The oligomerization (and particularly dimerization) of Hsp70 proteins plays an important role in their chaperoning activities. Here, we report that human stress-inducible Hsp70 possesses the highest propensity among analyzed Hsp70 homologs to form dimers in the presence of ATP. ATP-bound Hsp70 assembles in solution as an antiparallel dimer closely resembling the dimeric structures captured in DnaK and BiP crystals. ATP-dependent Hsp70 dimerization is necessary for efficient Hsp40 interaction and is differentially affected by TPR cochaperone binding.
Keywords: Chaperone, Structural Biology, Mass Spectrometry, Protein Conformation, Protein Structure, Protein-Protein Interactions, Allostery, Cochaperone
Graphical Abstract
Highlights
Hsp70 homologs differ in their oligomeric properties in the presence of ATP.
Human inducible Hsp70 forms ATP-dependent anti-parallel dimers with high propensity.
Dimerization of ATP-bound Hsp70 is required for effective Hsp70-Hsp40 interaction.
ATP-dependent interaction with Tomm34 TPR cochaperone disrupts Hsp70 dimer.
Abstract
Eukaryotic protein homeostasis (proteostasis) is largely dependent on the action of highly conserved Hsp70 molecular chaperones. Recent evidence indicates that, apart from conserved molecular allostery, Hsp70 proteins have retained and adapted the ability to assemble as functionally relevant ATP-bound dimers throughout evolution. Here, we have compared the ATP-dependent dimerization of DnaK, human stress-inducible Hsp70, Hsc70 and BiP Hsp70 proteins, showing that their dimerization propensities differ, with stress-inducible Hsp70 being predominantly dimeric in the presence of ATP. Structural analyses using hydrogen/deuterium exchange mass spectrometry, native electrospray ionization mass spectrometry and small-angle X-ray scattering revealed that stress-inducible Hsp70 assembles in solution as an antiparallel dimer with the intermolecular interface closely resembling the ATP-bound dimer interfaces captured in DnaK and BiP crystal structures. ATP-dependent dimerization of stress-inducible Hsp70 is necessary for its efficient interaction with Hsp40, as shown by experiments with dimerization-deficient mutants. Moreover, dimerization of ATP-bound Hsp70 is required for its participation in high molecular weight protein complexes detected ex vivo, supporting its functional role in vivo. As human cytosolic Hsp70 can interact with tetratricopeptide repeat (TPR) domain containing cochaperones, we tested the interaction of Hsp70 ATP-dependent dimers with Chip and Tomm34 cochaperones. Although Chip associates with intact Hsp70 dimers to form a larger complex, binding of Tomm34 disrupts the Hsp70 dimer and this event plays an important role in Hsp70 activity regulation. In summary, this study provides structural evidence of robust ATP-dependent antiparallel dimerization of human inducible Hsp70 protein and suggests a novel role of TPR domain cochaperones in multichaperone complexes involving Hsp70 ATP-bound dimers.
Protein homeostasis, including de novo protein synthesis surveillance, preprotein transport, misfolded protein degradation and aggregate dissolving, relies on the coordinated actions of the abundant Hsp70 and Hsp90 molecular chaperones (1, 2). Hsp70 proteins exhibit remarkable sequence and structural conservation across all kingdoms of life, suggesting conserved molecular mechanics (3, 4). In eukaryotes, Hsp70s have diversified into organelle-specific species including cytosolic Hsc70 and Hsp70 isoforms, and the endoplasmic reticulum (ER)1 isoform, BiP (5).
Hsp70 proteins contain two independent domains: an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD). The SBDβ subdomain forms a substrate-binding pocket with preference for hydrophobic polypeptide sequences (6, 7), whereas SBDα forms an α-helical lid that covers SBDβ through ionic contacts and helps to stabilize substrate binding (8, 9). The NBD and SBD are connected by a highly conserved hydrophobic linker (10). ATP coordination into the nucleotide-binding pocket of NBD leads to dramatic allosteric changes in Hsp70, characterized by NBD-SBDβ docking stabilizing the interdomain linker, accompanied by SBDα detachment from SBDβ and its docking onto the NBD (6, 11–14). The SBDβ domain in the ATP-bound conformation of Hsp70 has unfavorable kinetics of substrate binding, which leads to substrate release (15, 16). ATP hydrolysis triggers rearrangements in SBDβ reducing substrate on/off rates and therefore stabilizing substrate entrapment (17–19).
Two classes of cochaperones modulate the ATPase cycle of Hsp70 by accelerating ATP hydrolysis and ADP/ATP nucleotide exchange: Hsp40 homologs and nucleotide exchange factors (NEFs) (19, 20). Hsp40 proteins contain a J-domain (named after the bacterial Hsp40 protein DnaJ) that interacts with the Hsp70 interdomain linker at the NBD-SBDβ interface, stimulating the NBD catalytic center (18). The J-domain also helps to communicate the hydrolysis-stimulating role of the substrate, which is brought to Hsp70 by Hsp40 substrate-binding activity (17, 18). A structurally differing group of NEFs assists ADP to ATP exchange in the NBD of Hsp70 by convergent mechanics through opening of the nucleotide binding cleft (20). Interesting members of the NEF group are Hsp110 proteins, represented by yeast Sse1, which exhibit significant structural homology to the ATP-bound state of Hsp70 and mediate nucleotide exchange through heterodimerization with Hsp70 (14, 21).
Beside the ATPase regulating cochaperones, eukaryotes have evolved a large group of cochaperones containing tetratricopeptide repeat (TPR) domains that mediate their interaction with the conserved C-terminal EEVD motif of eukaryotic Hsp70 and Hsp90 (22). Apart from their TPR domains, these cochaperones bear additional domains with various functions diversifying molecular chaperones-mediated substrate processing (23).
Recently, dimerization and other oligomeric forms of Hsp70 family members in both conformational states, Apo/ADP- and ATP-bound, were described to play an important role in their chaperoning activities (24–28). Crystallographic studies revealed the presence of antiparallel ATP-bound dimers in the crystal structure of evolutionary distant Hsp70 orthologs DnaK (11, 13) and BiP (6, 12). The ATP-dependent dimerization of DnaK is necessary for its cooperation with DnaJ, however the dimeric state is very low in solution (25). In contrast, our previous isothermal titration calorimetry (ITC) analysis suggested that human inducible Hsp70 isoform exists predominantly as a dimer in an ATP supplemented buffer (29).
The aim of this work was to compare the levels of ATP-dependent dimerization in different Hsp70 proteins and to functionally and structurally characterize dimers of the human inducible Hsp70 isoform. Unlike DnaK and BiP, our data indicate that antiparallel dimerization of ATP-bound Hsp70 is highly pronounced in human Hsc70/Hsp70 cytosolic isoforms and is differentially regulated by interactions with Chip and Tomm34 TPR cochaperones.
EXPERIMENTAL PROCEDURES
Cloning and Protein Preparation
All coding sequences were cloned by Gateway recombination technology (Invitrogen, Carlsbad, CA). The full coding sequences of the E. coli dnaK (DnaK, P0A6Y8-1) and human HSPA1A (Hsp70, P0DMV8-1), HSPA8 (Hsc70, P11142-1) and HSPA5 (BiP, P11021-1, aa 19–654) genes and sequences coding for Hsp70 point mutants (I164D, T204A, D529A, N540A, E543A, N540A-E543A) were cloned into pDEST17 and pT7-SBP vectors containing an N-terminal His6 or SBP tag, respectively. The Hsp70 point mutations were prepared using QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The full coding sequences of the human DNAJB1 (Hsp40, P25685-1), TOMM34 (Tomm34, Q15785-1), BAG1 (Bag-1, Q99933-4) and STUB1 (Chip, Q9UNE7-1) genes were cloned into pDEST15, containing an N-terminal His6-GST tag cleavable by tobacco etch virus (TEV) protease. Genes were expressed in BL21(DE3) RIPL cells and purified as described (29). His6-tagged Hsp70 proteins were purified on HisTrap columns (GE Healthcare, Piscataway, NJ), dialyzed against buffer A (20 mm Tris, pH 7.2, 100 mm KCl) for 24 h, applied to HiTrap Q columns (GE Healthcare) and eluted by a linear gradient of buffer B (20 mm Tris, pH 7.2, 1 m KCl). Fractions containing His6-tagged proteins were concentrated and subjected to gel filtration using a HiPrep 16/60 Sephacryl S-100 HR column. An untagged stress-inducible Hsp70 protein was produced by TEV protease treatment of streptavidin matrix-bound SBP-tagged protein bearing TEV cleavage site between SBP tag and the protein sequence.
Cell Culture and Transfection
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 300 mg/l l-glutamine in a humidified atmosphere of 5% CO2 at 37 °C. Transient transfections of cells with the plasmids pCMV-N-HA-Hsp70 and pCMV-N-HA-Hsp70-N540A-E543A were performed by polyethylenimine (PEI). Cells were collected 24 h after transfection, lysed in 25 mm Hepes, pH 7.5, 75 mm KCl, 75 mm KAc, 2 mm MgCl2, and the lysates desalted by 7-kDa molecular mass cut-off Zeba spin desalting column (Thermo Fischer Scientific, Waltham, MA).
SBP Pull-down Assays
All proteins were exchanged into pull-down assay buffer (50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2) before mixing 70 pmol of various SBP-tagged Hsp70 constructs with streptavidin agarose beads for 30 min at 4 °C. After washing with assay buffer, 140 pmol of Tomm34 protein in buffer containing 0.2 mm ATP or no nucleotide was added to the beads for 1 h at 4 °C. Beads were washed before eluting with 2 mm biotin in assay buffer and analysis by SDS-PAGE.
Antibodies
Monoclonal antibodies to Tomm34, Hsp70 and Chip were previously prepared and characterized in-house. Blots were developed with rabbit anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (Dako, Santa Clara, CA).
Luciferase Refolding Assay
Luciferase refolding assay was performed as described previously with some changes (29). The refolding protein mixture contained 1 μm Hsp70 WT or its mutants, 2 μm Hsp40, 0.5 μm Bag-1 and 8 nm denatured luciferase in 25 mm Hepes, pH 7.2, 50 mm KAc, 2 mm DTT, 2 mm MgCl2. Luciferase activity was measured at 21 °C using an Infinite M1000 Pro (Tecan, Männedorf, Switzerland) at emission wavelengths of 560 nm and 100 ms integration time. The signal from samples with native luciferase was set as 100%. Luciferase activity of denatured luciferase only and the refolding activity of chaperone mixture without Hsp70 acted as negative controls.
Malachite Green ATPase Assay
Experiments used published protocols (29, 30). Hsp70 WT or its mutants (2 μm) were titrated with Hsp40 protein in buffer containing 50 mm Hepes, pH 7.5, 0.15 m KAc, 2 mm MgCl2, 2 mm ATP for 3 h at 37 °C. Malachite green reagent was added, and 34% sodium citrate was immediately added to halt the non-enzymatic hydrolysis of ATP. Samples were incubated for another 15 min before measuring absorbance at 620 nm. A phosphate standard curve was used to calculate pmol ATP/μm Hsp70/min.
