Wei et al. apply multi-layer proteomic profiling and systems biology approaches to define Wip1-deficient testis proteome and phosphoproteome landscapes, and they identify cell adhesion/tight junction, sperm motility, and inflammatory response pathways. These data establish the mechanism that proinflammatory cytokines may impair the blood-testis barrier dynamics by decreasing the expression of junction-associated proteins in Wip1 null testes, leading to subfertility and spermatogenesis defects.
Keywords: Phosphoproteome, iTRAQ, Molecular Biology, Knockouts, Quantification
Graphical Abstract
Highlights
Mice lacking Wip1 display spermatogenesis defects at the late-stage germ cell types.
Proteome and phosphoproteome profiling reveals impaired dynamics of testis junction.
Elevated levels of cytokines may lead to abnormal BTB structure and spermatogenesis.
Abstract
Mice lacking wild-type p53-induced phosphatase 1 (Wip1) display male reproductive defects including smaller testes, subfertility and spermatogenesis defects at the round- and elongating-spermatid stages. However, the molecular mechanisms underlying these abnormalities remain unclear. Here we examined the proteome and phosphoproteome of testes from Wip1-knockout mice using a quantitative proteomic approach. From a total of 6872 proteins and 4280 phosphorylation sites identified, 58 proteins and 159 phosphorylation sites were found to be differentially regulated compared with wild type mice. Pathway enrichment analyses revealed that these regulated proteins and phosphosites were mainly involved in adherens/tight junctions, apoptosis, inflammatory response, spermatogenesis, sperm motility, and cytoskeletal assembly and depolymerization. Wip1-knockout mice showed decreased expression of junction-associated proteins (occludin, ZO-1, and N-cadherin) and impaired integrity of the blood-testis barrier. In addition, Wip1 deficiency was associated with elevated levels of cytokines and germ cell apoptosis in the testis. These results suggest that proinflammatory cytokines may impair the blood-testis barrier dynamics by decreasing the expression of junction-associated proteins, which could lead to subfertility and spermatogenesis defects. Collectively, these findings help to explain the low reproductive function caused by Wip1 deletion and provide novel insights into our understanding of causes of male infertility.
Spermatogenesis comprises a complex process of mitosis, meiosis, and spermiogenesis in the mammalian testis (1). Maintaining the integrity of different types of cell-cell junctions is vital for the movement of germ cells across the blood-testis barrier (BTB)1 (2). The BTB is formed by tight junctions between adjacent Sertoli cells, dividing the seminiferous epithelium into distinct basal and adluminal compartments (3). The BTB is maintained collectively by different types of junctions, including claudins and occludin-based tight junctions, cadherin-based adherens junctions, and connexin-based gap junctions, all of which are in close structural and functional contact with each other (4, 5). The major functions of the BTB are to provide the appropriate microenvironment (structural support and nourishment) to develop germ cells and confer apical-basal polarity, sequester autoantigenic post-meiotic germ cells from the systemic circulation, prevent autoimmune recognition, and exclude potential cytotoxic substances (6, 7). In addition, a previous study reported that cytokine-mediated down-regulation of junction proteins resulted in impaired BTB integrity (8). Spermatogenesis may also be disrupted in the absence of the immunological and “fence” functions of the BTB, resulting in male subfertility or infertility (9, 10).
Wild-type (WT) p53-induced phosphatase 1 (Wip1) is a Mg2+/Mn2+ dependent serine/threonine phosphatase of the PP2C family that has been implicated in regulating several stress-induced and DNA damage-induced signaling networks. The gene encoding human Wip1, PPM1D, is highly expressed in testis (11). Wip1 phosphatase was originally studied as a potent regulator of tumorigenesis (12, 13), and emerging evidence has indicated roles for Wip1 in multiple physiological processes and pathophysiological conditions, such as cellular homeostasis (14), neutrophil migration and inflammation (15), B-cell development (16), bone marrow mesenchymal stem cells and macrophage migration (17, 18), autophagy, obesity, and atherosclerosis (19). Moreover, Wip1-knockout (Wip1-KO) mice presented with defects of the male reproductive organs and impaired spermatogenesis, eventually leading to reduced fertility (11, 20). Filipponi et al. confirmed that the activation of ataxia telangiectasia-mutated (ATM) kinase caused by damaged heterochromatin partially contributed to the testicular defects (20).
In the present study, we demonstrated that Wip1-deficient mice displayed subfertility and spermatogenesis defects at the round- and elongating-spermatid stages. We further studied the molecular mechanisms responsible for Wip1-related hypospermatogenesis using a proteomics approach to investigate differentially expressed testis proteins and proteins harboring differentially phosphorylated sites between wild-type and Wip1-deficient mice. Integrative proteomics and phosphoproteomics analyses suggested that impaired integrity of the BTB may be partially responsible for the defects in spermatogenesis and fertility in Wip1-deficient mice. These findings further our understanding of the mechanisms underlying the role of Wip1 in spermatogenesis and fertility, and offer a new perspective for future research into male fertility.
EXPERIMENTAL PROCEDURES
Mice and Breeding Assay
Wip1−/− (Wip1-KO) and Wip1+/+ (Wip1-WT) mice were kindly provided by the Key Laboratory of Human Disease Comparative Medicine of the Ministry of Public Health (Peking, Beijing, China) (11). Wip1+/− mice were generated by mating. All the CD1 female mice used in our study were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). Polymerase chain reaction (PCR) genotyping primers for Wip1+/+ and Wip1−/− were as follows: Wip1+/+ forward: 5′-GACAGTCCTGTGCCAAAATGCT-3′ and reverse 5′-GGTGACTTGATTGGTGGTGTAGA-3′; Wip1−/− forward: 5′-GCAGGGCTGTTTGTGGTGCT-3′ and reverse 5′-GCATGCTCCAGACTGCCTT-3′. All mice were bred and maintained under pathogen-free conditions with a 12-h light/dark cycle. All animal procedures were approved by the Animal Care and Use Committee of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences.
For breeding, 10-week-old Wip1+/+, Wip1+/−, and Wip1−/− male mice were housed singly with wild-type 8-week-old CD1 females. Copulatory plugs were monitored daily, and plugged females were moved to separate cages for monitoring of pregnancy. Females that were not pregnant within 2 weeks were replaced. Mating was carried out over 3 months, and pregnancy results and viable pups were recorded. If plugged female mice had not produced any pups by day 22 postcoitus, they were confirmed as not pregnant. Male mice were permitted to rest for 2 days, after which another female was put in this cage for mating. Each male mouse was used for four breeding cycles, as described above (21).
Histology, Immunostaining, and Terminal Deoxynucleotidyl Transferase Nick-end Labeling (TUNEL)
For histological examination, testes and epididymides were dissected out from mice, washed twice in phosphate-buffered saline (PBS), and fixed with Bouin's solution (Sigma-Aldrich, Darmstadt, Germany) overnight at 4 °C, dehydrated in graded concentrations of alcohol, and embedded in paraffin wax. Paraffin sections (5 μm) were cut and then stained with hematoxylin-eosin for morphological evaluation. Epididymal tissues were also examined by transmission electron microscopy, according to standard techniques. For immunostaining, deparaffinized and rehydrated slides were incubated with boiling 0.1 m sodium citrate (pH 6.0) for 10 min for antigen retrieval, and stained with respective antibodies: rabbit anti-mouse VASA homologue (MVH) (Abcam, Cambridge, UK), goat anti-protamine 2 (Prm2) (Santa Cruz Biotechnology, Danvers, MA), rabbit anti-Wilms tumor (WT1) (Abcam), rabbit anti-β-catenin (Abcam), rabbit anti-occludin (Abcam), rabbit anti-N-cadherin (Abcam), and rabbit anti-zona occludens-1 (ZO-1) (Cell Signaling Technology). Apoptotic cells were evaluated by TUNEL assay using dewaxed and rehydrated paraffin sections (5 μm). TUNEL-positive cells were detected using an In Situ Cell Death Detection Kit, POD (Roche, Basel, Switzerland), following the manufacturer's protocol. Apoptotic cells were stained brown and normal cells were stained blue-violet. Apoptotic cells were analyzed by random counting of TUNEL-positive cells in five visual fields/slide (each group, n = 4 repeats; each repeat, n = 3 slides) under a fluorescence microscope at ×400 magnification. All the images were taken under a fluorescence microscope (BX51, Olympus, Tokyo, Japan).
RNA Extraction and Real-time PCR
Testes were harvested in TRIzol reagent (1 ml reagent per 50–100 mg testis; Invitrogen ™, Rockford, IL). RNA was extracted using an RNA Extraction Kit (Tiangen, Beijing, China) according to the manufacturer's instructions, and its concentration and purity were determined using a Nano-100 micro-spectrophotometer (Allsheng, Hangzhou, China). First-strand cDNA was synthesized using a Reverse Transcription Kit (Thermo Fisher Scientific, Rockford, IL). Real-time PCR was performed with an ABI Prism 7500 device (Applied Biosystems, Foster City, CA) using SYBR Select Master Mix (Applied Biosystems) and the primers given in supplemental Table S1. Eight samples from different mice in each group were used. Quantifications were made in triplicate for each sample from individual testes. mRNA expression was analyzed using the comparative Ct method (ΔΔCT) with β-actin mRNA as the internal control.
BTB Integrity Assay
The integrity of the BTB was determined as described previously (22). For each mouse, 100 μl of 10 mg/ml fluorescein isothiocyanate (FITC)-conjugated dextran (Sigma-Aldrich) was injected into the caudal vein. Eight male mice from each group were euthanized about 30 min later, and their testes were removed immediately and embedded in OCT (Sakura Finetek, Torrance, CA). Frozen sections (about 7 μm) were cut using a cryostat microtome and examined under a fluorescence microscope (Olympus).
