This paper investigated the molecular pathways modulated in past P. falciparum-infected placentas. The proteome, phosphoproteome and glycoproteome analysis of P. falciparum-infected placentas that had developed placental malaria during pregnancy but had the parasites cleared by pharmacological treatment, revealed the activation of AKT and ERK signaling pathways and apoptosis. These molecular features open new perspectives towards novel therapeutic solutions.
Keywords: Malaria, Post-translational Modifications, Cell Death, Host-Pathogen Interaction, Infectious Disease, Parasite, Pregnancy, Placental Malaria
Graphical Abstract
Highlights
An integrated proteome, phosphoproteome and glycoproteome analysis was applied to past P. falciparum-infected placentas.
A total of 2946 proteins, 1733 N-linked glycosites and 4100 phosphosites were identified and quantified in the human placentas.
AKT and ERK signaling pathways are associated to an increased apoptosis in past P. falciparum-infected placentas.
Abstract
Malaria in pregnancy is a public health concern in malaria-endemic areas. Accumulation of maternal immune cells in the placenta and increased levels of inflammatory cytokines caused by sequestration of Plasmodium falciparum-infected erythrocytes have been associated to poor neonatal outcomes, including low birth weight because of fetal growth restriction. Little is known about the molecular changes occurring in a P. falciparum-infected placenta that has developed placental malaria during pregnancy but had the parasites cleared by pharmacological treatment (past infection). We conducted an integrated proteome, phosphoproteome and glycoproteome analysis in past P. falciparum-infected placentas aiming to find molecular changes associated with placental malaria. A total of 2946 proteins, 1733 N-linked glycosites and 4100 phosphosites were identified and quantified in this study, disclosing overrepresented processes related to oxidative stress, protein folding and regulation of apoptosis in past-infected placentas Moreover, AKT and ERK signaling pathways activation, together with clinical data, were further correlated to an increased apoptosis in past-infected placentas. This study showed apoptosis-related mechanisms associated with placental malaria that can be further explored as therapeutic target against adverse pregnancy outcomes.
Malaria is a protozoan disease transmitted by Anopheles mosquitoes, and one of the most important parasitic diseases in human beings (1). Malaria in pregnancy (MiP)1 (caused both by P. falciparum and P. vivax) can pose devastating consequences to a pregnant woman and the developing fetus (2), including maternal anemia and low birth weight because of preterm delivery and fetal growth restriction (3). Approximately 125 million pregnant women worldwide are exposed to the risks of MiP each year, resulting in 200,000 infant deaths (2).
Complications of MiP result mainly from the massive sequestration of the Plasmodium falciparum-infected erythrocytes (IE) in the placental intervillous blood spaces (4). The placenta provides an optimum environment to the development of the parasite subpopulations capable of binding to receptors, such as the chondroitin sulfate A (CSA), which is highly abundant in that organ and it is expressed on the surface of the syncytiotrophoblast; thus, being accessible as a binding receptor to infected red blood cells (5, 6). When placental malarial infection is poorly controlled, it results in chemokine release during the recruitment of the maternal monocytes to the intervillous blood spaces, which has been consistently associated with an increased risk for low birth weight (7). Other findings associated with placental malaria include the deposition of hemozoin—or malaria pigment, a product of the parasite hemoglobin digestion—in phagocytic leukocytes and within fibrin deposits in the intervillous space (3).
A pathologic classification scheme for placental malaria (PM) was first proposed by Garnham et al., and later slightly modified by Bulmer et al. (8) and Muehlenbachs et al. (9). Shortly, after an “active” form of infection characterized by the presence of parasites in the intervillous spaces with or without pigment in intervillous monocytes, the infection assumes a “chronic” form, when parasites are present in the intervillous spaces along with pigment as deposits or in macrophages within the fibrin, followed by a “past-chronic” form, when the pigment is present in the absence of parasites (8, 9). The placental malaria classification is useful as effective endpoints for interventional trials for prophylaxis or treatment drugs, as well as to predict clinical outcomes. For example, chronic infection has been most closely associated with decreased birth weight because of fetal growth restriction and lower maternal hemoglobin or severe anemia, whereas acute infection (especially with high parasitemia) has been more closely associated with preterm delivery (3). However, specific host cellular and molecular changes, as well as altered pathological pathways associated with past P. falciparum infected placentas are still not well elucidated.
Mass spectrometry-based proteomics analysis has the potential to identify large-scale changes in the protein expression and post-translational modification (PTM) patterns that can ultimately be associated to pathological manifestations (10). Proteomic approaches have been previously used to analyze changes in the protein expression occurring in the placental tissue during normal pregnancy (11), as well as in pathological conditions such as pre-eclampsia (12), fetal growth restriction (13), premature rupture of membranes (14) and gestational diabetes (15).
Our study is, to our knowledge, the first applying quantitative-based proteomics approaches to the deep characterization of the protein expression and PTMs in placental malaria models. First, we performed label-free proteomics in two placental malaria models from both mouse and human, to completely assess the biological process changes in infected placentas regardless stage of infection. Next, isobaric tag labeling followed by phosphopeptide and glycopeptides enrichment and high-resolution LC-MS/MS analyses were performed in past P. falciparum-infected and noninfected human placentas, disclosing altered pathways related to the oxidative stress generation and activation of the AKT and ERK, which were further confirmed by Western blotting. Moreover, we provided evidences that past-P. falciparum infected placentas present an increased expression of pro-apoptotic-related proteins BAX and cleaved caspase 3 and a decreased expression of anti-apoptotic-related proteins BCL2 and HSPB1. We also demonstrated that past-infected placentas have increased DNA fragmentation indicating that apoptosis is a processes occurring in placental malaria. Taken together, our findings support an apoptosis-related mechanism that can be further explored in larger cohort studies as a molecular change marker for placental malaria.
EXPERIMENTAL SECTION
Mice and Parasites
In this study, an experimental model of PM female C57BL/six pregnant(16). C57BL/6 (Wild-Type [WT]) from the animal breeding facility of the Institute of Biomedical Sciences at the University of São Paulo (ICB/USP) was used with ages from 8 to 10 weeks, which were bred and kept in a constant light-dark cycle (12 h/12 h) in conventional housing at animal facility of the ICB/USP Department of Parasitology. Mice received water and were fed with commercial NUVILAB CR-1 feed (Nuvital, Brazil) ad libitum. Erythrocytes infected with Plasmodium berghei NK65 constitutively expressing green fluorescent protein (GFP) were used to induce the placental malaria and to conduct in vitro experiments. Infection was daily monitored and parasitemia was assessed in blood smears stained with Giemsa, and afterward counted under a microscope. All procedures were performed in accordance with the national directives for animal welfare and were authorized by the ICB/USP ethics committee with protocol number 062 sheet 33 book 3.
Pregnancy Monitoring and Experimental Infection
Detection of a vaginal plug and measurement of the body weight were combined to set the gestation time, as previously described (16). Females (2 or 3) were placed with one male for 24 h and examined for the presence of the vaginal plug. The progression of pregnancy was monitored every other day based on the weight gain. Successful fertilization was confirmed between G10 and G13 when females exhibited in average 3 to 4g increase of their body weight. Therefore, the weight gain was considered an indication of pregnancy. Pregnant mice were infected intravenously (i.v.) on G13 with 1 × 105 iRBCs. G13 was determined as the optimal time point for the infection to enable the analysis of the malaria pathological features during the course of pregnancy and in the developing fetus, as infection at an earlier stage would not allow the pregnancy to reach its term, which is consistent with previous report (16). Pregnant mice underwent cesarean sections on G19, and their placentas were collected for histopathological analysis and protein extraction. Noninfected pregnant females were used as controls.
Study Design and Participants
A total of 600 pregnant women (noninfected and infected during pregnancy) of a Brazilian endemic region were enrolled through volunteer sampling, and followed until delivery, between January 2013 and April 2015. At the time of recruitment, socioeconomic and clinical data were collected, and peripheral blood was used to diagnose and/or confirm malaria infection. The women were followed by a trained nurse, which involved at least two domiciliary visits, in the second and third trimester, to monitor their clinical state, and collected a peripheral blood sample. For each episode of malaria, an additional blood sample was collected. At delivery, clinical data were collected from mother and newborn, as well as placental tissue and blood samples. All pregnant women who had malaria were treated with antimalarial drugs under medical prescription, according to the Brazilian Ministry of Health guidelines.
Women were excluded if they had a history during pregnancy of smoking and/or alcohol consumption, and who presented with infections (i.e. Toxoplasmosis, HIV, Hepatitis B and C virus, and Syphilis), and/or other comorbidities (i.e. hypertension, pre-eclampsia/eclampsia and diabetes mellitus), as well as women who had preterm delivery, stillbirth, and newborn with congenital malformation.
For this study, after applying the exclusion criteria, the selection of samples was only between uninfected pregnant women and pregnant women who presented P. falciparum infection during pregnancy. supplemental Fig. S1 illustrates the study design and patient selection used in this study.
Placenta and Blood Sample Collection and Storage
Immediately after removal/expulsion of the placenta, the maternal face of the placenta was identified, and one cotyledon was cut to collected placental blood sample into a tube containing heparin using vacuum tube for blood collection. Two maternal placental tissue samples of 2 cm3 were collected in two locations to one-third of the distance between the insertion of the cord and the edge of the placental disc. Subsequently, samples were stored in 10% buffered formalin and maintained at 4 °C for 24 h. After this time, the fragments were placed in another tube with 50 ml of clean buffered formalin to be sent to the laboratory for histological processing. In the same region of the cotyledons, one fragment of placental tissue of 1 cm3 was also collected, stored in RNAlater and maintained at −80 °C until analysis.
Screening of Malaria Infection and Sample Processing
All samples collected were screened for the presence of malaria parasites by microscopy and confirmed by real-time PCR (PET-PCR) (17). Peripheral and placental blood were collected in heparinized tubes, and then separated into plasma and whole blood cells by centrifugation. Total DNA was extracted from whole blood cells with a commercially available extraction kit (QIAmp DNA Mini Kit, Qiagen), according to manufacturer instructions. Thin and thick blood smears were Giemsa-stained.
Histopathology and Morphometric Assessment
Histopathological examination involved the use of placental tissue slides. Paraformaldehyde-fixed placentas were embedded in paraffin, sectioned at 5 μm, and mounted on silane-coated slides. These sections were dewaxed and rehydrated through descending concentrations of alcohol to distilled water, followed by Hematoxylin-Eosin (H&E) staining, and microscopical analysis using a Axio Imager M2 microscope coupled to a Axio Cam HRc camera (Carl Zeiss NTS GmBH, Oberkochen, BW, Germany). For the histological and morphometric analyses, the slides were coded and examined by two different observers in a blinded fashion.
The H&E-staining enabled assessing the syncytial nuclear aggregates, fibrinoid necrosis, and fibrin deposition (18). The presence of hemozoin was assessed through polarized light microscopy (19). The leukocyte (CD45), mononuclear inflammatory cells infiltrate (CD68), and the villous vascularity (CD31) were assessed by immunohistochemistry. The proliferation index was calculated through the quantitative image analysis of the anti-Ki-67/DAB staining (20).
Morphometric analyses of the mouse placentas were performed as previously described (21). In short, the vascular space was quantified by analyzing the hematoxylin-eosin (H&E)-stained sections of the placenta. For each section, three areas of the intervillous space were randomly selected for the image acquisition (200× magnification). The images were analyzed with the image processing and analysis program Image J (NIH, Bethesda, MD). Briefly, the images were subjected to an automated light analysis procedure in which noise removal was applied to ensure color and image quality standardization across the sections and specimens. The images were given a color threshold to cover the area corresponding to the blood space lumen. The percentage of coverage was calculated as the ratio between the number of pixels covered by the area defined by the threshold and the total number of pixels in the image. The blood vascular area in each placenta was assessed by the analysis of three nonconsecutive sections. The reported results correspond to single pregnant females, and they represent the average result from 3–9 placentas. Human barrier thickness was performed as previously described (18). Briefly, average distance between fetal vessel wall and villus outer membrane as measured by overlaying horizontal and vertical lines 4 mm apart on 5 random terminal villi at 63×. More details about the methods and staining used to quantify malaria-associated placental parameters can be found in supplemental Table S1.
