Abstract
Gene therapy represents an attractive alternative to treat hemophilia B. Here we established three hepatocyte-derived cell lines based on Huh7, PLC/PRF/5, and Hep3B cells stably carrying a mutated canine FIX (cFIXmut) transgene containing a single point mutation in the catalytic domain. Based on these in vitro models resembling a commonly used canine large animal model, the tetracycline-controlled transcriptional activator (Tet-on)-inducible CRISPR/Cas9 system and an optimized donor were used to correct mutated cFIX gene through homology-directed repair (HDR). For efficient delivery of designer nuclease and donor DNA, we produced a high-capacity adenovirus vector type 5 (HCAdV5) containing the Tet-on-inducible cFIX-specific CRISPR/Cas9 system and a single-stranded adeno-associated virus type 2 vector (ssAAV2) containing the modified donor. Moreover, we designed a single HCAdV5 delivering all components for HDR. Our amplification-refractory mutation system based on qPCR analysis (ARMS-qPCR) revealed that the single vector application in Huh7-cFIXmut cells resulted in up to 5.52% HDR efficiencies, which was superior to the two-vector strategy. Furthermore the single vector also resulted in increased phenotypic correction efficiencies assayed by ELISA. We conclude that HDR in combination with viral vector delivery holds great promise for the correction of mutated FIX in disease models.
Keywords: hemophilia B, gene therapy, homology directed repair, CRISPR/Cas9, HCAdV5, ssAAV2, single vector
Introduction
Hemophilia B, accounting for 20% of hemophilia cases, represents an X-linked genetic blood clotting disorder due to the deficiency of factor IX (FIX) induced by mutations in the FIX gene. Patients with functional plasma levels of FIX that are less than 1% of the normal value have a severe phenotype characterized by frequent spontaneous bleeding episodes that result in chronic, debilitating arthropathy and occasionally death.1 Fortunately, the increase of plasma FIX levels above 1% of physiologic levels can ameliorate the bleeding diathesis. Nowadays, the injection of FIX as a protein replacement strategy is used to control and prevent bleeding in hemophilia B patients. However, treating bleeding episodes only until hemostasis is achieved may not be sufficient to treat long-term effects, such as hemarthropathy.2 Furthermore, inhibitors and the high costs of frequent intravenous infusions of FIX impel more feasible treatments into consideration.
As an alternative, direct infusion of complementary FIX DNA into hemophilia B animals and patients through viral delivery was evaluated, and it has proven to be efficacious.3, 4 The clinical trials by Nathwani and colleagues5, 6, 7 showed long-term therapeutic FIX expression after gene transfer of an adeno-associated virus (AAV) vector based on serotype 8 (AAV8) for the treatment of 10 patients with severe hemophilia B. However, the duration of expression is still under consideration, as the AAV vector predominantly persists episomally in host cells. Since hepatocytes are quiescent and renewal is very slow in the recruited adult patients, long-term therapeutic effects are possible if no silencing of transgene expression occurs. Even liver-derived expression of hyperfunctional FIX-Padua reduced liver toxicity and showed no increased risk of thrombogenicity in hemophilia B mice and dogs,8 but the disease complexity and certain adverse events restrict its clinical usage.9, 10, 11 Recent studies demonstrated that recombinant AAV-mediated site-specific integration of human FIX (hFIX) cDNA into the albumin (Alb) locus expressed in hepatocytes successfully ameliorated the disease in mice, but a high vector dose might lead to increased off-target integration.12
As an attractive alternative to gene addition-based therapeutic strategies, gene correction approaches were implemented, which rely on nuclease-driven homology-directed repair (HDR). It was demonstrated in a murine model of hemophilia B that zinc-finger nuclease (ZFN)-based HDR could produce high levels of hFIX.13, 14 In our previous study, we performed HDR based on transcription activator-like effector nucleases (TALENs) and the CRISPR/Cas9 nuclease machinery to correct the mutated genomic canine FIX (cFIX) locus in vitro. We found that the CRISPR/Cas9 construct revealed higher activities than the TALEN pair15 on genomic levels, and, therefore, we decided to pursue the CRISPR/Cas9 approach based on the identical guide RNA (gRNA) sequences in the present study.
