. The entire experiment was repeated twice.
- Extracellular acidification rate (ECAR) of preadipocytes and adipocytes after 4 days of in vitro differentiation was determined by calculating the area under the curve (AUC) during measurements of basal respiration. The whole experiment was repeated three times. Data are shown as mean ± SEM of 3–7 cell lines per group.
- Basal respiration of preadipocytes and adipocytes after 4 days of in vitro differentiation was determined by calculating the area under the curve (AUC) during measurements of oxygen consumption rate (OCR). The whole experiment was repeated three times. Data are shown as mean ± SEM of 3–7 cell lines per group.
- Ratio of oxygen consumption rate (OCR) to extracellular acidification rate (ECAR) of preadipocytes and adipocytes after 4 days of in vitro differentiation. Data are shown as mean ± SEM of 3–7 cell lines per group.
- Representative Western blot analysis of three independent adipocyte cell lines after 6 days of in vitro differentiation for: pJnk T183/Y185 and total Jnk after treatment with 10 nM TNFα for 15 min (Inflammation); pAkt S473 and total Akt after treatment with 100 nM insulin for 15 min (Insulin Signaling); pStat5 Y694 and total Stat5 after treatment with 500 nM GH for 20 min (Growth Hormone).
Acidification of cell culture media is caused by both a plasma membrane H(+)‐ATPase pump and lactic acid production as an end product of glycolysis (Montcourrier et al, 1997). Consistent with the data showing different levels of pH in the culture media (Fig 2B), all of the Type 1 cell lines demonstrated high media acidification rates (Fig EV1B), whereas the Type 2 and Type 3 cells exhibited a mixture of high and low acidification rates. Seahorse flux analysis confirmed these differences with a 20–30% higher basal and maximal extracellular acidification rates (ECAR) in both Type 1 preadipocytes and adipocytes compared to those from Types 2 and 3 (Fig 4F). These changes occurred without significant changes in basal and maximal oxygen consumption rates (OCR) between adipocyte clusters. This resulted in a 20–30% lower ratio of OCR/ECAR in Type 1 preadipocytes and adipocytes compared to other subtypes, indicating a shift from oxidative to glycolytic metabolism (Fig 4G and H).
Numerous factors, including inflammatory cytokines, insulin, and growth hormone, affect adipocyte biology. To test whether exogenous hormones differentially affect these adipocyte subpopulations, three independent clonal preadipocyte lines from each subpopulation were differentiated, serum‐starved for 4 h, and stimulated with 20 ng/ml TNFα (TNFα), 100 nM insulin, or 400 ng/ml human growth hormone (GH) for 15 min. TNFα activated JNK signaling in both Type 1 and 2 adipocyte clones from as evidenced by robust phosphorylation of JNK on Thr183/Tyr185, whereas Type 3 clones were largely unresponsive (Figs 4I and EV2B). On the other hand, stimulation with insulin increased phosphorylation of AKT‐Ser473 in most robustly in Type 1 and 3 adipocytes, with 30–50% lower levels of AKT‐Ser473 phosphorylation observed in Type 2 clones although these differences did not quite reach statistical significance (Figs 4I and EV2C). By contrast, while growth hormone induced activation of Stat5‐Tyr694 tyrosine phosphorylation in all clones, stimulation was twofold higher in Type 2 cells compared to Types 1 and 3 (Figs 4I and EV2D). These differential responses were not due to differences in receptor abundance, since no significant differences in the expression of TNFα receptors, insulin receptors, or GH receptors were noted in the microarray analysis between the subtypes. Thus, these three subtypes of white preadipocytes/adipocytes possess not only differences in gene expression, but also significant metabolic differences as well as response to hormonal stimuli.
Utilizing lineage tracing models to differentially label adipocyte subpopulations
To determine whether the Type 1, 2, and 3 clonal cell lines might represent unique preadipocyte and adipocyte subpopulations in vivo, we crossed Wt1‐creERT2 (Zhou et al, 2008), Tagln‐cre (Zhang et al, 2006), or Mx1‐cre (Kuhn et al, 1995) mice to mice carrying the membrane Tomato/membrane Green (mT/mG) fluorescent protein reporter expressed in the Rosa26 locus (Muzumdar et al, 2007). This allows lineage tracing since in the absence of Cre expression, all cells express a cell membrane‐localized Tomato red, whereas in cells expressing the Cre recombinase, as well as all future cell lineages derived from these cells, there is gene rearrangement resulting in a loss of Tomato (red) fluorescent protein (mTFP) and expression of a membrane‐localized green fluorescent protein (mGFP). These membrane markers outline cell morphology and are distinguishable by FACS analysis, thus allowing identification of both preadipocytes and adipocytes marked by these transgenes. In these experiments, the Tagln‐cre was constitutively expressed, whereas the Wt1‐creERT2 was activated at e14.5 by administration of tamoxifen, and the Mx1‐cre was induced at birth by administration of Poly I:C. At 5–6 months of age, adipose tissue depots were collected from Wt1‐creERT2 ROSA26mT/mG, Tagln‐cre ROSA26mT/mG, and Mx1‐cre ROSA26mT/mG mice, digested with collagenase, and the SVF subjected to FACS separation to isolate the lineage‐marked preadipocytes (Rodeheffer et al, 2008). In this analysis, cells that were positive for markers of other committed lineages, i.e., endothelial cells (CD31), macrophages (CD45), and erythrocytes (Ter119), were removed to create a “lineage‐negative” population (Lin‐), and cells triple‐positive for CD29, CD34, and Sca1 in this population were considered to be preadipocytes and further sorted for Tomato or GFP fluorescence (Fig EV3A and B). Both the primary GFP+ and Tomato+ sorted cells from Wt1‐creERT2 ROSA26mT/mG and Tagln‐cre ROSA26mT/mG mice were able to undergo adipogenic differentiation, as assessed by Oil Red O staining of neutral lipids. The low numbers of primary GFP+ cells derived from Mx1‐cre ROSA26mT/mG did not allow us to confirm their adipogenic potential (Fig EV3A and B).
Figure EV3. FACS Sorting and Adipogenic Differentiation of Rosa26mT/mG preadipocytes.
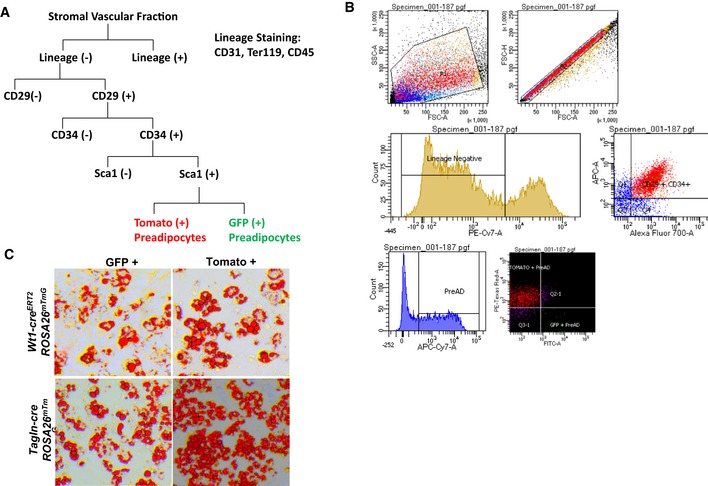
- Schematic of FACS sorting for GFP‐ and Tomato‐positive preadipocytes.
- Representative FACS plot of LacZ (+) APCs from the subcutaneous and perigonadal fat depots of Tbx15‐LacZ male mice at 6–8 weeks of age.
- Bright‐field image of primary mGFP‐ and mTomato‐positive preadipocytes after in vitro adipogenic differentiation. Neutral lipids were stained with Oil Red O, and original photographs were at 10× magnification (left panel).
Primary preadipocyte subtypes from display differential gene expression and metabolic properties
To confirm differences in the differential gene expression that we observed in preadipocytes subpopulations from the Immortomouse™ clonal cell lines, we performed qPCR analysis on mGFP‐ and mTomato‐positive preadipocytes isolated from the pooled visceral fat depots of Wt1‐creERT2;Rosa26mT/mG mice, the pooled subcutaneous fat depots of Mx1‐cre;Rosa26mT/mG, and both the pooled visceral and subcutaneous depots from Tagln‐cre;Rosa26mT/mG mice. Markers of Type 1 adipocytes, Wt1, Lrrn4, and Upk3b mRNA expression were 5‐ to 10‐fold higher in GFP+ compared to Tomato+ preadipocytes isolated from Wt1‐creERT2;Rosa26mT/mG mice, with no enrichment in expression of these markers in GFP+ preadipocytes from Tagln‐cre ROSA26mT/mG or Mx1‐cre ROSA26mT/mG mice (Fig 5A–C). Additionally, markers of Type 2 preadipocytes, including Tagln, Ctgf, and Krt19 mRNA, were significantly increased by 3‐ to ~50‐fold in GFP+ compared to Tomato+ preadipocytes isolated from either the visceral or subcutaneous fat of Tagln‐cre;Rosa26mT/mG mice, with no differences in these markers in GFP+ preadipocytes derived from Wt1‐creERT2;Rosa26mT/mG or Mx1‐cre;Rosa26mT/mG mice (Fig 5D–F). Finally, markers of Type 3 adipocytes, Mx1 and Casp1 mRNA, were increased ~5‐fold in GFP+ compared to Tomato+ preadipocytes isolated from Mx1‐cre;Rosa26mT/mG mice, while the expression of Cxcl12 mRNA remained unchanged. Cxcl12 mRNA expression was strongly enriched in GFP+ preadipocytes derived from Wt1‐creERT2;Rosa26mT/mG (Fig 5G–I). Thus, primary preadipocytes from our lineage tracing mouse models display gene expression profiles consistent with those observed in our clonal cell lines.