Peptide Binding
Fluorescein-NRLLLTG peptide binding experiments were performed in 50 mm Hepes, pH 7.5, 0.15 m KAc, 2 mm MgCl2, 0.01% Tween 20. To obtain equilibrium binding curves, mixtures containing 30 nm peptide were titrated with different concentrations of Hsp70 WT and mutant proteins and incubated overnight at 4 °C. Peptide binding kinetics and peptide release was analyzed using peptide and protein concentrations of 30 nm and 50 μm, respectively, where ATP (0.4 mm) was added after overnight incubation at 4 °C to measure release. To distinguish nonspecific binding, we used BSA with the same concentration as analyzed protein. All reactions were carried out in a total volume of 12 μl in 384-well black Nunc Plates (Thermo Fischer Scientific). Fluorescence polarization was measured at 21 °C using an Infinite M1000 Pro (Tecan) with excitation and emission wavelengths of 470 and 520 nm, respectively. Data were analyzed using GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego, CA).
Small-angle X-ray Scattering (SAXS)
The SAXS data sets were collected using the BioSAXS-1000, Rigaku at CEITEC (Brno, Czech Republic). Data were collected at 277 K with focused (confocal Max-Flux SAXS optic, Rigaku) Cu Kα X-ray (1.54 Å). Sample to detector (PILATUS 100K, Dectris) distance was 0.48 m covering a scattering vector (q = 4πsin(θ)/λ) range from 0.009 to 0.6 Å−1. Size exclusion buffer was used for the blank measurement. Hsp70-T204A protein samples were measured at 2.5 and 1.5 mg/ml. Six separate two-dimensional images were collected for buffer and sample (10 min exposure per image). Radial averaging, data reduction and buffer subtractions were performed using SAXSLab3.0.0r1, Rigaku. Six individual scattering curves (10 min exposures) were compared with check the radiation damage and averaged.
Integral structural parameters (supplemental Table S1) were determined using PRIMUS/qt ATSAS v.2.8.3 (31). For molecular weight estimations, an ab initio model was reconstructed using DAMMIF ATSAS v.2.8.3, where the annealing mode was set to “fast,” whereas all other parameters were kept at default. The ab initio modeling for superimposition with atomic models was performed by DAMMIN ATSAS v.2.8.3, where the computation mode was set to “slow,” point symmetry of the particle to “P2” and all other parameters were kept at default. Evaluation of solution scattering and fitting to experimental scattering curves was performed using CRYSOL ATSAS v.2.8.3, where automatic constant subtraction was allowed; other parameters were kept at default. Superimposition of the atomic and ab initio models was performed by SUPCOMB ATSAS v.2.8.3. UCSF Chimera was used for graphical representation (32).
Surface Plasmon Resonance (SPR)
SPR was carried out on a Biacore T200 system (GE Healthcare). Biotinylated Hsp40 (EZ-Link™ Thermo Fischer Scientific) was immobilized on Series S Sensor chip SA (GE Healthcare) for about 460 response units. All measurements were performed at 21 °C in SPR buffer (25 mm Hepes, pH 7.5, 100 mm KAc, 2 mm MgCl2, 0.005% Tween 20). Each Hsp70 protein (1 μm) was pre-incubated with or without 0.2 mm ATP and injected for 20 min at 10 μl/min. The first channel on the sensor was coated with biotin and used as a background control. The data were plotted after subtracting this background control.
Analytical Size Exclusion Chromatography (SEC)
Separations by SEC were carried out using Superdex 200 Increase 10/300 GL (GE Healthcare) pre-equilibrated with 25 mm Hepes, pH 7.5, 150 mm KAc, 2 mm MgCl2. 75 μl of Hsp70 proteins (40 μm) pre-incubated with/without 0.2 mm ATP for 20 min at 21 °C was injected and isocratically eluted at 0.3 ml/min. In experiments testing the influence of Hsp40 on Hsp70 dimers, Hsp40 (10 μm) was added at the end of the Hsp70 incubation with ATP and incubated for 5 min. In experiments analyzing the effect of TPR domain cochaperones on Hsp70 dimerization, Tomm34 or Chip proteins were mixed with Hsp70 in equimolar ratio (40 μm each) in the presence/absence of 0.2 mm ATP. Incubation time, injection volume and flow rate were as described above. From each run, 300 μl fractions corresponding to protein peaks were collected and analyzed using gel electrophoresis and Coomassie staining. Apparent molecular weights of proteins and protein complexes were determined using calibration curves obtained from a mixture of Thyroglobulin, Apoferritin, Amylase, Alcohol Dehydrogenase, Albumin and Carbonic Anhydrase.
Desalted lysates of HEK293 cells with overexpressed HA-tagged Hsp70 WT or N540A-E543A mutant were incubated with/without 2 mm ATP for 20 min at 21 °C before loading on columns pre-equilibrated with cell lysis buffer 25 mm Hepes, pH 7.5, 75 mm KCl, 75 mm KAc, 2 mm MgCl2. The injection volume was 200 μl. 500 μl fractions isocratically eluted at flow rate of 0.3 ml/min were collected and analyzed by gel electrophoresis and Western blotting. All analyses were performed at least three times.
Chemical Cross-linking
Chemical cross-linking was performed using disuccinimidyl adipate (DSA, Creative Molecules, Vancover, Canada). Hsp70, its dimerization mutants or homologs DnaK, Hsc70 and BiP (40 μm each) in 50 mm Hepes, pH 7.5, 150 mm KAc, 2 mm MgCl2 were incubated in the presence or absence of ATP (0.2 mm) for 20 min at 21 °C. Ten molar excess of the cross-linking agent was added and incubated for 30 min at 21 °C. 5 μg protein samples were resolved by LDS-PAGE (pre-cast 4–12% gel, NuPage) and visualized by Coomassie staining.
For glutaraldehyde chemical cross-linking, Hsp70, Tomm34 or both (40 μm each) in 25 mm Hepes, pH 7.5, 150 mm KAc, 2 mm MgCl2 were incubated for 20 min at 21 °C with or without ATP (0.2 mm). Glutaraldehyde was added (final concentration 0.5 mm) and the reactions stopped after 10 min with Tris, pH 8 (final concentration 80 mm). Samples were diluted 100x in 2× CSB loading buffer and 5 μl separated by SDS-PAGE, blotted and probed with Tomm34 or Hsp70 antibody. Lysates of HEK293 cells with overexpressed HA-tagged Hsp70 WT or N540A-E543A mutant were desalted into 25 mm Hepes, pH 7.5, 75 mm KCl, 75 mm KAc, 2 mm MgCl2 and incubated with/without 2 mm ATP for 20 min at 21 °C before mixing with glutaraldehyde (final concentration 0.5 mm) for 10 min at 21 °C. The reactions were stopped by adding Tris, pH 8 (final concentration 80 mm), samples were supplemented with 2× CSB loading buffer and analyzed by SDS-PAGE and Western blotting with Hsp70 antibody. All electrophoresis-based experiments were performed at least three times.
Native Electrospray Ionization Mass Spectrometry (ESI-MS)
Hsp70 variants (40 μm) were incubated in 25 mm Hepes, pH 7.5, 150 mm KAc, 2 mm MgCl2 with or without 0.2 mm ATP for 20 min, transferred into 200 mm ammonium acetate pH 7.5 using Zeba Spin columns (0.5 ml, 7-kDa cut off) and diluted to 20 μm protein concentration. To study Hsp70-cochaperone interactions, we pre-incubated Hsp70 with or without ATP as described above, mixed with the cochaperone in equimolar ratio, transferred into ammonium acetate and performed native ESI-MS analysis on Waters Synapt G2Si. Samples were electrosprayed from home-made gold-coated borosilicate spraying tips. Two sets of parameters differing in the trap collision energy (1V and 70V) were used. Although low collisional activation led to better transmission of dimeric molecules, higher collisional activation provided better resolution and can be used for MW estimation. Other parameters were: sampling cone voltage 50V, source offset 20V, trap gas 4 ml/min and source temperature 30 °C. Data were analyzed in MassLynx 4.1.
Molecular Modeling
The homology model of dimeric human stress-inducible Hsp70 was built using Modeler 9.17 (33). The D and F chains of BiP (PDB code 5E84) with sequence identity 64% with human stress-inducible Hsp70, were used as template.
Experimental Design and Rationale for Hydrogen/Deuterium Exchange (HDX) Mass Spectrometry
Hsp70 and its point mutants N540A, E543A and N540A-E543A were pre-incubated in the presence or absence of 5-fold molar excess of ATP for 30 min in 50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2. Deuterium labeling was initiated by 5-fold dilution into D2O-based 50 mm Hepes, pD 7.5, 0.1 m KAc, 2 mm MgCl2. After 20 s to 2 h at 21 °C, exchange was quenched by 8-fold dilution in 1 m glycine, pH 2.3. Each sample was injected into LC-system and processed as described (34) with minor modifications: desalting lasted 3 min, analytical column used was ZORBAX 300SB-C18, 0.5 × 35 mm, 3.5 μm (Agilent Technologies) with flow rate 20 μl. Gradient was shortened to 7 min, finishing at 25% solvent B. MS used 15 T FT-ICR (SolariX XR, Bruker Daltonics, Bremen, Germany). Data were acquired with ftmsControl 2.1.0 and HyStar 4.0, processed in DataAnalysis 4.1 and analysis of deuterated samples used an in-house program Deutex (unpublished). Peptides arising from pepsin digestion were identified in a separate LC-MS/MS run, where the gradient elution was performed for 35 min. Each full MS scan was followed by six MS/MS scans of the top most intense ions. Mascot generic files were generated in Data Analysis 4.1 using two peak-picking algorithms - FTMS and SNAP. These data were searched by MASCOT 2.5 against a database containing Hsp70 WT and mutant sequences and against the pepsin sequence. Search parameters were: no enzyme specificity, no modification considered, precursor tolerance 3 ppm (although most hits were below 1 ppm), fragment ion tolerance 0.05-Da. All assignments were checked manually. Based on replicate measurements (wild-type in triplicate, mutants in duplicate, all independent preparations), the average standard deviation of deuterium determination was < 0.17-Da. Differences greater than 0.35-Da in deuterium incorporation between samples were considered significant (35). Fully deuterated controls were performed for all protein versions and the data were corrected for back-exchange as described (36).
Software
Multiple sequence alignments were performed using DNASTAR Lasergene Suite (www.dnastar.com). Molecular structures were rendered using PyMOL (www.pymol.org).
RESULTS
Hsp70 and Hsc70 Exhibit Increased Propensity to Form ATP-induced Oligomers/Dimers
To evaluate the propensity of different Hsp70 homologs to undergo ATP-specific oligomerization, we performed analytical size-exclusion chromatography (SEC) of DnaK, Hsp70, Hsc70 and BiP pre-incubated in the absence (Apo form) or presence of ATP (Fig. 1A). DnaK eluted as a predominant peak with an apparent molecular weight (MW, all SEC-derived MW are considered apparent, see Experimental Procedures) of 115 kDa under both conditions. In the presence of ATP, a small population of DnaK eluted earlier, with MW 245 kDa, which might represent ATP-bound DnaK dimers (25). Although Hsp70 eluted as a predominant peak with MW 95 kDa and a minor peak with MW 210 kDa in the Apo form, pre-incubation with ATP led to its redistribution to a predominant peak of MW 160 kDa and a minor peak at 80 kDa. Similarly, Hsc70 Apo form shifted from the dominant peak at MW 90 kDa and a minor peak of 210 kDa to two major peaks with MW of 135 and 85 kDa and an early peak of 315 kDa after ATP addition. BiP in the Apo form eluted in three peaks with MW 335, 225 and 100 kDa. Incubation with ATP decreased its oligomerization, evidenced by BiP redistribution to a dominant peak of MW 100 kDa. This behavior of BiP in SEC experiments was reported earlier (28). It is noteworthy that the low MW peaks of Hsp70, Hsc70 and BiP eluted slightly later in the presence of ATP. This observation reflects ATP-induced docking of NBD and SBD domains leading to protein compaction (10, 28). Surprisingly, similar behavior was not observed for ATP-bound DnaK. This might be explained by the insufficient SEC separation of the docked and undocked DnaK molecules, or by a more continuous distribution of DnaK molecules between docked/undocked/intermediate states in the presence of ATP (11, 37, 38). To verify whether our purified proteins are conformationally active, we tested their ability to release substrate peptide in the presence of ATP (supplemental Fig. S1A). All proteins exhibited decreased peptide binding after ATP addition, confirming their allosteric activity. These results show that these Hsp70 homologs have varying propensities to oligomerize in the Apo form (with BiP being the most oligomeric, Fig. 1A) and especially in the ATP-bound state, where Hsp70 and Hsc70 exhibited the most pronounced ability to self-oligomerize. To exclude the possibility that ATP-induced oligomerization of Hsp70 protein was caused by the affinity purification tag (His6), we performed SEC after tag cleavage by TEV protease (supplemental Fig. S1B). The Apo form of tag-free Hsp70 shifted from MW 90 kDa to 130 kDa after ATP addition, indicating that oligomerization is His6-independent. As the yield of the tag-free proteins is low, we used tagged proteins in the next experiments.