The CRISPR/Cas9 machinery probably represents one of the most popular genome-editing technologies, which can efficiently induce DNA double-strand breaks (DSBs) at the desired genomic target locus.16, 17, 18 The CRISPR/Cas9 system initiates either error-prone non-homologous end joining (NHEJ) or error-free HDR in the presence of a repair template.19, 20 Recently, non-viral and early generation adenoviral delivery of the CRISPR/Cas9 machinery along with the homologous donor sequence was utilized to correct mutated FIX in a respective mouse model.21, 22 In one of these studies, it was concluded that non-viral vector delivery is superior to early generation adenoviral vector delivery of the CRISPR/Cas9 machinery.21
Here we explored advanced high-capacity adenoviral vectors based on human adenovirus type 5 (HCAdV5), for delivery of the CRISPR/Cas9 technology, in combination with the single-stranded adeno-associated virus type 2 vector (ssAAV2), for delivery of the donor DNA. HCAdV vectors show an improved toxicity profile and are less immunogenic to early generation adenoviral vectors deleted for one or more early adenovirus genes.23 Because of the high similarity between the hFIX and cFIX genes and the fact that FIX is primarily expressed in and released from liver cells, we generated three liver cell lines stably expressing mutated cFIX under the control of the liver-specific human α1-antitrypsin (hAAT) promoter. The mutated cFIX transgene corresponds to the mutation observed in a canine model for hemophilia B carrying a single missense mutation (G1477A on the nucleotide level), resulting in a substitution of glutamic acid (Glu) for glycine (Gly) at position 418 in the catalytic domain of the protein.24 This mutation results in a complete absence of circulating FIX in affected animals. This fact makes it ideal for gene therapy, including HDR, because genotypic and phenotypic correction can be simply detected. Vectors and gene correction strategies can first be explored in vitro and then translated and explored in this large animal model, which is a precondition to perform clinical trials. Here, we explored the two-vector strategy, in which we used HCAdV5 and ssAAV2 vectors as vehicles to deliver doxycycline (DOX)-induced CRISPR/Cas9 and a modified cFIX donor sequence, respectively. Moreover, we established a single-vector strategy employing HCAdV5 to deliver all components for HDR. The single-vector strategy for HDR of mutated cFIX in liver cells revealed higher repair efficiencies than the two-vector strategy.
Results
Generation and Characterization of In Vitro Hemophilia B Models for Phenotypic Correction
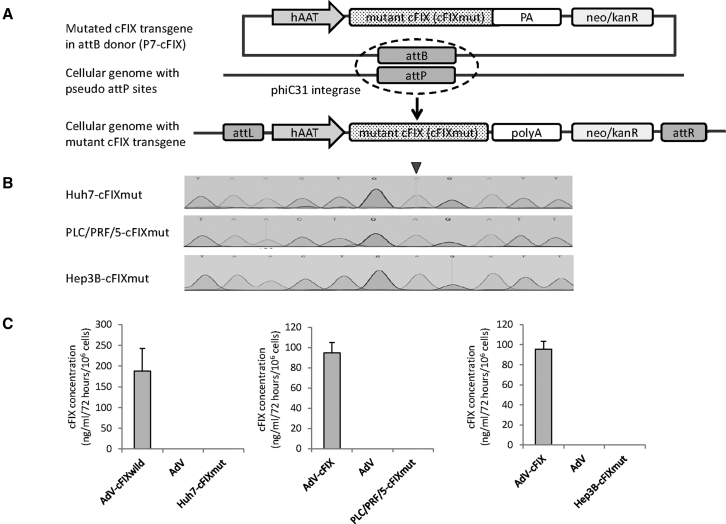
Based on the hemophilia B canine model, hepatocyte-derived Huh7, PLC/PRF/5, and Hep3B cells stably carrying the mutated cFIX-coding sequence (cFIXmut) with a single missense mutation (G1477A on the nucleotide level) were established using the phiC31 integration system.25 The transgene for somatic integration expresses cFIXmut under the control of the hepatocyte-specific hAAT promoter in plasmid P7-cFIX (Figure 1A). At 3 weeks post-co-transfection and selection with G418, single-cell clones were picked and cultured in 24-well plates. Subsequently, whole genomic DNA was extracted, and stable cell clones were checked for positive integration events of the cFIXmut transgene via Sanger sequencing (Figure 1B). Sequencing results of final cell lines Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut revealed integration of the mutant cFIX-encoding transgene. To further evaluate these cell lines, we measured integrated cFIX cDNA copy numbers in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells using qPCR. Results revealed that 2.3, 1.4, and 0.8 copies of the target sequence were integrated into the different cell lines, respectively.