Angiogenic Factors and Leptin Measurement
Angiopoietins 1 and 2 (ANG-1 and ANG-2), TEK receptor tyrosine kinase (TIE-2), vascular endothelial growth factor A (VEGFA), VEGFR1/FLT1, VEGFR2/FLK1 receptors, and the leptin hormone were measured in placental plasma (1:20 dilution for all factors) by using the DuoSet ELISA development kits (R&D), according to the directives provided by the manufacturer.
Measurement of Cytokines/Anaphylatoxins by Bead Array
The cytokines (IL-12p70, TNF, IL-10, IL-6, IL-1b, and IL-8), and complement proteins (C3a, C4a, and C5a) were measured in the placental plasma, using a CBA human inflammatory kit and CBA human anaphylatoxin kit (BD Biosciences), respectively, according to manufacturer protocol. Samples were analyzed in a two-laser BD FACSCalibur flow cytometer with a CellQuest version 5.2 software (BD Biosciences), and the concentrations were computed by using the FCAP array software version 3.0.1 (BD Biosciences).
Statistical Analysis of Clinical Data
Data were analyzed by using the Stata 14.2 (StataCorp) and GraphPad Prism 6.0 software. The variables were summarized by using the mean and S.D., medians and interquartile ranges (IQR), or by using frequencies and percentages. Differences in the mean values between groups were assessed with the Mann-Whitney U-tests accordingly. Categorical data and proportions were analyzed by using chi-square tests. All p values were 2-sided, with 0.05 significance level.
t test followed by Benjamini-Hochberg correction was applied to assess the differences in the mean values between groups for angiogenic factors, leptin, cytokines and anaphylotoxins. Corrected p value (q-value) <0.1 was considered as significant.
Ethical Considerations
Ethical clearance was provided by the research committee of the University of São Paulo (Plataforma Brasil, CAAE: 03930812.8.0000.5467), according to the Resolution no. 466/12 from the Brazilian National Health Committee. All the participants in the study provided a written informed consent.
Placenta Proteins Extraction
Pieces of mouse placentas were homogenized in liquid nitrogen by using mortar and pestle and resuspended in a PBS extraction buffer containing 8 m urea, 10 mm DTT and mixture inhibitor protease. Later, the human placenta tissues (∼50 mg), stored in RNAlater, were washed in 80% acetonitrile (cold) to remove the RNAlater and resuspended in an extraction buffer containing 6 m urea, 10 mm DTT extraction buffer containing 6 m urea, 10 mm DTT, 1 mm NaF, 1 mm Sodium orthovanadate and a mixture inhibitor protease.
Human placenta tissues were lysed through a 2-min shaking at 30Hz with a 5 mm stainless steel bead in a TissueLyser (Qiagen, Chadstone, VIC, Australia). Protein concentration was determined by Qubit fluorimetric detection method. Proteins were reduced by adding DTT to a final 10 mm concentration and 30-min incubation. Prior to the digestion by the addition of IAA to a final 40 mm concentration and the 30min incubation in the dark at room temperature, proteins were alkylated. To quench the reaction, DTT was added to a final concentration of 5 mm. Trypsin was added (1:50, w/w), and the mixture was incubated overnight at 37 °C. Resulting peptide mixtures were desalted with hydrophilic-lipophilic-balanced solid phase extraction (SPE) (Waters) and peptides eluted in 1 ml of 50% (v/v) acetonitrile (ACN), 1% (v/v) trifluoroacetic acid (TFA) and 70% ACN/1% TFA. Samples were dried in a vacuum concentrator and reconstituted in the appropriate buffer for glycopeptides and phosphopeptide enrichment steps.
TMT Labeling
A total of 100 μg of each placenta samples was diluted to a final concentration of 100 mm of TEAB labeled with TMT10plex Mass Tag labeling (Thermo), according to the instructions provided by the manufacturer. Tubes containing the different isobaric chemical tags (0.8 mg each) were added to 41 μl anhydrous acetonitrile at room temperature. Reagents were dissolved by 5 min, vortexing and solutions were gathered by centrifugation. Protein samples were labeled by adding 41 μl of TMT isobaric tag, followed by a 1h-incubation step at room temperature. To quench the reaction, 5% hydroxyl-amine (8 μl per sample) was added, followed by a 15min incubation at RT. Samples were finally pooled, divided into two aliquots (∼500 μg each) and purified on an Oligo R3 reversed phase micro-column. TMT-labeled samples were mixed in equal amounts and divided in two fractions for glycopeptides and phosphopeptides enrichment procedures.
Enrichment of Sialic Acid Containing Glycopeptides and Phosphopeptides by TiO2 Chromatography
One of the aliquots from the placenta TMT-labeled samples was submitted to a sialic acid enrichment containing glycopeptides by using TiO2 (22, 23). Samples were resuspended in 1 ml loading buffer (1 m glycolic acid, 80% ACN, 5% TFA), incubated with TiO2 beads (GL Sciences, Japan, 5 μm using a total of 0.6 mg TiO2 beads per each 100 μg peptides) and shaken at room temperature for 15min in batch format. Suspension was centrifuged at 1000 × g for 1min and supernatant loaded onto a second TiO2 batch (containing half the amount of TiO2 as used in the beginning); next, it was shaken at room temperature for 15 min. The two TiO2 batches were washed in 100 μl loading buffer and centrifuged at 1000 × g for 1 min. Supernatant was removed and beads were washed in 100 μl washing buffer 1 (80% ACN, 1% TFA) and centrifuged at 1000 × g for 1 min. Supernatant was removed and beads were washed in 100 μl washing buffer 2 (20% ACN, 0.2% TFA) and centrifuged at 1000 × g for 1 min. Supernatant was removed and beads were dried in a vacuum centrifuge for 5 min. Bound peptides were eluted in 100 μl of 1% ammonium hydroxide during 15 min, vortexed, and then centrifuged at 1000 × g for 1min. Eluted peptides were dried by vacuum centrifugation to produce the enriched sialylated glycopeptide and the phosphopeptide fraction.
Glycopeptide Enrichment by Using HILIC
The second placenta TMT-labeled sample aliquot was reconstituted in 100 μl 80% (v/v) acetonitrile and 1% (v/v) trifluoroacetic acid. Peptides were loaded onto an in-house PolyHYDROXYETHYL A™ HILIC resin (PolyLC Inc) packed onto a C8 disk (Empore) in a p200 pipette tip (24). The flow-through and washing fraction (80% (v/v) acetonitrile and 1% (v/v) trifluoroacetic acid) were collected. The enriched glycopeptides were eluted in 50 μl 0.1% (v/v) TFA followed by 50 μl of 25 mm NH4HCO3 and finally 50 μl of 50% (v/v) acetonitrile. The three eluted fractions were combined, dried by vacuum centrifugation and purified on an Oligo R3 reversed phase micro-column.
PNGase F deglycosylation
A TiO2 or HILIC-enriched glycopeptides (from protein trypsin digestion and endogenous glycopeptides) aliquot was resuspended in 50 mm TEAB, pH 8.0 and deglycosylated with 500U of PNGase F (New England Biolabs, Ipswich, MA) and 0.1U Sialidase A (Prozyme, Hayward, CA) for 12 h at 37 °C. After incubating, peptides were purified on an Oligo R3 reversed phase micro-column, dried by vacuum centrifugation and reconstituted in 50 μl of 0.1% formic acid prior to LC-MS/MS analysis.
Hydrophilic Liquid Interaction Chromatography (HILIC) Fractionation
The formerly N-linked sialylated glycopeptides and phosphopeptides enriched by TiO2 as well as the flow through (nonmodified peptides) were resuspended in 90% ACN, 0.1% TFA and injected onto an in-house packed TSKgel Amide-80 HILIC (Tosoh Bioscience, 5 μm) 320 μm × 170 mm μHPLC column by using the Agilent 1200 micro-HPLC instrument (22). The samples were soon suspended in solvent B (90% ACN, 0.1% TFA) by adding 10% TFA followed by water, and the acetonitrile was finally added slowly to the aqueous solution to prevent peptide precipitation by the high acetonitrile concentration. Peptides were loaded onto a 320 ID peak HILIC column and eluted in 6 μl/min by decreasing the solvent B concentration (100–60% ACN, 0.1% TFA in water) for 42 min. Fractions were automatically collected in a 96-well plate in 1min intervals after the UV detection at 210 nm, dried by vacuum centrifugation, and stored at −20 °C until performing the LC-MS/MS analysis. The fractions were combined up to a total of 8–16 fractions, according to the UV detection. All fractions were dried by vacuum centrifugation.
Mass Spectrometry Analysis
Label-free Mouse Placenta Samples
Peptides were separated by nano-LC-MS/MS on an in-house packed 17 cm × 100m Reprosil-Pur C18-AQ column (3 μm; Dr. Maisch GmbH, Germany) by using an Easy-LC nano-HPLC (Proxeon, Odense, Denmark). The HPLC gradient was 0–34% solvent B (A 0.1% formic acid; B 90% ACN, 0.1% formic acid) for 50min. at a 250 nL/min flow. Mass spectrometric analysis was performed by using a Thermo Orbitrap Fusion (Q-OTqIT, Thermo). Survey scans of peptide precursors from 400 to 1400 m/z were performed at 120K resolution (at 200 m/z) with a 5 × 105-ion count target. Tandem MS was performed by isolation at 1.6 Th with the quadrupole, HCD fragmentation with normalized collision energy of 38. The MS2 ion count target was set at 10e5, and the max injection time was 60 ms. The length of the dynamic exclusion was set to 10 s with a 10 ppm tolerance around the selected precursor and its isotopes. The instrument run at its maximum speed mode in 3s cycles. Two independent experiments were performed with two technical replicates.
Label-free and TMT-labeled Human Placenta Samples
The peptides (resuspended in 0.1% FA) were automatically injected and loaded on a ReproSil-Pur C18 AQ (Dr. Maisch, Ammerbuch-Entringen, Germany) in-house packed trap column (2 cm x 100 μm inner diameter; 5 μm). Peptides were separated at 250 nl/min on an analytical ReproSil-Pur C18 AQ (Dr. Maisch, Ammerbuch-Entringen, Germany) packed in house column (17 cm × 75 μm; 3 μm) by reversed phase chromatography, which was operated on an EASY-nanoLC system (Thermo Fisher Scientific, Odense, Denmark). Mobile phase was 95% ACN/0.1% FA (B) and water/0.1% FA (A). Gradient was from 3% to 28% solvent B in 52 min, 28–47% B in 5min, 45–100% B in 5min and 8min at 100% B. The nanoLC was connected online to a Q Exactive HF Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in positive ion mode and using data-dependent acquisition. The Orbitrap acquired the full MS scan with an automatic gain control (AGC) target value of 3 × 106 ions and a maximum filling time of 100ms. Each MS scan was acquired at 120,000 full width half maximum (FWHM) high-resolution at m/z 200 in the Orbitrap with 400–1600 Da mass range. The 10 most abundant peptide ions were selected from MS for higher energy collision-induced dissociation (HCD) fragmentation (collision energy: 29V) in case they were at least doubly charged. Fragmentation was performed at high resolution (60,000 FWHM) for a 1 × 105 target and 200 ms maximum injection time by using an isolation a 1.2 m/z window and a 30 s. dynamic exclusion. Raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride) partner repository with the data set identifier PXD008079.