Figure 5. Primary preadipocyte and adipocyte subtypes exhibit differential gene expression profiles and phenotypic characteristics.
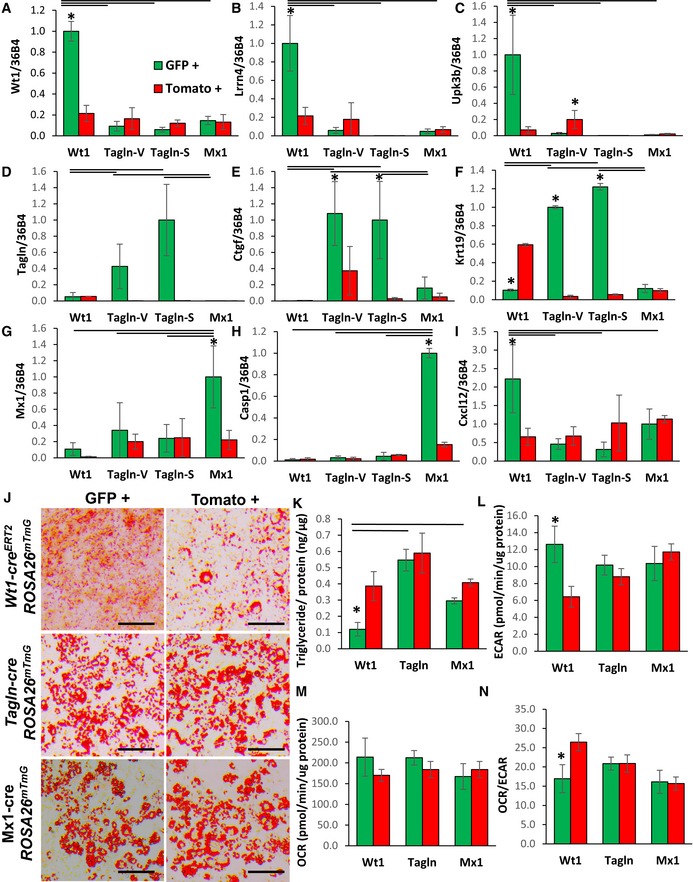
-
A–IExpression of Wt1, Lrrn4, Upk3b, Tagln, Ctgf, Krt19, Mx1, Casp1, and Cxcl12 in primary preadipocytes isolated from white adipose tissue of 5‐ to 6‐month‐old Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG male mice. mGFP‐ and mTomato‐positive preadipocytes were isolated from the pooled visceral fat depots (perigonadal, perirenal, mesenteric, and pericardial) of Wt1‐creERT2;Rosa26mT/mG mice, the pooled subcutaneous fat depots (subcutaneous and scapular white) of Mx1‐cre;Rosa26mT/mG, and both the pooled visceral (Tagln–V) and subcutaneous depots (Tagln‐S) from Tagln‐cre;Rosa26mT/mG mice. Data are shown as mean ± SEM of 4‐6 mice/group.
-
JBright‐field image of immortalized mGFP‐ and mTomato‐positive preadipocytes after in vitro adipogenic differentiation. Neutral lipoids were stained with Oil Red O, and original photographs were at 10× magnification (left panel). Scale bar = 100 μm.
-
KTriglyceride content of immortalized mGFP‐ and mTomato‐positive preadipocytes after 6 days of differentiation. Data are shown as mean ± SEM of 5–8 replicates, and the entire experiment was repeated with three independent cell lines.
-
LBasal extracellular acidification rate (ECAR) of immortalized mGFP‐ and mTomato‐positive preadipocytes was determined by calculating the area under the curve (AUC) in basal conditions. The whole experiment was repeated twice. Data are shown as mean ± SEM of three cell lines per group.
-
MBasal respiration of preadipocytes was of immortalized mGFP‐ and mTomato‐positive preadipocytes determined by calculating the area under the curve (AUC) during measurements of oxygen consumption rate (OCR). The whole experiment was repeated twice. Data are shown as mean ± SEM of three cell lines per group.
-
NRatio of basal oxygen consumption rate (OCR) to basal extracellular acidification rate (ECAR) of immortalized mGFP‐ and mTomato‐positive preadipocytes. Data are shown as mean ± SEM of three cell lines per group.
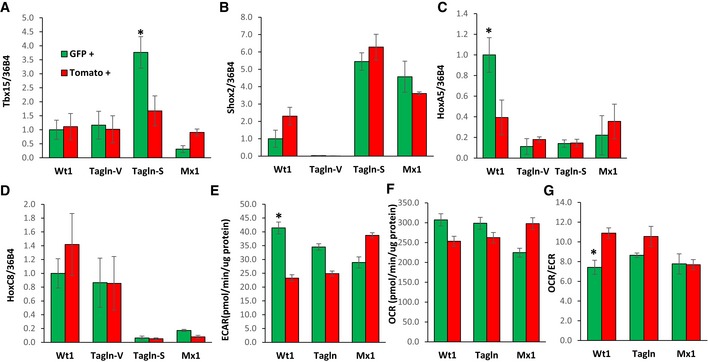
Interestingly, the expression of the developmental genes Tbx15 and HoxA5 was highly enriched within specific subpopulations. Tbx15 mRNA expression, which we previously showed marks a subset of cells in the subcutaneous adipose tissue (Lee et al, 2017a), was ~3‐fold higher in GFP+ preadipocytes derived from the subcutaneous fat of Tagln‐cre;Rosa26mT/mG. HoxA5 mRNA expression was ~2.5‐fold higher in GFP+ preadipocytes from the visceral fat of Wt1‐creERT2;Rosa26mT/mG compared to the corresponding Tomato+ controls. In contrast, the mRNA expression of Shox2 was higher in preadipocytes derived from the subcutaneous fat compared to visceral fat‐derived preadipocytes, while HoxC8 mRNA expression was higher in preadipocytes derived from visceral fat compared to those derived from subcutaneous fat, but did not vary within these three adipocyte subpopulations (Fig EV4A–D).
Figure EV4. Gene expression and maximal metabolic rates of primary preadipocyte subpopulations.
-
A–DExpression levels of Shox2, Tbx15, HoxC8, and HoxC5 mRNA were compared using quantitative real‐time PCR (qPCR) in primary preadipocytes isolated from white adipose tissue of 5‐ to 6‐month‐old Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG male mice. mGFP‐ and mTomato‐positive preadipocytes were isolated from the pooled visceral fat depots (perigonadal, perirenal, mesenteric, and pericardial) of Wt1‐creERT2;Rosa26mT/mG mice, the pooled subcutaneous fat depots (subcutaneous and scapular white) of Mx1‐cre;Rosa26mT/mG, and both the pooled visceral (Tagln–V) and subcutaneous depots (Tagln‐S) from Tagln‐cre;Rosa26mT/mG mice. Data are shown as mean ± SEM of 4‐6 mice/group.
-
EMaximal extracellular acidification rate (ECAR) of immortalized mGFP‐ and mTomato‐positive preadipocytes was determined by calculating the area under the curve (AUC) during measurements of maximal extracellular acidification rate. The whole experiment was repeated twice. Data are shown as mean ± SEM of three cell lines per group.
-
FMaximal respiration of preadipocytes was of immortalized mGFP‐ and mTomato‐positive preadipocytes determined by calculating the area under the curve (AUC) during measurements maximal of oxygen consumption rate (OCR). The whole experiment was repeated twice. Data are shown as mean ± SEM of three cell lines per group.
-
GRatio of maximal oxygen consumption rate (OCR) to maximal extracellular acidification rate (ECAR) of immortalized mGFP‐ and mTomato‐positive preadipocytes.