Fig. 1.
Homologs of Hsp70 differ in their propensity to dimerize in the presence of ATP. A, Bacterially purified DnaK, Hsp70, Hsc70 and BiP proteins (40 μm) were pre-incubated with or without ATP (0.2 mm, 20 mins, 21 °C) before separation by analytical SEC. Apparent MW of eluting peaks is indicated (see Experimental procedures). B, Hsp70 homologs (40 μm) were pre-incubated with or without ATP for 20 mins before addition of chemical cross-linker (10 molar excess of DSA). The cross-linked complexes were separated by LDS-PAGE (pre-cast gradient (4–12%) gel, NuPage). The molecular weight standard is indicated. C, Native ESI-MS spectra of Hsp70 and Hsc70 proteins (20 μm) were acquired after their pre-incubation without (Apo) or with ATP (0.2 mm). The charged states corresponding to monomers and dimers are labeled with single and double dots, respectively.
To explore the presence and the nature of the oligomers in the Apo and ATP state further, we performed chemical cross-linking (DSA) of Hsp70 proteins incubated with or without ATP and analyzed the resulting complexes by SDS-PAGE (Fig. 1B). In agreement with SEC profiles, we observed ATP-dependent assembly of Hsp70 and Hsc70 proteins. Conversely, ATP decreased the amount of BiP oligomeric structures. The MW of Hsp70 and Hsp70 ATP-induced oligomers is ∼150 kDa, strongly suggesting dimer formation. As with SEC data, ATP-dependent dimers of DnaK were not detectable.
Next, we verified the molecular masses of Hsp70 and Hsc70 protein assemblies formed after incubation with ATP by native electrospray ionization mass spectrometry (24, 27, 39, 40). Here, we used a lower protein concentration (20 μm) than for SEC (40 μm) to avoid spray instability and subsequent irreproducibility of data (higher concentrations caused frequent plugging of the ESI emitters). A decrease in monomer/dimer ratio was observed between Apo- and ATP treated Hsp70 (Fig. 1C), although the amount of dimer in the ATP state does not reflect ratios from SEC. This can be caused by lower gas-phase stability, poorer transmission of the dimer and/or by the lower protein concentration (20 μm) than used for SEC (40 μm). A similar picture was observed for Hsc70, corroborating the SEC results. The 315 kDa peak in Hsc70 SEC analysis was not detected by native ESI-MS, possibly because of its gas-phase instability. These results indicate that Hsp70 homologs differ in their oligomeric properties in solution in the presence of ATP, with human stress-inducible Hsp70 predominantly forming a dimeric population.
ATP-induced Dimerization of the Human Stress-inducible Hsp70 Is a Time and Concentration Dependent Process Requiring Intact ATP-bound Conformation of the Protein
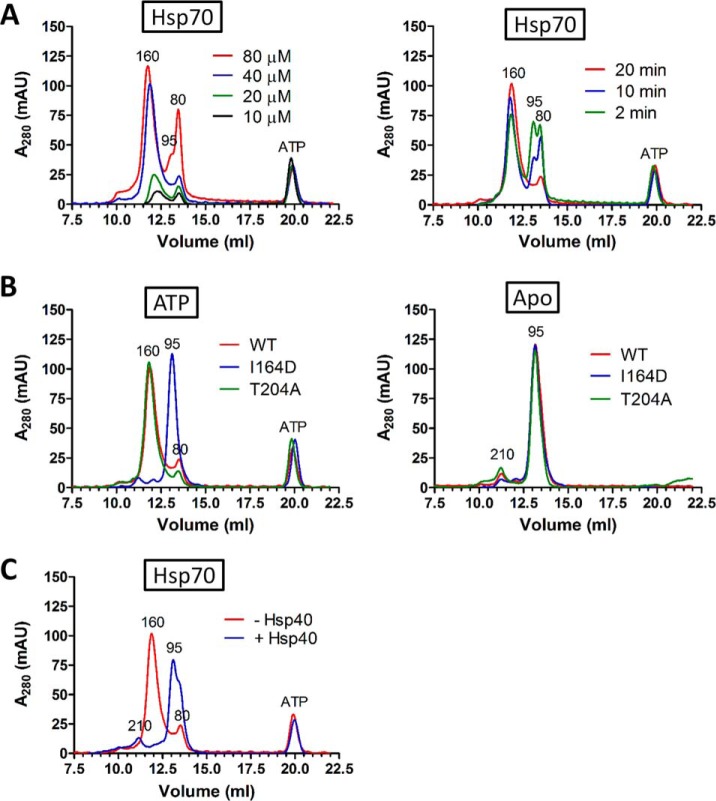
In these experiments we focused on the human inducible isoform of Hsp70. To characterize ATP-dependent dimerization, we measured the concentration and time dependence of this process by SEC (Fig. 2A). Under a constant ATP concentration (200 μm), Hsp70 dimers are detectable from 10 μm up to 80 μm protein concentration, with 40 μm showing the highest dimer to monomer peak intensities ratio. When incubated with ATP for increasing time intervals (2, 10, 20 min), Hsp70 dimers formed in a time-dependent manner, demonstrating that ATP-triggered Hsp70 dimer-monomer association/dissociation equilibrium is concentration- and time-dependent.
Fig. 2.
ATP-induced dimerization of Hsp70 requires its intact NBD-SBDβ docked conformation. A, Hsp70 was mixed at different concentrations (10, 20, 40, 80 μm; 20 min incubation) or for different time intervals (2, 10, 20 min) with ATP (0.2 mm) at 21 °C before separation by SEC. B, Hsp70 and its mutant forms (I164D, T204A) (40 μm) were incubated without (Apo) or with ATP (0.2 mm, 20 min) at 21 °C before SEC. C, Hsp70 (40 μm) was incubated with ATP (0.2 mm, 21 °C) for 20 min. Hsp40 (10 μm) was added or not for 5 min and samples were separated by SEC. Apparent MW of eluting peaks is indicated (see Experimental procedures).
To evaluate whether the ATP-induced conformation of Hsp70 is necessary for its dimerization, we tested the behavior of the Hsp70 I164D mutant, which binds ATP but is not able to reach NBD-SBD docked conformation (Fig. 2B) (29). The elution profiles of I164D under both nucleotide conditions show a dominant peak at MW 95 kDa, indicating that this protein is not able to form ATP-dependent dimers and remains monomeric. Therefore, the ATP-dependent dimerization of Hsp70 requires its ATP-induced conformation. Next, we tested T204A mutant whose intrinsic ATPase activity is low, suggesting it might establish ATP-bound dimers with higher rate and/or stability (29, 41). The T204A mutant elution profile was almost identical to WT, with only a slight increase in dimer to monomer peak intensities, implying that dimerization is not affected by the intrinsic ATP conversion rate under our experimental setup. Finally, we probed the behavior of Hsp70 ATP-bound dimers in the presence of the ATPase-activating cochaperone, Hsp40 (Fig. 2C). A 5 min incubation of preformed Hsp70 dimers with substoichiometric amounts of Hsp40 led to the complete disappearance of the ATP-dependent dimer peak, with a concomitant increase of the 95 kDa peak belonging to the monomeric Apo/ADP state of Hsp70. A minor early eluting peak of Hsp70 Apo state dimer (apparent MW 210 kDa) also appeared. These data support the requirement of ATP-induced Hsp70 conformation for dimerization and indicate that Hsp40 can recognize and stimulate the ATPase activity of dimeric Hsp70.
ATP-dependent Hsp70 Dimer Assembles In An Antiparallel Fashion Resembling the Dimers Captured In the Crystal Structures of DnaK and BiP
The conformational requirements for Hsp70 ATP-induced dimer formation suggest that the dimer might acquire a quaternary structure observed in the DnaK (11, 13) and BiP crystal packings (12), with two ATP-bound protomers assembled in an antiparallel orientation (Fig. 3A). Two residues (DnaK numbering N537, D540) in an α-helical bundle subdomain of DnaK are important for its ATP-dependent anti-parallel dimerization and are conserved in human Hsp70 (human numbering N540 and E543) (25). We rationalized that if these residues retained their role in human Hsp70 ATP-dependent dimerization, their mutation would disturb dimerization. Indeed, SEC of N540A, E543A and N540A-E543A in Apo and ATP-bound state showed decreasing levels of dimeric forms in the order N540A>E543A>N540A-E543A, with the double mutant being unable to reach the dimeric structure (Fig. 3B). We saw a similar trend by chemical cross-linking and native ESI-MS (Fig. 3C and 3D). In native ESI-MS, the dimer of T204A mutant in ATP-bound state exhibited higher stability in the gas phase than WT and we therefore analyzed the dimerization interface mutant N540A-E543A also on the background of T204A mutation (T204A-N540A-E543A, supplemental Fig. S2A). This measurement further supported the importance of residues N540 and E543 for Hsp70 ATP-dependent dimerization. Additionally, we detected a minor population of Hsp70 dimers in Apo state, which is in concordance with current knowledge and our SEC data (Fig. 3D) (24). To exclude that diminished dimerization of mutants is caused by their impaired allosteric activity upon ATP binding, we analyzed WT and mutant proteins with or without ATP by HDX mass spectrometry (Fig. 3E and supplemental Fig. S2B). The ATP-induced deuteration changes are similar in WT and dimerization mutants, highlighted by protection of the NBD and opening of SBDβ. More precisely, the peptides covering allosterically important regions involved in linker-mediated domain docking (11–13, 29) are protected in all proteins in the presence of ATP: NBD docking interface (covered by peptides 143–150, 150–167 and 168–180), SBDβ docking interface (413–427, 478–486) and linker region (394–401). These results indicate that the allosterically induced ATP-bound conformation is preserved in the N540A, E543A and N540A-E543A mutants. To independently verify the mutants′ ability to reach the ATP-bound state, we tested their interaction with Tomm34, which we have shown to be ATP-dependent (supplemental Fig. S2C) (29). All proteins increasingly interacted with Tomm34 in the presence of ATP, supporting their conformational activity.
Fig. 3.