Protein Identification and Quantification
Raw files were imported into a MaxQuant version 1.5.3.30 for the protein identification and quantification (25). For protein identification in MaxQuant, the database search engine Andromeda (26) was used to search for the MS/MS spectra against a database composed by the Uniprot Mouse Protein Database (April 15, 2015 release; 45,185 entries) and Uniprot Plasmodium berghei (strain Anka) Protein Database (April 13, 2015 release; 11,656 entries) or Reviewed Uniprot Human Protein Database (April 15, 2015 release; 45,185 entries) with a 4.5 ppm tolerance level for MS, and 20ppm for MS/MS. Enzyme specificity was set to fully specific trypsin with maximum two missed cleavages. Carbamidomethylation of cysteine (57.021464 Da) was set as fixed modification, and the oxidation of methionine (15.994915 Da) and N-terminal acetylation protein (42.010565 Da), deamidation (0.984015 Da) and phosphorylation at S/T/Y (79.966330) were selected as variable modifications. The minimum peptide length was set to 7 amino acids. For label-free quantification, it was used the 'match between runs' feature of the MaxQuant that enables to identify the transfer between samples based on the accurate mass and retention time applied with a 0.7 min match time window and 20 min alignment time window. For the TMT-labeling quantification, the ion MS2: 10plex TMT reporter was included in the search parameters. All identifications were filtered to achieve a 1% protein-false discovery rate (FDR).
Label-free protein quantification was based on the maxquant label-free algorithm by using both unique and razor peptides for the protein quantification; at least 2 ratio counts were required for a protein quantification to be considered valid. Protein abundance was calculated based on the normalized spectral protein intensity (LFQ intensity) (27). Statistical analyses of the proteome data were performed by using the Perseus v.1.5.4.1 available in the MaxQuant environment. First, proteins identified in the reverse database, potential contaminants and proteins only identified per site were excluded for further analysis. LFQ intensities were log2 transformed and the mean two technical replicates values for each independent experiment was calculated. The t test analysis was applied both in the past infected and noninfected groups with p value set to p < 0.05. For the TMT labeled samples, the reporter ion intensities from redundant peptides were summed. Quantitation of each identified formerly N-glycopeptide, phosphopeptide or nonmodified peptides was determined by the normalized reporter ion intensities by log2 and then subtracting them by the log2 mean of each sample. Student t test with p < 0.05 was applied to identify the regulated glycopeptides or nonmodified peptides before further bioinformatics analyses. Annotated MS/MS spectra can be found in MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the following search keys: ipfwhr5osl, hwm0mqbx2s, dkscsltllo, jwreohtivy, rnb08ldiee. Details about each data set can be found in supplemental Table S4, S5, and S7–S9.
Bioinformatics Analysis
Protein-protein interaction networks were construed by using the STRING software, which were visualized and edited with the Cytoscape 2.8.2 (21). The Enrichr software was used to perform the Kinase Enrichment Analysis (KEA), which is a gene-set library of the human or mouse kinases and their known substrates collected from literature reports provided by six kinase-substrate databases: HPRD, PhosphoSite, PhosphoPoint, Phospho.Elm NetworKIN, and MINT. The Kinase Enrichment Analysis (http://www.maayanlab.net/KEA2/) was also performed by using the regulated phosphosites with site score probability >0.9.
Phosphorylation motif analyses were performed with the web-based motif-x tool (http://motif-x.med.harvard.edu/) (28). Prealigned sequences of peptides ±6 residues around the serine phosphorylation-sites identified were analyzed against the IPI mouse database as background with the “occurrence” parameter at 20 and “significance” parameter at 0.000001.
Functional annotation enrichment analysis of regulated glycopeptides and nonmodified peptides were performed by using the DAVID Gene Functional Classification Tool (18) with significant enrichment set as p < 0.05.
Western Blot Analysis
The lysis and protein extraction of the placenta tissue that generated the lysate used for the Western blot analysis are described in the “Placenta proteins extraction” section. Lysate of human placenta containing 50 μg of proteins were separated by discontinuous electrophoresis (4% polyacrylamide stacking gel and 12% polyacrylamide resolving gel). Proteins were transferred to PVDF membranes, incubated with a solution of Blocking Buffer (Thermo Fisher Scientific, Waltham, MA USA) and BSA 5% 1:1, and then incubated with the following antibodies: p-AKT (S473, Cell Sig., 1:500), AKT (Cell Sig, 1:500), p-ERK (Y204, Santa Cruz, 1:1000), ERK1/1 (Cell Sig., 1:1000), p-S6 (S235/236, Cell Sig, 1:1000), S6 (Cell Sig, 1:1000), p-4EBP1 (S65, Cell Sig, 1:1000), 4EBP1 (Cell Sig, 1:1000), cleaved caspase-3 (Cell Sig, 1:1000), BAX (Cell Sig, 1:1000), BCL2 (Cell Sig, 1:500), HSPB1 (Cell Sig, 1:1000), p-HSBP1(S15 and S82 Cell Sig, 1:1000) and TUB (Sigma-Aldrich, 1:10.000) at 4 °C overnight. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (Vector Laboratories, Burlingame, CA). Image acquisition was performed by using the Alliance chemiluminescence detection system (UVITEC, Cambridge, UK). Quantitative densitometry was carried out using the ImageJ software (National Institute of Health [NIH]). The volume density of the chemiluminescent bands was calculated as integrated optical density×mm2 after background correction. The subsequent densitometry analysis of bands was performed through the Image J Software or Image Lab. Phosphorylation levels were calculated by normalizing the intensity of the phosphorylated by the nonphosphorylated form. Each ratio was normalized by the mean of noninfected ratios, generating the “folds versus noninfected” values which were used for statistical comparison. All the results were analyzed for Gaussian distribution and passed the normality test. The statistical differences between group means were tested by unpaired two-tailed t test. A value of p < 0.05 was considered as statistically significant. For each type of tissue -infected or noninfected- n = 5 independent experiments were performed in duplicate.
Evaluation of Apoptosis
Apoptosis was assessed using paraffin blocks of tissue arrays sectioned at 5 μm intervals, mounted on slides and processed for immunohistochemical staining. DNA fragmentation was evaluated using DeadEnd™ Colorimetric TUNEL (Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling) System (Promega, Wisconsin) according to manufacturer's instructions. A placenta section was used as negative control by suppression of enzyme in reaction. Data are presented as fold-change in relation to the mean of the control group.
Quantitative Analysis of Immunoreactivity
The immunoreactivity images were acquired using an Imager.D2 Optical Microscope equipped with Axiocam Color and ZEN software (Zeiss Group, Baden-Württemberg, DE). Quantification was performed on the villous area from 24 past-infected and 32 noninfected placentas using a 40X objective. Three images from each placenta were selected and analyzed through the ImageJ program (National Institutes of Health, Bethesda, MD). Briefly, the density of DAB (3, 3-diaminobenzidine) staining and the tissue area in each field were measured using the color deconvolution plugin. The immunoreactivity results were normalized by the tissue area present in the respective field, and then divided by the mean value obtained in the controls.
Experimental Design and Statistical Rationale
For the label-free experiments depicted in Fig. 2, four mice (two infected and two un-infected) and 10 human samples (5 controls and 5 past-infected) were run. Each replicate represents a biological replicate. The statistical significance is reported in supplemental Tables S4 and S5. The TMT-based isobaric labeling quantification was performed with five biological replicates for the past-infected and five for the noninfected human placentas. The statistical significance is reported in supplemental Tables S7, S8, and S9. Western blotting orthogonal confirmation was performed on five biological replicates for the past-infected and five for the noninfected human placentas as depicted in Figs. 6, 7, and supplemental Fig. S7. Validation of the apoptosis-induced mechanism was performed on 24 past-infected and 32 noninfected human placentas considered as biological replicates, Fig. 7.
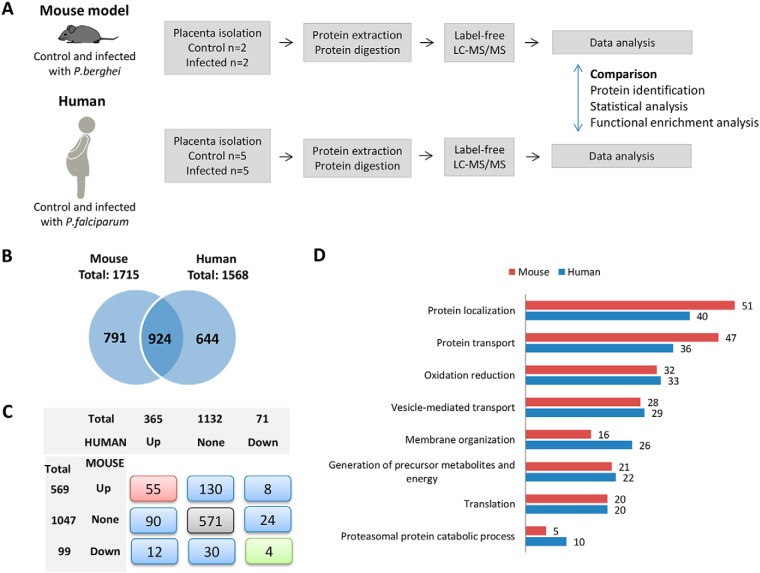
Fig. 2.
Comparative label-free proteomics in mouse and human infected and noninfected placentas disclosed shared biological processes associated with placental malaria. A, Translational-comparative approach workflow for label-free mass spectrometry-based proteomics in two malaria-infected placenta models: human and mouse. B, Distribution of proteins identified in placentas isolated from mouse (control and P. berghei-infected groups) and human (control and P. falciparum-infected groups). C, Distribution and overlap of regulated and nonregulated protein coding genes identified in placentas isolated from mouse and human. D, Common enriched biological processes based on gene ontology biological process terms in mouse and human infected placentas followed by the number of up-regulated proteins identified by LC-MSMS related to each process (Benjamini, p < 0.05).
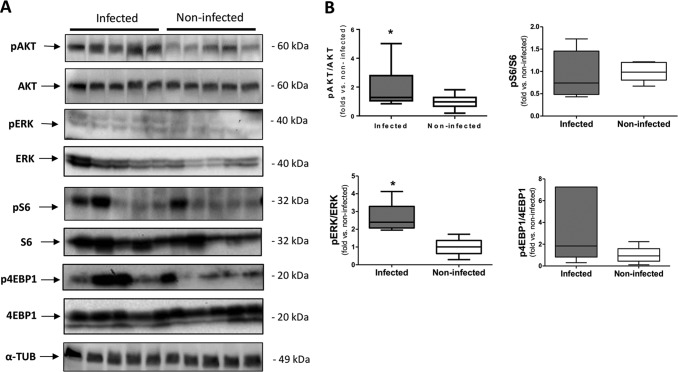
Fig. 6.
Immunoblotting validation of the predicted-activated pathways in malaria past-infected placentas. A, Representative western blots of phospho: total expression levels of AKT (S473), ERK(Y204), S6 (S235/236) and 4EBP1 (S65). α-tubulin was used as loading control. The same cohort of samples from the mass spectrometry analysis was used for this assay. B, Densitometry analysis showed that the phosphorylation levels of AKT (S473) and ERK (Y204) were significantly higher in the past-infected placentas compared with noninfected placentas (n = 5 infected and 5 noninfected, Student's t test *p < 0.05).