To investigate the metabolism of the preadipocytes subtypes derived from our lineage tracing mouse models, GFP+ and Tomato+ preadipocytes were immortalized by transducing cells with a lentivirus expressing an SV40 large T antigen. While all of preadipocytes retained the ability to differentiate and accumulate neutral lipid, as assessed by Oil Red O staining (Fig 5J), quantitation of triglyceride content after differentiation revealed that similar to the Type 1 clonal cell lines, GFP+ adipocytes from Wt1‐creERT2;Rosa26mT/mG mice accumulated ~60% less triglyceride than Tomato+ adipocytes from the same mice, or compared to GFP+ adipocytes from Tagln‐cre;Rosa26mT/mG or Mx1‐cre;Rosa26mT/mG mice (Fig 5K). Also similar to the Type 1 clonal cell lines, GFP+ adipocytes from Wt1‐creERT2;Rosa26mT/mG mice exhibited twofold higher basal and maximal extracellular acidification rates (ECAR) compared to the corresponding Tomato+ preadipocytes. These changes occurred without significant changes in basal and maximal oxygen consumption rates (OCR) and resulted in 30–40% lower ratios of basal and maximal OCR/ECAR in GFP+ preadipocytes derived from Wt1‐creERT2;Rosa26mT/mG mice compared to paired Tomato+ preadipocytes. On the other hand, no differences were observed between GFP+ and Tomato+ preadipocytes derived from Tagln‐cre;Rosa26mT/mG or Mx1‐cre;Rosa26mT/mG mice (Figs 5L–N and EV4E–G). Taken together, these data demonstrate that GFP+ preadipocytes derived from Wt1‐creERT2 ROSA26mT/mG, Tagln‐cre ROSA26mT/mG, and Mx1‐cre ROSA26mT/mG mice largely recapitulate both the gene expression and phenotypic differences observed in the clonal preadipocytes cell lines and thus represent the Type 1, 2, and 3 preadipocytes, respectively.
Preadipocyte subpopulations are distributed in a depot‐specific manner in vivo
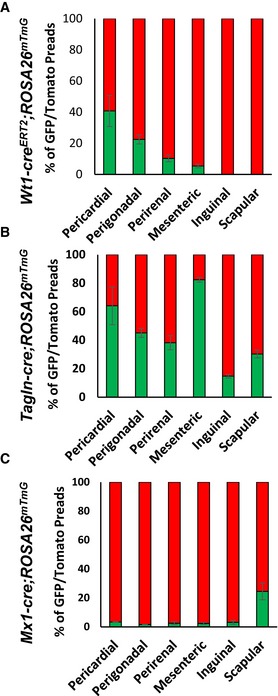
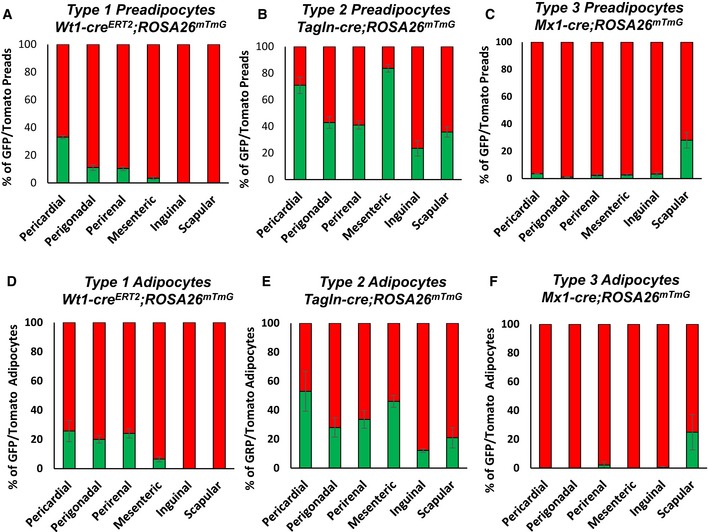
Quantitation of preadipocytes from the different fat depots indicates the depot‐dependent contribution of these subpopulations to the total preadipocyte population. Type 1 preadipocytes, represented by GFP‐positive preadipocytes from male Wt1‐creERT2 ROSA26mT/mG, were found in all visceral WAT depots, but in different relative abundance. Thus, pericardial fat had the highest percentage (40.7 ± 9.9%) followed by perigonadal (22.5 ± 2.9%), perirenal (10.3 ± 2.1%), and mesenteric (5.4 ± 1.7%) far. By contrast, Type 1 preadipocytes were virtually absent in both subcutaneous inguinal and scapular WAT (< 0.1%) (Fig 6A). Type 2 preadipocytes, represented by GFP‐positive preadipocytes from male Tagln‐cre;ROSA26mT/mG mice, on the other hand, were present in all adipose tissue depots, although again the proportions of these cells varied between adipose tissue depots with highest levels in mesenteric (82.3 ± 1.8%) WAT followed by pericardial (64.1 ± 13.2%), perigonadal (45.1 ± 3.2%), perirenal (38.2 ± 4.9%), subcutaneous scapular WAT (30.3 ± 2.7%) and inguinal WAT (14.8 ± 1.2%) (Fig 6B). Finally, Type 3 preadipocytes, represented by GFP‐positive preadipocytes from male Mx1‐cre;ROSA26mT/mG mice, were present in high numbers in subcutaneous scapular WAT (24.5 ± 2.8%) and present in low numbers (~2–3%) in preadipocytes from all other depots (Fig 6C). Female mice Wt1creERT2 ROSA26mT/mG, Tagln‐cre ROSA26mT/mG, and Mx1‐cre ROSA26mT/mG mice displayed a similar preadipocyte distribution as male mice (Fig EV5A–C).
Figure 6. Preadipocytes subpopulations are distributed in a depot‐specific manner in vivo .
-
A–CNumber of mGFP‐ and mTomato‐positive preadipocytes isolated by FACS from the indicated white adipose depots from 5‐ to 6‐month‐old male Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG mice. Data are shown as mean ± SEM of 4–9 mice.
Figure EV5. Preadipocytes and adipocyte subpopulations in female mice.
-
A–CNumber of mGFP‐ and mTomato‐positive preadipocytes isolated by FACS from each of the indicated white adipose depots from 5‐ to 6‐month‐old female Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG mice. Data are shown as mean ± SEM of 4–9 mice.
-
D–FQuantitation of mGFP‐ and mTomato‐positive adipocytes from each of the indicated white adipose depots from 5‐ to 6‐month‐old female Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG mice. Adipocytes were counted from four non‐overlapping images/depot/mouse. Data are shown as mean ± SEM of 4–9 mice.
Adipocytes subpopulations are differentially distributed in an adipose depots in vivo
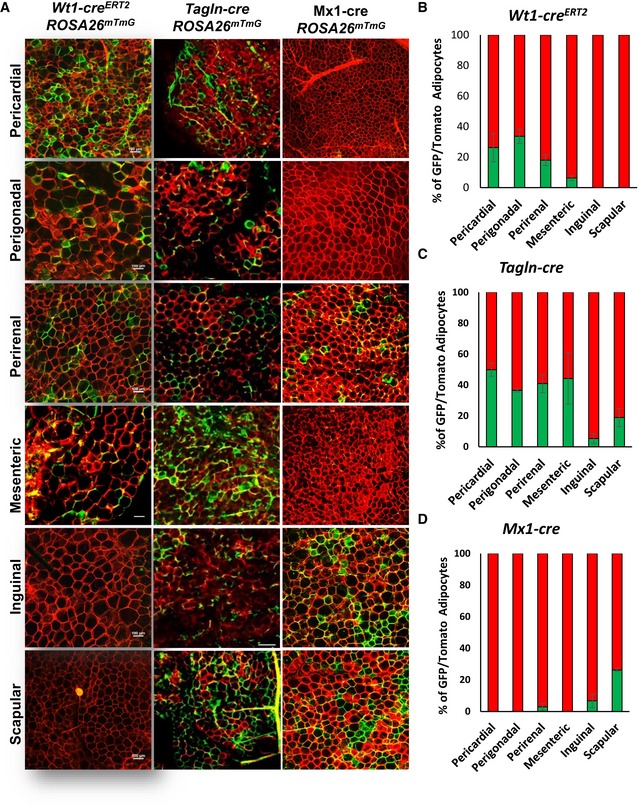
While adipocytes in any depot are ultimately derived from preadipocytes in that depot, the number of adipocytes of each type could or could not parallel the number of preadipocytes depending on rates of differentiation and cell turnover in vivo. Thus, we also assessed the distribution of GFP+ adipocytes in male Wt1‐creERT2 ROSA26mT/mG, Tagln‐cre ROSA26mT/mG, and Mx1‐cre ROSA26mT/mG mice in the different fat depots from mice at 5–6 months of age, when all depots are well represented. Consistent with their representation in preadipocytes, Wt1‐GFP+ adipocytes were found in only visceral and pericardial adipose tissue depots with the highest level in perigonadal fat (33.8 ± 4.9%), followed by pericardial fat (26.2 ± 5.9%), perirenal fat (18.3 ± 4.1%), and mesenteric fat (6.3 ± 1.7%), with no GFP+ cells in subcutaneous adipose tissue depots (Fig 7A and B). Type 2 adipocytes, marked by Tagln‐driven GFP, were present in all adipose tissue depots, but highest in pericardial (49.8 ± 4.2%) fat, followed by mesenteric (44.2 ± 16.6%), perirenal (41.0 ± 5.9%), and perigonadal fat (36.5 ± 1.0%) with much lower levels in scapular (19.0 ± 5.8%) and inguinal (5.3 ± 3.2%) WAT (Fig 7A and C). Type 3 adipocytes, marked by expression of Mx1, were present in highest numbers in scapular white fat (26.2 ± 4.2%), with small numbers also found in the inguinal (6.5 ± 2.5%) and perirenal (2.1 ± 1.5%) fat (Fig 7A and D), but not in other depots. Again, female mice Wt1creERT2 ROSA26mT/mG, Tagln‐cre ROSA26mT/mG, and Mx1‐cre ROSA26mT/mG mice showed a similar adipocyte distribution as male mice (Fig EV5D–F).