Tissue Lysis, Protein Extraction, Digestion, Labeling, and Peptide Fractionation
Frozen testis samples were ground and lysed in 10 ml buffer (2 m thiourea, 8 m urea, 0.1% CHAPS, with one tablet of protease inhibitor (cOmplete Mini EDTA-free mixture, Roche Applied Science, Indianapolis, IN) and one tablet of phosphatase inhibitor mixture (PhosSTOP, Roche Applied Science)), and subsequently homogenized by TissueLyser LT (Qiagen, Duesseldorf, Germany) (settings: 3 × 60s; 50 oscillation/s) with a steel ball. The homogenate was cleared using centrifugation at 12,000 rpm for 10 min at 4 °C. Supernatants were collected, and protein concentration was quantified by the Bradford assay with BSA protein as standard. In detail, the proteins in lysis buffer were reduced in 50 mm DTT for 1 h at 56 °C and subsequently alkylated in the dark with 5 mm iodoacetamide at room temperature for 30 min. For subsequent digestion with trypsin, the solution was diluted with 25 mm ammonium bicarbonate (PH 8.5) and trypsin was added (1:50, w/w) and digestion was performed for 6h and the second trypsin digestion was performed at 37 °C overnight. After trypsin digestion, 5% of peptides were used for whole-proteome analysis, and the remaining 95% were used for phosphopeptide enrichment and further phosphoproteome analysis. For proteome analysis, peptides were labeled with each component of the 8-plex isobaric tags for relative and absolute quantification (iTRAQ) reagent kit (AB SCIEX, Foster city, CA), according to the manufacturer's instructions. In detail, two iTRAQ labels, 119 and 121 (pooled extracts from three KO and three WT testis tissues), were chosen as reference mixtures. Labels 113, 114, 115, 116, 117, and 118 were used for the relative abundance of three KO and three WT testis tissues included in two technical replicate experiments. Label 119/121 was used for assessing the systematic error. Labels 119 and 121 were technique replicates of the pooled extracts from all testis tissues. Therefore, their theoretical log2 transformed ratio (log2 (label 119/121)) should be 0. Experimentally, the average log2 transformed ratio is 0, and the standard deviation (S.D.) is 0.126. Eventually, mean+2SD (1.283) and mean-2SD (0.779) was used to decide the fold change threshold. Additionally, to identify more proteins, reverse-phase chromatography was applied using a RIGOL l-3000 system (Beijing RIGOL Technology Co., Ltd., Beijing, China) to separate the mixed peptides. Peptide mixtures were reconstituted in 100 μl of mobile phase A (2% acetonitrile in ddH2O, pH 10) and centrifuged at 14,000 rpm for 20 min prior to loading onto the column (Durashell-C18, 4.6 × 250 mm, 5 μm, 100 Å, Agela). After loading, the peptides were eluted at a flow rate of 700 μl/min by stepwise injection of mobile phase B (98% acetonitrile in ddH2O, pH 10). Ten fractions were collected by measuring the absorbance at 214 nm. Each fraction was desalted with a C18 column (Waters, Milford, MA) and vacuum-dried prior to liquid chromatography tandem-mass spectrometry (LC-MS/MS).
Phosphopeptide Enrichment
Phosphopeptide enrichment was carried out using a Phosphopeptide Enrichment Kit (Pierce, Thermo Fisher Scientific). Briefly, the dried peptides from each sample were dissolved in 150 μl of binding buffer. TiO2 beads were washed twice with washing buffer, and a total of 1 mg of tryptic peptides solution was incubated with an appropriate amount (tryptic peptide: TiO2 = 1:1, w/w) of TiO2 beads by end-over-end rotation at room temperature for 30 min. The phosphopeptide-bound beads were collected by brief centrifugation, washed twice with 500 μl washing buffer, and transferred to a C18 StageTip (Thermo Fisher Scientific) placed on the top of a 1.5-ml centrifuge tube. The StageTip was centrifuged to remove the wash buffer completely and phosphopeptides were collected from the resin with elution buffer. The eluents were dried and stored at −80 °C until further LC-MS/MS analysis.
LC-MS/MS Analysis Based On Q-Exactive Mass Spectrometry
The peptide mixtures were analyzed using a Q-Exactive mass spectrometer fitted with an EASY-nLC 1000 System (Thermo Fisher Scientific). The dried peptides and phosphopeptides were reconstituted in 20 μl of 0.1% formic acid, loaded on an EASY-Spray column (12 cm × 75 μm, 3 μm C18 resin (Dr. Maisch GmbH, Ammerbuch, Germany)) interfaced with a Q-Exactive MS (Thermo Fisher Scientific). Peptide and phosphopeptide mixtures were eluted using 100% ddH2O, 0.1% formic acid (buffer A), and 100% acetonitrile, 0.1% formic acid (buffer B) under a 113 min gradient with a flow rate of 300 nL/min (8–30% buffer B for 77min, 30–50% buffer B for 10min, 50–95% buffer B for 15min, and finally 5% buffer B for 11min). The eluted peptides and phosphopeptides were analyzed on a Q-Exactive mass spectrometer (Thermo Fisher Scientific) with MS survey scan (m/z range 400–1800) (70,000 resolution, 3 × 106 AGC target, and 20 ms maximal ion time) and MS/MS spectra (17,500 resolution, high-energy collision dissociation, 2m/z isolation window, 27 normalized collision energy, 2 × 105 AGC, 100 ms maximal ion time, 30 s dynamic exclusion, and top number 20).
Mass Spectrometry Data Analysis
The RAW mass spectrometry files were processed using Maxquant software (version 1.5.2), based on Andromeda as a search engine against the UniProt mouse protein database (UniProt 2016_05_28; 16,793 sequences) combined with the target-decoy and contaminants database searching strategy. For proteome analysis, the parameters for database searching were set as follows: fully tryptic enzyme specificity; two maximum missed cleavages; iTRAQ 8-plex labels at the N-terminal and lysine residues and carbamidomethylation of cysteine (Cys) residues were specified as a fixed modification; ion mass tolerances was set by Andromeda (20 ppm. first search, 4.5 ppm. main search); oxidation of methionine and protein N-terminal acetylation were allowed as variable modifications (23). For the phosphoproteome, spectra were searched with an initial maximal mass deviation of 6 ppm and 20 ppm for precursor and fragment ions, static mass shift for Cys, dynamic mass shift for oxidation (Met), N-acetylation and phospho-S (serine) T (threonine) Y (tyrosine), two maximal missed cleavages, three maximal modification sites, and the assignment of a, b, and y ions. Trypsin with full enzyme specificity and only peptides with a minimum length of 6 amino acids were selected. The minimum Maxquant score for phosphorylation sites was 40. The false discovery rate was evaluated by searching against the databases with the reversed amino acid sequences. We required a maximum FDR of 1% for identification of protein, peptide, and phosphorylation sites.
For proteome analysis, at least one unique peptide was considered for the identification and quantification of the proteins (24–26). The quantitative analysis was carried out on the log2-values of the measured reporter intensities and the data were normalized based on the median using perseus software (version 1.5.5). A two-sample t test was performed within Perseus 1.5.5 software. Proteins with p values <0.05 and fold-change ratios ≥1.283 or ≤0.779 were considered as differentially expressed proteins (DEPs). For phosphoproteome analysis, quantification was done at phosphosite level by calculating its intensity values. Further data processing was done with the Perseus software (version 1.5.5). Phosphoproteomics data were log2-transformed, and the following filtered steps were applied: only phosphosites belonging to class I phosphorylation sites (localization probability >0.75 and score difference >5). Additionally, phosphorylation sites were quantified at a maximum of one missing quantification value within the four replicates. Data were normalized to the median of each sample and the missing values were replaced with random numbers drawn from normal distribution of each sample. Changes in phosphosites between knockout group and wild type group were determined by a two-sample t test with p < 0.05 and a minimum of a 1.5-fold difference (27). Western-blotting was used to measure the phosphorylation status of β-catenin to validate the phosphoproteome regulation.
Functional Analysis
Identified DEPs and proteins harboring differentially phosphorylated sites were functionally annotated using the Gene Ontology (GO) Consortium (http://www.geneontology.org) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (http://www.kegg.jp). To understand these DEPs and proteins harboring differentially phosphorylated sites in terms of the published literature, interactions among them in relation to function and biological pathways were determined using ingenuity pathway analysis (IPA) (http://www.ingenuity.com). Interactions among all the related differential proteins and proteins harboring differentially phosphorylated sites were subsequently analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) 10.5 database (http://string-db.org).
Western Blotting
Testis lysates were run on sodium dodecyl sulfate-polyacrylamide gels and immunoblotting was performed according to standard techniques. The primary antibodies used were as follows: rabbit anti-β-catenin, rabbit anti-N-cadherin, rabbit anti-occludin, rabbit anti-Bcl-2, rabbit anti-F4–80, rabbit anti-Bax, rabbit anti-caspase-3, rabbit anti-renalase, rabbit anti-prostaglandin D synthase (Ptgds), and rabbit anti-Myd88 (all Abcam); mouse anti-Wip1, mouse anti-caspase-9, rabbit anti-interleukin (IL)-1β, rabbit anti-tumor necrosis factor (TNF)-α, rabbit anti-ZO-1, rabbit anti-β-actin, and rabbit anti-p-β-catenin (Q-R-R-T-Sp-M-G-G-T-Q, Phospho-S552 (all Cell Signaling Technology); rabbit anti-spink2 (Lifespan, Salt lake city, UT). Immunoblots were repeated at least three times for each sample from three KO and WT mice, respectively. The visualized bands were quantified by calculating net light density value with Gel Image System 1D software (v4.2), based on a Tanon-5200 Chemiluminescent Imaging System (Tanon, Shanghai, China).
Experimental Design and Statistical Rationale
Ten-week-old male WT and Wip1-KO mice on a C57BL/6 background were sacrificed, intact testis were dissected, snap-frozen with liquid nitrogen, and stored at −80 °C until use. With proteome, two sets of 8-plex iTRAQ experiments were performed as technical replicates. Each 8-plex iTRAQ experiment contains three independent biological replicates of Wip1-WT, three independent biological replicates of Wip1-KO, and two technical replicates of pooled sample as references. With phosphoproteome, four independent biological experiments (in total four Wip1-WT and Wip1-KO testis tissue samples) without technical replication were performed. The results from the replicates were combined when analyzing the data with the Perseus software (version 1.5.5).