Figure 1.
Construction of Stable Mutated Liver Cells Using phiC31 Integrase
(A) The vector containing the attB site, neo/kanR, and the mutated canine factor IX (cFIX) expression cassette and the phiC31 expression plasmid were co-transfected into liver-derived cell lines (Huh7 cells, PLC/PRF/5 cells, and Hep3B cells). In the presence of phiC31 integrase, an attB-containing donor can be unidirectionally integrated into a target cell genome, resulting in an integrated transgene flanked by attL and attRR sites. After selection with G418 for 3 weeks and sequence verification, the stable mutated cFIX cells (Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut) were prepared for homology-directed repair (HDR) correction experiments. hAAT, human alpha-1-antitrypsin promoter; PA, polyadenylation signal; neo, neomycin; kanR, kanamycin resistance. (B) The cFIX fragment of stable cell clones was checked using Sanger sequencing. The black arrow indicates the point mutation G1477A in cFIX. (C) The supernatants of cFIX stable cell clones were checked using ELISA. Data points represent SEM of three independent experiments performed in triplicates. AdV-cFIX, supernatant of cells infected with adenovirus vector carrying a wild cFIX transgene; AdV, supernatant of cells infected with an irrelevant adenovirus vector; Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut, the supernatants of the respective cFIX stable cell clones.
Next we tested the expression of cFIX protein in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cell lines by performing an ELISA using the supernatant of cultured cells. As a positive control, parental Huh7, PLC/PRF/5, and Hep3B cells were infected with a previously described HCAdV encoding wild-type cFIX.26 As shown in Figure 1C, wild-type cFIX was released from cells infected with cFIX-encoding HCAdV into the supernatant and was measurable by ELISA (Figure 1C). Without additional infection with the cFIX-encoding HCAdV, cFIX protein was undetectable in the supernatant (Figure 1C). This indicated that these cell lines (Huh-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut) can be used as an in vitro model for phenotypic correction and that the ELISA setup can differentiate between cFIX and hFIX.