Conserved N540 and E543 residues are required for stable ATP-induced dimerization of Hsp70. A, Crystal structure of DnaK ATP-bound dimer (PDB code 4jne) with N537 and E540 residues highlighted in red (N540, E543 numbering in human inducible Hsp70). The NBD domain is depicted in yellow, SBDβ in marine and SBDα in green. B, Hsp70 and N540A, E543A and N540A-E543A (40 μm) were incubated without (Apo) or with ATP (0.2 mm, 20 min) at 21 °C before SEC. C, Hsp70 WT, N540A, E543A and N540A-E543A proteins (40 μm) were pre-incubated with or without ATP for 20 min before addition of chemical cross-linker (10 molar excess of DSA). The cross-linked complexes were separated by LDS-PAGE (pre-cast gradient (4–12%) gel, NuPage). The molecular weight standard is indicated. D, Native ESI-MS spectra of Hsp70 WT, N540A, E543A and N540A-E543A proteins (20 μm) were acquired after their pre-incubation without (Apo) or with ATP. The charged states corresponding to monomers and dimers are labeled with single and double dots, respectively. E, Deuteration level (exchanged deuterons, D) differences of Hsp70 WT, N540A, E543A and N540A-E543A peptides in ATP-bound and nucleotide-free state (Apo) after 1200 s (for other incubation times see supplemental Fig. S2B) incubation in deuterated buffer. Numbers at the left indicate the Hsp70 peptide fragments; schematic representation at the left shows Hsp70 domain constitution; L, interdomain linker; T, C-terminal tail. The asterisks indicate peptides covering allosterically important regions of Hsp70 molecule. Dashed lines in the graph indicate significance level of 0.35 Da (see Experimental procedures).
To evaluate the overall shape of Hsp70 ATP-dependent dimers in solution more closely, we performed small-angle X-ray scattering (SAXS) (Fig. 4 and supplemental Table S1), using T204A mutant to prevent ATP hydrolysis during sample preparation and handling. T204A protein was pre-incubated with ATP and its dimeric form was separated by SEC before SAXS at two protein concentrations (1.5 mg/ml, 2.5 mg/ml). The data were fitted to the theoretical scattering functions based on the atomic coordinates of various Hsp70 dimeric crystal structures or of our Hsp70 homology-based ATP-dependent dimer model (see Experimental Procedures). The best fit to the experimental data was obtained for Hsp70 dimer model (1.5 mg/ml, χ2 = 1.2; 2.5 mg/ml, χ2 = 1.1, Fig. 4A and 4B), supporting the proposed human Hsp70 ATP-dependent dimer model (Fig. 4C) over the crystal structures of DnaK and BiP dimers (PDB: 4b9q (chains AC), 5e84 (chains DF)). Taken together, these analyses suggest that the ATP-bound monomers of inducible human Hsp70 protein assemble in solution as an antiparallel dimer in a similar orientation as DnaK and BiP dimers in their crystal structures (11–13).
Fig. 4.

ATP-bound Hsp70 monomers assemble as antiparallel dimers in solution as determined by small-angle X-ray scattering. A, χ2-ranked fitting of Hsp70 dimeric crystal structures (PDB codes with indicated chains) and homology-based Hsp70 dimer model (see Experimental Procedures) to the SAXS experimental data obtained for Hsp70-T204A mutant at two concentrations. Fitting to the experimental scattering curves was performed using CRYSOL ATSAS v.2.8.3. B, CRYSOL fit of Hsp70 ATP dimer atomic model. Simulated scattering data of dimeric Hsp70 atomic model (red) is fitted to experimentally obtained solution scattering showing overall good fit with χ2 = 1.1. C, Atomic model of Hsp70 ATP-dependent dimer, developed as described in Experimental Procedures. NBD is highlighted in yellow, SBDβ in marine and SBDα in green. N- and C-terminal unstructured regions of the model are omitted from the figure. The image was created in PyMOL.
Hsp70 ATP-dependent Dimers Assemble Through Conserved Interfaces
To pinpoint the structural rearrangements introduced to the monomeric (Apo form) and dimeric (ATP-bound) Hsp70 states by N540A, E543A and N540A-E543A mutations, we compared the HDX rate of these mutants with the WT protein (Fig. 5 and supplemental Fig. S3), an approach that has been applied previously to study protein homodimerization (42–44). We observed significant destabilization of the SBD-αAB helices covered by peptides 519–529, 530–542, 533–542 in Apo N540A and N540A-E543A mutants, with the latter exhibiting more pronounced changes (Fig. 5A and 5C, and supplemental Fig. S3B). Because these peptides either contain or are in close vicinity to the substituted N540 and E543 residues, the detected structural changes are most likely because of local disturbances caused by the mutations, although the E543A mutation itself does not have an effect on this region. E543A and N540A-E543A showed increased deuterium incorporation in the loop connecting αC and αD helices and the αD helix (peptide 582–601) under both nucleotide conditions (Fig. 5A and 5C), suggesting an important role of E543 in the stability of this region independently of conformation. As the αC/αD helices form a compact α-helical structural unit together with the αB helix (Fig. 5B), the loosening of the αD helix might explain the additive effect of E543A in destabilizing the αA/αB helices in the N540A-E543A mutant (Fig. 5A) (9). Because N540 and E543 together with D529 are involved in positioning the α-helical lid-SBDβ (Fig. 5B) (8, 9, 16) and D529 is present in the region destabilized by N540A and N540A-E543A substitutions (peptide 519–529), we expect the mutants to have defects in the kinetics of substrate binding (see below, Fig. 6B).
Fig. 5.
Hsp70 ATP-dependent dimers assemble through NBD-NBD and NBD-SBDα interfaces. A, Deuteration level (exchanged deuterons, D) differences between Hsp70 WT and N540A, E543A and N540A-E543A peptides in nucleotide-free state (Apo) measured after 120 s (for other incubation times see supplemental Fig. S3B) incubation in deuterated buffer. The graph is labeled as in Fig. 3E. Red dots indicate peptides highlighted in part B and C of this figure. B, Crystal structure of human SBD (PDB code 4po2). SBDα (lid) helices and the lid-SBDβ positioning ionic/polar contacts are indicated. SBDβ is labeled in marine and SBDα in green. Regions covered by peptides highlighted by red dots in A are red. C, Deuteration kinetics of indicated peptides from Apo and ATP-bound Hsp70 proteins. D, Deuteration level (exchanged deuterons, D) differences between Hsp70 WT and N540A, E543A and N540A-E543A in ATP-bound state measured after 20 s (for other incubation times see supplemental Fig. S3B) incubation in deuterated buffer. The graph is labeled similarly as in Fig. 3E. Red dots indicate peptides highlighted (red/pink) on the atomic structures in E, E. 48–68, 268–282, 294–302, 519–529 and 530–542 peptides highlighted in D were projected onto the homology-based Hsp70 ATP-bound dimer atomic model (see Experimental Procedures) and the corresponding peptides (see supplemental Fig. S4C) in DnaK were projected onto the crystal structure of DnaK ATP-bound dimer (PDB code 4jne). The peptides are colored in red or pink in the respective protomer. The atomic structures are shown in both ribbon and surface representations. Surface representation was also manually separated for clarity (bottom structures). NBD is highlighted in yellow, SBDβ in marine and SBDα in green. N- and C-terminal unstructured regions of the model are omitted from the figure. The image was created in PyMOL.
Fig. 6.
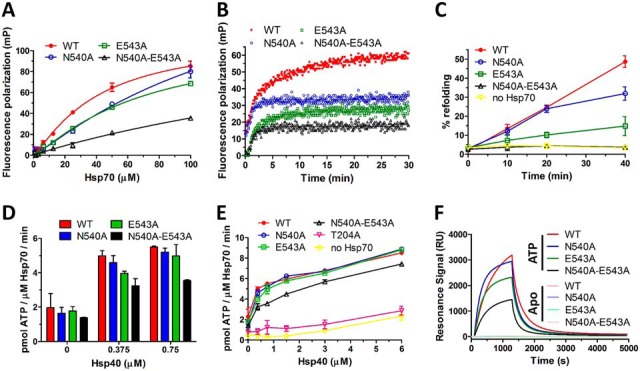
E543A and N540A-E543A mutants have severely impaired substrate binding/refolding activities and lower interaction with Hsp40. A, Equilibrium binding curves of F-NRLLLTG peptide binding to Hsp70 WT and mutants under nucleotide-free conditions. Fluorescence polarization was determined at 30 nm peptide and increasing Hsp70 concentrations. Error bars represent S.D.; n = 3 independent experiments. B, Kinetics of F-NRLLLTG interaction with WT and mutant Hsp70s under nucleotide- free conditions determined by fluorescence polarization. Protein and peptide concentrations were 50 μm and 30 nm, respectively. C, Firefly luciferase was chemically denatured, mixed with Hsp70 WT or mutants (1 μm), Hsp40 (2 μm), Bag-1 (0.5 μm) and ATP (2 mm) and recovered luminescence was measured. Error bars represent S.D.; n = 3 independent experiments. D, ATPase activity of Hsp70 WT and mutants at lower Hsp40 concentrations, for full results see E. E, ATPase activity of Hsp70 WT and mutants (2 μm) was tested at various Hsp40 concentrations in malachite green assay. Error bars represent S.D.; n = 3 independent experiments. F, SPR measurement of Hsp70 WT and mutants binding to Hsp40 under nucleotide-free conditions (Apo) or in the presence of ATP.