Data were analyzed using GraphPad Prism (version 6) statistical software (GraphPad software; San Diego, CA) and the results were presented as mean ± S.E. (S.E.) of at least three separate experiments if not specified otherwise and compared with two-tailed unpaired Student's t test between the Wip1-KO and Wip1-WT groups. The significance level was indicated as ***p < 0.001, **p < 0.01, *p < 0.05, p > 0.05 not significant (ns).
RESULTS
Knockout of Wip1 Results in Reduced Fertility in Mice
Based on the normally high expression of Wip1 in testis and spermatozoa (11, 20), we monitored the fertility of Wip1-KO male mice by mating different Wip1 genotypes with WT CD1 females. The pregnancy rates (84.31% versus 97.73% or 97.50%, p < 0.001) and average litter sizes (11.21 ± 0.65 versus 12.70 ± 0.35 or 12.44 ± 0.39, p < 0.05) were significantly reduced in Wip1-KO males compared with WT or heterozygous males (Table I).
Table I. Fertility data for different Wip1 males mated with WT females.
| Genotype of mice |
No. of male mice | No. of mating females | No. of plugged females | Pregnancy rate (No. of litter/mating) | Average litter size | |
|---|---|---|---|---|---|---|
| Male | Female | |||||
| +/+ | +/+ | 11 | 44 | 44 | 97.73% (43/44) | 12.70 ± 0.35 (546/43) |
| +/− | +/+ | 10 | 40 | 40 | 97.50% (39/40) | 12.44 ± 0.39 (467/39) |
| −/− | +/+ | 15 | 60 | 51 | 84.31%*** (43/51) | 11.21 ± 0.65* (482/43) |
The presented errors are shown in S.E.
*p < 0.05, and
***p < 0.001, Wip1−/− vs. Wip1+/+ or Wip1+/−.
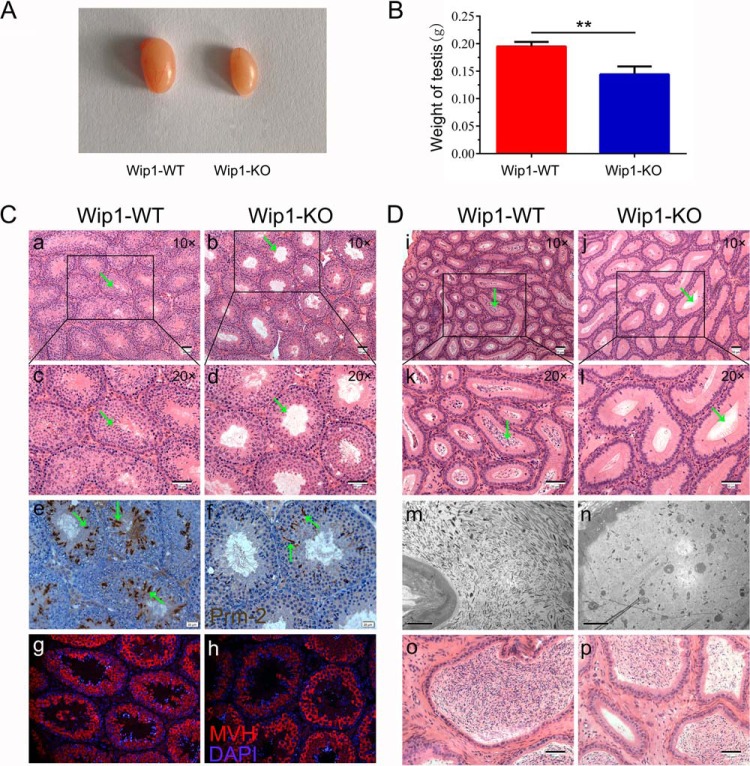
In accordance with previous observations, gross examination of testis size revealed a 26% decrease in testis weight in Wip1-KO compared with WT males (Fig. 1A, 1B). Wip1-KO testes displayed abnormal architecture in histological sections, with many tubules lacking late-stage germ cell types (Fig. 1Ca-d; green arrows). We also investigated the stages of spermatogenesis affected by Wip1 deletion by immunostaining. Consistent with the histological sections, expression of Prm2 (specifically expressed in spermatids) was significantly reduced in Wip1-KO males compared with control males (Fig. 1Ce-f; green arrows). However, there were no abnormalities in MVH protein levels in primordial germ cells derived from Wip1-KO and WT males (Fig. 1Cg-h). Moreover, the epididymal caput and cauda in Wip1-KO males often exhibited abnormal tubules with an absence of spermatozoa (Fig. 1Di-l, o-p; green arrows). Furthermore, transmission electron microscopy of epididymides (Fig. 1Dm-n) verified that the sperm count was markedly reduced in Wip1-KO compared with WT mice. Subfertility of Wip1-KO males could thus be attributed to a reduced numbers of sperm produced by the null testes and defects in the epididymis.
Fig. 1.
Wip1 deficiency disrupts spermatids development in the testis and reduces sperm count in the epididymis in mice. A, Overall testis sizes in 10-week-old Wip1-KO and WT mice. B, Weights of testes harvested from adult mice were reduced in Wip1-KO mice. Histological analyses of testes from 10-week-old Wip1-KO and WT mice at (C, a-b) low magnification (×10) and at (C, c-d) high magnification (×20). Immunohistochemical images of testis sections from 10-week-old WT and Wip1-KO mice stained with (C, e-f) anti-Prm2 antibody and (C, g-h) anti-MVH antibody. Hematoxylin-eosin (H&E)-stained sections of caput epididymides from 10-week-old WT and Wip1-KO mouse testes at (D, i-j) low magnification (×10) and at (D, k-l) high magnification (×20). (D, m-n) Transmission electron micrographs of caput epididymides from 10-week-old WT and Wip1-KO mice. (D, o-p) H&E-stained sections of cauda epididymides from 10-week-old WT and Wip1-KO mice. Data are expressed as mean ± S.E. **p < 0.01. Scale bars: 50 μm (Ca-d, Cg-h, Di-l, Do-p); 20 μm (Ce-f, Dm-n).
Integrative Whole Proteome and Phosphoproteome Profiling Identifies Cell Adhesion/Tight Junction Pathways
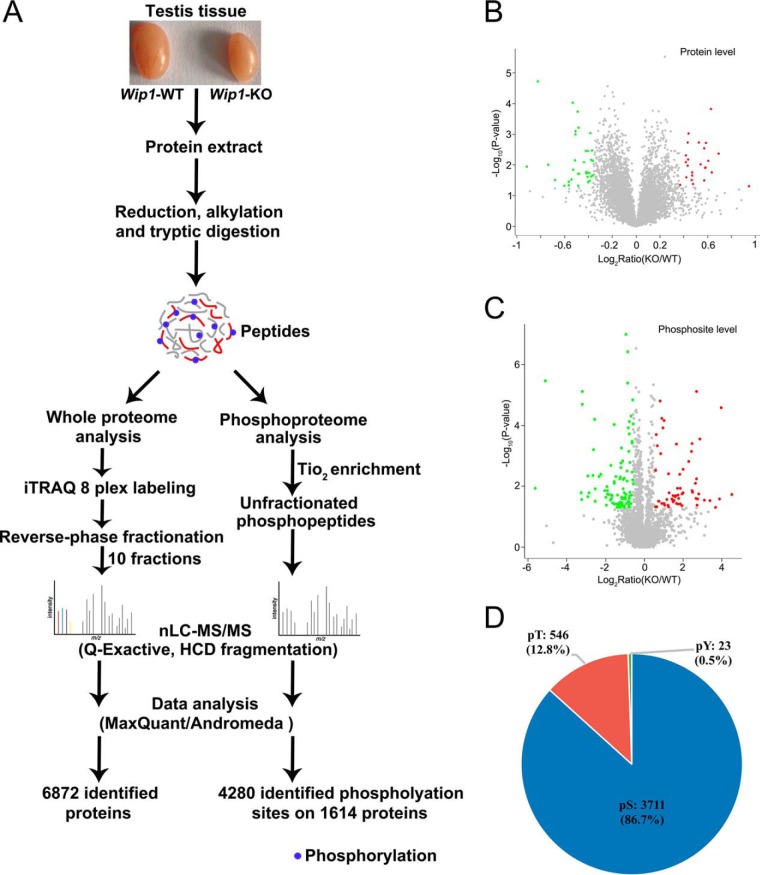
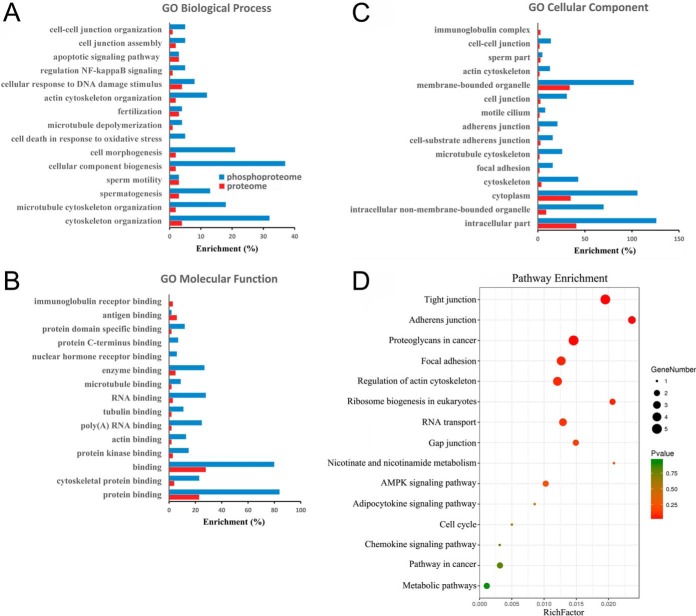
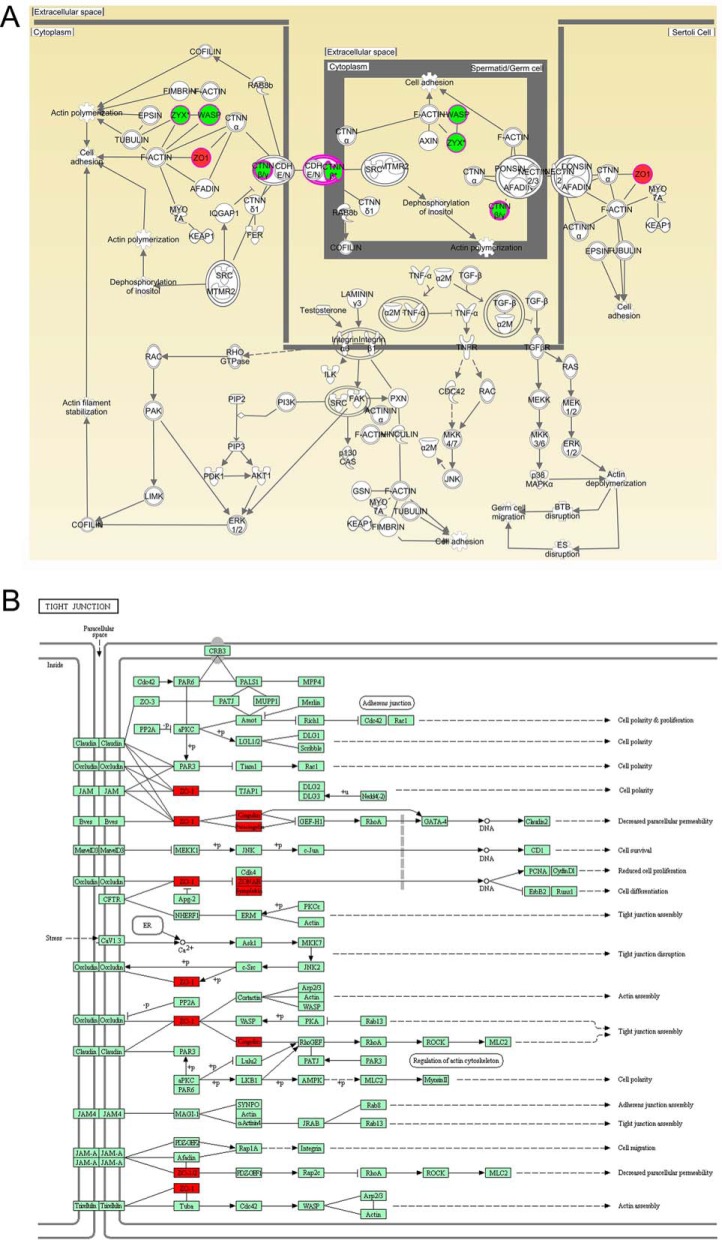
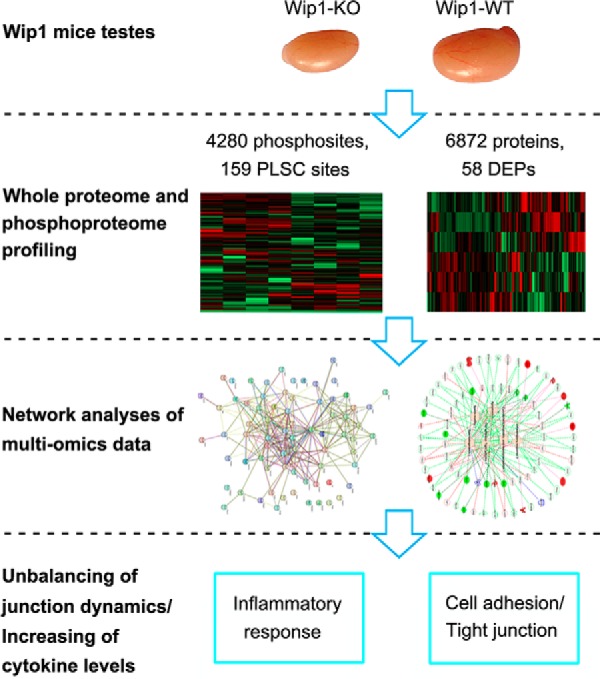
We employed a quantitative proteomics approach to analyze the global protein expression profiles and phosphorylation events between WT and KO mouse testes. After dissection, the testis tissues were processed as schematically described in Fig. 2A and in Experimental Procedures. In total, we identified 6872 proteins and 4280 phosphorylation sites on 1614 proteins (supplemental Tables S2 and S3, <1% FDR). The 4280 phosphorylation sites were composed of 3711 phosphorylated serine (pS), 546 phosphorylated threonine (pT), and 23 phosphorylated tyrosine (pY) residues (Fig. 2D). Among these, 23 DEPs and 60 phosphosites on 54 proteins were upregulated, and 35 DEPs and 99 phosphosites on 90 proteins were down-regulated in Wip1 null testes, with p values that are significant (Fig. 2B, 2C, supplemental Tables S2 and S3). In addition, we compared the phosphoproteome data of specific phosphopeptide against the related protein expression data from proteomic study (supplemental Table S7). In total, 1431 identified proteins were overlapped between phosphoproteome and proteome profiling data. Among these, for most differentially regulated phosphorylation sites, the corresponding protein expression difference was no significant from the proteome profiling data. Of the 58 DEPs, only 2 were also identified as differentially regulated phosphoproteins (supplemental Table S7). To gain