Molecular Design of CRISPR/Cas9 and gRNA Expression Cassettes and Donor DNA
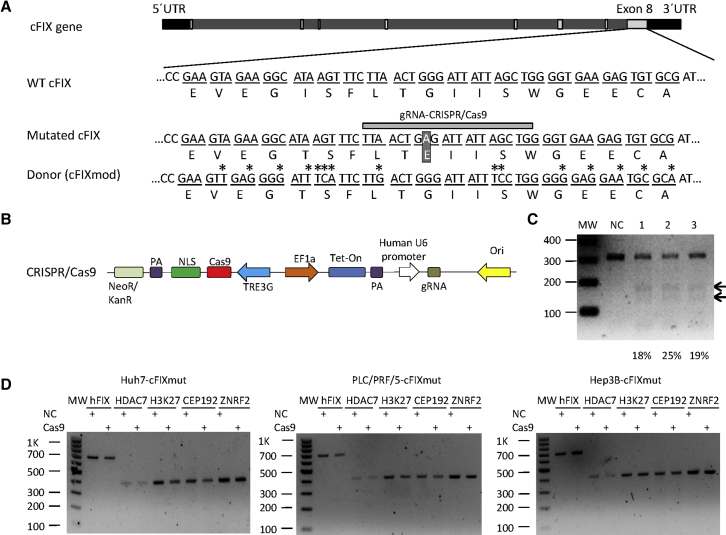
Evans and colleagues24 identified and characterized a hemophilia B canine model that carries a natural single missense mutation (G1477A) (Figure 2A), resulting in the substitution of glutamic acid for glycine-418 in the serine protease domain of the molecule and a complete lack of cFIX protein expression. Therefore, the gene correction of Gly418Glu in the cFIX gene represents an attractive site for exploring gene correction approaches to treat hemophilia B caused by mutated FIX. To guarantee that the donor sequence was gRNA resistant, we mutated three bases in the target sequence of the gRNA recognition region (Figure 2A), which we described in detail in our previous publication.15 An alternative solution to render the donor DNA gRNA resistant would be mutating the PAM sequence through synonymous substitution. However, applying this strategy was impossible for the experimental setup used in this project, because the PAM sequence was represented in our target sequence in two codons, TGG and GGT. TGG was the only codon for tryptophan and GGT represented the codon for glycine. Glycine would be converted to other amino acids if the first base or/and the second base is/are mutated.
Figure 2.
Construction of Non-viral Homology-Directed System for Correcting Mutated Canine Coagulation Factor IX
(A) Schematic outline of the cFIX target locus. The top panel shows the cFIX gene, including the UTRs (black bars), introns (bright gray bars), and exons (light gray bars). Nucleotide and amino acid sequences of wild-type cFIX (WT cFIX) and a major disease causing cFIX mutation (G1477A, mutated cFIX) are shown (white letters in dark square). The bottom panel shows the mutated donor sequence (cFIXmod) used in this study (mutations are marked with an asterisk), which results in the identical amino acid sequence as WT cFIX. The gray horizontal bar schematically shows the guide RNA (gRNA)-binding site used in this study (gRNA-CRISPR/Cas9). Differences in the donor and mutated cFIX sequences are marked with stars. (B) Schematic outline of the DNA sequences contained in the Tet-on-inducible CRISPR/Cas9 for cutting the cFIX-mutated strand and the donor DNA that was transfected as PCR product (cFIXmod). NLS, nuclear localization signal; PA, polyadenylation signal; TREG3, TRE3G promoter; EF1a, human elongation factor-1 alpha promoter; gRNA, guide RNA; Tet-on, tetracycline-controlled transcriptional activation. (C) CRISPR/Cas9 nuclease activity measured by T7E1 assay after transfection with the CRISPR/Cas9-encoding plasmid and the donor DNA. MW, molecular-weight size marker; NC, negative control referring to the mixture of untreated Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells; 1, CRISPR/Cas9-treated Huh7-cFIXmut cells; 2, CRISPR/Cas9-treated PLC/PRF/5-cFIXmut cells; 3, CRISPR/Cas9-treated Hep3B-cFIXmut cells. (D) CRISPR/Cas9 nuclease off-target analysis of the top 5 predicted off-target sites. These were the histone deacetylase 7 gene (HDAC7), DNA-packaging protein Histone H3 (H3K27), the centrosomal protein of 192 kDa (CEP192), and the Zinc and Ring Finger 2 gene (ZNRF2), which were analyzed in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells by T7E1 assay after transfection with the CRISPR/Cas9-encoding plasmid.