Inspection of the ATP-bound state of Hsp70 dimerization mutants revealed several peptides with differential deuteration levels at early incubation times of the experiment (Fig. 5D 5E, and supplemental Fig. S3). These peptides can be divided into four groups according to deuteration rate under nucleotide-free and ATP-bound conditions: i) peptides with similarly increased deuteration under both nucleotide conditions, indicating ATP-independent structural changes; ii) peptides exhibiting elevated deuteration in the Apo state with further increased deuteration upon ATP addition; iii) peptides with increased deuteration only in the ATP-bound state with no correlation to the degree of dimerization deficiency introduced by N540A, E543A and N540A-E543A mutations; iv) peptides increasingly deuterated only in the presence of ATP, with E543A and N540A-E543A mutants being more affected than N540A mutant. Group i) is represented by the single peptide 582–601, whose role has been described above. Group ii) encompasses peptides 519–529, 530–542 and 533–542. Peptide 519–529 covers a region that does not show detectable ATP-induced changes in WT protein (supplemental Fig. S3A). The presence of ATP increases the degree of destabilization induced in this site only in the N540A-E543A mutant (Fig. 5C and supplemental Fig. S3A). Moreover, the E543A mutation does not affect deuteration of the 519–529 peptide in the Apo form but leads to elevated deuteration in the ATP-bound state (Fig. 5C and supplemental Fig. S3A). That the E543A and N540A-E543A mutations are more deleterious for Hsp70 ATP-dependent dimerization than N540A (Fig. 3B, 3C and 3D) indicates a role for the 519–529 region in dimerization. The αB helix covered by peptides 530–542 and 533–542 reacts to ATP addition by increased deuterium uptake in WT protein (supplemental Fig. S3A). Destabilization of this part of the protein detected in the Apo forms of N540A and N540A-E543A mutants is more pronounced in their ATP-bound states, particularly for N540A-E543A (Fig. 5C and supplemental Fig. S3A). Like 519–529, deuteration of 530–542 and 533–542 peptides in E543A is elevated only in its NBD-SBDβ docked conformation (Fig. 5C and supplemental Fig. S3A). As the quaternary structure of Hsp70 ATP-dependent dimers in solution resembles the crystal packing of BiP ATP-bound dimers (Fig. 4) (12), we assume that increased deuteration rates of 530–542 and 533–542 peptides in the ATP-bound conformation of dimerization mutants reflect both mutation-induced loosening of the α-helical region structure (Fig. 5B) and dimer destabilization because of elimination of the conserved N540 and E543A residues mediating the intermolecular NBD-SBDα contacts (Fig. 3A) (12). Group (iii) is represented by 487–510 peptide (supplemental Fig. S3A). The deuteration profile of this peptide probably reflects N540A-induced structural changes in the ATP-bound state of Hsp70 protomer. Peptides belonging to group iv) include 48–68, 268–282, 294–302, 353–368, 547–564 and 550–576 (Fig. 5D and 5E, and supplemental Fig. S3A). The differential deuteration of these peptides in ATP-bound forms of E543A and N540A-E543A mutants might mirror their role in the dimerization interface. When projected onto the surface of DnaK (13) and Hsp70 dimer (homology-based model, see Experimental Procedures), group (4) peptides 48–68, 268–282 and 294–302 localize to the dimer interface together with 519–529 and 530–542 (Fig. 5E). The same is true for the dimer contacts in BiP and for the interface of Hsc70 NDB with Sse1 (supplemental Fig. S4A) (45). The interface regions identified for Hsp70 are not compatible with Sse1 dimer crystal structure (supplemental Fig. S4A) (14). However, although the regions covered by 353–368 and 547–564/550–576 peptides do not match any protein-protein contacts in dimeric Hsp70, they form an interface when projected onto the Sse1 dimer (supplemental Fig. S4B). Next, we searched for the residues mediating the intermolecular contacts in the respective dimeric structures (supplemental Fig. S4A, S4C). The intermolecular NBD-SBDα contacts are conserved in all of the analyzed proteins except the Sse1 dimer. However, this interface is present in Sse1-Hsc70 NBD heterodimers (supplemental Fig. S4A). Interestingly, although the intermolecular NBD-NBD contacts localize to the aligned peptide sequences, the residues mediating the interactions differ in Hsp70 isoforms (supplemental Fig. S4A, S4C). This suggests that differences in the NBD-NBD interface contribute to Hsp70 isoforms' varying dimerization propensities (Fig. 1A). Moreover, we observed that the highly conserved D529 (Hsp70 numbering) residue participates in intramolecular contacts involving NBD-NBD interface residues in all the analyzed proteins (supplemental Fig. S4A). To test the role of D529 in Hsp70 ATP-dependent dimers, we analyzed the dimerization properties of D529A mutant (supplemental Fig. S5A and S5B). This mutant can dimerize, although the ATP-bound monomer-dimer equilibrium is impaired compared with WT protein and the D529A substitution destabilized the SBDα subdomain αAB helixes in both conformational states (supplemental Fig. S5C). Because the αB helix contains N540 and E543 residues, the D529A-induced structural rearrangements in this region might account for the dimerization imbalance.
Taken together, the antiparallel Hsp70 ATP-dependent dimer assembles in solution through the interfaces captured in the previously solved crystal structures of Hsp70 isoforms (DnaK, BiP, Sse1-Hsc70 NBD) (12, 25, 45), although the identity of the residues mediating the interaction is isoform-specific. Moreover, our HDX data suggest that Hsp70 ATP-dependent dimers might fluctuate between BiP-like (12) and Sse1-like (14) dimer organizations.
Dimerization of ATP-bound Hsp70 Is Required for Effective Hsp70-Hsp40 Interaction and Hsp40-mediated Hsp70 Chaperoning Activity
The structural changes introduced to Hsp70 by mutating N540 and E543 impair its substrate binding capacity (Fig. 6A and 6B). To assess the chaperoning activity of dimerization deficient mutants, we measured their ability to refold denatured luciferase in vitro with Hsp40 and Bag1 cochaperones as the ATPase activating and nucleotide-exchange factors (Fig. 6C) (19, 46, 47). E543A and N540A-E543A mutants had severely impaired refolding capacity, whereas the N540A mutant allowed luciferase refolding, albeit to a lower degree than WT protein. Next, we determined Hsp40-stimulated ATPase activity. Although the ATPase activity measured at low Hsp40 concentrations (Fig. 6D) mirrored the dimerization defects of N540A, E543A and N540A-E543A proteins, at higher Hsp40 concentrations N540A and E543A mutants reached ATPase levels comparable to WT protein (Fig. 6E). The N540A-E543A mutant exhibited significantly lower activity across all Hsp40 concentrations. These results indicate that these mutants are compromised in their ability to associate and functionally cooperate with Hsp40 either during in vitro refolding or concentration-dependent ATPase stimulation (25). To test mutant protein binding to Hsp40 directly, we used an established surface plasmon resonance (SPR) assay (Fig. 6F) (25, 48). We did not detect interaction between Hsp40 and the Hsp70 variants under nucleotide-free conditions. Addition of ATP led to a high binding signal for WT protein. The binding curves of N540A, E543A and N540A-E543A mutants reflected their ability to form dimers (Fig. 3), with N540A-E543A having considerably diminished affinity to Hsp40. These data suggest that the ATP-dependent dimerization of human inducible Hsp70 is required for its efficient interaction with Hsp40, like the DnaK-DnaJ interaction (25).
Tomm34 and Chip Cochaperones Bind to Hsp70 ATP-induced Dimers With Different Outcome
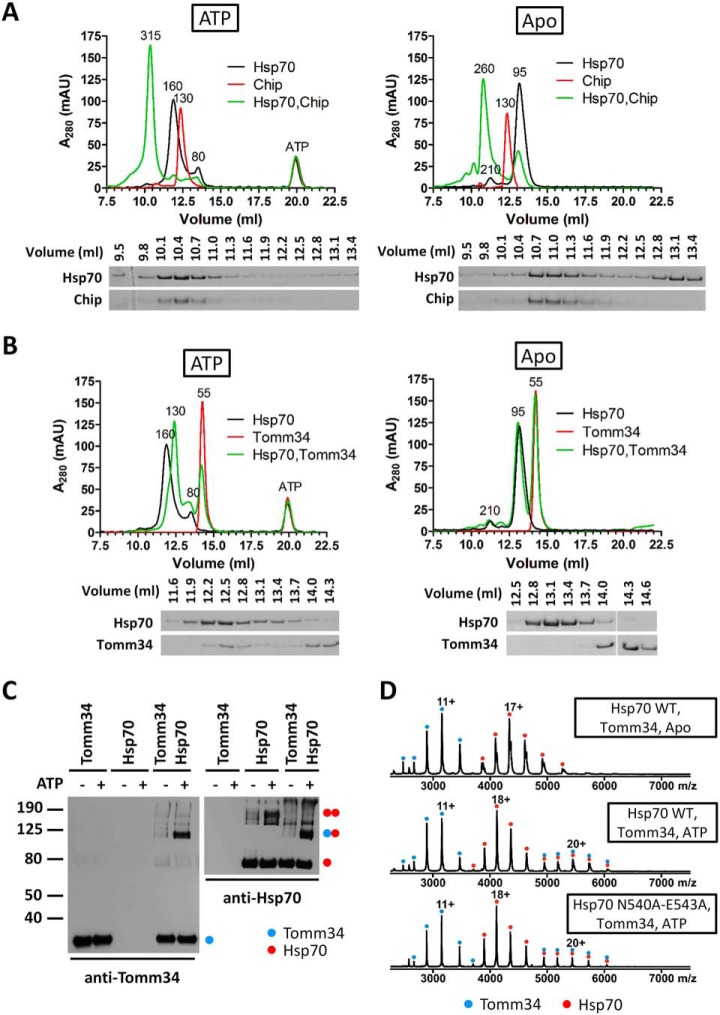
To delineate the behavior of Hsp70 ATP-dependent dimers further, we evaluated engagement in multichaperone complexes by analyzing the binding of ATP-induced Hsp70 dimers to the TPR cochaperones Tomm34 (34 kDa) (29) and Chip (35 kDa) (49, 50). As a negative control we used Hsp70 lacking its C-terminal PTIEEVD motif that mediates the interaction with TPR domains (supplemental Fig. S6A, S6B) (22). In SEC, Chip eluted as a single peak with MW 130 kDa under both nucleotide conditions (Fig. 7A). In accordance with these data, it is known that Chip forms dimers that aberrantly separate in SEC as larger oligomers because of their non-globular shape (49, 51). We also verified Chip by native ESI-MS, which clearly showed a MW corresponding to dimeric molecules (supplemental Fig. S6C). The separation of Chip/Hsp70 mixtures under nucleotide-free conditions revealed that Chip was completely saturated by Hsp70 and the elution volume of the resulting complex corresponded to 260 kDa suggesting a 2:1 stoichiometry (Chip2:Hsp70). Because the proteins were mixed in equimolar concentrations, a fraction of Hsp70 remained free as evidenced by the presence of the 95 kDa elution peak. The addition of ATP to the Chip/Hsp70 mixture led to the formation of one predominant peak with MW 315 kDa indicating the presence of a Chip2:Hsp702 complex. As controls, we did not detect interactions between Chip and Hsp70ΔPTIEEVD with or without nucleotide addition (supplemental Fig. S6A). However, Hsp70ΔPTIEEVD was able to assemble as dimers in the presence of ATP. These data indicate that Chip dimers form a stable complex with ATP-induced Hsp70 dimers. This is supported by native ESI-MS (supplemental Fig. S6D). In the Apo state, complexes corresponding to Chip2:Hsp70 and to a lesser extent Chip2:Hsp702 was also detected. On incubation with ATP, we detected only Chip2:Hsp702 complexes and Hsp70 dimers because of disruption of the larger MW complexes during ESI-MS. Moreover, the Hsp70 N540A-E543A mutant did not form ATP-induced Chip2:Hsp702 complexes, exhibiting similar behavior to Hsp70 WT/Chip mixtures in the Apo state.
Fig. 7.
ATP-dependent dimer of Hsp70 associates with Chip and Tomm34 TPR domain cochaperones through different mechanisms. A and B, Hsp70 WT and Chip or Tomm34 were mixed (both at 40 μm) and pre-incubated with or without ATP (0.2 mm, 20 min, 21 °C) before separation by analytical SEC. Apparent MW of eluting peaks is indicated (see Experimental Procedures). Proteins in the separated fractions were analyzed by SDS-PAGE and Coomassie staining. C, Hsp70, Tomm34 or both (40 μm each) were mixed in the presence or absence of ATP (0.2 mm) for 20 min at 21 °C. Glutaraldehyde was added (0.5 mm final concentration) for 10 mins and the reactions stopped with Tris, pH 8 (80 mm final concentration). Samples were diluted in 2× CSB loading buffer, separated by SDS-PAGE, blotted and probed by Tomm34 or Hsp70 antibodies. Blue and red dots, respectively, indicate positions of Tomm34 and Hsp70 monomers, homodimers and heterodimers. D, Native ESI-MS spectra of Hsp70 WT or N540A-E543A mutant mixtures with Tomm34 (20 μm) were acquired after pre-incubation without (Apo) or with ATP. The charged states corresponding to Tomm34/Hsp70 monomers and heterodimers are labeled with single and double dots, respectively.