insights into the biological functions regulated in Wip1 knockout progression, we characterized the DEPs and PLSC phosphoproteins by GO enrichment in relation to biological process, molecular function, and cellular component (supplemental Table S4). GO enrichment analysis showed that the three categories covering most of the differentially expressed terms were mainly related to adherens/tight junctions, spermatogenesis, apoptotic process, immune response, and microtubule/cytoskeleton organization (Fig. 3A–3C, Tables II and III). KEGG pathway enrichment analysis indicated that most of the DEPs were related to immune response, amino acid metabolism, and tight junction, similar to the GO term enrichments (Tables II and III, supplemental Fig. S1 and supplemental Table S5). KEGG pathway analysis of these PLSC phosphoproteins showed tight junction and adherens junction as the two most predominant pathways, involving seven related phosphoproteins (ZO-1 (Tjp1), ZONAB (Ybx3), Cingulin (Cgn), Paracingulin (Cgnl1), Symplekin (Sympk), β-catenin (Ctnnb1), and WASP (Wasl) (Fig. 3D, Fig. 4B, Table II, supplemental Fig. S2, and supplemental Table S6). The germ cell-Sertoli cell junction signaling pathway is presented in Fig. 4A. In this pathway, Wip1 deficiency might induce down-regulation of β-catenin phosphorylation and broken adherens junctions between germ cells and Sertoli cells, eventually resulting in actin deploymerization, BTB disruption, and germ cell migration.
Fig. 2.
Whole proteome and phosphoproteome analysis of Wip1-deficient mice testis. A, Schematic diagram of proteome and phosphoproteome analysis. After dissection, the testis from WT and Wip1-KO mice were lysed in 8 m urea and digested with trypsin. Peptides were either directly labeled with iTRAQ-8plex or enriched using TiO2 beads prior to mass spectrometry analysis. Proteomics and phosphoproteomics data were then qualitatively, quantitatively and functionally analyzed. B, Volcano plot of the quantified proteins in all biological replicates (the proteins that showed significant down- and upregulated after statistical analysis are reported in green and red). C, Volcano plot of the quantified phosphosites in all biological replicates after TiO2 enrichment (the phosphosites that showed significant down- and upregulated after statistical analysis are reported in green and red). D, Distribution of the phosphorylated serine, threonine, and tyrosine residues among the identified phosphorylation sites.
Fig. 3.
Integrative analysis of Wip1-deficient proteome and phosphoproteome identifies cell adhesion/tight junction pathways. A-C, Functional enrichment of DEPs and PLSC phosphoproteins by the Gene Ontology (GO) database. A, Biological process analysis; B, molecular function analysis; C, cellular component analysis. Red and blue columns represent the proteome and phosphoproteome, respectively. D, Bubble graph describing the distribution of PLSC phosphoproteins in a classified KEGG pathway. Bubble size represents the number of PLSC phosphoproteins involved. Gradation from green to red indicates a decreasing p value (increasing significance).
Table II. Proteins and phosphoproteins involved in cell adhesion/tight junction pathways showing altered expression in Wip1-KO mice.
| Accession number | Protein description | Phosphorylation site | p value | Fold change |
KO |
WT |
|---|---|---|---|---|---|---|
| (KO/WT) | S.D. | S.D. | ||||
| Q91Z83 | Myosin-7 (Myosin heavy chain 7) | — | 0.027 | 1.644 | 0.358 | 0.061 |
| Q02248 | Catenin beta-1 | S (552) | 0.021 | 0.580 | 0.532 | 0.137 |
| P39447 | Tight junction protein ZO-1 | S (617) | 0.018 | 6.995 | 0.490 | 0.032 |
| P23242 | Gap junction alpha-1 protein | S (306) | 0.006 | 0.418 | 0.097 | 0.019 |
| Q62523 | Zyxin | S (336) | 0.049 | 0.380 | 0.228 | 0.046 |
| Q91YD9 | Neural Wiskott-Aldrich syndrome protein | Y (253) | 0.002 | 0.436 | 0.030 | 0.038 |
| Q9WTQ5 | A-kinase anchor protein 12 | T (635) | 0.016 | 0.105 | 0.023 | 0.559 |
| Q9WTQ5 | A-kinase anchor protein 12 | S (733) | 0.018 | 23.134 | 1.797 | 0.073 |
| P40124 | Adenylyl cyclase-associated protein 1 | S (34) | 7.58E-06 | 0.109 | 0.024 | 0.036 |
| Q61165 | Sodium/hydrogen exchanger 1 | S (790) | 0.013 | 0.270 | 0.763 | 0.116 |
| P14602 | Heat shock protein beta-1 (HspB1) | S (86) | 0.039 | 0.387 | 0.252 | 0.092 |
| P14602 | Heat shock protein beta-1 (HspB1) | S (15) | 3.84E-07 | 0.552 | 0.349 | 0.870 |
| P17095 | High mobility group protein HMG-I/HMG-Y | S (36) | 0.033 | 0.414 | 0.211 | 0.008 |
| Q8K4L3 | Supervillin | S (857) | 0.048 | 0.485 | 0.157 | 0.045 |
| Q8K4L3 | Supervillin | S (220) | 0.002 | 0.592 | 0.022 | 0.165 |
| Q8K4L3 | Supervillin | S (227) | 0.002 | 0.592 | 0.022 | 0.165 |
| Q61235 | Beta-2-syntrophin | S (373) | 0.043 | 0.487 | 0.101 | 0.060 |
| O70318 | Band 4.1-like protein 2 | S (379) | 0.005 | 0.489 | 0.043 | 0.044 |
| Q80X82 | Symplekin | S (1256) | 4.03E-06 | 0.556 | 0.113 | 0.172 |
| Q8BRT1 | CLIP-associating protein 2 | S (376) | 0.018 | 3.890 | 0.196 | 0.048 |
| Q99KG5 | Lipolysis-stimulated lipoprotein receptor | S (407) | 0.002 | 0.302 | 0.118 | 0.108 |
| P59242 | Cingulin | S (149) | 0.044 | 0.413 | 0.290 | 0.084 |
| Q6AW69 | Cingulin-like protein 1 | S (284) | 0.026 | 0.511 | 0.098 | 0.046 |
| Q9JKB3 | Y-box-binding protein 3 | S (2) | 0.025 | 3.422 | 0.162 | 0.916 |
Table III. Proteins and phosphoproteins involved in inflammatory response pathway with alterant levels in Wip1-KO mice.