To construct a controllable HDR system, we introduced the tetracycline-controlled transcriptional activator (Tet-on) 3G tetracycline-inducible expression system to regulate CRISPR/Cas9 expression, rendering gene editing dependent on the addition of DOX (Figure 2B). To avoid that the corrected cFIX was recognized by gRNA and cut by CRISPR/Cas9, we recently designed a modified donor sequence, cFIXmod, through synonymous base pair substitution,15 resulting in a DNA sequence with altered nucleotides but the identical translation (Figure 2A). Based on the same study, we previously screened two gRNA candidates for the mutated cFIX target sequence and the more efficient one was employed in this study.15 We treated the Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells with the Tet-on-inducible CRISPR/Cas9 plasmid, and we used the T7 endonuclease 1 (T7E1) assay to analyze the nuclease-cutting efficacy. The insertion or deletion (indel) frequency was quantified with Image Lab 5.2, and the results showed that CRISPR/Cas9 treatment resulted in cutting of the target locus in all the three cell lines (Figure 2C). Possible off-target sites were predicted for the cFIX gRNA using the COSMID software,27 and the top five sites were verified with the Off-spotter online tool28 and the Cas-OFFinder29 CRISPR/Cas9 off-target predictive tools. Identified off-target sites were the histone deacetylase 7 gene (HDAC7), a DNA-packaging protein Histone H3 gene (H3K27), the centrosomal protein of 192-kDa gene (CEP192), and the Zinc and Ring Finger 2 gene (ZNRF2). After transfection of Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells with the Tet-on-inducible CRISPR/Cas9 plasmid and the gRNA cassette, the T7E1 assay showed undetectable indels at the identified off-target sites (Figure 2D).
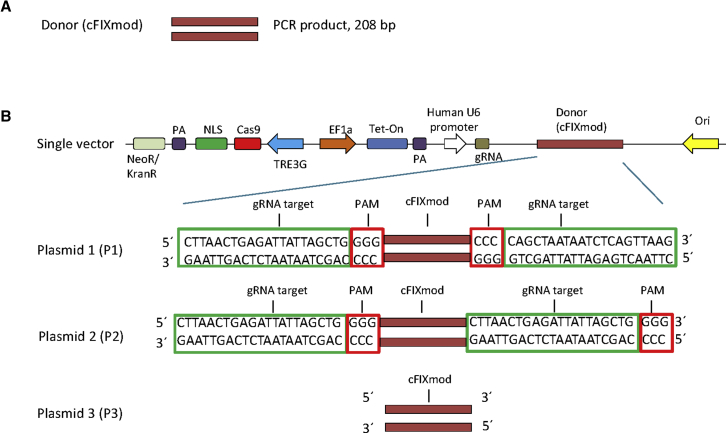
To deliver the donor DNA for HDR, we pursued two strategies. In non-viral approaches, we used a PCR product containing the cFIXmod sequence of 208 bp in length (Figure 3A). To reduce complexity of the system, we also designed an approach that was based on a single molecule carrying all components for HDR (Figure 3B). This included CRISPR/Cas9, the gRNA expression cassette, and the donor DNA. To optimize the system, we constructed three molecular setups of single vectors that delivered all components for HDR: plasmid 1 (P1), plasmid 2 (P2), and plasmid 3 (P3) (Figure 3B). For P1 and P2, the donor was flanked by the gRNA recognition sequence to achieve release of the donor from the plasmid. The difference between them was that the gRNA recognition sequences in P1 were in the opposite orientation, while the gRNA recognition sequences of P2 flanked the donor DNA in the identical orientation. The donor contained in P3 lacked the flanking gRNA recognition sequence.
Figure 3.
Design of Donor DNA Sequences for the Two-Vector and the Single-Vector Systems
(A) Donor DNA used for the two-vector system. A PCR product of 208 bp (cFIXmod) covering the mutated cFIX location was used. (B) Schematic diagram of single vectors containing the Tet-on-inducible CRISPR/Cas9 for cutting the cFIX-mutated strand and the donor DNA. Three molecular setups of single vectors, which delivered all components for HDR, were designed: plasmid 1 (P1), plasmid 2 (P2), and plasmid 3 (P3). For P1 and P2, the donor is flanked by the gRNA recognition sequence. Note that the gRNA recognition sequences in P1 are in the opposite orientation while the gRNA recognition sequences of P2 flank the donor DNA in the identical orientation. The donor contained in plasmid P3 lacks the gRNA recognition sequence. NLS, nuclear localization signal; PA, polyadenylation signal; TREG3, TRE3G promoter; EF1a, human elongation factor-1 alpha promoter; gRNA, guide RNA; Tet-on, tetracycline-controlled transcriptional activator.