Tomm34 eluted as a single peak of 55 kDa representing its monomeric structure (Fig. 7B, see also Fig. 7C and 7D). The elution profiles of Tomm34/Hsp70 and Tomm34/Hsp70ΔPTIEEVD in the absence of ATP were almost identical, with clearly separated peaks for individual proteins (Fig. 7B and supplemental Fig. S6B). This was expected because Tomm34-Hsp70 interaction is both ATP- and PTIEEVD-dependent (29). Accordingly, Hsp70ΔPTIEEVD dimers formed in the presence of ATP did not interact with Tomm34 (supplemental Fig. S6B). Separation of Tomm34/Hsp70 mixture in the presence of ATP resulted in a dominant peak with MW corresponding to 130 kDa and a concomitant decrease in ATP-dependent Hsp70 dimers and Tomm34 peak intensities. Conversely, we observed an increase in the Hsp70 ATP-bound monomer peak (80 kDa) (Fig. 7B). SDS-PAGE revealed that the 130 kDa peak contains Tomm34 and Hsp70. The apparent MW of the 130 kDa peak suggests the stoichiometry of the Tomm34/Hsp70 complex to be 1:1. We also chemically cross-linked the individual Tomm34 or Hsp70 proteins or their complex and analyzed the resulting assemblies by Western blotting using Tomm34 and Hsp70 specific antibodies (Fig. 7C). The ATP-dependent Hsp70 dimer (migrating at approx. 150 kDa) dissociates upon interaction with Tomm34 and the resulting Tomm34:Hsp70 complex has 1:1 stoichiometry according to its migration at approx. 110 kDa in the denaturing gel. An ATP-dependent 1:1 complex between Hsp70 WT/N540A-E543A and Tomm34 was also detected by native ESI-MS, indicating that stable Hsp70 ATP-dependent dimerization is not required for the Hsp70-Tomm34 interaction (Fig. 7D). Taken together, the ATP-induced Hsp70 dimer can complex with TPR cochaperones. However, although Hsp70 homodimers remain intact upon interaction with Chip, they are destabilized during ATP-dependent interaction with Tomm34 and form a 1:1 heterodimer.
ATP-dependent Hsp70 Dimers Are Distributed In High-molecular Weight Protein Complexes Ex Vivo
To gain insight into the potential role of ATP-dependent Hsp70 dimer formation in the cellular proteome, we compared the distribution of HA-tagged WT and N540A-E543A proteins in cell lysates fractionated by SEC (Fig. 8A). Firstly, lysates were processed by preparative gel filtration to remove low-molecular weight compounds including ATP. Before SEC, the processed lysates were incubated in the presence or absence of ATP for 20 min. In the absence of ATP, both WT and N540A-E543A proteins were evenly distributed in the early fractions (elution volume 7.5–12 ml) and accumulated in the later fractions (elution volume 12.5–13.5 ml) that correspond to their low oligomeric/monomeric forms (see Fig. 3B). Pre-incubation with ATP led to N540A-E543A becoming mainly low oligomeric, with almost complete loss in the early fractions. In contrast, WT Hsp70 retained its uniform distribution in the early fractions with slightly increased population of low oligomeric species in the presence of ATP. To probe the conformational status of Hsp70 in cell lysates we also tested the level of Tomm34 in SEC fractions (29). Tomm34 shifted to earlier fractions after ATP pre-incubation. Of note, Tomm34 coeluted with the low oligomeric fractions of WT and N540A-E543A Hsp70, indicating the presence of 1:1 complexes. This result shows that both Hsp70 variants were in NBD-SBDβ docked conformation and interacted with Tomm34 in the ATP-treated samples. Analogously, we performed glutaraldehyde cross-linking of desalted total cell lysates from the cells overexpressing HA-tagged WT and N540A-E543A proteins and separated the cross-linked complexes by SDS-PAGE (Fig. 8B). We observed an increase in the level of higher-molecular weight protein complexes containing HA-Hsp70 WT in the presence of ATP. On the contrary, the multiprotein complexes of HA-tagged N540A-E543A captured in the absence of ATP rapidly disassembled in the presence of ATP. These observations suggest that the ATP-dependent dimerization properties of Hsp70 are necessary for its participation in higher-molecular weight protein complexes in cells.
Fig. 8.
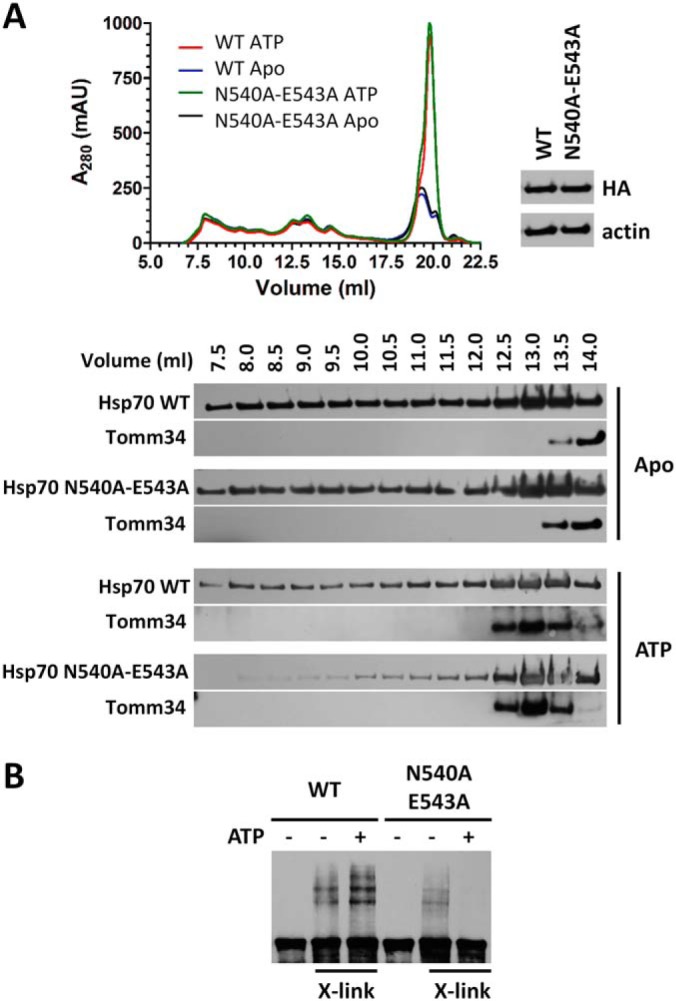
ATP-dependent dimerization of Hsp70 is required for its participation in high molecular weight complexes detected ex vivo. A, Desalted lysates from HEK293 cells overexpressing HA-tagged Hsp70 WT or N540A-E543A mutant to the same level (evaluated by Western blotting using anti-HA antibody) were incubated with/without 2 mm ATP for 20 min at 21 °C before SEC. The presence of Hsp70 and Tomm34 proteins in the separated fractions was tested by Hsp70/Tomm34 antibodies. B, Desalted HEK293 cell lysates with overexpressed HA-tagged Hsp70 WT or N540A-E543A were incubated with/without 2 mm ATP for 20 min at 21 °C before mixing with glutaraldehyde (0.5 mm final concentration) for 10 min at 21 °C. The reaction was stopped by adding Tris, pH 8 (80 mm final concentration), samples were supplemented with 2× CSB loading buffer and analyzed by SDS-PAGE and Western blotting with Hsp70 antibody.
DISCUSSION
Formation and disassembly of a small population of ATP-dependent DnaK dimers plays a critical role in DnaJ-regulated DnaK ATPase activity (25, 26). Moreover, conservancy of residues involved in mediating homo-dimerization of ATP-bound DnaK in its eukaryotic homologs supports a functional role of the ATP-dependent dimer (52). Here, we demonstrated that human Hsc70 and Hsp70 proteins form ATP-dependent dimers in solution with markedly higher efficiency compared with DnaK (Fig. 1A). The higher affinity of self-interactions between ATP-bound Hsp70 monomers versus Apo monomers is demonstrated by efficient ATP-bound dimer formation at low concentrations of Hsp70 (10 μm) compared with the more than 200 μm of Apo Hsp70 needed to assemble into dimers (Fig. 2A) (24, 53). Interestingly, Hsc70 was shown to follow ATP-dependent monomerization similarly to BiP in number of previous studies using SEC and SAXS (54–57). The discordance in the results might be caused by the fact that Hsc70 in those studies was purified from animal tissues and/or separated on Superose 12 SEC column. It has been demonstrated that posttranslational modifications and buffer ionic-strength influence Hsp70 ATP-dependent dimerization (27) and that Superose 12 ionically interacts with separated proteins complicating the interpretation (58). Alternatively, the presence of Hsp40 contamination in purified Hsc70 proteins would lead to rapid Hsc70 ATP-bound dimer dissociation after ATP addition (Fig. 2C). Hsp40 was also reported to promote ATP-dependent dimerization/oligomerization of Hsp70/Hsc70 purified from animal tissues (59) or insect cells (27), indicating that the role of Hsp70 posttranslational modification might be decisive for its oligomerization properties. In addition, an SAXS study describing the structures of recombinant bacterially purified Hsc70 in ADP- and ATP-bound conformations revealed ATP-induced compaction of Hsc70 reflecting NBD-SBDβ docking in monomers (57). However, the authors later discovered that the expression construct they used contained a mutation causing E543K substitution (60). As we now demonstrate, E543 is crucial for ATP-dependent Hsp70 dimerization.
The structural analyses (SAXS, HDX, Fig. 4 and 5, and supplemental Fig. S3) with dimerization-deficient mutants of human Hsp70 (N540A, E543A, N540A-E543A, Fig. 3) revealed that Hsp70 predominantly assembles in solution as an antiparallel dimer, closely resembling the crystal packing of its homologs (6, 11–13, 45). A recent study suggested the presence of antiparallel human Hsp70 dimers in multichaperone complexes (27), however their subunit orientation in the ATP-bound state inferred from chemical cross-linking does not satisfy the topology of antiparallel Hsp70 ATP-dependent dimers captured in DnaK/BiP structures and described here (6, 11–13). This discrepancy could stem from the use of Hsp70 purified from eukaryotic expression system by Morgner et al. (27) (i.e. post-translationally modified) or be caused by the error-prone intra- and inter-molecular cross-links differentiation because of the homo-dimeric nature of Hsp70 (61). Although the residues mediating NBD-SBDα contacts in the antiparallel dimer are conserved, the NBD-NBD interfaces show lower conservation, as documented by inspection of crystal structures and our homology-based Hsp70 dimer model (supplemental Fig. S4). Therefore, NBD-SBDα contacts might serve as an elemental dimer interface that is accompanied by homolog-specific NBD-NBD interactions allowing for functional differentiation of Hsp70 dimeric species. Mechanistically, stable dimerization of ATP-bound Hsp70 may prevent Hsp70 monomers from premature association with Hsp110 and Bag NEFs as documented by evidence that Hsp110/BAG interfaces with Hsp70 NBD largely overlap with NBD-NBD contacts in the Hsp70 ATP-dependent dimer (47, 62, 63). Accordingly, although Bag1M protein increased the dissociation rate of ADP analog from Hsc70, its role in the dissociation of ATP analog was negligible (64).
Contrary to previous results that DnaK dimerization mutants N537A and D540A did not perturb substrate binding equilibria (25), N540, E543A and N540A-E543A substitutions in human Hsp70 severely compromised its substrate binding thermodynamics (Fig. 6A) because of a large destabilization of the Hsp70 SBDα region (Fig. 5 and supplemental Fig. S3). The faster kinetics of substrate binding for the dimerization mutants (Fig. 6B) resembles the behavior of lidless DnaK and the D540A-E548A DnaK mutant (8, 16). Some DnaK/Hsp70 C-terminal truncation mutants exhibit intramolecular binding of the destabilized SBDα hydrophobic motifs into the substrate binding site of SBDβ (65, 66). This self-binding increases the intrinsic ATPase rate of these proteins (67–69). We do not expect the dimerization mutants to follow this mechanism because their basal ATPase activity is comparable to WT protein (Fig. 6D). Therefore, the elution profile of the predominantly monomeric N540A-E543A mutant (Fig. 3B) showing a dominant peak at MW 95 kDa in the presence of ATP is likely to mirror complex processes involving both impaired ATP-bound monomer/dimer equilibrium and destabilization of the SBDα subdomain (Fig. 5), rather than increased ATP hydrolysis by this mutant.