Figure 7. Adipocyte subpopulations are distributed in a depot‐specific manner in vivo .
-
ARepresentative images of whole‐mount preparations of the indicated white adipose tissue depots of 5‐ to 6‐month‐old Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG male mice. The photographs were taken at 10× magnification. Scale bar = 100 μm.
-
B–DQuantitation of mGFP‐ and mTomato‐positive adipocytes from each of the indicated white adipose depots from 5‐ to 6‐month‐old male Wt1‐creERT2;Rosa26mT/mG, Tagln‐cre;Rosa26mT/mG, and Mx1‐cre;Rosa26mT/mG mice. Adipocytes were counted from four non‐overlapping images per depot per mouse. Data are shown as mean ± SEM of 4–9 mice.
By adding the percentages of each subtype, we estimate that these three lineages describe > 90% of preadipocytes in the pericardial and mesenteric depots; about 40% of preadipocytes in the perigonadal, perirenal, and scapular adipose depots; but only about 30% of preadipocytes in the inguinal adipose tissue depot (Fig 8A and B). Similarly, these three subtypes describe ~60–70% of mature adipocytes in the visceral (perigonadal, perirenal, pericardial, and mesenteric) WAT depots, ~50% of adipocytes in scapular white fat and ~20% of cells in the inguinal fat pad. However, ~30–40% of adipocytes in visceral depots arise from as‐of‐yet unidentified lineages, and over 50% of adipocytes from the inguinal and scapular white depots, respectively, remain undefined by these models (Fig 8C and D). Taken together, these data demonstrate that white adipose tissue is comprised of at least three definable adipocyte precursor subpopulations that give rise to discrete adipocyte populations in vivo. These subpopulations of adipocytes and preadipocytes are distributed in a depot‐specific manner. Thus, each white adipose tissue depot is comprised of different proportions of distinct adipocyte subpopulations, which differ in their precursors, as well as their molecular and functional phenotypes (Fig 8E).
Figure 8. Distribution analysis of adipocyte subpopulations within adipose tissue depots.
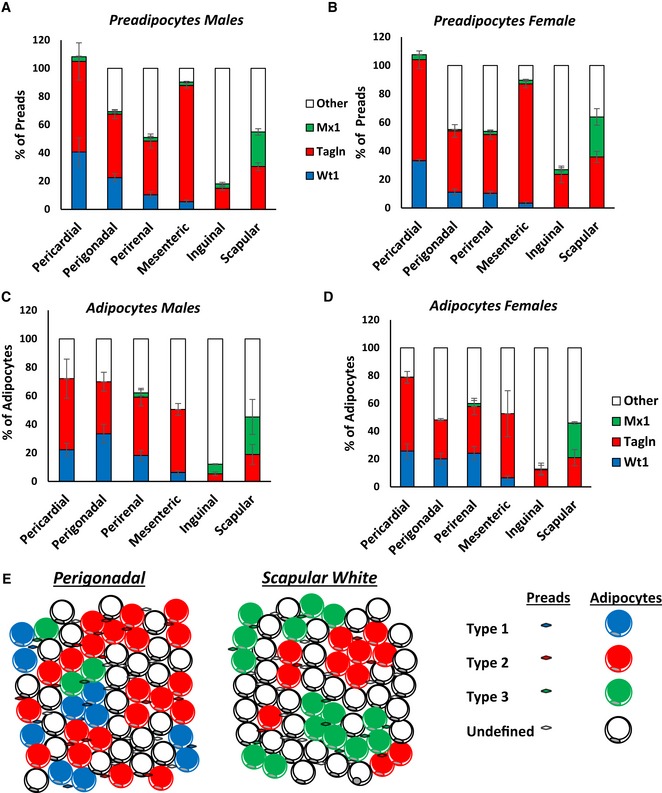
-
A, BNumber of mGFP‐positive preadipocytes isolated by FACS from each of the indicated white adipose depots from 5‐ to 6‐month‐old Wt1‐creERT2, Tagln‐cre, and Mx1‐cre crossed to Rosa26mT/mG mice from the experiment shown in Fig 6B. Data are shown as mean ± SEM of 4–9 mice.
-
C, DNumber of mGFP‐positive adipocytes from each of the indicated white adipose depots from 5‐ to 6‐month‐old Wt1‐creERT2, Tagln‐cre, and Mx1‐cre crossed to Rosa26mT/mG mice. Four separate non‐overlapping images/depot/mouse from the experiment shown in Fig 6C. Data are shown as mean ± SEM of 4–9 mice.
-
EModel of adipose tissue heterogeneous within a single fat depot. Each adipose tissue depot is comprised of a mix of Types 1–3 and uncharacterized preadipocytes and adipocytes.
Discussion
Over the past decade, it has become clear that the risk of metabolic disease in individuals with obesity is in part determined by differences in the amount and physiological effects of adipocytes accumulated in visceral versus subcutaneous white adipose depots (Tran et al, 2008; Smith & Kahn, 2016; Gaggini et al, 2017), as well as relative abundance and activity of brown, beige, and white fat in the individual or animal (Chondronikola et al, 2016; Cohen & Spiegelman, 2016; Kusminski et al, 2016). In this study, we have explored the important question as to how much of the effect of adipocytes in different white fat depots might be due to intrinsic differences in adipocytes in these depots, as well as heterogeneity of the white adipocyte population itself. To answer this question, we have created novel clonal white adipocyte precursor cell lines from a mouse containing a temperature‐sensitive Sv40T antigen. We find that these clonal white adipose cell lines represent at least three distinct adipocyte precursor subpopulations which have unique metabolic properties, gene expression profiles, and responses to stimulation by insulin, inflammatory cytokines, and GH. Lineage tracing studies followed by FACS analysis of preadipocyte populations indicate that these subpopulations retain the gene expression and phenotypic differences observed in the clonal cell lines. Furthermore, these three subtypes are present at different levels in pericardial, mesenteric, inguinal, perigonadal, perirenal, and scapular white adipose tissue depots in vivo. More importantly, these cells contribute to heterogeneity in the mature adipocytes in each fat depot.
This heterogeneity in white adipocytes and preadipocytes results in creating a unique cellular composition in each visceral and subcutaneous depot. Thus, in the three classical visceral depots studied in mice, Type 1 adipocytes, i.e., cells marked by history of expression of Wilms’ tumor 1 (Wt1), were only present in visceral depots, but were present at high levels in visceral depots below the diaphragm, such as perigonadal fat (34%), and above the diaphragm, such as pericardial fat (26%). and lowest level (~6%) in mesenteric fat, with no contribution to either subcutaneous adipose depot studied. On the other hand, Type 2 adipocytes, marked by a history of Tagln expression, were highest in pericardial fat (~50%), a less well‐studied WAT depot and one that was not even used for isolation of the original preadipocytes clones. Type 2 adipocytes were also found in mesenteric, perirenal, and perigonadal fat, and represented about 19% of adipocytes in the subcutaneous inguinal fat pad. Type 3 adipocytes, i.e., those marked by history of Mx1 expression, were highest in number in scapular white fat (~25%), another depot not utilized in the selection of the original defining clones, with smaller numbers in the inguinal and perirenal fat. The relative ratio of preadipocytes from different subpopulations largely matches the ratio of the adipocytes from these subpopulations in the different adipose tissue depots. The similarity in distribution of preadipocytes and adipocytes indicates the ability of preadipocytes from different subpopulations to undergo adipogenic differentiation and their turnover rate once formed are likely similar.