| Accession number | Protein description | Phosphorylation site | p value | Fold change |
KO |
WT |
|---|---|---|---|---|---|---|
| (KO/WT) | S.D. | S.D. | ||||
| Q9DAC0 | Chemokine-like factor superfamily member 2B | — | 0.009 | 0.714 | 0.142 | 0.106 |
| P28654 | Decorin | — | 0.009 | 1.343 | 0.157 | 0.012 |
| P22366 | Myeloid differentiation primary response protein MyD88 | — | 0.017 | 1.551 | 0.244 | 0.138 |
| P01878 | Ig alpha chain C region | — | 0.007 | 1.447 | 0.173 | 0.048 |
| P01872 | Ig mu chain C region | — | 0.016 | 1.455 | 0.221 | 0.077 |
| P01863 | Ig gamma-2A chain C region | — | 7.44E-05 | 1.908 | 0.048 | 0.083 |
| Q80V62 | Fanconi anemia group D2 protein homolog (FACD2) | — | 0.044 | 1.291 | 0.189 | 0.112 |
| Q6NS57 | Mitogen-activated protein kinase-binding protein 1 | S (1252) | 0.004 | 2.359 | 0.048 | 0.059 |
| Q9JKB3 | Y-box-binding protein 3 | S (2) | 0.025 | 3.422 | 0.162 | 0.916 |
| Q923E4 | NAD-dependent protein deacetylase sirtuin-1 | S (532) | 6.41E-05 | 0.167 | 0.035 | 0.189 |
| Q62523 | Zyxin | S (336) | 0.049 | 0.380 | 0.228 | 0.046 |
| P23242 | Gap junction alpha-1 protein (Connexin-43) | S (306) | 0.006 | 0.418 | 0.097 | 0.019 |
| Q91YD9 | Neural Wiskott-Aldrich syndrome protein | Y (253) | 0.002 | 0.436 | 0.030 | 0.038 |
| P14602 | Heat shock protein beta-1 (HspB1) | S (86) | 0.039 | 0.387 | 0.252 | 0.092 |
| P14602 | Heat shock protein beta-1 (HspB1) | S (15) | 3.84E-07 | 0.552 | 0.349 | 0.870 |
| Q6NZM9 | Histone deacetylase 4 (HD4) | S (629) | 0.019 | 0.524 | 0.121 | 0.070 |
| Q9D0J8 | Parathymosin | S (2) | 0.035 | 0.451 | 0.202 | 0.165 |
Fig. 4.
Functional annotations of PLSC phosphoproteins by IPA and KEGG databases in absence of Wip1. A, Some PLSC phosphoproteins were predicted to participate in the germ cell-Sertoli cell junction pathway in Wip1-deficient testes by IPA. B, KEGG pathway analysis of these PLSC phosphoproteins showed tight junction as the most predominant pathways, involving five related phosphoproteins (ZO-1 (Tjp1), ZONAB (Ybx3), Cingulin (Cgn), Paracingulin (Cgnl1), and Symplekin (Sympk).
We applied comprehensive bioinformatics analysis to assess the integrated functions and processes of these DEPs and PLSC phosphoproteins. The significant functions and processes related to these proteins and phosphoproteins analyzed in IPA pertained to a network consisting of 20 proteins and 58 phosphoproteins involved in cell adhesion/tight junction, apoptosis, inflammatory response, spermatogenesis, sperm motility, and cytoskeletal assembly and deploymerization (supplemental Fig. S3A). Furthermore, proteins are known to perform multiple functions within complex networks, and we therefore obtained more detailed information about these six processes by searching for interactions between DEPs and PLSC phosphoproteins using the STRING database (supplemental Fig. S3B). Taken together, the GO and KEGG term enrichments and IPA analysis unanimously indicated that DEPs and PLSC phosphoproteins participated in the cell adhesion/tight junction processes.
Knockout of Wip1 Impairs Junction-related Protein Expression and BTB Integrity
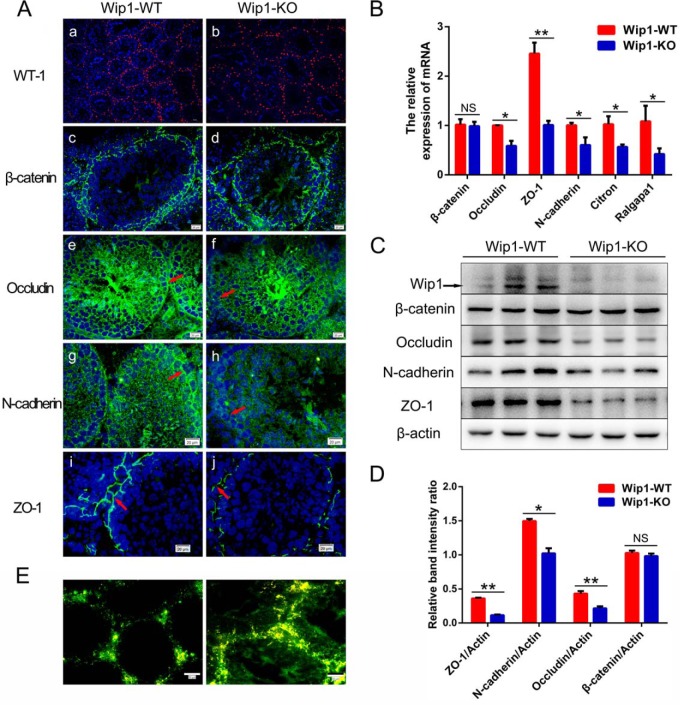
The distribution and expression levels of BTB-component proteins were investigated to validate the altered cell adhesion/tight junction signaling pathways. We investigated the immunoreactivities of Sertoli cell marker and adherens/tight junction proteins. The localizations of these proteins in the testis were in accordance with previous results (28). Immunostaining of Sertoli cells with anti-rabbit WT1 showed that the number of Sertoli cells in the seminiferous tubules was unaltered in Wip1-KO mouse testes (Fig. 5Aa-b). Tight junction and adherens junction proteins (occludin, ZO-1, N-cadherin) showed intermittent, diffuse staining and weaker fluorescence intensity in Wip1-KO mouse testes compared with WT testes (Fig. 5Ae-j). mRNA levels of tight junction and adherens junction genes (occludin, ZO-1, N-cadherin, citron) were significantly downregulated in Wip1-KO compared with WT testes (Fig. 5B). In addition, the GTPase-related gene, Ralgapa1, which is involved in the recruitment of cadherins to the adherens junction, was also significantly reduced (Fig. 5B). Moreover, occludin, N-cadherin, and ZO-1 protein levels were significantly decreased by 1.9-fold, 1.5-fold, and 3.0-fold, respectively (Fig. 5C, 5D). However, there was no change in β-catenin mRNA and protein levels and immunoreactivity in Wip1-KO mouse testes (Fig. 5). Finally, FITC-labeled dextran was used as a tracer to evaluate the BTB integrity. FITC green fluorescence was mainly distributed at the periphery of the seminiferous tubules and failed to enter the adluminal compartment of the seminiferous epithelium in WT testes (Fig. 5E). In contrast, after Wip1 knockout, dextran permeated beyond the BTB and entered the adluminal compartment, indicating the impaired integrity of the BTB and abnormal spermatogenesis (Fig. 5E). Collectively, these findings uniformly indicated that disruption of BTB integrity was partially responsible for the subfertility associated with Wip1 deficiency.
Fig. 5.
Wip1 deficiency disrupts the expression of junction-associated proteins and the integrity of BTB. (A, a-d) Immunofluorescence images of testis sections from 10-week-old WT and Wip1-KO mice stained with anti-WT1 and anti-β-catenin body. (A, e-j) Immunoreactivity of junction-associated proteins (occludin, N-cadherin, ZO-1) was altered in the seminiferous tubules of Wip1-deficient mice. Distribution of occludin (red arrows; e-f), N-cadherin (red arrows; g-h), and ZO-1 (red arrows; i-j) in the seminiferous epithelium of WT and Wip1-KO mice (n = 4 per group) counterstained with DAPI. Scale bars: 50 μm (a-b); 20 μm (c-j). B, Relative mRNA expression levels of junction-associated components in WT and Wip1-KO mouse testes. C, Relative protein expression levels of junction-associated components in WT and Wip1-KO mouse testes. β-actin served as a loading control. D, Quantified relative band intensity ratio of C. E, In vivo functional assay (FITC-labeled dextran tracer) to determine BTB integrity in WT and Wip1-KO mouse testes. In testes sections from WT mice, FITC green fluorescence was only visible in the interstitial spaces and basal compartment, whereas in sections of testes from Wip1-KO mice, FITC green fluorescence was observed in the interstitial spaces and basal compartment, and in the lumen of the seminiferous tubules. Scale bars: 50 μm. Data are expressed as mean ± S.E. *p < 0.05, **p < 0.01, NS: nonsignificant.