Our data showed that the degree of dimerization defects in the analyzed mutants was mirrored by their decreased interaction with Hsp40 and refolding activity (Fig. 6CDEF). Dimerization deficient E543A and N540A-E543A mutants did not refold denatured luciferase even at a saturating concentration of Hsp40 (2 μm, Fig. 6C). Because N540A-E543A binds substrates very weakly (Fig. 6A and 6B), its inactivity in the refolding assay cannot be clearly distinguished from its dimerization and Hsp40 binding defects. However, although N540A and E543A have comparable defects in substrate binding activity (Fig. 6A), N540A mediates a significantly higher level of luciferase refolding (Fig. 6C). In addition, N540A mutation allows a significant level of Hsp70 dimerization and Hsp40 interaction (Fig. 3B, 3C, 3D, Fig. 6F). Therefore, we speculate that Hsp40-mediated substrate transfer to ATP-bound Hsp70 requires chaperone transient dimerization. Evidence exists suggesting that substrate binding to SBDβ needs to be followed by SBDα closing over SBDβ for effective Hsp40/substrate-stimulated ATPase activity of Hsp70 (11, 26). Thus, transient stabilization of the SBDα subdomain of an Hsp70 protomer by its docking onto the NBD domain of the partner Hsp70 molecule might provide a time window for productive Hsp40-mediated substrate positioning into SBDβ before the SBDα subdomain closes over SBDβ. This hypothesis is consistent with the paradoxical observation that the DnaJ-DnaK interaction relies on both ATP-dependent DnaK dimerization (25) and dimer dissociation allowing free movement of the SBDα subdomain (26). Additionally, our SEC analysis of Hsp70 ATP-dependent dimers coincubated with Hsp40 (Fig. 2C) revealed complete dimer dissociation, however, the resulting monomers separated as both ATP-bound and ADP/Apo forms (apparent MW 80 and 95 kDa, respectively) indicating that Hsp40 dimer interaction with Hsp70 ATP-bound dimer does not activate the ATPase activity of both Hsp70 protomers. This observation suggests that Hsp702:Hsp402 interaction is asymmetrical.
Hsp70 isolated from cells is commonly observed in larger protein complexes (70–72). Our ex vivo experiments indicated that ATP-dependent Hsp70 dimerization is vital for Hsp70 participation in these high molecular weight complexes (Fig. 8A). Furthermore, antiparallel dimeric Hsp70 species were detected in in vitro reconstituted complexes involving Hsp90, Hop, Hsp40, and GR (Glucocorticoid receptor) (27), supporting the functional role of Hsp70 dimerization in multichaperone assemblies. The level of inducible Hsp70 is elevated under proteotoxic conditions and chronically increased in some pathologies, including cancer (73–78). Given that intracellular concentrations of Hsp70 can reach up to tens of μm under stress conditions (79, 80), the physiological importance of ATP-dependent Hsp70 dimers for overcoming proteotoxic stress becomes a likely hypothesis.
Considering the recently discovered regulatory role of ATP-induced dimerization for Hsp70 chaperoning activity (Hsp40 binding) (25, 26) and multichaperone complex formation (27), differential effects of TPR cochaperone binding to Hsp70 ATP-bound dimers (Fig. 7) are likely to be important for overall activity. Association of Hsp70 ATP-bound dimers with Chip may facilitate spatial coordination of Hsp40/Hsp70-mediated substrate processing and ubiquitination (50, 81). Conversely, the inhibitory role of Tomm34 on Hsp70-mediated refolding activity (29) can be attributed to its ability to dissociate Hsp70 ATP-bound dimers (Fig. 7B and 7C) necessary for substrate capture during effective Hsp70-Hsp40 interaction (18, 25, 26, 82, 83). We have previously suggested that the additional Tomm34-Hsp70 binding site is located between residues 533–543 in the SBDα subdomain (29). This region contains both residues (N540, E543) required for the formation of Hsp70 antiparallel dimers. It is yet to be determined whether Tomm34 occupies the 533–543 region directly or modulates its availability for Hsp70 homodimerization through allosteric changes in the SBDα subdomain. Nevertheless, ATP-dependent Hsp70 dimer dissociation by Tomm34 provides structural support for our previously suggested model of Tomm34-mediated pre-protein shuttling between Hsp70 and Hsp90 enabling its cytosolic transport in semifolded but aggregation-proof form (29). In addition, our results support the notion that TPR domain cochaperone binding to EEVD motif of Hsp70 does not follow a uniform scaffolding mechanism and suggest that the interaction is cochaperone specific (50).
The thermodynamics of Hsp70-Tomm34 interactions determined by our previous ITC analysis (29) is likely to reflect multiple equilibria transitions (e.g. Hsp70 ATP-bound monomer/dimer equilibrium), complicating data interpretation. Hsp70:Tomm34 complex stoichiometry was determined to be 2:1, whereas the current study shows that Tomm34 associates with ATP-bound Hsp70 monomer (Fig. 7B, 7C and 7D). A plausible explanation for this discrepancy would be that Hsp70 dimer dissociation by Tomm34 binding to one of the protomers renders the other protomer incapable of further association with Tomm34.
In conclusion, ATP-dependent antiparallel Hsp70 dimerization is an evolutionary conserved mechanism for Hsp70 activity regulation that has been adapted by different Hsp70 homologs with various propensities. In contrast to bacterial DnaK and human BiP, human stress-induced Hsp70 is largely dimeric in the presence of ATP in vitro and ex vivo, which might reflect the specific role of stress-induced Hsp70 in the maintenance of cellular proteostasis under proteotoxic conditions. Moreover, Hsp70 ATP-bound dimers are used either as a scaffold for TPR domain cochaperone binding (Chip) or, conversely, cochaperone binding (Tomm34) leads to rapid disassembly, which may explain the inhibitory effect of Tomm34 on the Hsp70 chaperone system (29).
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/archive/) partner repository with the dataset identifier PXD010069. Small angle scattering datasets, atomic model and fits have been deposited to the Small Angle Scattering Biological Data Bank (www.sasbdb.org) as entry SASDDN6.
Supplementary Material
Acknowledgments
We thank Dr. P. J. Coates for critical reading of the manuscript.
Footnotes
* This work was mainly supported by Czech Science Foundation (16-20860S), additional support was provided by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR, LO1413, LQ1604 and LO1509), by the Ministry of Health of the Czech Republic - conceptual development of research organization (MMCI, 00209805) and EU CZ.1.05/1.1.00/02.0109 and OPPK CZ.2.16/3.1.00/24023. CIISB research infrastructure project LM2015043 funded by MEYS CR is gratefully acknowledged for the financial support of the measurements at the CF X-ray Diffraction and Bio-SAXS, CEITEC and at the Center of Molecular Structure, BioCeV. Part of the work was carried out with the support of Core Facility Biomolecular Interactions and Crystallization of CEITEC - Central European Institute of Technology under CEITEC - open access project LM2011020 funded by MEYS CR under the activity “Projects of major infrastructures for research, development and innovations”.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- ADP
- adenosine diphosphate
- ATP
- adenosine triphosphate
- Bag-1
- Bcl2-associated athanogene 1
- Chip
- carboxyl terminus of Hsp70-interacting protein
- ESI-MS
- electrospray ionization mass spectrometry
- GST
- glutathione S-transferase
- HDX
- hydrogen/deuterium exchange
- Hsc
- heat shock cognate
- Hsp
- heat shock protein
- NBD
- nucleotide-binding domain
- NEF
- nucleotide exchange factor
- PDB
- protein data bank
- SAXS
- small-angle X-ray scattering
- SBD
- substrate-binding domain
- SBP
- streptavidin-binding peptide
- SEC
- size exclusion chromatography
- SPR
- surface plasmon resonance
- TEV
- tobacco etch virus
- Tomm34
- translocase of outer mitochondrial membrane 34
- TPR
- tetratricopeptide repeat
- WT
- wild type.