In addition to this heterogeneity, our data demonstrate that cell lines created from the SVF of different fat depots of the Immortomouse retain depot‐specific aspects of their gene expression signature. Thus, cell lines derived from subcutaneous depots retain high levels of Shox2 and Tbx15, while visceral depots retain high expression of HoxC8 and HoxA5, both characteristics of the adipocytes and preadipocytes in these depots (Gesta et al, 2006; Macotela et al, 2012). Interestingly, we find that HoxA5 is highly expressed in Type 1 preadipocytes, while Tbx15 is highly expressed in Type 2 preadipocytes derived from the subcutaneous depot. On the other hand, the expression of other depot‐specific markers, including Shox2 and HoxC8, is not different between the adipocyte subpopulations. These data suggest that some, but not all, depot‐specific differences in gene expression may reflect the differential cellular contribution of the subpopulations to specific adipose tissue depots.
One unexpected finding from these studies was that while the adipocyte precursor pool from perigonadal (visceral) fat pad completely failed to differentiate in vitro into mature adipocytes, over 35% of the clonal precursors derived by FACS separation from this fat pad did differentiate in vitro. Other attempts to utilize Immortomouse to develop adipocyte cell lines, especially in those derived from the visceral adipose depots, also exhibited poor adipogenic differentiation (Church et al, 2015). These data suggest that there is a population of cells in the SVF of perigonadal adipose tissue that exerts an anti‐adipogenic effect on other cells in this depot. This might represent the CD142+ population that was recently discovered to have anti‐adipogenic properties (Schwalie et al, 2018) or some other negatively regulatory cell type. Even after separation of these cells into clonal cell lines, there is a difference in adipogenic potential depending on depot of origin. For example, about ~70% of clonal cell lines derived from the inguinal depot are adipogenic compared to only ~35% of clonal cell lines derived from the perigonadal depot.
Of the three types of preadipocytes/adipocytes identified here, only the Type 1 cells, marked by expression of Wilms’ tumor 1 (Wt1), have been previously described (Lansley et al, 2011; Chau et al, 2014) as a subpopulation of white adipocytes in visceral depots arising from mesothelial cells progenitors. In our study, we show that this preadipocyte/adipocyte subpopulation is uniquely characterized by highly glycolytic metabolism and highly responsive to the cytokine TNFα. The glycolytic nature of Type 1 adipocytes is independent of that linked to expression of the developmental gene Tbx15, which we have previously shown marks a glycolytic subpopulation of subcutaneous white adipocytes (Lee et al, 2017a) and also marks glycolytic fibers in skeletal muscle (Lee et al, 2015). Indeed, whereas Wt1 expression is common in cells of the perigonadal visceral fat pad, Tbx15 is expressed in very few cells of these cells. Indeed, Tbx15 expression is expressed only at low levels in primary Type 1 preadipocytes and is more highly expressed in subcutaneous Type 2 preadipocytes. Another feature of Type 1 adipocytes is that they have reduced triglyceride accumulation after in vitro differentiation. These differences may reflect the reduced insulin‐induced glucose uptake and de novo lipogenesis observed in this subtype of adipocyte.
Type 2 preadipocytes/adipocytes, marked in vivo by history of Tagln expression, are widely distributed and contribute to varying degrees to all adipose tissue depots. Tagln, also known as smooth muscle protein 22‐alpha, is a TGFβ‐inducible gene that regulates cytoskeletal organization and has a positive role in the adipogenic, osteogenic, and myogenic differentiation of bone marrow‐derived stem cells (Elsafadi et al, 2016). Tagln has been shown to be expressed in expressed in vascular and visceral smooth muscle, as well as pericytes and fibroblasts, and is regarded an early marker of smooth muscle differentiation (Li et al, 1996; Klein et al, 2011). In this regard, it is interesting that perivascular cells or pericytes have been identified as an important source of white adipocyte precursors (Tang et al, 2008; Jiang et al, 2014), as well as a subpopulation of brite adipocytes (Long et al, 2014; Berry et al, 2016). Furthermore, perivascular multipotent stem cells express high levels of Tagln that are further induced by TGFβ1 treatment (Klein et al, 2011). In contrast to previous reports which show no contribution of Tagln lineage cells to white adipocyte development (Berry et al, 2016), our data clearly demonstrate that Tagln‐cre contributes widely to white preadipocyte and adipocyte populations. One potential reason for this difference is that we utilized a mouse model in which Cre was knocked into the endogenous Tagln locus (Zhang et al, 2006), whereas the previous study used a transgenic mouse carrying Cre driven by a 2.8 kb piece of the Tagln promoter (Holtwick et al, 2002), which may not include all of the promoter or enhancer elements necessary for Tagln expression in WAT preadipocytes.
Type 3 preadipocytes and adipocytes are marked by expression of Mx1 and are most abundant in scapular white adipose tissue, with small numbers of cells in the inguinal and perirenal adipose tissue depots, i.e., primarily in subcutaneous depots with a limited presence in visceral fat. Induction of Mx1‐cre with Poly I:C has been shown to rapidly induce recombination of floxed genes in hematopoietic lineages (Kuhn et al, 1995), and it has been suggested that some bone marrow‐derived hematopoietic cells may serve as progenitors for a small subpopulation of adipocytes in the visceral depots (Majka et al, 2010; Gavin et al, 2016). This seems unlikely to account for the results in the present study, since the largest number of Type 3 adipocytes was observed in the scapular white fat, a non‐visceral depot. Interestingly, Type 3 adipocytes also have high expression of Casp1 and Cxcl12. Although expression of Casp1 and Cxcl12 is correlated with insulin resistance and macrophage infiltration (Stienstra et al, 2010; Kim et al, 2014), we find that Type 3 adipocytes are insulin sensitive and display an attenuated response to TNFα compared to Type 1 and 2 adipocytes.
Previous studies have demonstrated that subcutaneous, but not visceral, WAT adipocytes are almost completely derived from a lineage marked by recombination by a paired related homeobox transcription factor 1 (Prx1) driven cre construct (Sanchez‐Gurmaches et al, 2015). Since both Type 2 and 3 adipocytes are found in the subcutaneous WAT depots, our data indicate that within the subcutaneous WAT, adipocytes are derived from both a Prx1‐cre‐positive and Tagln‐ or Mx1‐positive lineage. On the other hand, since Type 1–3 adipocytes are also found in visceral WAT, adipocytes from Wt1‐, Tagln‐, or Mx1‐positive lineages can also derive from Prx1‐cre‐negative lineages. Indeed, our microarray analysis indicates that Type 1–3 adipocytes display a wide range of Prx1 expression levels. Sanchez‐Gurmaches and Guertin have also shown that scapular white subcutaneous and perirenal WAT can also be found to be derived from a lineage that is both myogenic factor 5 (Myf5) and paired box 3 (Pax3) positive (Sanchez‐Gurmaches & Guertin, 2014). Type 2 adipocytes are also found within both depots, while Type 1 adipocytes are present in perirenal WAT and Type 3 adipocytes are present in scapular WAT, suggesting that at least a subset of Type 1–3 preadipocytes and adipocytes may also be derived from these Myf5/Pax3‐positive lineages. Taken together with our data, these other studies are beginning to highlight the developmental complexities which underlie WAT formation.
In addition to lineage differences, the WAT adipocyte subpopulations we have identified have clear phenotypic and functional differences, despite the fact that they are similar in mRNA expression of the key adipokines, adiponectin, and leptin, Type 1 adipocytes and their precursors display highly glycolytic metabolism. In addition, in comparison with Type 1 cells, Type 2 and 3 adipocytes display increased insulin‐mediated glucose uptake and tend to have increased de novo lipogenesis. In addition, stimulation of Stat5‐Tyr694 phosphorylation by growth hormone is highest in Type 2 adipocytes. Interestingly, both fat‐specific knockout of the insulin receptor and over‐expression of growth hormone unmask intrinsic heterogeneity of white adipose tissue leading to a bimodal distribution of adipocyte cell size (Bluher et al, 2004; Troike et al, 2017). Since insulin and GH regulate lipogenesis and lipolysis to positively and negatively regulate adipocyte size, respectively, we hypothesize that the differential responses may, at least in part, contribute to these changes in size distribution. Although we have focused on white fat, studies have demonstrated that brown and bright adipocytes also exhibit intrinsic heterogeneity in terms of thermogenic capacity (Xue et al, 2015) and gene expression (Lee et al, 2017b). Thus, taken together, it appears that white, brite, and brown adipose tissue are composed of populations that are phenotypically, developmentally, and metabolic heterogeneous.
While all three types of precursor cells can differentiate into adipocytes, they can also differentiate into osteoblasts and chondrocytes when provided with optimized media and growth factors. Thus, all three types of cells are pluripotential mesenchymal precursors, and not exclusively committed preadipocytes. On the other hand, we were unable to induce myogenic differentiation of any of these clonal adipocyte lines, suggesting there is some level of lineage restriction/specification. Interestingly, despite their adipogenic potential, none of the clonal cell lines showed a significant induction of a thermogenic gene expression profile in response to either BMP7 or forskolin treatment, indicating that these cell lines are white, and not beige or brite cells. This occurs despite the fact that immortalization of preadipocytes with Sv40 T antigen tends to promote a brown adipocyte metabolic program by inhibiting retinoblastoma protein function (Hansen et al, 2004). However, in the Immortomouse clonal cell lines, Sv40 T antigen is only expressed and stable under permissive conditions (33°C; in the presence of IFNγ), and differentiation and analysis of these cells were performed under non‐permissive conditions for T antigen expression (37°C; in the absence of IFNγ). Indeed, previous experiments have demonstrated that the inducible expression of T antigen tsA58 in the Immortomouse™ clonal cell lines does not induce a brown adipocyte phenotype (Morganstein et al, 2008).