Knockout of Wip1 Increases Cytokine Levels and Germ Cell Apoptosis in Testes
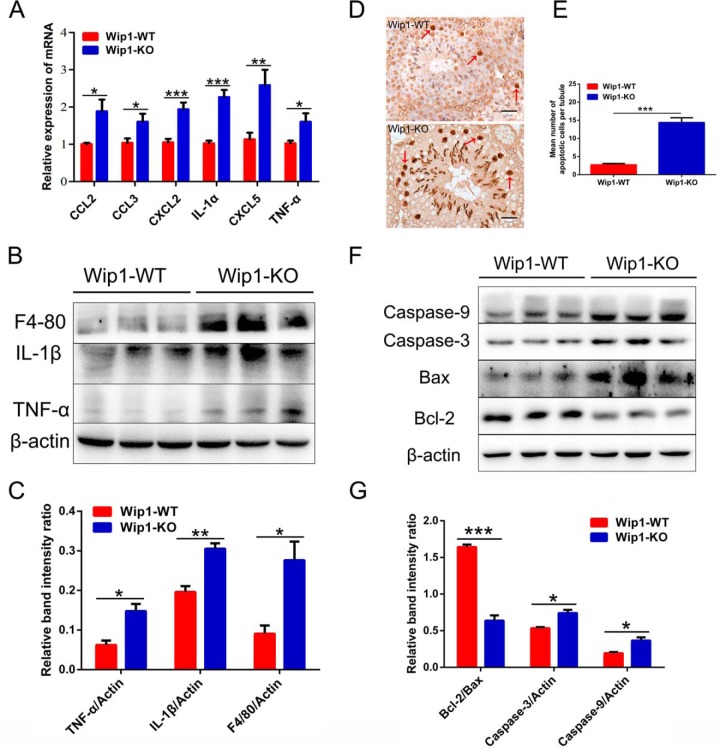
Several studies have shown that increased cytokine expression disrupted BTB integrity by inducing the down-regulation of junction-associated proteins (2, 29). Based on the results of omics analyses, we showed that the expression levels of cytokines and chemokines, including TNF-α, IL-1α, IL-1β, CCL2 (MCP-1), CCL3 (MIP-1α), CXCL2 (MIP-2α), and CXCL5, were significantly increased in Wip1-KO mouse testes (Fig. 6A–6C), whereas the murine macrophage marker F4–80 was also significantly enhanced, associated with the increased expression of chemokines (Fig. 6A–6C).
Fig. 6.
Wip1-deficient mice exhibit elevated cytokine levels and germ cell apoptosis in testis. A, Relative mRNA expression levels of cytokines and chemokines in WT and Wip1-KO mouse testes. B, Relative protein expression levels of cytokines and murine macrophage marker (F4–80) in WT and Wip1-KO mouse testes. β-actin served as a loading control. C, Quantified relative band intensity ratio of B. D, TUNEL-positive cells in WT and Wip1-KO mouse testis sections appeared brown (red arrows). E, Mean number of apoptotic cells per tubule based on mean ± S.E. for four animals in each group. F, Relative protein expression levels of apoptosis-related components in WT and Wip1-KO mouse testes. β-actin served as a loading control. G, Quantified relative band intensity ratios of Bcl-2/Bax, caspase-3/actin, and caspase-9/actin were calculated by densitometric scanning of band intensity. Data are expressed as mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 50 μm.
As indicated above, Wip1-KO testes demonstrated disruption of BTB integrity and increased expression of cytokines and chemokines, indicative of germ cell apoptosis (30, 31). We therefore examined if depletion of Wip1 provoked testicular germ cell apoptosis using TUNEL assay. Few apoptotic germ cells were detected in WT mouse testes, but these were obviously elevated in Wip1-KO testes (Fig. 6D; red arrows). Wip1-deficient testes showed a significant increase (5.30-fold, p < 0.001) in the mean number of apoptotic germ cells in the seminiferous tubules compared with WT testes (Fig. 6E). We confirmed the effects of Wip1 depletion on the key proteins and enzymes in the apoptotic pathway by immunoblotting. Wip1-KO mouse testes presented significantly reduced expression levels of Bcl-2 and elevated levels of Bax, caspase-3, and caspase-9 (Fig. 6F). The Bcl-2/Bax, caspase-3/actin, and caspase-9/actin ratios are shown in Fig. 6G. These results suggest that disruption of BTB integrity induced germ cell apoptosis and spermatids loss in the testis, eventually leading to reduced fertility.
DEPs and PLSC Phosphoproteins Were Verified By Western Blotting
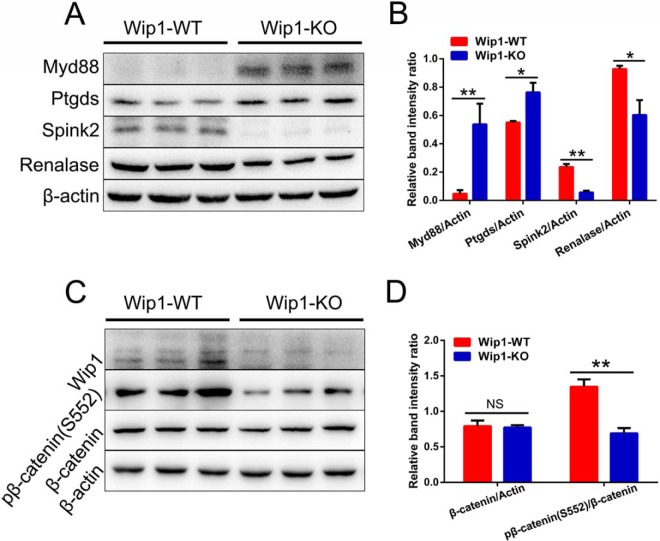
We verified the accuracy of the quantitative proteomic and phosphoproteomic results by Western blotting of four DEPs and one PLSC phosphoprotein, including Myd88, Ptgds, Spink2, Renalase, and pβ-catenin (S552), which were related to inflammatory response, spermatogenesis, sperm motility, and cell adhesion. The Western blotting results were in accordance with the proteomic and phosphoproteomic results, thus validating the reliability of the omics data (Fig. 7).
Fig. 7.
Confirmation of differential expression of proteins and PLSC phosphoproteins. A, Expression levels of Myd88, Ptgds, Spink2, and Renalase in WT and Wip1-KO mice were confirmed by Western blotting. β-actin served as a loading control. B, Quantified relative band intensity ratio of A. C, Expression levels of total and phosphorylated β-catenin in WT and Wip1-KO mouse were confirmed by Western blotting. β-actin served as a loading control. D, Quantified relative band intensity ratios of β-catenin/actin and p-β-catenin (S552)/β-catenin were calculated by densitometric scanning of band intensity. Data are expressed as mean ± S.E. *p < 0.05, **p < 0.01, NS: nonsignificant.
DISCUSSION
Wip1 has been studied extensively in relation to multiple aspects of tumorigenesis, stress response, DNA damage response, and inflammatory response, as well as cellular homeostasis, cell development and migration, obesity, and atherosclerosis. Two of the most apparent phenotypes of Wip1-deficient mice include defects in testis development, spermatogenesis, and fertility (11, 20). Wip1-deficient males have profoundly reduced fertility, presumably related to the aberrant architecture of the seminiferous tubules, as well as a paucity of spermatids in the testis. Moreover, we confirmed that Wip1-KO mice have significantly reduced numbers of mature sperm in the epididymis. We therefore concluded that Wip1 plays an indispensable role in the development of the male reproductive structures.
Although Filipponi et al. reported that testicular defects in Wip1-deficient mice were partially because of the activation of ATM by damaged heterochromatin (20), the effects have not been extensively examined at the protein level. We therefore performed proteomics and phosphoproteomics analyses to clarify the molecular mechanisms underlying the spermatogenesis and fertility defects in Wip1-deficient mice, and identified 58 DEPs and 141 PLSC phosphoproteins, 20 and 58 of which have previously been reported to potentially participate in the processes of cell adhesion/tight junctions, apoptosis, inflammatory response, spermatogenesis, sperm motility, and cytoskeletal assembly and depolymerization. Furthermore, we predicted the interaction networks among these DEPs and PLSC phosphoproteins using the STRING database and investigated their functions and roles in Wip1 knockout progression by performing GO and KEGG pathway enrichment analyses. Integrative GO analysis of the DEPs and PLSC phosphoproteins revealed significant enrichment of proteins mainly involved in adherens/tight junctions, apoptotic process, immune response, spermatogenesis, and microtubule/cytoskeleton organization. Most of the DEPs were related to immune response, amino acid metabolism, and tight junction according to KEGG pathway enrichment analysis. The KEGG pathway describing tight junction and adherens junction were the two most highly enriched in terms of PLSC phosphoproteins in Wip1-KO compared with WT mice, including seven related proteins, (ZO-1 (Tjp1), ZONAB (Ybx3), Cingulin (Cgn), Paracingulin (Cgnl1), Symplekin (Sympk), β-catenin (Ctnnb1), and WASP (Wasl). Further analysis of PLSC phosphoproteins by IPA identified the germ cell-Sertoli cell junction signaling pathway in Wip1-deficient testes. We therefore speculated that the phosphorylation level of β-catenin might function as a central regulator of adherens junction required for spermatogenesis and fertility. Chang et al. reported that spermatid-specific deletion of β-catenin resulted in a significantly reduced sperm count, increased germ cell apoptosis, and impaired fertility, and attributed this to the role of β-catenin in the adherens junctions between Sertoli cells and elongating spermatids (32). Wnt signaling is activated by Wnt ligands and increased β-catenin protein stability (33), and has been shown to be important for testis development and spermatogenesis (34). Down-regulation of p-β-catenin (S552) might thus affect Wnt activity and adherens junctions between germ cells and Sertoli cells, eventually resulting in spermatogenesis defects and BTB disruption. This hypothesis was partially validated by Cho et al., who found that Wip1 enhanced the Wnt response by directly dephosphorylating NLK and increasing β-catenin-TCF/LEF1 interactions (35). Integrative whole proteome and phosphoproteome profiling thus revealed that spermatogenesis and fertility defects in Wip1-deficient mice may be associated with abnormal cell adhesion/tight junction processes.