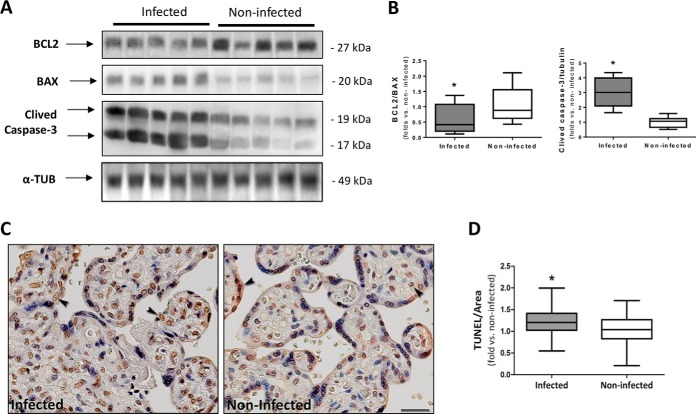
Fig. 7.
Higher apoptosis induction in past P. falciparum-infected placentas. A, Western blot analysis of BCL2, BAX, and Caspase-3 levels in past infected and noninfected placentas. α-tubulin was used as loading control and for normalization. The same cohort of sample from the mass spectrometry analysis was used for this assay. B, The BCL2/Bax and Caspase-3/Tubulin ratios were calculated by densitometry analysis (n = 5 infected and 5 noninfected, Student t test *p < 0.05). C, Apoptotic nuclei (TUNEL stain brown) in past-infected and noninfected placentas. Arrowheads show positive nuclei examples. The images in each panel were acquired at 400× magnification and the scale bar represents 25 μm. Mayer's hematoxylin counterstaining. Independent cohort of samples was used for this assay. D, Quantification of TUNEL immunostaining intensity (pixels/μm2) per tissue area is represented by number of fold-increase to noninfected mean (n = 24 infected and 32 noninfected placentas, Mann Whitney test *p < 0.05).
RESULTS
Samples and Characteristics of the Study
This study was performed with a cohort of ten placentas from which five were obtained from women with no recorded evidence of any Plasmodium infection during pregnancy (noninfected), and five from women with microscopically and molecularly (RT-PCR) documented P. falciparum during pregnancy (past infected). The details of the study design and patient selection is shown in supplemental Fig. S1. Table I shows the gestational week when the mother was infected and developed malaria as well as the level of parasitemia. At delivery, parasitemia was negative in all infected mothers. Hemozoin was observed in three out of five infected placentas (Table I).
Table I. Human placenta samples used in this study and time and parasitemia of infection during pregnancy.
| Samples | 1st infection | 2nd infection | 3rd infection | RT-PCR for P. falciparum | Hemozoin in placental tissue |
|---|---|---|---|---|---|
| 7 | 24 (37733.7) | - | - | Positive | No |
| 17 | 29 (2698.1) | 33 (6.3) | - | Positive | Yes |
| 124 | 14 (1754.0) | 29 (10340.0) | 32 (46.6) | Positive | Yes |
| 289 | 27 (4503.0) | - | - | Positive | Yes |
| 417 | 25 (1499.0) | - | - | Positive | No |
| 194 | - | - | - | Negative | No |
| 301 | - | - | - | Negative | No |
| 459 | - | - | - | Negative | No |
| 476 | - | - | - | Negative | No |
| 484 | - | - | - | Negative | No |
Time of infection expressed in weeks. Values of parasitemia presented in 103 DNA copies, obtained by PET-PCR quantification.
No significant differences as to the characteristics of the mothers: maternal age, one pregnancy (primigravidae), maternal weight gain, % hematocrit, hemoglobin (g/dl), and placental weight were observed (p > 0.05) The newborn outcome had significant lower Rohrer index (p = 0.028), head circumference (p = 0.008), and chest circumference (p = 0.011) in the past malaria-infected group compared with the noninfected group supplemental Table S2).
The placentas of women who experienced a past P. falciparum infection during pregnancy had a significant increase in sVEGFR-2 and Tie2 levels, and a decrease in the sFlt-1 and leptin levels (t test followed by Benjamini Hochberg, q<0.1 supplemental Table S3). There was no significant difference between both groups of women as to the placental histopathology parameters (syncytial nuclear aggregates, fibrin deposition score, fibrinoid necrosis, proliferation index, leukocytes infiltrated (CD45+) and mononuclear cells infiltrated (CD68+)) (supplemental Table S3). Similarly, placental plasma levels of TNFα, IL1B, IL6, IL8, IL10, IL12 cytokines and C3a, C4a and C5a anaphylotoxins did not vary between women with past Plasmodium falciparum infection during pregnancy and those without (supplemental Table S3).
Comparative Proteomics Between Mouse and Human Models for Placental Malaria Discloses Shared Biological Processes Occurring in Infected Placentas
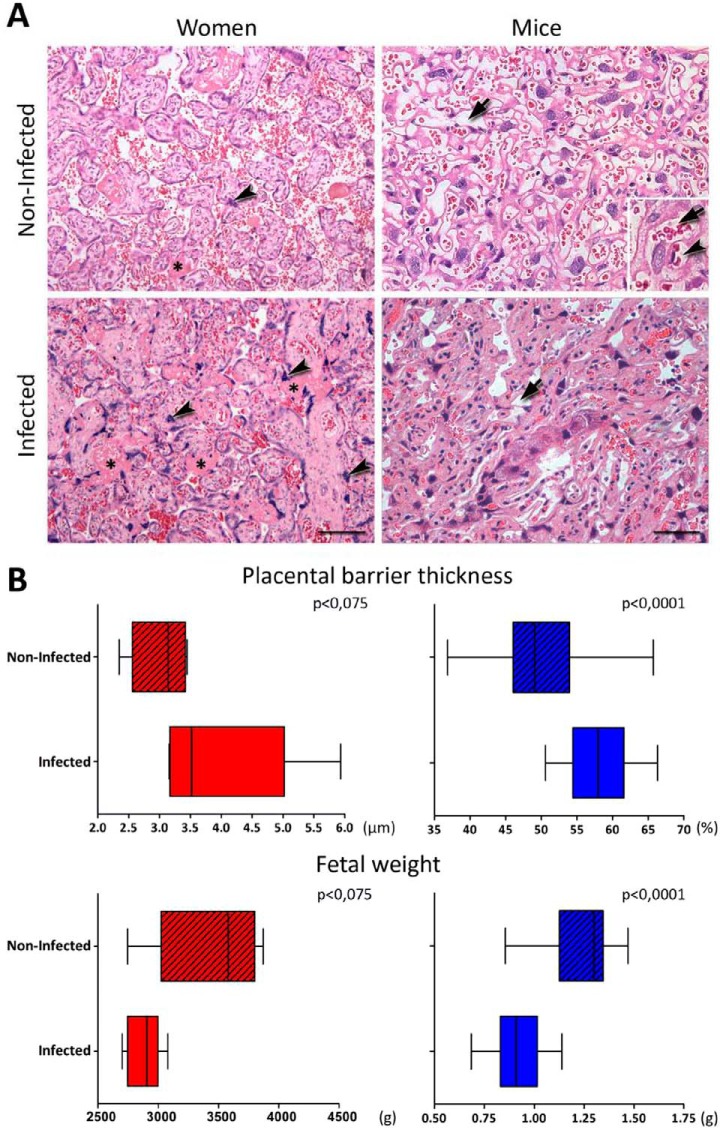
In the first stage of our approach, we aimed to identify the overall biological process changes occurring in malaria-infected placentas isolated from mice and human. In both species, the placental tissue of infected pregnant revealed some abnormalities compared with noninfected. We observed extensive areas with fibrin deposits and syncytial knots proliferations, besides an increase of the placental barrier thickening and disorganization of the labyrinthine zone (Fig. 1A). The infection also contributes considerably to low birth weight, which is seen in both humans and mice (Fig. 1B). Subsequently, label-free proteomic analyses were applied in two biological models both from human (n = 5 past-infected, n = 5 noninfected) and mouse malaria-infected placentas (n = 2 infected, n = 2 noninfected). At delivery, a biopsy of the placenta was isolated from both models with the respective control. The proteins were extracted and digested with trypsin and the resulting peptides were submitted to LC-MS/MS analysis. After protein identification and quantification, a comparison was performed in the placentas from mice actively infected with P. berghei and noninfected and in human past infected with P. falciparum and noninfected (Fig. 2A).
Fig. 1.
Features of placental malaria (PM) in women and mice. A, HE-stained placentas from women and mice showing differences in the thickness of the placental barrier. In human placentas images, the asterisks point to fibrin deposits and the arrowheads indicates presence of syncytial knots, which are more frequent in the presence of the infection. In the images of mice placental the arrows highlight the vascular spaces of the labyrinth region. The insert shows the placental barrier, where the maternal vascular space (arrow) and the fetal capillary light (arrowhead) are highlighted. B, Graphic representations of placental thickness (n = 5 infected and 5 non-infected human and n = 5 infected and 5 non-infected mouse) and fetal weight (n = 5 infected and 5 non-infected human and n = 5 infected and 5 non-infected mouse) of infected and non-infected from human (red) and mouse (blue) species evidencing a decrease in both infected groups. The human placental barrier thickness was quantified by the average distance between the fetal vessel wall and villus outer membrane, and the mice placental area occupied by blood sinusoids was quantified in relation to the total placental area. Both procedures are described in Materials and Methods. Scale bar represents 100 μm (Women) and 50 μm (Mice). Mann Whitney Test was used.
A total of 1715 and 1568 proteins were identified with less than 1% FDR in the placenta tissue from pregnant mice and human, respectively, with an overlap of ∼40% (924/2359 proteins) (Fig. 2B and supplemental Table S4 and S5). The Pearson correlation coefficient between the two data sets was 0.643 (supplemental Fig. S2A). Similar dynamic range was observed between the two proteomes, as shown by scatter plot graph in supplemental Fig. S2B. We also compared the distribution of abundance of protein by using the common or exclusive proteins identified in mouse and/or human data sets. The distribution of proteins identified in common between the two data sets is represented by a decreased frequency from high to less abundant proteins, whereas the distribution of exclusive proteins identified in each data set is represented by an increased frequency from high to less abundant proteins (supplemental Fig. S2C).
Next, differentially regulated proteins between infected and noninfected placentas were determined by applying the t test analysis in the log2-transformed LFQ intensities separately in data sets from mouse or human. We also considered regulated proteins those identified exclusively in each condition (infected or noninfected) (supplemental Table S4 and S5). Fig. 2C shows the distribution and overlapping of significantly regulated proteins (up or down) or nonregulated proteins upon infection in mice and human placenta data sets. Most of the proteins showed the same direction of regulation both in mice and in human data sets (55 up-regulated, 571 nonregulated and 4 down regulated) and 20 proteins showed opposite directions.
Differentially regulated proteins identified in infected and noninfected placentas were analyzed as to over-represented biological process by using the gene ontology (GO) terms. We observed shared significant enriched process associated to up-regulated proteins identified both in mice and human infected placentas (supplemental Table S6) and the top 8 most commonly significant enriched biological processes are shown in Fig. 2D. Significant altered processes (p < 0.05) were observed especially related to the protein metabolism (protein localization, transport and proteasomal protein catabolic process) and energy (generation of precursor metabolites and energy), which involved from 5 to 51 up-regulated proteins identified in infected human or mouse placentas (Fig. 2D). We also performed the same enrichment analysis by using down-regulated proteins identified in infected mouse and human compared with noninfected placentas, but only intracellular transport was retrieved as a shared enriched biological process from down-regulated proteins in the mouse or human data sets (supplemental Table S6).