Whereas the three white adipocyte subtypes we have identified describe 60–70% of mature adipocytes in the pericardial and three visceral adipose tissue depots, they are still 30–40% not identified by expression of these markers, and the percentage of undefined cells is more than 50% in the subcutaneous adipose depots. Likewise, even utilizing a combination of three independent clustering algorithms to identify preadipocyte subtypes, ~30% of the cell lines were left unclustered, consistent with the presence of additional adipocyte subtypes. Some of this may be related to methodological limitations of classification, especially in the in vivo lineage tracing using Wt1‐CreERT2 and Mx1‐Cre to mark Type 1 and 3 adipocytes, since the cre recombinase in both of these lines required induction early in development. On the other hand, it seems quite likely that there are other subtypes of white preadipocytes/adipocytes which could be distinguished using other criteria and combinations of gene expression. Identifying these additional adipocyte lineages will be of importance in future studies.
In summary, taken together, our results demonstrate that white adipose tissue is comprised of at least three distinct adipocyte subpopulations that arise from different populations of adipocyte precursor cells and have distinct molecular markers, differences in metabolism, and differential responses to exogenous stimuli. These different types of adipocytes are found in each adipose depot in different numbers and therefore contribute to the differing biology of individual adipose depots. In analogy to the study other biological cell types, such as lymphocytes or skeletal muscle fibers, studies of adipocyte biology need to take into account the effects different adipocyte types. More importantly, further investigation of these adipocyte subpopulations may provide unique ways to manipulate the physiology and new targets in the treatment of obesity and related disorders.
Materials and Methods
Animals and diets
C57BL/6 mice (Jackson Labs) and the Immortomouse (Jat et al, 1991) were housed under a 12‐h light/dark cycle in a temperature‐controlled room and allowed ad libitum access to water and food containing 22% calories from fat, 23% from protein, and 55% from carbohydrates (Mouse Diet 9F 5020; PharmaServ). Animal care and study protocols were approved by the Animal Care Committee of Joslin Diabetes Center and were in accordance with the National Institutes of Health guidelines.
Wt1‐creERT2 (Zhou et al, 2008), Tagln‐cre (Zhang et al, 2006), Mx1‐cre (Kuhn et al, 1995), and Rosa26mT/mG (Muzumdar et al, 2007) on a C57BL/6 background (ordered from Jackson Labs) were housed at 22°C under a 14‐h light, 10‐h dark cycle, 3–4 mice per cage, with ad libitum access to water and standard laboratory chow (ProLab RMH 3000). All experiments were approved by the Ohio University Institutional Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines.
Culture and differentiation of clonal cell lines
Immortomouse™ clonal preadipocyte cell lines were created from the stromovascular fraction (SVF) of the subcutaneous and perigonadal fat of a single 8‐week‐old male mouse. The SVF cells were isolated by collagenase digestion of subcutaneous and perigonadal fat, plated and grown at 33°C in preadipocyte growth media (Macotela et al, 2012) supplemented with 10 units/ml interferon‐gamma (PeproTech). After two passages, 190 individual cells were sorted without gating using an Aria flow cytometer (BD Biosciences), and one cell was placed in each well. Cells were grown at 33°C with 10 units/ml interferon γ into clonal cell lines. All clonal preadipocyte cell lines were treated with 25 μM plasmocin (InvivoGen) for 2 weeks to eliminate mycoplasma and tested regularly for mycoplasma contamination. The adipogenic capacity of cell lines was assessed by lipid accumulation by Oil Red O staining after the induction of differentiation.
For differentiation, cells were seeded at 20,000 cells per well in 48‐well plates. After cells reached confluence, the medium was replaced by differentiation medium (growth medium with no growth factors, but with 2% FBS, 1 μM dexamethasone, 0.5 μM isobutylmethylxanthine, 100 nM insulin, and 1 μM rosiglitazone). After 2 days, the medium was replaced with growth medium containing 2% FBS and 100 nM insulin for 2 more days, then for 2–3 days with growth medium with 2% FBS alone additional days to allow for differentiation.
Gene expression by microarray analysis and bioinformatics
RNA was extracted and biotin‐labeled complementary RNA (cRNA) was prepared from confluent cultures of the 24 clonal cell lines depicted in Fig EV1B. Complementary RNA was hybridized to Affymetrix M430 2.0 arrays, and microarray analysis was performed on globally scaled data (MAS 5.0). Normalized microarray data were analyzed with R/Bioconductor (Gentleman, 2005). Differential expression was calculated with the limma package (Ritchie et al, 2015), hierarchical clustering used the hclust function, and affinity propagation used the APCluster package (Bodenhofer et al, 2011).
Analysis of gene expression by quantitative PCR and western blotting
Total RNA was isolated using an RNeasy mini kit (Qiagen). 3 μg of total RNA was reverse transcribed in 100 μl using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A portion (5 μl) of diluted (1/5) reverse transcription reaction was amplified with specific primers (300 nM each) in a 10 μl PCR using a SYBR green PCR master mix (Applied Biosystems). Analysis of individual gene expression was carried out in an ABI Prism 7900HT sequence detector with initial denaturation at 95°C for 10 min, followed by 40 PCR cycles, each cycle consisting of 95°C for 15 s, 60°C for 1 min, and 72°C for 1 min, and SYBR green fluorescence emissions were monitored after each cycle. For each gene, mRNA expression was calculated relative to 36B4 expression. Amplification of specific transcripts was confirmed by the melting‐curve profiles (cooling the sample to 68°C and heating slowly to 95°C with measurement of fluorescence) at the end of each PCR. The primers used for quantitative PCR are described in Table EV1.
Proteins were extracted from cells, subjected to SDS–PAGE, and transferred to polyvinylidene fluoride membranes. The blots were blocked and probed with antibodies as specified in Table EV2.
Functional analysis of clonal cell lines
Triglyceride quantitation
Triglycerides were quantified using the Sigma‐Aldrich Triglyceride Quantitation Kit according to the manufacturer's instructions.
Glucose uptake
Preadipocytes and differentiated adipocytes were serum deprived in DMEM for overnight and then starved in KRH buffer supplemented with 2 mM sodium pyruvate for a further 2 h. The cells were then incubated in the presence or absence of insulin for 15 min. Glucose uptake was initiated by addition of 2‐[3H] deoxyglucose at a concentration of 0.1 mM 2‐deoxyglucose in 1 μCi 2‐[3H] deoxyglucose in KRH buffer and incubated for 4 min at room temperature. Glucose uptake measurements have been corrected for nonspecific diffusion (5 μM cytochalasin B).
Lipogenesis
Adipocytes were serum deprived in DMEM for overnight and then starved in KRH supplemented with 0.1% free fatty acid BSA (final volume = 500 μl) in the absence (basal) or presence of insulin for 15 min. D‐glucose (final concentration = 5 mM) containing 1 μCi/ml of 14C‐D‐glucose for 2 h. Adipocytes were then lysed with 0.1% SDS, and total lipids were extracted by adding 0.5 ml of Dole solution, followed by addition of 0.5 ml heptane. 0.5 ml was then transferred to a scintillation vial, dried overnight, and the lipid content was weighed. 5 ml of scintillation liquid was added, and the radioactivity was counted. The 14C‐D‐glucose incorporated into lipid was then normalized against lipid content.
Lipolysis
Differentiated adipocytes were serum deprived in DMEM for 2 h and then starved in KRH buffer supplemented with 2 mM sodium pyruvate for a further 2 h. The cells were washed carefully and incubated in KRH buffer supplemented with 2 mM sodium pyruvate in the presence or absence of 1 μM isoproterenol for 2 h. Glycerol release was measured by colorimetric reaction as previously described (Lee et al, 2013).
Metabolic analysis
Oxygen consumption and extracellular acidification rates were measured using the XF24 Extracellular Flux Analyzer from Seahorse Bioscience. One hour before the experiment, cells were washed and incubated in 630 μl of non‐buffered (without sodium carbonate) DMEM (4.5 g/l glucose) pH 7.4 at 37°C in a non‐CO2 incubator. Three replicates per cell type were included in the experiment, and four wells evenly distributed within the plate were used for correction of temperature variations. During the time course of the experiment, oxygen concentration and media acidification were measured over time periods of 2 min at 6‐min intervals, consisting of a 2 min of mixing period and a 4‐min waiting period. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) over the 2‐min measurement period were calculated using the Fixed Delta technique for determining the slope. Maximal ECAR was measured in the presence of 1.0 μM oligomycin. Maximal OCR was measured in the presence of 1.0 μM carbonyl cyanide‐4‐(trifluoromethoxy)phenylhydrazone (FCCP).