Based on the omics data, we explored the distribution and expression levels of tight junction and adherens junction (apical ectoplasmic specialization) proteins and BTB integrity in Wip1-deficient testes. Wip1 deficiency decreased expression and immunoreactivity of levels of tight junction-related proteins (occludin and ZO-1) and apical ectoplasmic specialization protein (N-cadherin) in the testes, with similar reductions in mRNA expression. Adherens junctional complexes, such as ectoplasmic specializations, facilitate cellular interactions that are critical for both adhesion and signaling between Sertoli cells and germ cells (36). Previous studies indicated that Sertoli cell-germ cell adherens junctions undergo extensive restructuring to promote germ cell maturation and spermation (36, 37). Elongating/elongated spermatids adhere to Sertoli cells via the apical ectoplasmic specialization, a testis-specific actin-rich cell junction (38). Recent studies suggested that disruption of Sertoli cell-spermatid adhesion at the apical ectoplasmic specialization resulted in impaired functioning of basal ectoplasmic specialization/tight junctions at the BTB (39). This suggests that disruption of the apical ectoplasmic specialization/tight junction because of Wip1 deletion may contribute to spermatid sloughing and oligospermia in pathological conditions associated with reduced fertility potential. The production and distribution of tight junction and ectoplasmic specialization proteins are tightly associated with the assembly of a functional BTB (40). Based on these results, we sought to determine if the BTB was compromised in Wip1-deficient testes using a FITC-dextran tracer and showed that the BTB became inefficient and was unable to prevent the diffusion of substances into the lumen of the seminiferous tubules. Disruption of the BTB can result in hypospermatogenesis and subsequent subfertility or infertility (9). Overall, the present results indicated that Wip1 deficiency could damage the normal BTB structure and function by impairing adherens/tight junctions, leading to spermatids loss and subfertility.
The mechanisms responsible for the disruption of cell adhesion and the BTB remain unclear. In light of our omics results, we detected the expression levels of some cytokines and chemokines, and showed that mRNA levels of TNF-α, IL-1α, CCL2 (MCP-1), CCL3 (MIP-1α), CXCL5, and CXCL2 (MIP-2α) were all significantly increased in Wip1-KO compared with WT mouse testes, whereas protein levels of TNF-α and IL-1β were also elevated. Cytokine-mediated down-regulation of junction proteins increases BTB permeability (8). Michelle et al. reported that TNF-α inhibited the steady-state protein levels of occludin, ZO-1, and N-cadherin, resulting in the disruption of BTB dynamics (29). TNF-α may thus act as a crucial regulator of junction dynamics and be involved in the impairment of Sertoli cell-germ cell adhesion. CCL2 (MCP-1), CCL3 (MIP-1α), CXCL2 (MIP-2α), and CXCL5 not only recruit macrophages, but also promote tissue-resident macrophage proliferation and increase the inflammatory response (41). Wip1 deficiency significantly increased the protein expression of F4–80 in the testis, which could be achieved by increased expression of chemokines. Disruption of BTB integrity can result in germ cell apoptosis, whereas TNF-α can also trigger germ cell apoptosis (30, 31). The death signal triggers the Bcl-2 family (e.g. Bax and Bcl-2) or can directly induce an increase in mitochondrial membrane permeability, activate the cysteine protease, and eventually lead to apoptosis (42). Our data showed that Wip1 depletion induced obvious apoptotic morphological changes and increased TUNEL-positive cells, as well as enhancing protein levels of caspase-9, caspase-3, and pro-apoptotic Bax, while decreasing levels of anti-apoptotic Bcl-2 in the testes. Proinflammatory cytokines may be related to testicular germ cell apoptosis and impaired BTB dynamics, both of which partially contribute to subfertility and spermatogenesis defects in Wip1-deficient males.
In summary, knockout of Wip1 resulted in reduced fertility and spermatogenesis defects at the round- and elongating-spermatid stages. Integrative proteomics and phosphoproteomics analyses indicated that DEPs and PLSC phosphoproteins were mainly involved in cell adhesion/tight junctions, apoptosis, inflammatory response, spermatogenesis, and cytoskeletal assembly and depolymerization. Based on verification of the omics results, we propose that changes in BTB integrity and BTB-associated junction proteins may represent one pathway responsible for the low male reproductive function in Wip1-deficient mice (Fig. 8).
Fig. 8.
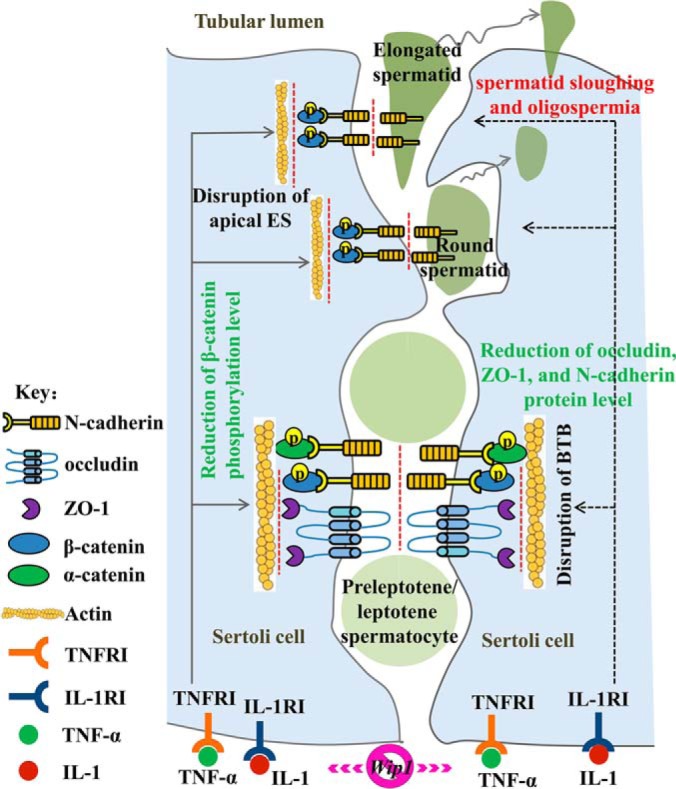
Schematic model of the proposed mechanisms responsible for the subfertility and spermatogenesis defects induced by Wip1 deficiency. Proinflammatory cytokines impairs the BTB and apical ectoplasmic specialization dynamics by decreasing the expression of junction-associated proteins (occludin, ZO-1, N-cadherin, and pβ-catenin (S552)), which effect could be partially responsible for the spermatid sloughing and oligospermia and subfertility in Wip1-deficient mice.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD009220. Project name: Integrative proteomic and phosphoproteomic profiling of testis from Wip1-knockout mice: insights into mechanisms of reduced fertility; Project accession: PXD009220.
Supplementary Material
Acknowledgments
We thank Dr. Bing Pan of the School of Basic Medical Sciences, Peking University for providing excellent technical support and for editing the English text of a draft of this manuscript.
Footnotes
* This research was supported by the National Natural Science Foundation of China (31330074, 31572378), the Major National Scientific Research Projects (2015CB943101), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (ASTIP-IAS05), and the Special Fund of Chinese Central Government for Basic Scientific Research Operations in Commonweal Research Institutes (2016ywf-yb-4).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- BTB
- blood–testis barrier
- Wip1
- wild-type p53-induced phosphatase 1
- WT
- wild-type
- KO
- knockout
- TUNEL
- terminal deoxynucleotidyl transferase nick-end labeling
- iTRAQ
- isobaric tags for relative and absolute quantification
- DEPs
- differentially expressed proteins
- GO
- gene ontology
- KEGG
- kyoto encyclopedia of genes and genomes
- IPA
- ingenuity pathway analysis
- STRING
- search tool for the retrieval of interacting genes
- FDR
- false discovery rate
- PLSC
- phosphorylation level significantly changed.