Integrative Glycoproteomic and Phosphoproteomic Analysis Reveals Altered Pathways Associated with Past-Stage Placental Malaria
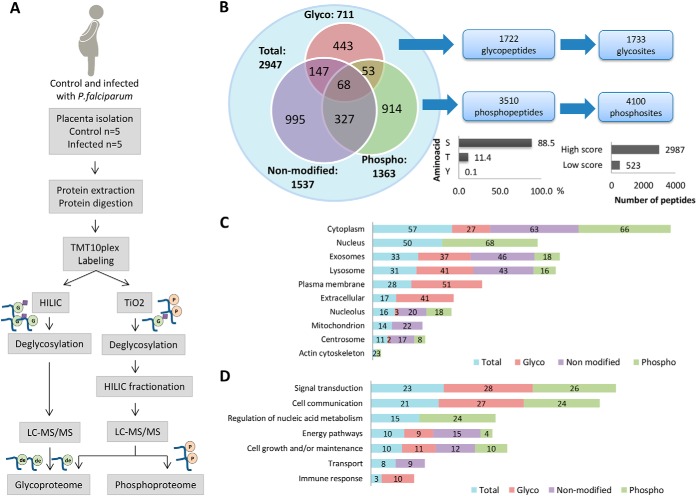
In the second stage of our approach, we aimed to characterize the altered pathways occurring in past P. falciparum-infected human placentas. In this regard, isobaric labeling-based quantitative proteomics combined to glycopeptide and phosphopeptide enrichment, using HILIC and/or TiO2, were performed to evaluate post-translational modifications, as well as protein expression in past-infected and noninfected placentas (Fig. 3A).
Fig. 3.
Integrated analyses of proteome, phosphoproteome and glycoproteome using isobaric labeling strategy in past P. falciparum infected and noninfected human placentas. A, Proteomics workflow for enrichment, identification, and quantification of phosphopeptides and glycopeptides in past P. falciparum infected and noninfected human placentas. B, Overall distribution of proteins identified in the proteome, phosphoproteome and glycoproteome fractions and total number of glycopeptides/glycosites and phosphopeptides/phosphosites identified by LC-MS/MS. The bar graphs represent % phosphosites in S, T or Y (left) and the number of phosphosites with high score (>0.99) and low score (<0.99). C, Percentages of proteins identified in the proteome, phosphoproteome and glycoproteome located in the indicated ten major organelles based on their gene ontology cellular components D, and seven major gene ontology biological processes.
By using such approach, we were able to identify 2946 proteins encompassing 1537 proteins, 711 glycoproteins and 1363 phosphoproteins, altogether reporting the largest protein data set so far reported in human placenta (Fig. 3B and supplemental Table S7–S9). By combining the glycopeptides identified using HILIC and TiO2, we attained 1733 glycosites from 1722 unique glycopeptides (Fig. 3B and supplemental Table S8 and S9) with a 41% overlapping and 53% in the glycopeptides and glycoprotein level, between both enrichment strategies, respectively (supplemental Fig. 3A and 3B). Furthermore, a total of 4100 phosphosites from 3510 unique phosphopeptides were identified, which were mainly localized at serine (88.5%), followed by threonine (11.4%) and tyrosine (0.1%) (Fig. 3B). We also grouped the phosphopeptides according to the localization score probability in high (≥0.99) and low score (<0.99) attaining a total amount of 2987 (73%) and 523 (27%) of high and low localization probability score peptides, respectively (Fig. 3B and supplemental Table S8). Reproducibility among biological replicates was assessed through the Pearson correlation analysis for the nonmodified, TiO2 and HILIC fractions. High correlation coefficient among biological replicates was observed, especially for the nonmodified fraction, where both in past infected and noninfected group the Pearson correlation was >0.94 in every comparison. As to the HILIC fraction, the correlation among biological replicates was >0.93 in the noninfected group, but an increase was observed in the past infected group, where some comparisons showed a <0.9 coefficient. The same was observed in TiO2 fraction, in which the past infected and noninfected groups presented a coefficient ranging from 0.86 to 0.95 (supplemental Fig. S4).
Furthermore, the distribution of proteins identified in each data set (glycoproteome, phosphoproteome and proteome) was compared with the cellular location and biological processes. We observed that glycoproteins are mainly located in the plasma membrane and extracellular component, whereas the phosphoproteins appear mainly in the cytoplasm and nucleus (Fig. 3C). Although glycoproteins and phosphoproteins had different cellular location, they shared common biological processes such as signal transduction and cell communication (Fig. 3D), which were not enriched in the nonmodified peptide data set. We also observed that specific biological processes were enriched in the phosphoprotome and glycoproteome data sets as for instance the “nucleic acid metabolism regulation” and the “immune response,” respectively. Moreover, the proteins identified in the nonmodified data set were particularly enriched for the “transport” process (Fig. 3D).
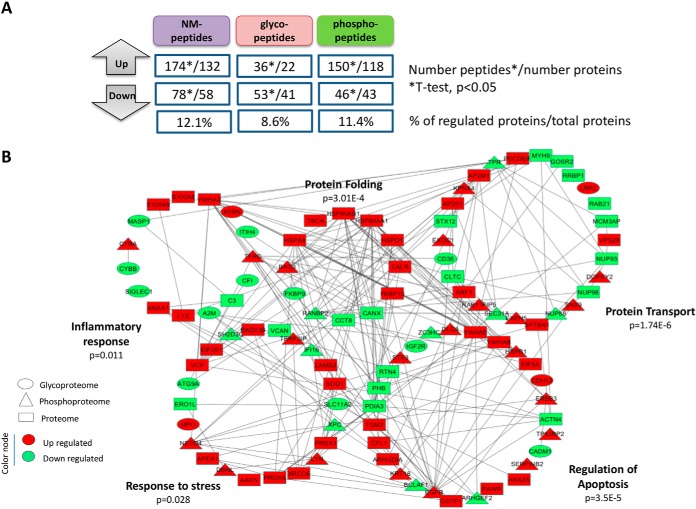
To assess specific changes in the glycopeptides, phosphopeptides and nonmodified peptides levels, a t test analysis was performed in the normalized reporter ion intensities from past-infected and noninfected conditions (supplemental Table S7–S9). For the glycopeptide data set, we filtered only the peptides bearing the deamidation site on the N-linked glycosylation motif, NXS/T/C (where X is not proline) (supplemental Table S8 and S9). Fig. 4A and supplemental Table S10 show the number of significantly regulated nonmodified-, glyco-, phospho-peptides in past-infected compared with noninfected placentas (t test, p < 0.05), together with the number of proteins that carried those regulated peptides. The percentage of regulation related to the total number of proteins identified in each data set is also shown in Fig. 4A. In addition, heatmaps and principal component analysis (PCA) were performed to visualize protein expression patterns and sample clusterization by using the differentially regulated nonmodified/glyco/phospho-peptides (supplemental Fig. S5). Interestingly, when nonmodified peptides or glycopeptides could separate between past-infected and noninfected groups (supplemental Fig. S5). However, when using phosphopeptides, an overlap between past-infected and noninfected groups was observed, indicating that phosphorylation events is likely more affected by intra-individual variability (supplemental Fig. S5). We combined the list of regulated proteins identified in all data sets and performed a protein-protein network analysis in order to visualize the interactions between the differentially regulated proteins in an overrepresented biological process context (Fig. 4B).
Fig. 4.
Analysis of differentially regulated nonmodified peptides, glycopeptides and phosphopeptides using the normalized TMT report ion intensities from past P. falciparum infected and noninfected placentas. A, Overall distribution of the number of differentially (up and down) nonmodified peptides/proteins, glycopeptides/glycoproteins and phosphopeptides/phosphoproteins in past malaria-infected compared with noninfected placentas (n = 5 infected and 5 noninfected, Student's t test, p < 0.05). The % of regulated proteins/glycoproteins/phosphoproteins is displayed relatively to the total number of proteins/glycoproteins/phosphoproteins identified in each data set. B, Network models delineating complementary coverage of protein networks provided by the integration of regulated proteins identified in the proteome, glycoproteome and phosphoproteome fractions.
The list of differentially regulated proteins was submitted to functional enrichment analysis by using biological process gene ontology (GO) terms. The complete list of enriched biological process GO terms and pathways is available in the supplemental Table S11. We selected five significant enriched biological processes that could be involved in placental malaria to be highlighted in the network: protein transport, regulation of apoptosis, protein folding, inflammatory response and cellular response to the stress encompassing the 41, 39, 14, 26, and 22 regulated proteins, respectively. We also detailed the level of regulation (glycosylation, phosphorylation or nonmodified) and the direction of the regulation (up and down upon P. falciparum infection) within each pathway. The final network shown in Fig. 4B demonstrates that altered biological processes may result from different regulating levels not only in the protein expression level, but also at PTM level.
Phosphoproteomics Discloses Altered Pathways in Past P. falciparum-infected Placentas
From the phosphoproteome data set, 111 phosphoproteins were recorded as “kinases” and 36 as “phosphatase” by using the gene ontology (GO) terms for molecular function (supplemental Fig. S6A). We also identified 26 and 27 phosphoproteins recorded as protein kinases and phosphatases, respectively in the proteome data set. Considering the total of phosphatase/kinase identified in the phosphoproteome and proteome data set, 146/52 was attained, having 25 shared in both data sets (supplemental Fig. S6A). Among the identified kinases/phosphatases, 17 were regulated at phosphorylation level (PAK1, ALPPL2, DUSP3, STK3, PRPF4B, LYN, MARK2/MARK4/MARK1/MARK3, IGF2R, EGFR, CAMK2G, ERBB3, NRK, PRKCD, AAK1, MARK2, MTMR3 and MARK3) and 7 were regulated at the proteome level (CAMK2G, CPPED1, MYLK, HSP90AA1, CAB39, HSP90AB1, ALPP).
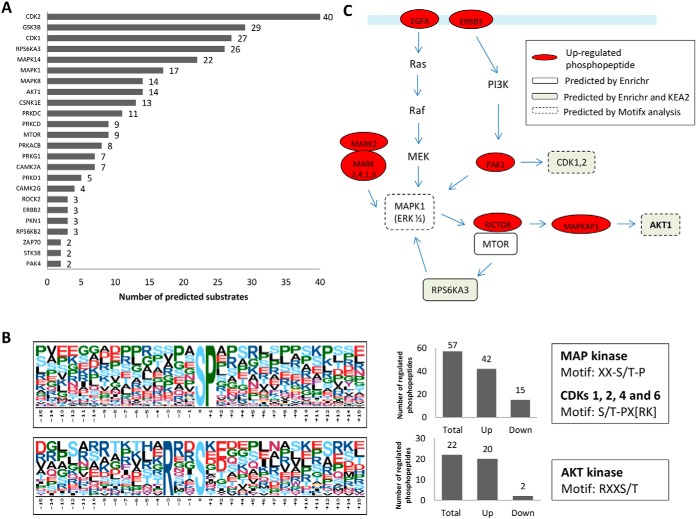
To explore the kinome involved in the differentially regulated phosphopeptides, a kinase enrichment analysis was performed by using the Enrichr software. A significant overrepresentation of substrates for 24 kinases was observed and represented together with the number of phosphoproteins identified by LC-MS/MS related to each kinase in Fig. 5A. In addition, protein-protein interaction analysis showed that 21 out of 24 predicted kinases are linked in the same network (supplemental Fig. S6B). From the 24 significant enriched kinases (adjusted p value <0.05), 12 were identified by LC-MS/MS in the phosphoproteome (MTOR, CSNK1E, PAK4, AKT1, GSK3B, ROCK2, PRKCD, CAMK2G, RPS6KA3, PRKDC, ERBB2) and/or proteome (ROCK2, CAMK2G, CDK2, PRKDC) data sets, but none of them were differentially regulated between past infected and noninfected placentas. We performed an additional analysis by using the KEA2 software from an input set of regulated phosphosites identified with the site probability score >0.9. From the input list, 21 phosphosites had a kinase matching, and from them, 11 were shared with the Enrichr prediction kinase (CAMK2A, MAPK14, CDK1, CSNK1E, PRKCD, PRKDC RPS6KA3, MAPK8, AKT1, GSK3B and CDK2). Additionally, the overrepresented motif analysis showed two enriched phosphorylation motifs “SP” and “RXXS”, which were previously described as recognition motifs for CDKs 1, 2, 4 and 6 Kinase (29, 30), MAP kinase (31) and AKT Kinase (32), thus confirming the kinome data (Fig. 5B). We combined the results from predicted kinases with the significant regulated phosphopeptides identified by the LC-MSMS to create a mechanistic model of altered pathways and biological process occurring in placental malaria (Fig. 5C).