REFERENCES
- 1. Calamini B., and Morimoto R. I. (2012) Protein homeostasis as a therapeutic target for diseases of protein conformation. Curr. Topics Med. Chem. 12, 2623–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young J. C., Agashe V. R., Siegers K., and Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 3. Young J. C. (2010) Mechanisms of the Hsp70 chaperone system. Biochim. Biol. Cell. 88, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boorstein W. R., Ziegelhoffer T., and Craig E. A. (1994) Molecular evolution of the HSP70 multigene family. J. Mol. Evolution 38, 1–17 [DOI] [PubMed] [Google Scholar]
- 5. Radons J. (2016) The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones 21, 379–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang J., Zong Y., Su J., Li H., Zhu H., Columbus L., Zhou L., and Liu Q. (2017) Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat. Communications 8, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., and Zuiderweg E. R. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Structural Biol. 7, 298–303 [DOI] [PubMed] [Google Scholar]
- 8. Fernandez-Saiz V., Moro F., Arizmendi J. M., Acebron S. P., and Muga A. (2006) Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J. Biol. Chem. 281, 7479–7488 [DOI] [PubMed] [Google Scholar]
- 9. Zhang P., Leu J. I., Murphy M. E., George D. L., and Marmorstein R. (2014) Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS ONE 9, e103518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., and Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 [DOI] [PubMed] [Google Scholar]
- 12. Yang J., Nune M., Zong Y., Zhou L., and Liu Q. (2015) Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23, 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Q., and Hendrickson W. A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhuravleva A., and Gierasch L. M. (2015) Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. U.S.A. 112, E2865–E2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buczynski G., Slepenkov S. V., Sehorn M. G., and Witt S. N. (2001) Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J. Biol. Chem. 276, 27231–27236 [DOI] [PubMed] [Google Scholar]
- 17. Mayer M. P., Schroder H., Rudiger S., Paal K., Laufen T., and Bukau B. (2000) Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nature structural biology 7, 586–593 [DOI] [PubMed] [Google Scholar]
- 18. Kityk R., Kopp J., and Mayer M. P. (2018) Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol. Cell 69, 227–237 e224 [DOI] [PubMed] [Google Scholar]
- 19. Yu H. Y., Ziegelhoffer T., and Craig E. A. (2015) Functionality of Class A and Class B J-protein cochaperones with Hsp70. FEBS Lett. 589, 2825–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bracher A., and Verghese J. (2015) The nucleotide exchange factors of Hsp70 molecular chaperones. Front Mol. Biosci. 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yam A. Y., Albanese V., Lin H. T., and Frydman J. (2005) Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280, 41252–41261 [DOI] [PubMed] [Google Scholar]
- 22. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., and Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 23. Allan R. K., and Ratajczak T. (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aprile F. A., Dhulesia A., Stengel F., Roodveldt C., Benesch J. L., Tortora P., Robinson C. V., Salvatella X., Dobson C. M., and Cremades N. (2013) Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS ONE 8, e67961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarbeng E. B., Liu Q., Tian X., Yang J., Li H., Wong J. L., Zhou L., and Liu Q. (2015) A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J. Biol. Chem. 290, 8849–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Q., Li H., Yang Y., Tian X., Su J., Zhou L., and Liu Q. (2017) A disulfide-bonded DnaK dimer is maintained in an ATP-bound state. Cell Stress Chaperones 22, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morgner N., Schmidt C., Beilsten-Edmands V., Ebong I. O., Patel N. A., Clerico E. M., Kirschke E., Daturpalli S., Jackson S. E., Agard D., and Robinson C. V. (2015) Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Reports 11, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Preissler S., Chambers J. E., Crespillo-Casado A., Avezov E., Miranda E., Perez J., Hendershot L. M., Harding H. P., and Ron D. (2015) Physiological modulation of BiP activity by trans-protomer engagement of the interdomain linker. Elife 4, e08961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Durech M., Trcka F., Man P., Blackburn E. A., Hernychova L., Dvorakova P., Coufalova D., Kavan D., Vojtesek B., and Muller P. (2016) Novel entropically driven conformation-specific interactions with Tomm34 protein modulate Hsp70 protein folding and ATPase activities. Mol. Cell. Proteomics 15, 1710–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang L., Bertelsen E. B., Wisen S., Larsen E. M., Zuiderweg E. R., and Gestwicki J. E. (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 372, 167–176 [DOI] [PubMed] [Google Scholar]
- 31. Franke D., Petoukhov M. V., Konarev P. V., Panjkovich A., Tuukkanen A., Mertens H. D. T., Kikhney A. G., Hajizadeh N. R., Franklin J. M., Jeffries C. M., and Svergun D. I. (2017) ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallography 50, 1212–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Computational Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 33. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 34. Kadek A., Kavan D., Felice A. K., Ludwig R., Halada P., and Man P. (2015) Structural insight into the calcium ion modulated interdomain electron transfer in cellobiose dehydrogenase. FEBS Lett. 589, 1194–1199 [DOI] [PubMed] [Google Scholar]
- 35. Jensen P. F., Larraillet V., Schlothauer T., Kettenberger H., Hilger M., and Rand K. D. (2015) Investigating the interaction between the neonatal Fc receptor and monoclonal antibody variants by hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteomics 14, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., and Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schlecht R., Erbse A. H., Bukau B., and Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
- 38. Wieteska L., Shahidi S., and Zhuravleva A. (2017) Allosteric fine-tuning of the conformational equilibrium poises the chaperone BiP for post-translational regulation. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ebong I. O., Morgner N., Zhou M., Saraiva M. A., Daturpalli S., Jackson S. E., and Robinson C. V. (2011) Heterogeneity and dynamics in the assembly of the heat shock protein 90 chaperone complexes. Proc. Natl. Acad. Sci. U.S.A. 108, 17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stengel F., Baldwin A. J., Painter A. J., Jaya N., Basha E., Kay L. E., Vierling E., Robinson C. V., and Benesch J. L. (2010) Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. U.S.A. 107, 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Z., Hartl F. U., and Bracher A. (2013) Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 20, 929–935 [DOI] [PubMed] [Google Scholar]
- 42. Woodcock J. M., Goodwin K. L., Sandow J. J., Coolen C., Perugini M. A., Webb A. I., Pitson S. M., Lopez A. F., and Carver J. A. (2018) Role of salt bridges in the dimer interface of 14–3-3zeta in dimer dynamics, N-terminal alpha-helical order, and molecular chaperone activity. J. Biol. Chem. 293, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haladova K., Mrazek H., Jecmen T., Halada P., Man P., Novak P., Chmelik J., Obsil T., and Sulc M. (2012) The combination of hydrogen/deuterium exchange or chemical cross-linking techniques with mass spectrometry: mapping of human 14–3-3zeta homodimer interface. J. Structural Biol. 179, 10–17 [DOI] [PubMed] [Google Scholar]
- 44. Heintz U., and Schlichting I. (2016) Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence. Elife 5, e11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., and Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takayama S., Bimston D. N., Matsuzawa S., Freeman B. C., Aime-Sempe C., Xie Z., Morimoto R. I., and Reed J. C. (1997) BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 16, 4887–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., and Moarefi I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 48. Mayer M. P., Laufen T., Paal K., McCarty J. S., and Bukau B. (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289, 1131–1144 [DOI] [PubMed] [Google Scholar]
- 49. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., and Pearl L. H. (2005) Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 50. Zhang H., Amick J., Chakravarti R., Santarriaga S., Schlanger S., McGlone C., Dare M., Nix J. C., Scaglione K. M., Stuehr D. J., Misra S., and Page R. C. (2015) A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure 23, 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nikolay R., Wiederkehr T., Rist W., Kramer G., Mayer M. P., and Bukau B. (2004) Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 279, 2673–2678 [DOI] [PubMed] [Google Scholar]
- 52. Malinverni D., Marsili S., Barducci A., and De Los Rios P. (2015) Large-scale conformational transitions and dimerization are encoded in the amino-acid sequences of Hsp70 chaperones. PLoS Computational Biol. 11, e1004262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trcka F., Durech M., Man P., Hernychova L., Muller P., and Vojtesek B. (2014) The assembly and intermolecular properties of the Hsp70-Tomm34-Hsp90 molecular chaperone complex. J. Biol. Chem. 289, 9887–9901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao B., Eisenberg E., and Greene L. (1996) Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J. Biol. Chem. 271, 16792–16797 [DOI] [PubMed] [Google Scholar]
- 55. Benaroudj N., Triniolles F., and Ladjimi M. M. (1996) Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSC70. J. Biol. Chem. 271, 18471–18476 [DOI] [PubMed] [Google Scholar]
- 56. Buxbaum E., and Woodman P. G. (1996) Binding of ATP and ATP analogues to the uncoating ATPase Hsc70 (70 kDa heat-shock cognate protein). Biochem. J. 318 (Pt 3), 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilbanks S. M., Chen L., Tsuruta H., Hodgson K. O., and McKay D. B. (1995) Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry 34, 12095–12106 [DOI] [PubMed] [Google Scholar]
- 58. Lee S. C., and Whitaker J. R. (2004) Are molecular weights of proteins determined by superose 12 column chromatography correct? Journal of agricultural and food chemistry 52, 4948–4952 [DOI] [PubMed] [Google Scholar]
- 59. King C., Eisenberg E., and Greene L. (1995) Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J. Biol. Chem. 270, 22535–22540 [DOI] [PubMed] [Google Scholar]
- 60. Ha J. H., Hellman U., Johnson E. R., Li L., McKay D. B., Sousa M. C., Takeda S., Wernstedt C., and Wilbanks S. M. (1997) Destabilization of peptide binding and interdomain communication by an E543K mutation in the bovine 70-kDa heat shock cognate protein, a molecular chaperone. J. Biol. Chem. 272, 27796–27803 [DOI] [PubMed] [Google Scholar]
- 61. Lima D. B., Melchior J. T., Morris J., Barbosa V. C., Chamot-Rooke J., Fioramonte M., Souza T., Fischer J. S. G., Gozzo F. C., Carvalho P. C., and Davidson W. S. (2018) Characterization of homodimer interfaces with cross-linking mass spectrometry and isotopically labeled proteins. Nat. Protocols 13, 431–458 [DOI] [PubMed] [Google Scholar]
- 62. Arakawa A., Handa N., Ohsawa N., Shida M., Kigawa T., Hayashi F., Shirouzu M., and Yokoyama S. (2010) The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure 18, 309–319 [DOI] [PubMed] [Google Scholar]
- 63. Williamson D. S., Borgognoni J., Clay A., Daniels Z., Dokurno P., Drysdale M. J., Foloppe N., Francis G. L., Graham C. J., Howes R., Macias A. T., Murray J. B., Parsons R., Shaw T., Surgenor A. E., Terry L., Wang Y., Wood M., and Massey A. J. (2009) Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 52, 1510–1513 [DOI] [PubMed] [Google Scholar]
- 64. Brehmer D., Rudiger S., Gassler C. S., Klostermeier D., Packschies L., Reinstein J., Mayer M. P., and Bukau B. (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Structural Biol. 8, 427–432 [DOI] [PubMed] [Google Scholar]
- 65. Umehara K., Hoshikawa M., Tochio N., and Tate S. I. (2018) Substrate binding switches the conformation at the lynchpin site in the substrate-binding domain of human Hsp70 to enable allosteric interdomain communication. Molecules 23, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang H., Kurochkin A. V., Pang Y., Hu W., Flynn G. C., and Zuiderweg E. R. (1998) NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry 37, 7929–7940 [DOI] [PubMed] [Google Scholar]
- 67. Swain J. F., Schulz E. G., and Gierasch L. M. (2006) Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J. Biol. Chem. 281, 1605–1611 [DOI] [PubMed] [Google Scholar]
- 68. Sun L., Edelmann F. T., Kaiser C. J., Papsdorf K., Gaiser A. M., and Richter K. (2012) The lid domain of Caenorhabditis elegans Hsc70 influences ATP turnover, cofactor binding and protein folding activity. PLoS ONE 7, e33980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Freeman B. C., Myers M. P., Schumacher R., and Morimoto R. I. (1995) Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14, 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Faou P., and Hoogenraad N. J. (2012) Tom34: a cytosolic cochaperone of the Hsp90/Hsp70 protein complex involved in mitochondrial protein import. Biochim. Biophys. Acta 1823, 348–357 [DOI] [PubMed] [Google Scholar]
- 71. Chavez J. D., Schweppe D. K., Eng J. K., and Bruce J. E. (2016) In Vivo Conformational Dynamics of Hsp90 and Its Interactors. Cell Chem. Biol. 23, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klucken J., Shin Y., Masliah E., Hyman B. T., and McLean P. J. (2004) Hsp70 Reduces alpha-Synuclein Aggregation and Toxicity. J. Biol. Chem. 279, 25497–25502 [DOI] [PubMed] [Google Scholar]
- 73. Sherman M. Y., and Gabai V. L. (2015) Hsp70 in cancer: back to the future. Oncogene 34, 4153–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yaglom J. A., Wang Y., Li A., Li Z., Monti S., Alexandrov I., Lu X., and Sherman M. Y. (2018) Cancer cell responses to Hsp70 inhibitor JG-98: Comparison with Hsp90 inhibitors and finding synergistic drug combinations. Sci. Reports 8, 3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kundrat L., and Regan L. (2010) Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49, 7428–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kawazoe Y., Nakai A., Tanabe M., and Nagata K. (1998) Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur. J. Biochem. 255, 356–362 [DOI] [PubMed] [Google Scholar]
- 77. Woo J. A., Liu T., Zhao X., Trotter C., Yrigoin K., Cazzaro S., Narvaez E., Khan H., Witas R., Bukhari A., Makati K., Wang X., Dickey C., and Kang D. E. (2017) Enhanced tau pathology via RanBP9 and Hsp90/Hsc70 chaperone complexes. Hum. Mol. Genet. 26, 3973–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lackie R. E., Maciejewski A., Ostapchenko V. G., Marques-Lopes J., Choy W. Y., Duennwald M. L., Prado V. F., and Prado M. A. M. (2017) The Hsp70/Hsp90 Chaperone Machinery in Neurodegenerative Diseases. Frontiers Neurosci. 11, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. De Los Rios P., and Barducci A. (2014) Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. Elife 3, e02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nollen E. A., Brunsting J. F., Song J., Kampinga H. H., and Morimoto R. I. (2000) Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell Biol. 20, 1083–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang H. T., Zeng L. F., He Q. Y., Tao W. A., Zha Z. G., and Hu C. D. (2016) The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim. Biophys. Acta 1863, 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jin Y., Awad W., Petrova K., and Hendershot L. M. (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27, 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Petrova K., Oyadomari S., Hendershot L. M., and Ron D. (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/archive/) partner repository with the dataset identifier PXD010069. Small angle scattering datasets, atomic model and fits have been deposited to the Small Angle Scattering Biological Data Bank (www.sasbdb.org) as entry SASDDN6.