Preadipocyte isolation
Preadipocytes were isolated as previously described (Rodeheffer et al, 2008). SVF was prepared from each fat pad by collagenase treatment. SVF was resuspended in erythrocyte lysis buffer, pelleted by centrifugation, and labeled with appropriate antibodies (Table EV2) in staining solution (HBSS 10% FBS). After staining, cells were filtered through a 40 μm cell strainer and single cells were sorted according to the expression of surface markers (CD31−CD45−Ter119−CD29+CD34+Sca1+) with an Aria flow cytometer (BD Biosciences). GFP+ and Tomato+ preadipocytes were immortalized by transduction with lentiviral particles and immortalized by infection with a pBABE retrovirus encoding an SV40 T antigen followed by selection with 2 g/ml of puromycin. All immortalized preadipocyte cell lines were treated with 25 μM plasmocin (InvivoGen) for 2 weeks to eliminate mycoplasma and tested regularly for mycoplasma contamination.
Statistics, sample size, and data analysis
Normal distribution was assessed by the Kolmogorov–Smirnov test (P > 0.05). All differences were analyzed by ANOVA, or Student's t‐test if normally distributed. A Wilcoxon T‐test was used for data that were not normally distributed. Results were considered significant if P < 0.05. Sample sizes for the in vitro studies (ranging from three to seven cell lines/group) were determined by the number of independent cell lines available, and for in vivo studies, analysis started with four to six mice per group. In case sufficient statistical power was not achieved, we performed additional experiments when possible. There were no excluded data in the analysis. There was no blinding, nor randomization.
Author contributions
KYL designed and performed experiments, analyzed data, and wrote the paper; QL designed and performed experiments and analyzed data; RS designed and performed experiments; JMD designed experiments and analyzed data; SU designed and performed experiments; CRK designed experiments, analyzed data, and wrote the paper. All authors discussed the results and implications and commented on the manuscript at all stages. CRK is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Acknowledgements
We are grateful to Christie Penniman (Joslin Diabetes Center) for animal care, Girijesh Buruzula and Joyce LaVecchio (DRC Flow Cytometry Core), the Ohio University Microscopy Core, and Michele Pate (Ohio University Flow Cytometry Core) for technical assistance. This work was supported by NIH grant R01 DK082655, DK036836, DK007260 (CRK), the Joslin DRC P30 DK036836, the Joslin training grant T32 DK007260, an American Diabetes Association mentor‐based award (CRK), the Mary K. Iacocca Professorship (CRK), start‐up funds from Ohio University College of Osteopathic Medicine (KYL), the American Diabetes Association Junior Faculty Development Award 1‐17‐JDF‐055 (KYL), iMed the initiative for personalized medicine of the Helmholtz Association (SU), and the project Ageing and Metabolic Programming (AMPro) (SU).
The EMBO Journal (2019) 38: e99291
Contributor Information
Kevin Y Lee, Email: leek2@ohio.edu.
C Ronald Kahn, Email: c.ronald.kahn@joslin.harvard.edu.
Data availability
Microarray data deposited at Gene Expression Omnibus (Accession #: GSE110531).
References
- Berry DC, Jiang Y, Graff JM (2016) Mouse strains to study cold‐inducible beige progenitors and beige adipocyte formation and function. Nat Commun 7: 10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M, Patti ME, Gesta S, Kahn BB, Kahn CR (2004) Intrinsic heterogeneity in adipose tissue of fat‐specific insulin receptor knock‐out mice is associated with differences in patterns of gene expression. J Biol Chem 279: 31891–31901 [DOI] [PubMed] [Google Scholar]
- Bodenhofer U, Kothmeier A, Hochreiter S (2011) APCluster: an R package for affinity propagation clustering. Bioinformatics 27: 2463–2464 [DOI] [PubMed] [Google Scholar]
- Boquest AC, Shahdadfar A, Fronsdal K, Sigurjonsson O, Tunheim SH, Collas P, Brinchmann JE (2005) Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Mol Biol Cell 16: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau YY, Bandiera R, Serrels A, Martinez‐Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, Stimson RH, Walker BR, Chapuli RM, Schedl A, Hastie N (2014) Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol 16: 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondronikola M, Volpi E, Borsheim E, Porter C, Saraf MK, Annamalai P, Yfanti C, Chao T, Wong D, Shinoda K, Labbe SM, Hurren NM, Cesani F, Kajimura S, Sidossis LS (2016) Brown adipose tissue activation is linked to distinct systemic effects on lipid metabolism in humans. Cell Metab 23: 1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Brown M, Rodeheffer MS (2015) Conditional immortalization of primary adipocyte precursor cells. Adipocyte 4: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Spiegelman BM (2016) Cell biology of fat storage. Mol Biol Cell 27: 2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Kahn CR (2010) The role and importance of brown adipose tissue in energy homeostasis. Curr Opin Pediatr 22: 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankel SN, Fadnes DJ, Stavrum AK, Stansberg C, Holdhus R, Hoang T, Veum VL, Christensen BJ, Vage V, Sagen JV, Steen VM, Mellgren G (2010) Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS ONE 5: e11033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsafadi M, Manikandan M, Dawud RA, Alajez NM, Hamam R, Alfayez M, Kassem M, Aldahmash A, Mahmood A (2016) Transgelin is a TGFbeta‐inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleston organization. Cell Death Dis 7: e2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer‐Lorente R, Bejar MT, Badimon L (2014) Notch signaling pathway activation in normal and hyperglycemic rats differs in the stem cells of visceral and subcutaneous adipose tissue. Stem Cells Dev 23: 3034–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich‐Horvat P, Liu CY, Vasan RS, Murabito JM, Meigs JB, Cupples LA, D'Agostino RB Sr, O'Donnell CJ (2007) Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation 116: 39–48 [DOI] [PubMed] [Google Scholar]
- Frey B, Dueck D (2007) Clustering by passing messages between data points. Science 315: 972–976 [DOI] [PubMed] [Google Scholar]
- Gaggini M, Carli F, Gastaldelli A (2017) The color of fat and its central role in the development and progression of metabolic diseases. Horm Mol Biol Clin Invest 31: 1868–1883 [DOI] [PubMed] [Google Scholar]
- Gao H, Volat F, Sandhow L, Galitzky J, Nguyen T, Esteve D, Astrom G, Mejhert N, Ledoux S, Thalamas C, Arner P, Guillemot JC, Qian H, Ryden M, Bouloumie A (2017) CD36 is a marker of human adipocyte progenitors with pronounced adipogenic and triglyceride accumulation potential. Stem Cells 35: 1799–1814 [DOI] [PubMed] [Google Scholar]
- Gavin KM, Gutman JA, Kohrt WM, Wei Q, Shea KL, Miller HL, Sullivan TM, Erickson PF, Helm KM, Acosta AS, Childs CR, Musselwhite E, Varella‐Garcia M, Kelly K, Majka SM, Klemm DJ (2016) De novo generation of adipocytes from circulating progenitor cells in mouse and human adipose tissue. FASEB J 30: 1096–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R (2005) Bioinformatics and computational biology solutions using R and Bioconductor
- Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR (2006) Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA 103: 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliemann J, Vinten J (1974) Lipogenesis and insulin sensitivity of single fat cells. J Physiol 236: 499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, Petrovic N, Enerback S, Nedergaard J, Cinti S, te Riele H, Kristiansen K (2004) Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA 101: 4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J, Kuhn M (2002) Smooth muscle‐selective deletion of guanylyl cyclase‐A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci USA 99: 7142–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen I, Heymsfield SB, Allison DB, Kotler DP, Ross R (2002) Body mass index and waist circumference independently contribute to the prediction of nonabdominal, abdominal subcutaneous, and visceral fat. Am J Clin Nutr 75: 683–688 [DOI] [PubMed] [Google Scholar]
- Jat PS, Noble MD, Ataliotis P, Tanaka Y, Yannoutsos N, Larsen L, Kioussis D (1991) Direct derivation of conditionally immortal cell lines from an H‐2Kb‐tsA58 transgenic mouse. Proc Natl Acad Sci USA 88: 5096–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Berry DC, Tang W, Graff JM (2014) Independent stem cell lineages regulate adipose organogenesis and adipose homeostasis. Cell Rep 9: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori‐Katayama M, Kaiho A, Ishizu Y, Okamura‐Oho Y, Hino O, Abe M, Kishimoto T, Sekihara H, Nakamura Y, Suzuki H, Forrest AR, Hayashizaki Y (2011) LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS ONE 6: e25391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Yoon JH, Ghim J, Yea K, Song P, Park S, Lee A, Hong CP, Jang MS, Kwon Y, Park S, Jang MH, Berggren PO, Suh PG, Ryu SH (2014) CXCL12 secreted from adipose tissue recruits macrophages and induces insulin resistance in mice. Diabetologia 57: 1456–1465 [DOI] [PubMed] [Google Scholar]