Fig. 5.
Insights into activated pathways and kinases provided by the phosphoproteomics data in past malaria-infected placentas. A, Kinase/substrate prediction analysis using the KEA algorithm within the Enrichr software. The graph shows significant enriched kinases (p < 0.05) and the number of substrates related to each kinase which were identified by LC-MS/MS with significant changes in phosphorylation levels. B, Phosphorylation motifs serine-phosphorylation sites identified by LC-MS/MS were extracted and pre-aligned sequences of peptides ±15 residues around the identified phosphorylation sites were analyzed against the IPI human database as background with the web-based motif-x tool with default settings. The numbers of sequences with the respective phosphorylation motifs as well as the direction of the regulation are shown in the bar graphs (right). C, Proposed model integrating predicted kinases, motif-x analysis and regulated phosphopeptides. White and gray rectangular marked proteins are kinases predicted by Enrichr or Enrichr/KEA2, respectively, and the red circled marked proteins were identified by LC-MSMS with at least one up-regulated phosphopeptide.
Up-regulated phosphopeptides from EGFR, ERBB4, MARK2, MARK2,4,1,3 and PAK1 identified in the phosphoproteomics data set may be linked to the mTOR pathway via activation of ERK1/2 (MAPK1), which was predicted by Enrichr and motif analysis. We also identified an up-regulation of RICTOR phosphopeptide, which is a subunit of mTORC2, a protein complex nucleated by the quinase mTOR that phosphorylates AKT1 at Ser473 enhancing its kinase activity, such a response also predicted by KEA2, Enrichr and motif analysis (Fig. 5C).
P. falciparum Induces Phosphorylation-driven Activation of AKT and ERK1/2 in Past Infected Placentas
With the aim to confirm the predicted AKT and ERK activation, we measured the phosphorylated contents of AKT (S473) and ERK1/2 (Y204) in protein extracts from both past P. falciparum infected and noninfected placentas. A significant increase in the phosphorylated content of AKT (S473) and ERK1/2 (Y204) (Y705) was observed in past-infected placentas (Fig. 6).
We also evaluated whether the activity of mTORC1 was also increased in past-infected placentas as a result from the activation of its upstream inducer AKT by measuring the phosphorylated content of several mTORC1 downstream substrates. As illustrated in Fig. 6, no significant difference was observed in the phosphorylation levels of S6 (S235/236) and 4EBP1 (S65) in past-infected placentas compared with noninfected ones.
Past-P. falciparum Infected Placentas Showed Increased Apoptosis Compared with Noninfected Placentas
Based on the over-represented “regulation of apoptosis” and “response to stress”, biological processes that were highlighted in the protein-protein network (Fig. 4B), we assessed placenta apoptotic signaling pathway. A significant impairment of the BCL2/BAX ratio, an elevation of cleaved-caspase-3 levels (Fig. 7A and 7B) and an increase of TUNEL staining (Fig. 7C and 7D) were observed in the past-infected placentas compared with noninfected ones. These results indicate that the apoptosis pathway is triggered by a past P. falciparum infection (Fig. 7).
Moreover, up-regulation of HSPB1 phosphorylation was also identified in the past-infected placentas and within “regulation of apoptosis” network (Fig. 4B and supplemental Table S10). Because HSPB1 function has previously been associated to expression and phosphorylation at the S15 and S82 sites (33, 34), the HSPB1 expression and phosphorylation levels in past-infected and noninfected placentas were evaluated. The results evidenced that past-infected placentas present a decrease in the HSPB1 protein expression levels and a slight increase in the phosphorylation level at the HSPB1 S82 (supplemental Fig. S7).
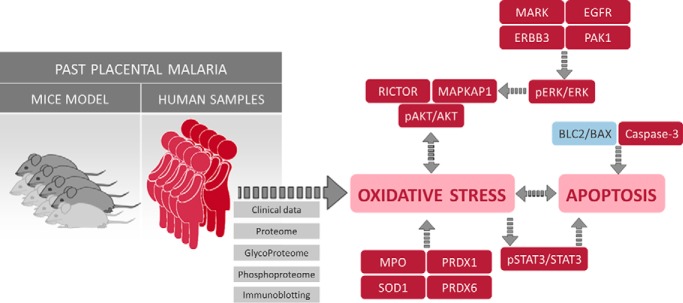
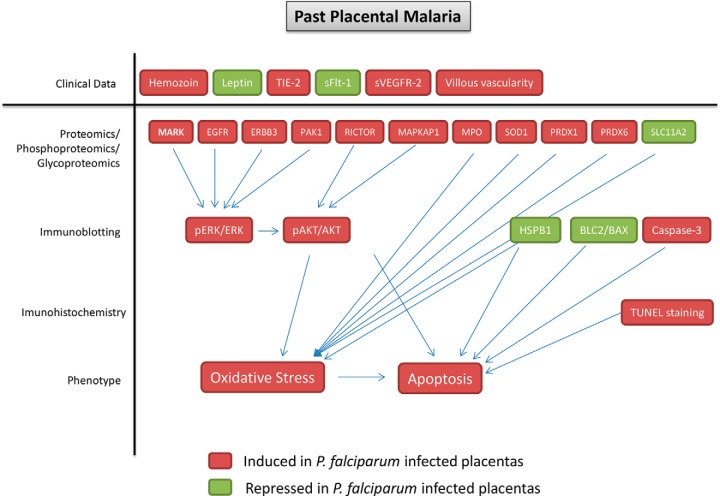
Additionally, a schematic visualization was supplied that summarizes the findings of this study (Fig. 8). The results from proteomics, phosphoproteomics and glycoproteomics were integrated with clinical data, immunoblotting and immunohistochemistry assay to show that placentas subjected to past P. falciparum infection present late consequences characterized by activation of ERK/AKT pathways and induction of oxidative stress and apoptosis.
Fig. 8.
Summary of the findings provided by clinical data, proteomics/phosphoproteomics/glycoproteomics data, immunoblotting and immunohistochemistry showing oxidative stress and apoptosis as biological events related to past placental malaria.
DISCUSSION
Placenta is a unique transitional tissue during pregnancy supporting the fetus nutritionally and metabolically (35). Therefore, disturbances on the maternal-fetal interface may alter the growth and development of the embryo in utero causing adverse outcomes in the newborn (36).
P. falciparum-infected placentas are characterized by accumulation of maternal immune cells (predominantly monocytes/macrophages and neutrophils) and increased levels of inflammatory cytokines (4). It has been proposed that intrauterine growth restriction and low birth weight result from an impaired maternal-fetal circulation, because of placental abnormalities caused by the infection, including deposition of hemozoin, placental sequestration of parasitized erythrocytes and increased amount of monocytes in the intervillous space causing hypoxia and inflammation (1). These events are associated with the active/chronic form of malaria infection, when the parasite and/or immune cells are present and levels of cytokines are high (9). However, little is known about the molecular taking place in the placentas as late consequences of a past malaria infection during pregnancy, i.e. a condition where the mother developed placental malaria but was treated and had the parasite cleared (9).
This is the first study providing a deep characterization of protein expression and PTMs in past malaria-infected and noninfected placentas. Using label-free proteomics, 1715 proteins were identified in the placenta of mice, and 1568 proteins were identified in the human placenta. In addition, a data set of 2947 proteins, 1733 glycosites and 4100 phosphosites were described by combining peptide isobaric labeling followed by phosphopeptide and glycopeptides enrichment and LC-MS/MS analysis in human placenta. Until this study, Wang et al. provided the highest number, in terms of protein identification by the LC-MSMS in human placentas (12, 37) with 2636 proteins, 1027 N-glyco- and 2094 phosphosites identified in the placentas from normal and preeclamptic pregnancies. All these data sets demonstrate the importance and strength of mass spectrometry-based proteomics strategies to expand both the database and knowledge of protein expression and PTMs associated with normal and pathological processes in human placentas.
In this study, we focused in past placental malaria, in which parasites were not detected and no significant evidence of inflammation was observed at the time of the delivery and placenta collection (supplemental Table S3). Our purpose was to understand specific molecular changes in the proteome, phosphoproteome and glycoproteome that occur in past P. falciparum-infected placentas.
Firstly, we made a comparison of the regulated proteins identified from two complementary malaria-infected mouse and human placenta models to acquire insights about the biological process associated to Plasmodium sp. infection. It is important to mention that P. berghei-infected mice reproduced the pathogenesis of placental malaria in pregnant human (21, 38). Moreover, infection of pregnant C57BL/6 females with NK65 P. berghei parasites consists of a robust experimental model to identify host molecular elements involved in PM pathogenesis (39). Therefore, by comparing two biological models we expected to increase the relevance and confidence of the altered processes associated to placental malaria, regardless model and stage of infection. Both showed that malaria-infected placentas have altered biological processes related to protein metabolism, including: protein location and transport, ubiquitin-dependent protein catabolism, proteasomal-mediated protein catabolism and protein folding, as well as process related to energy generation (generation of precursor metabolites and energy derivation by oxidation of organic compounds). It is well known that fetal growth is directly related to maternal nutrient availability and ability to transport these nutrients from maternal circulation to the fetus (35, 36). The rupture of amino acid transport in malaria-infected placentas with intervillositis was recently correlated with reduced birth weight (40). Moreover, protein folding and protein catabolic process (ubiquitin-dependent protein catabolism, proteasomal protein catabolism) are also important biological processes related to cell metabolism, survival, and adaptation to cell stress (41, 42). We also observed differences in mouse and human placenta responses to malaria infection not only in protein-specific regulation, but also in an overrepresented biological process, especially in the data set of down-regulated proteins in both mouse and human. Those differences can be attributed not only to the biological model, but also to differences in the stage of infection, in which the active form was observed in mouse infected-placentas, which is characterized by the presence of the parasite; a condition not observed in the past infected human placentas.
In the second part of this study, we performed phosphoproteome and glycoproteome analysis to explore novel mechanisms associated to past placental malaria, which could be linked to previous results attained by comparing the two infected-placenta models, as wells as to the clinical outcomes reported in these samples data set.
By integrating different levels of protein information, including protein abundance and PTMs, improved coverage of proteins related to cell signaling and communication, as well as different cellular organelles, such as the nucleus in the phosphoproteome data and plasma membrane/extracellular proteins in the glycoproteome data was obtained. It was previously demonstrated that such integrative strategy improves the interpretation of disease-related networks, which is essential to draw reliable hypothesis and conclusions regarding disease-related mechanism and altered pathways (43).
Enriched biological processes related to protein folding and transport were found in the integrated-proteomics data set to be altered in past-infected human placentas, confirming the previous label-free proteomic comparison between mouse and human placental malaria models. Additionally, processes linked to inflammation, response to stress and regulation of apoptosis were found to be significantly overrepresented in the data set of regulated proteins/phosphoproteins/glycoproteins, supporting the notion that P. falciparum promotes changes in biological processes associated to cell stress responses in placenta.
By using predicted kinases enrichment analysis and integrated protein-protein network analysis, we found alterations in mTOR, ERK and AKT1 signaling pathways in infected placentas. These results together with the two-plasmodium infected-placentas models were a starting point to investigate and validate the phenotype related to apoptosis, oxidative stress and mTOR, ERK, and AKT1 signaling in past-infected and noninfected placentas.