REFERENCES
- 1. Guo X., and Jian Shen Zhengrong Xia Rui Zhang Ping Zhang Chun Zhao Jun Xing Ling Chen Wen Chen Min Lin Ran Huo Bing Su Zuomin Zhou Sha J. (2010) Proteomic Analysis of Proteins Involved in Spermiogenesis in Mouse. J. Proteome Res. 9, 1246–1256 [DOI] [PubMed] [Google Scholar]
- 2. Yao P. L., Lin Y. C., and Richburg J. H. (2010) Mono-(2-ethylhexyl) phthalate-induced disruption of junctional complexes in the seminiferous epithelium of the rodent testis is mediated by MMP2. Biol. Reprod. 82, 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rode K., Sieme H., Richterich P., and Brehm R. (2015) Characterization of the equine blood-testis barrier during tubular development in normal and cryptorchid stallions. Theriogenology 84, 763–772 [DOI] [PubMed] [Google Scholar]
- 4. Pelletier R. M., and Byers S. W. (1992) The blood-testis barrier and Sertoli cell junctions: structural considerations. Microsc. Res. Tech. 20, 3–33 [DOI] [PubMed] [Google Scholar]
- 5. Wong E. W. P., Mruk D. D., and Cheng C. Y. (2008) Biology and regulation of ectoplasmic specialization, an atypical adherens junction type, in the testis. BBA-Biomembranes 1778, 692–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mruk D. D., and Cheng C. Y. (2004) Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr. Rev. 25, 747–806 [DOI] [PubMed] [Google Scholar]
- 7. Wong E. W., Mruk D. D., and Cheng C. Y. (2008) Biology and regulation of ectoplasmic specialization, an atypical adherens junction type, in the testis. Biochim. Biophys. Acta 1778, 692–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lui W. Y., and Lee W. M. (2009) Molecular mechanisms by which hormones and cytokines regulate cell junction dynamics in the testis. J. Mol. Endocrinol. 43, 43–51 [DOI] [PubMed] [Google Scholar]
- 9. Qiu L., Zhang X., Zhang Y., Gu J., Chen M., Zhang Z., Wang X., and Wang S. L. (2013) Sertoli cell is a potential target for perfluorooctane sulfonate-induced reproductive dysfunction in male mice. Toxicol. Sci. 135, 229–240 [DOI] [PubMed] [Google Scholar]
- 10. Cheng C. Y., and Mruk D. D. (2012) The blood-testis barrier and its implications for male contraception. Pharmacol. Rev. 64, 16–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J., Nannenga B., Demidov O. N., Bulavin D. V., Cooney A., Brayton C., Zhang Y., Mbawuike I. N., Bradley A., Appella E., and Donehower L. A. (2002) Mice Deficient for the Wild-Type p53-Induced Phosphatase Gene (Wip1) Exhibit Defects in Reproductive Organs, Immune Function, and Cell Cycle Control. Mol. Cell. Biol. 22, 1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buss M. C., Remke M., Lee J., Gandhi K., Schniederjan M. J., Kool M., Northcott P. A., Pfister S. M., Taylor M. D., and Castellino R. C. (2015) The WIP1 oncogene promotes progression and invasion of aggressive medulloblastoma variants. Oncogene 34, 1126–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emelyanov A., and Bulavin D. V. (2015) Wip1 phosphatase in breast cancer. Oncogene 34, 4429–4438 [DOI] [PubMed] [Google Scholar]
- 14. Park H. K., Panneerselvam J., Dudimah F. D., Dong G., Sebastian S., Zhang J., and Fei P. (2011) Wip1 contributes to cell homeostasis maintained by the steady-state level of Wtp53. Cell cycle 10, 2574–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun B., Hu X., Liu G., Ma B., Xu Y., Yang T., Shi J., Yang F., Li H., Zhang L., and Zhao Y. (2014) Phosphatase Wip1 negatively regulates neutrophil migration and inflammation. J. Immunol. 192, 1184–1195 [DOI] [PubMed] [Google Scholar]
- 16. Yi W., Hu X., Chen Z., Liu L., Tian Y., Chen H., Cong Y. S., Yang F., Zhang L., Rudolph K. L., Zhang Z., Zhao Y., and Ju Z. (2015) Phosphatase Wip1 controls antigen-independent B-cell development in a p53-dependent manner. Blood 126, 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang Y., Liu L., Sheng M., Xiong K., Huang L., Gao Q., Wei J., Wu T., Yang S., Liu H., Mu Y., and Li K. (2015) Wip1 knockout inhibits the proliferation and enhances the migration of bone marrow mesenchymal stem cells. Exp. Cell Res. 334, 310–322 [DOI] [PubMed] [Google Scholar]
- 18. Tang Y., Pan B., Zhou X., Xiong K., Gao Q., Huang L., Xia Y., Shen M., Yang S., Liu H., Tan T., Ma J., Xu X., Mu Y., and Li K. (2017) Wip1-dependent modulation of macrophage migration and phagocytosis. Redox Bio. 13, 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Guezennec X., Brichkina A., Huang Y. F., Kostromina E., Han W., and Bulavin D. V. (2012) Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab. 16, 68–80 [DOI] [PubMed] [Google Scholar]
- 20. Filipponi D., Muller J., Emelyanov A., and Bulavin D. V. (2013) Wip1 controls global heterochromatin silencing via ATM/BRCA1-dependent DNA methylation. Cancer Cell 24, 528–541 [DOI] [PubMed] [Google Scholar]
- 21. Chen Q., Peng H., Lei L., Zhang Y., Kuang H., Cao Y., Shi Q. X., Ma T., and Duan E. (2011) Aquaporin3 is a sperm water channel essential for postcopulatory sperm osmoadaptation and migration. Cell Res. 21, 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao X., Mruk D. D., Tang E. I., Massarwa R., Mok K. W., Li N., Wong C. K., Lee W. M., Snapper S. B., Shilo B. Z., Schejter E. D., and Cheng C. Y. (2014) N-wasp is required for structural integrity of the blood-testis barrier. PLoS Genet. 10, e1004447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 24. Corradini E., Vallur R., Raaijmakers L. M., Feil S., Feil R., Heck A. J., and Scholten A. (2014) Alterations in the cerebellar (Phospho)proteome of a cyclic guanosine monophosphate (cGMP)-dependent protein kinase knockout mouse. Mol. Cell. Proteomics 13, 2004–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zanivan S., Meves A., Behrendt K., Schoof E. M., Neilson L. J., Cox J., Tang H. R., Kalna G., van Ree J. H., van Deursen J. M., Trempus C. S., Machesky L. M., Linding R., Wickstrom S. A., Fassler R., and Mann M. (2013) In vivo SILAC-based proteomics reveals phosphoproteome changes during mouse skin carcinogenesis. Cell Rep. 3, 552–566 [DOI] [PubMed] [Google Scholar]
- 26. Huang H., Larsen M. R., Palmisano G., Dai J., and Lametsch R. (2014) Quantitative phosphoproteomic analysis of porcine muscle within 24 h postmortem. J. Proteomics 106, 125–139 [DOI] [PubMed] [Google Scholar]
- 27. Huang H., Haar Petersen M., Ibanez-Vea M., Lassen P. S., Larsen M. R., and Palmisano G. (2016) Simultaneous Enrichment of Cysteine-containing Peptides and Phosphopeptides Using a Cysteine-specific Phosphonate Adaptable Tag (CysPAT) in Combination with titanium dioxide (TiO2) Chromatography. Mol. Cell. Proteomics 15, 3282–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chojnacka K., Bilinska B., and Mruk D. D. (2017) Annexin A2 is critical for blood-testis barrier integrity and spermatid disengagement in the mammalian testis. Biochim. Biophys. Acta 1864, 527–545 [DOI] [PubMed] [Google Scholar]
- 29. Li M. W., Xia W., Mruk D. D., Wang C. Q., Yan H. H., Siu M. K., Lui W. Y., Lee W. M., and Cheng C. Y. (2006) Tumor necrosis factor {alpha} reversibly disrupts the blood-testis barrier and impairs Sertoli-germ cell adhesion in the seminiferous epithelium of adult rat testes. J. Endocrinol. 190, 313–329 [DOI] [PubMed] [Google Scholar]
- 30. Yao P. L., Lin Y. C., and Richburg J. H. (2009) TNF alpha-mediated disruption of spermatogenesis in response to Sertoli cell injury in rodents is partially regulated by MMP2. Biol. Reprod. 80, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hou W. G., Zhao J., Li Z., Li W., Li T., Xiong L. Z., and Zhang Y. Q. (2012) Effects of electromagnetic pulse irradiation on the mouse blood-testicle barrier. Urology 80, 225 e221–226 [DOI] [PubMed] [Google Scholar]
- 32. Chang Y. F., Lee-Chang J. S., Harris K. Y., Sinha-Hikim A. P., and Rao M. K. (2011) Role of beta-catenin in post-meiotic male germ cell differentiation. PloS one 6, e28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clevers H., and Nusse R. (2012) Wnt/beta-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 34. Golestaneh N., Beauchamp E., Fallen S., Kokkinaki M., Uren A., and Dym M. (2009) Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 138, 151–162 [DOI] [PubMed] [Google Scholar]
- 35. Cho S. J., Cha B. S., Kwon O. S., Lim J., Shin D. M., Han D. W., Ishitani T., Jho E. H., Fornace A. J., and Cha H. J. (2017) Wip1 directly dephosphorylates NLK and increases Wnt activity during germ cell development. Biochim. Biophys. Acta 1863, 1013–1022 [DOI] [PubMed] [Google Scholar]
- 36. Mruk D. D., and Cheng C. Y. (2004) Cell-cell interactions at the ectoplasmic specialization in the testis. Trends Endocrin. Met. 15, 439–447 [DOI] [PubMed] [Google Scholar]
- 37. Yan H. H., Mruk D. D., Wong E. W., Lee W. M., and Cheng C. Y. (2008) An autocrine axis in the testis that coordinates spermiation and blood-testis barrier restructuring during spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 8950–8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grove B. D., and Vogl A. W. (1989) Sertoli cell ectoplasmic specializations: a type of actin-associated adhesion junction? J. Cell Sci. 93, 309–323 [DOI] [PubMed] [Google Scholar]
- 39. Yan H. H., and Cheng C. Y. (2006) Laminin alpha 3 forms a complex with beta3 and gamma3 chains that serves as the ligand for alpha 6beta1-integrin at the apical ectoplasmic specialization in adult rat testes. J. Biol. Chem. 281, 17286–17303 [DOI] [PubMed] [Google Scholar]
- 40. Wong E. W., Mruk D. D., and Cheng C. Y. (2008) Biology and regulation of ectoplasmic specialization, an atypical adherens junction type, in the testis. Biochim. Biophys. Acta 3, 692–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X. X., Ying P., Diao F., Wang Q., Ye D., Jiang C., Shen N., Xu N., Chen W. B., Lai S. S., Jiang S., Miao X. L., Feng J., Tao W. W., Zhao N. W., Yao B., Xu Z. P., Sun H. X., Li J. M., Sha J. H., Huang X. X., Shi Q. H., Tang H., Gao X., and Li C. J. (2013) Altered protein prenylation in Sertoli cells is associated with adult infertility resulting from childhood mumps infection. J. Exp. Med. 210, 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao J., Zhai L., Liu Z., Wu S., and Xu L. (2014) Leptin level and oxidative stress contribute to obesity-induced low testosterone in murine testicular tissue. Oxid. Med. Cell. Longev. 2014, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD009220. Project name: Integrative proteomic and phosphoproteomic profiling of testis from Wip1-knockout mice: insights into mechanisms of reduced fertility; Project accession: PXD009220.