- Klaus S (2004) Adipose tissue as a regulator of energy balance. Curr Drug Targets 5: 241–250 [DOI] [PubMed] [Google Scholar]
- Klein D, Weisshardt P, Kleff V, Jastrow H, Jakob HG, Ergun S (2011) Vascular wall‐resident CD44+ multipotent stem cells give rise to pericytes and smooth muscle cells and contribute to new vessel maturation. PLoS ONE 6: e20540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Kusminski CM, Bickel PE, Scherer PE (2016) Targeting adipose tissue in the treatment of obesity‐associated diabetes. Nat Rev Drug Discovery 15: 639–660 [DOI] [PubMed] [Google Scholar]
- Lansley SM, Searles RG, Hoi A, Thomas C, Moneta H, Herrick SE, Thompson PJ, Newman M, Sterrett GF, Prele CM, Mutsaers SE (2011) Mesothelial cell differentiation into osteoblast‐ and adipocyte‐like cells. J Cell Mol Med 15: 2095–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Gesta S, Boucher J, Wang XL, Kahn CR (2011) The differential role of Hif1beta/Arnt and the hypoxic response in adipose function, fibrosis, and inflammation. Cell Metab 14: 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Yamamoto Y, Boucher J, Winnay JN, Gesta S, Cobb J, Bluher M, Kahn CR (2013) Shox2 is a molecular determinant of depot‐specific adipocyte function. Proc Natl Acad Sci USA 110: 11409–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Singh MK, Ussar S, Wetzel P, Hirshman MF, Goodyear LJ, Kispert A, Kahn CR (2015) Tbx15 controls skeletal muscle fibre‐type determination and muscle metabolism. Nat Commun 6: 8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Sharma R, Gase G, Ussar S, Li Y, Welch L, Berryman DE, Kispert A, Bluher M, Kahn CR (2017a) Tbx15 defines a glycolytic subpopulation and white adipocyte heterogeneity. Diabetes 66: 2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kim SN, Kwon HJ, Granneman JG (2017b) Metabolic heterogeneity of activated beige/brite adipocytes in inguinal adipose tissue. Sci Rep 7: 39794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miano JM, Cserjesi P, Olson EN (1996) SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ Res 78: 188–195 [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, Castellot JJ Jr, Rosen ED, Spiegelman BM (2014) A smooth muscle‐like origin for beige adipocytes. Cell Metab 19: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macotela Y, Emanuelli B, Mori MA, Gesta S, Schulz TJ, Tseng YH, Kahn CR (2012) Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes 61: 1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka SM, Fox KE, Psilas JC, Helm KM, Childs CR, Acosta AS, Janssen RC, Friedman JE, Woessner BT, Shade TR, Varella‐Garcia M, Klemm DJ (2010) De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proc Natl Acad Sci USA 107: 14781–14786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcourrier P, Silver I, Farnoud R, Bird I, Rochefort H (1997) Breast cancer cells have a high capacity to acidify extracellular milieu by a dual mechanism. Clin Exp Metastasis 15: 382–392 [DOI] [PubMed] [Google Scholar]
- Morganstein DL, Christian M, Turner JJ, Parker MG, White R (2008) Conditionally immortalized white preadipocytes: a novel adipocyte model. J Lipid Res 49: 679–685 [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L (2007) A global double‐fluorescent Cre reporter mouse. Genesis 45: 593–605 [DOI] [PubMed] [Google Scholar]
- Ramseyer VD, Granneman JG (2016) Adrenergic regulation of cellular plasticity in brown, beige/brite and white adipose tissues. Adipocyte 5: 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M, Phipson B, Wu D, Hu Y, Law C, Shi W, Smyth G (2015) Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 43: e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM (2008) Identification of white adipocyte progenitor cells in vivo . Cell 135: 240–249 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM (2006) Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444: 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salans LB, Dougherty JW (1971) The effect of insulin upon glucose metabolism by adipose cells of different size. Influence of cell lipid and protein content, age, and nutritional state. J Clin Invest 50: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Gurmaches J, Guertin DA (2014) Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat Commun 5: 4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Gurmaches J, Hsiao WY, Guertin DA (2015) Highly selective in vivo labeling of subcutaneous white adipocyte precursors with Prx1‐Cre. Stem Cell Rep 4: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen P, van Marken Lichtenbelt WD, Spiegelman BM (2015) The future of brown adipose tissues in the treatment of type 2 diabetes. Diabetologia 58: 1704–1707 [DOI] [PubMed] [Google Scholar]
- Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, Wolfrum C, Deplancke B (2018) A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559: 103–108 [DOI] [PubMed] [Google Scholar]
- Seydoux J, Muzzin P, Moinat M, Pralong W, Girardier L, Giacobino JP (1996) Adrenoceptor heterogeneity in human white adipocytes differentiated in culture as assessed by cytosolic free calcium measurements. Cell Signal 8: 117–122 [DOI] [PubMed] [Google Scholar]
- Smith U, Kahn BB (2016) Adipose tissue regulates insulin sensitivity: role of adipogenesis, de novo lipogenesis and novel lipids. J Intern Med 280: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, Middelbeek RJ, Townsend KL, Lee MY, Takahashi H, So K, Hitchcox KM, Markan KR, Hellbach K, Hirshman MF, Tseng YH, Goodyear LJ (2015) A novel role for subcutaneous adipose tissue in exercise‐induced improvements in glucose homeostasis. Diabetes 64: 2002–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JM (2012) The fat controller: adipocyte development. PLoS Biol 10: e1001436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R, Joosten LA, Koenen T, van Tits B, van Diepen JA, van den Berg SA, Rensen PC, Voshol PJ, Fantuzzi G, Hijmans A, Kersten S, Muller M, van den Berg WB, van Rooijen N, Wabitsch M, Kullberg BJ, van der Meer JW, Kanneganti T, Tack CJ, Netea MG (2010) The inflammasome‐mediated caspase‐1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metab 12: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM (2008) White fat progenitor cells reside in the adipose vasculature. Science 322: 583–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Giorgadze N, Pirtskhalava T, Thomou T, DePonte M, Koo A, Forse RA, Chinnappan D, Martin‐Ruiz C, von Zglinicki T, Kirkland JL (2006) Fat depot‐specific characteristics are retained in strains derived from single human preadipocytes. Diabetes 55: 2571–2578 [DOI] [PubMed] [Google Scholar]
- Tchkonia T, Lenburg M, Thomou T, Giorgadze N, Frampton G, Pirtskhalava T, Cartwright A, Cartwright M, Flanagan J, Karagiannides I, Gerry N, Forse RA, Tchoukalova Y, Jensen MD, Pothoulakis C, Kirkland JL (2007) Identification of depot‐specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am J Physiol Endocrinol Metab 292: E298–E307 [DOI] [PubMed] [Google Scholar]
- Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia‐Martin R, Grinspoon SK, Gorden P, Kahn CR (2017) Adipose‐derived circulating miRNAs regulate gene expression in other tissues. Nature 542: 450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Yamamoto Y, Gesta S, Kahn CR (2008) Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab 7: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troike KM, Henry BE, Jensen EA, Young JA, List EO, Kopchick JJ, Berryman DE (2017) Impact of growth hormone on regulation of adipose tissue. Compr Physiol 7: 819–840 [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Scherer PE (2006) Adipose tissue‐derived factors: impact on health and disease. Endocr Rev 27: 762–778 [DOI] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR (2008) New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlamov O, Chu M, Cornea A, Sampath H, Roberts CT Jr (2015) Cell‐autonomous heterogeneity of nutrient uptake in white adipose tissue of rhesus macaques. Endocrinology 156: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM (2012) Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150: 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Lynes MD, Dreyfuss JM, Shamsi F, Schulz TJ, Zhang H, Huang TL, Townsend KL, Li Y, Takahashi H, Weiner LS, White AP, Lynes MS, Rubin LL, Goodyear LJ, Cypess AM, Tseng YH (2015) Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med 21: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Gesta S, Lee KY, Tran TT, Saadatirad P, Kahn CR (2010) Adipose depots possess unique developmental gene signatures. Obesity 18: 872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhong W, Cui T, Yang M, Hu X, Xu K, Xie C, Xue C, Gibbons GH, Liu C, Li L, Chen YE (2006) Generation of an adult smooth muscle cell‐targeted Cre recombinase mouse model. Arterioscler Thromb Vasc Biol 26: e23–e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera‐Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT (2008) Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Data Availability Statement
Microarray data deposited at Gene Expression Omnibus (Accession #: GSE110531).