Mammalian target of rapamycin (mTOR) is a serine/threonine quinase that acts as the catalytic core of two protein complexes mTORC1 and mTORC2, which are composed by different proteins, undergo distinct mechanisms of regulation and control diverse cellular processes (44). Our phosphoproteomics and predicted kinase data are indicative of likely activation mTORC2-AKT signaling pathway in past-infected placenta. Indeed, key components of mTORC2 namely RICTOR and MAPKAP1 (also known as mSIN1), which are essential for complex stability and activity (45), were identified by LC-MSMS as significantly up-regulated phosphopeptides in past-malaria-infected placentas. Corroborating those findings, mTORC2 downstream substrate AKT1 was found to be significantly enriched in kinase enrichment analysis, as well as significantly overrepresented in a motif analysis using regulated phosphosites showing “RXXS” AKT motif. Confirming those predictions, we found in past-infected placentas a significant increase in the phosphorylated content of AKT (S473), such a phosphorylation that is catalyzed by mTORC2. Interestingly, phosphorylation of MAPKAP at T86 was previously shown to increase mTORC2 phosphorylation of AKT (46), being therefore a candidate mechanism to explain this phenotype.
In addition to AKT, past-infected placenta also featured an activation of ERK signaling pathway as evidenced by its significant enrichment in kinase enrichment analysis, overrepresentation in motif analysis using regulated phosphosites showing “XX-S/T-P” MAP kinase motif ERK (Y204) and increased phosphorylated content (Y204) in immunoblotting. Despite well-established role of both ERK and AKT as upstream activators of mTORC1, no changes were detected in the activity of this complex as evaluated by the absence of differences in the phosphorylated contents of mTORC1 downstream products phosphoS6 (S235/236) and phospho 4EBP1 (S65) in past-infected placentas. In contrast to our findings, a previous study by Dimasuay et al. had demonstrated that the activity of both mTOR complexes is decreased in placental malaria with intervillositis, as evidenced by the reduced content of mTORC1 downstream products phosphoS6 and phospho4EBP1 and mTORC2 downstream product phosphoAKT S473. In addition, they also showed that mTOR inhibition was associated to high levels of the cytokines IL-1β, IL-6, IL-8, TNF and IL-10 which are secreted by monocytes exposed to infected erythrocytes (40). A possible reason to explain the discrepancy in mTORC1 and 2 activities between both studies relies in the fact that, in contrast to Dimasuay et al. (40), our experiments were performed in past-infected placentas where no parasites or intervillositis were found (supplemental Table S2). Additionally, Festuccia et al., showed that deletion of RICTOR in macrophages exacerbates cytokine secretion induced by LPS. Therefore, mTORC2-AKT activation may be a mechanism to reduce the production of proinflammatory cytokines and control the inflammation (47).
Full AKT activation involves its phosphorylation in two residues: threonine 308 (Thr308) in the activation loop by phosphoinositide-dependent kinase (PDK)1, and in serine 473 (Ser473) in the hydrophobic motif by mTORC2 (48). Los et al. reviewed the dual role of AKT both in survival and in deadly response (49). Sadidi et al. showed that the activated AKT assumes a pro-survival role in cells exposed to oxidative insults (50). However, Nogueira et al. demonstrated that cells with enhanced AKT activity can be selectively killed because of their hypersensitivity to ROS (51). Moreover, a pro-apoptotic activity of AKT was reported by Maddika et al., where the apoptin-induced apoptosis depends on the abnormal phosphatidylinositol 3-kinase (PI3-kinase)/AKT activation, resulting in the activation of the cyclin-dependent kinase CDK2 (52). Interestingly, we also identified in our phosphoproteomics data that CDK2 is a significantly overrepresented kinase in the past-infected placentas (Fig. 4).
Apoptosis, defined as programmed cell death, may be initiated by several stimuli, including hypoxia and oxidative stress (53). This process is associated to placental dysfunction and pregnancy complications, such as early loss of pregnancy (54), pre-eclampsia (55), and intrauterine growth restriction (IUGR) (56). Aiming to perform a deep investigation of the “regulation of apoptosis,” which was retrieved by a biological process enrichment analysis using differentially regulated proteins, we assessed by immunoblotting BCL2/BAX ratio, as well as cleaved caspase-3 in past-P. falciparum infected and noninfected placentas. We observed in past-P. falciparum infected, but not control placentas, a significant reduction in BCL2/BAX ratio and an increase in both cleaved caspase-3 content and DNA fragmentation by TUNEL staining, indicating an activation of apoptosis. Corroborating these findings, Sharma et al., demonstrated that pregnant mice infected with Plasmodium berghei presented increased levels of malondialdehyde, an index of lipid peroxidation and oxidative stress, decreased activity of the antioxidant enzyme catalase and enhanced apoptosis (57). Sarr et al. showed that apoptosis is increased in P. chabaudi-infected pregnant aborting mice but the apoptosis markers were restricted to maternal blood inflammatory cells and junctional zone trophoblasts (58). In the same study by Sarr, apoptosis markers were also evaluated in Plasmodium falciparum-infected placental human tissue sections. TUNEL and cleaved CASP3 staining were confined to leukocytes infiltrating the intervillous space (58). In contrast to our findings, Crocker et al. (59) showed malaria-infected and noninfected placentas had no significant difference in apoptosis measurement. In that same study, when infected placentas were subdivided into placentas with active and past infection, no difference was observed between active and noninfected samples, but significant decrease of apoptosis was observed in past infected compared with noninfected placentas. This discrepancy can be attributed to the placental malaria models differences, besides the extension of inflammatory cells present in infected placenta and the cell population stained for apoptosis and considered for comparison.
Recently, Liu et al. (60), proposed that hemozoin, a pigment produced by the Plasmodium parasites by the degradation of hemoglobin, promotes apoptosis in trophoblast cells by activating the STAT3/caspase-3/PARP signaling pathway. Additionally, in mosquito cells, hemozoin was shown to play a role inducing NO synthase expression, as well as AKT and ERK activation (61) and, in macrophages, hemozoin induces a powerful synergistic effect on the IFN-gamma-inducible NO generation via ERK- and NF-kappaB-dependent pathways (62).
Oxidative stress is an important mechanism induced by the human host in response to infections, and in the case of malaria, it can handle the death of the parasites (63). Oxidative stress-mediated apoptosis was observed in infected-hepatocytes (64), as well as in endothelial cells incubated with sera from malaria infected patients (65). Sarr et al., showed that P. chabaudi -infected pregnant mice had increased lipid peroxidation and increased antioxidant gene expression. P. chabaudi -infected pregnant mice treated with superoxide dismutase mimetic, showed improved embryo survival and pregnancy outcomes (66). Increased levels of proteins involved in the response to stress were identified by LC-MS/MS in past-infected placentas, such as MPO, SOD1, PRDX1 and PRDX5, which can be associated with the increased apoptosis phenotype.
This study showed apoptosis-related mechanisms, including oxidative stress generation and activation of AKT and ERK pathways as a feature associated with past placental malaria. Further larger cohort studies are required in order to determine whether the apoptosis markers could be used to predict the clinical outcomes related to placental malaria.
DATA AVAILABILITY
Raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride) partner repository with the dataset identifier PXD008079. Annotated MS/MS spectra can be found in MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the following search keys: ipfwhr5osl, hwm0mqbx2s, dkscsltllo, jwreohtivy, rnb08ldiee.
Supplementary Material
Acknowledgments
We thank the women from “Alto do Juruá” valley who agreed in participating in the study, as well as the nurses and technicians from the Hospital da Mulher e da Crianc̨a do Juruá and Gerência de Endemias/SESACRE team for their invaluable assistance. We also thank the directory of Santa Casa de Misericórdia de Cruzeiro do Sul, and Universidade Federal do Acre for the support. Additionally, we thank Bernardo Paulo Albe and Erika Paula Machado Peixoto for their technical assistance. Also, we thank Alexandre Macedo de Oliveira from Centers for Disease Control and Prevention (CDC) for his ongoing support of our study and the past and present participating investigators from Prof. Claudio R.F. Marinho laboratory for assistance during fieldwork and scientific input. We thank Venkatachalam Udhayakumar, Luciana Flannery and Naomi Lucchi from Malaria Laboratory Research and Development Unit at CDC for all the support on the establishment and training of the PET-PCR technique, which was funded by the U.S. Agency for International Development (USAID) through the Amazon Malaria Initiative (AMI).
Footnotes
 This article contains supplemental material. The authors declare that they have no conflict of interest.
This article contains supplemental material. The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- MiP
- malaria in pregnancy
- IE
- infected erythrocytes
- CSA
- chondroitin sulfate A
- HiLic
- hydrophilic liquid interaction chromatography.
REFERENCES
- 1. White N. J., Pukrittayakamee S., Hien T. T., Faiz M. A., Mokuolu O. A., and Dondorp A. M. (2014) Malaria. Lancet 383, 723–735 [DOI] [PubMed] [Google Scholar]
- 2. Dellicour S., Tatem A. J., Guerra C. A., Snow R. W., and ter Kuile F. O. (2010) Quantifying the number of pregnancies at risk of malaria in 2007: a demographic study. PLoS Med. 7, e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rogerson S. J., Hviid L., Duffy P. E., Leke R. F., and Taylor D. W. (2007) Malaria in pregnancy: pathogenesis and immunity. Lancet. Infectious Diseases 7, 105–117 [DOI] [PubMed] [Google Scholar]
- 4. Brabin B. J., Romagosa C., Abdelgalil S., Menendez C., Verhoeff F. H., McGready R., Fletcher K. A., Owens S., D'Alessandro U., Nosten F., Fischer P. R., and Ordi J. (2004) The sick placenta-the role of malaria. Placenta 25, 359–378 [DOI] [PubMed] [Google Scholar]
- 5. Fried M., and Duffy P. E. (1996) Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272, 1502–1504 [DOI] [PubMed] [Google Scholar]
- 6. Viebig N. K., Levin E., Dechavanne S., Rogerson S. J., Gysin J., Smith J. D., Scherf A., and Gamain B. (2007) Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PloS One 2, e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abrams E. T., Brown H., Chensue S. W., Turner G. D., Tadesse E., Lema V. M., Molyneux M. E., Rochford R., Meshnick S. R., and Rogerson S. J. (2003) Host response to malaria during pregnancy: placental monocyte recruitment is associated with elevated beta chemokine expression. J. Immunol. 170, 2759–2764 [DOI] [PubMed] [Google Scholar]
- 8. Bulmer J. N., Rasheed F. N., Francis N., Morrison L., and Greenwood B. M. (1993) Placental malaria. I. Pathological classification. Histopathology 22, 211–218 [DOI] [PubMed] [Google Scholar]
- 9. Muehlenbachs A., Fried M., McGready R., Harrington W. E., Mutabingwa T. K., Nosten F., and Duffy P. E. (2010) A novel histological grading scheme for placental malaria applied in areas of high and low malaria transmission. J. Infectious Dis. 202, 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson J. M., Vandre D. D., and Ackerman W. E. (2009) Placental proteomics: a shortcut to biological insight. Placenta 30, S83–S89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gharesi-Fard B., Zolghadri J., and Kamali-Sarvestani E. (2015) Proteome differences in the first- and third-trimester human placentas. Reproductive Sci. 22, 462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F., Shi Z., Wang P., You W., and Liang G. (2013) Comparative proteome profile of human placenta from normal and preeclamptic pregnancies. PloS One 8, e78025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miao Z., Chen M., Wu H., Ding H., and Shi Z. (2014) Comparative proteomic profile of the human placenta in normal and fetal growth restriction subjects. Cell. Physiol. Biochem. 34, 1701–1710 [DOI] [PubMed] [Google Scholar]
- 14. Chang A., Zhang Z., Zhang L., Gao Y., Zhang L., Jia L., Cui S., and Wang P. (2013) Proteomic analysis of preterm premature rupture of membranes in placental tissue. Arch. Gynecol Obstetrics 288, 775–784 [DOI] [PubMed] [Google Scholar]
- 15. Liu B., Xu Y., Voss C., Qiu F. H., Zhao M. Z., Liu Y. D., Nie J., and Wang Z. L. (2012) Altered protein expression in gestational diabetes mellitus placentas provides insight into insulin resistance and coagulation/fibrinolysis pathways. PloS One 7, e44701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barboza R., Reis A. S., da Silva L. G., Hasenkamp L., Pereira K. R., Camara N. O., Costa F. T., Lima M. R., Alvarez J. M., Boscardin S. B., Epiphanio S., and Marinho C. R. (2014) MyD88 signaling is directly involved in the development of murine placental malaria. Infection Immunity 82, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lucchi N. W., Narayanan J., Karell M. A., Xayavong M., Kariuki S., DaSilva A. J., Hill V., and Udhayakumar V. (2013) Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PloS One 8, e56677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Souza R. M., Ataide R., Dombrowski J. G., Ippolito V., Aitken E. H., Valle S. N., Alvarez J. M., Epiphanio S., and Marinho C. R. (2013) Placental histopathological changes associated with Plasmodium vivax infection during pregnancy. PLoS Neglected Tropical Diseases 7, e2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romagosa C., Menendez C., Ismail M. R., Quinto L., Ferrer B., Alonso P. L., and Ordi J. (2004) Polarisation microscopy increases the sensitivity of hemozoin and Plasmodium detection in the histological assessment of placental malaria. Acta Tropica 90, 277–284 [DOI] [PubMed] [Google Scholar]
- 20. Tuominen V. J., Ruotoistenmaki S., Viitanen A., Jumppanen M., and Isola J. (2010) ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 12, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neres R., Marinho C. R., Goncalves L. A., Catarino M. B., and Penha-Goncalves C. (2008) Pregnancy outcome and placenta pathology in Plasmodium berghei ANKA infected mice reproduce the pathogenesis of severe malaria in pregnant women. PloS One 3, e1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palmisano G., Lendal S. E., Engholm-Keller K., Leth-Larsen R., Parker B. L., and Larsen M. R. (2010) Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat. Protocols 5, 1974–1982 [DOI] [PubMed] [Google Scholar]
- 23. Larsen M. R., Jensen S. S., Jakobsen L. A., and Heegaard N. H. (2007) Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell. Proteomics 6, 1778–1787 [DOI] [PubMed] [Google Scholar]
- 24. Mysling S., Palmisano G., Hojrup P., and Thaysen-Andersen M. (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 29. Lowe E. D., Tews I., Cheng K. Y., Brown N. R., Gul S., Noble M. E., Gamblin S. J., and Johnson L. N. (2002) Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry 41, 15625–15634 [DOI] [PubMed] [Google Scholar]
- 30. Endicott J. A., Noble M. E., and Tucker J. A. (1999) Cyclin-dependent kinases: inhibition and substrate recognition. Curr. Opinion Structural Biol. 9, 738–744 [DOI] [PubMed] [Google Scholar]
- 31. Yang S. D., and Huang T. J. (1994) Identification of -R-X-(X)-S/T-X3-S/T- as consensus sequence motif for autophosphorylation-dependent protein kinase. J. Biol. Chem. 269, 29855–29859 [PubMed] [Google Scholar]
- 32. Obata T., Yaffe M. B., Leparc G. G., Piro E. T., Maegawa H., Kashiwagi A., Kikkawa R., and Cantley L. C. (2000) Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275, 36108–36115 [DOI] [PubMed] [Google Scholar]
- 33. Theriault J. R., Lambert H., Chavez-Zobel A. T., Charest G., Lavigne P., and Landry J. (2004) Essential role of the NH2-terminal WD/EPF motif in the phosphorylation-activated protective function of mammalian Hsp27. J. Biol. Chem. 279, 23463–23471 [DOI] [PubMed] [Google Scholar]
- 34. Rogalla T., Ehrnsperger M., Preville X., Kotlyarov A., Lutsch G., Ducasse C., Paul C., Wieske M., Arrigo A. P., Buchner J., and Gaestel M. (1999) Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J. Biol. Chem. 274, 18947–18956 [DOI] [PubMed] [Google Scholar]
- 35. Brett K. E., Ferraro Z. M., Yockell-Lelievre J., Gruslin A., and Adamo K. B. (2014) Maternal-fetal nutrient transport in pregnancy pathologies: the role of the placenta. Int. J. Mol. Sci. 15, 16153–16185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kidima W. B. (2015) Syncytiotrophoblast Functions and Fetal Growth Restriction during Placental Malaria: Updates and Implication for Future Interventions. BioMed Res. Int. 2015, 451735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang F., Wang L., Shi Z., and Liang G. (2013) Comparative N-glycoproteomic and phosphoproteomic profiling of human placental plasma membrane between normal and preeclampsia pregnancies with high-resolution mass spectrometry. PloS One 8, e80480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marinho C. R., Neres R., Epiphanio S., Goncalves L. A., Catarino M. B., and Penha-Goncalves C. (2009) Recrudescent Plasmodium berghei from pregnant mice displays enhanced binding to the placenta and induces protection in multigravida. PloS One 4, e5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodrigues-Duarte L., de Moraes L. V., Barboza R., Marinho C. R., Franke-Fayard B., Janse C. J., and Penha-Goncalves C. (2012) Distinct placental malaria pathology caused by different Plasmodium berghei lines that fail to induce cerebral malaria in the C57BL/6 mouse. Malaria J. 11, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dimasuay K. G., Aitken E. H., Rosario F., Njie M., Glazier J., Rogerson S. J., Fowkes F. J., Beeson J. G., Powell T., Jansson T., and Boeuf P. (2017) Inhibition of placental mTOR signaling provides a link between placental malaria and reduced birthweight. BMC Med. 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregersen N., and Bross P. (2010) Protein misfolding and cellular stress: an overview. Methods Mol. Biol. 648, 3–23 [DOI] [PubMed] [Google Scholar]
- 42. Kaushansky A., and Kappe S. H. (2015) Host ER stress during malaria parasite infection. EMBO Reports 16, 883–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park J. M., Park J. H., Mun D. G., Bae J., Jung J. H., Back S., Lee H., Kim H., Jung H. J., Kim H. K., Lee H., Kim K. P., Hwang D., and Lee S. W. (2015) Integrated analysis of global proteome, phosphoproteome, and glycoproteome enables complementary interpretation of disease-related protein networks. Sci. Reports 5, 18189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weichhart T., Hengstschlager M., and Linke M. (2015) Regulation of innate immune cell function by mTOR. Nat. Rev.. Immunol. 15, 599–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., and Su B. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 46. Yang G., Murashige D. S., Humphrey S. J., and James D. E. (2015) A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Reports 12, 937–943 [DOI] [PubMed] [Google Scholar]
- 47. Festuccia W. T., Pouliot P., Bakan I., Sabatini D. M., and Laplante M. (2014) Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PloS One 9, e95432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 49. Los M., Maddika S., Erb B., and Schulze-Osthoff K. (2009) Switching Akt: from survival signaling to deadly response. BioEssays 31, 492–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sadidi M., Lentz S. I., and Feldman E. L. (2009) Hydrogen peroxide-induced Akt phosphorylation regulates Bax activation. Biochimie 91, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nogueira V., Park Y., Chen C. C., Xu P. Z., Chen M. L., Tonic I., Unterman T., and Hay N. (2008) Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 14, 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maddika S., Panigrahi S., Wiechec E., Wesselborg S., Fischer U., Schulze-Osthoff K., and Los M. (2009) Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin's anticancer toxicity. Mol. Cell. Biol. 29, 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharp A. N., Heazell A. E., Crocker I. P., and Mor G. (2010) Placental apoptosis in health and disease. Am. J. Reproductive Immunol. 64, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hempstock J., Jauniaux E., Greenwold N., and Burton G. J. (2003) The contribution of placental oxidative stress to early pregnancy failure. Human Pathol. 34, 1265–1275 [DOI] [PubMed] [Google Scholar]
- 55. Heazell A. E., Buttle H. R., Baker P. N., and Crocker I. P. (2008) Altered expression of regulators of caspase activity within trophoblast of normal pregnancies and pregnancies complicated by preeclampsia. Reproductive Sci. 15, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 56. Levy R., Smith S. D., Yusuf K., Huettner P. C., Kraus F. T., Sadovsky Y., and Nelson D. M. (2002) Trophoblast apoptosis from pregnancies complicated by fetal growth restriction is associated with enhanced p53 expression. Am. J. Obstetrics Gynecol. 186, 1056–1061 [DOI] [PubMed] [Google Scholar]
- 57. Sharma L., Kaur J., and Shukla G. (2012) Role of oxidative stress and apoptosis in the placental pathology of Plasmodium berghei infected mice. PloS One 7, e32694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sarr D., Bracken T. C., Owino S. O., Cooper C. A., Smith G. M., Nagy T., and Moore J. M. (2015) Differential roles of inflammation and apoptosis in initiation of mid-gestational abortion in malaria-infected C57BL/6 and A/J mice. Placenta 36, 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crocker I. P., Tanner O. M., Myers J. E., Bulmer J. N., Walraven G., and Baker P. N. (2004) Syncytiotrophoblast degradation and the pathophysiology of the malaria-infected placenta. Placenta 25, 273–282 [DOI] [PubMed] [Google Scholar]
- 60. Liu M., Hassana S., and Stiles J. K. (2016) Heme-mediated apoptosis and fusion damage in BeWo trophoblast cells. Sci. Reports 6, 36193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Akman-Anderson L., Olivier M., and Luckhart S. (2007) Induction of nitric oxide synthase and activation of signaling proteins in Anopheles mosquitoes by the malaria pigment, hemozoin. Infection Immunity 75, 4012–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jaramillo M., Gowda D. C., Radzioch D., and Olivier M. (2003) Hemozoin increases IFN-gamma-inducible macrophage nitric oxide generation through extracellular signal-regulated kinase- and NF-kappa B-dependent pathways. J. Immunol. 171, 4243–4253 [DOI] [PubMed] [Google Scholar]
- 63. Percario S., Moreira D. R., Gomes B. A., Ferreira M. E., Goncalves A. C., Laurindo P. S., Vilhena T. C., Dolabela M. F., and Green M. D. (2012) Oxidative stress in malaria. Int. J. Mol. Sci. 13, 16346–16372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guha M., Kumar S., Choubey V., Maity P., and Bandyopadhyay U. (2006) Apoptosis in liver during malaria: role of oxidative stress and implication of mitochondrial pathway. FASEB J. 20, 1224–1226 [DOI] [PubMed] [Google Scholar]
- 65. Hemmer C. J., Lehr H. A., Westphal K., Unverricht M., Kratzius M., and Reisinger E. C. (2005) Plasmodium falciparum Malaria: reduction of endothelial cell apoptosis in vitro. Infection Immunity 73, 1764–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sarr D., Cooper C. A., Bracken T. C., Martinez-Uribe O., Nagy T., and Moore J. M. (2017) Oxidative stress: A potential therapeutic target in placental malaria. ImmunoHorizons 1, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride) partner repository with the dataset identifier PXD008079. Annotated MS/MS spectra can be found in MS-Viewer (http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the following search keys: ipfwhr5osl, hwm0mqbx2s, dkscsltllo, jwreohtivy, rnb08ldiee.