Genetic Correction of Mutated cFIX through Non-viral Gene Transfer
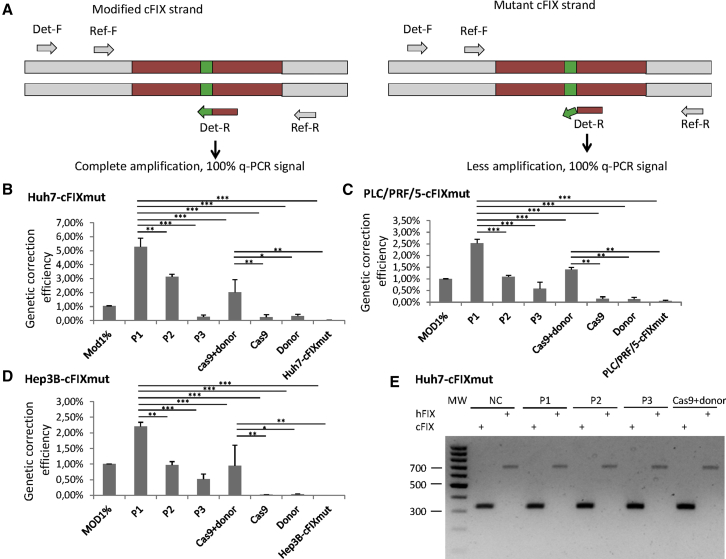
Next we conducted HDR with the Tet-on-inducible single-plasmid system (P1, P2, and P3) and the two-component system (CRISPR/Cas9 plasmid and the cFIXmod PCR product) in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells. At 72 h post-transfection, we measured gene correction efficiencies using our previously established amplification-refractory mutation system based on qPCR analysis (ARMS-qPCR) approach.30 The ARMS-qPCR contains two pairs of primers, the reference primer pair and the detection primer pair (Figure 4A). The reference primers bind to the common region of all samples, and, therefore, the PCR reaction is not influenced by HDR. For the detection PCR primer pair, the forward primer binds the common region while the reverse primer binds to the mutation site. After transfection of different single- and two-component systems using constructs displayed in Figures 2B and 3, we observed that HDR efficiencies in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells using the P1 single-plasmid strategy showed highest correction frequencies of 5.28%, 2.53%, and 2.21%, respectively (Figures 4B–4D). The other single vectors (P2 and P3) and the two-vector system also revealed detectable but lower HDR frequencies (Figures 4B–4D).
Figure 4.
Gene Correction of Mutated cFIX through Naked DNA
(A) The principle of the amplification-refractory mutation system qPCR (ARMS-qPCR) assay. Two qPCRs of each sample are performed. Primers of the reference PCR (Re-F,-R) amplify the upstream area of the HR region. The forward detection primer (Det-F) binds the upstream area of the HR region, and the reverse primer (Det-R) specifically binds to the HR cassette. (B–D) Naked DNA-mediated HDR events in mutated cFIX stable cell lines measured via ARMS-qPCR. Mod1%, Cas9 + donor, Cas9, Donor, and cell-cFIXmut display controls containing 1% modified template, cells transfected with CRISPR/Cas9 and optimized donor sequence, cells transfected with CRISPR/Cas9, cells transfected with optimized donor sequence, and cFIX stable cell lines Huh7-cFIXmut (B), PLC-PRF-5-cFIXmut (C), and Hep3B-cFICmut (D). (E) Indels measured by T7E1 assay after transfection with the non-viral homology-directed repair plasmids at the endogenous hFIX locus and the uncorrected cFIX transgene in Huh7-cFIXmut cells. MW, molecular weight markers; NC, negative control, PLC-cFIXmut or Hep3B-cFIXmut without treatment. P1, P2, P3, and Cas9 + donor display cells treated with plasmid 1, plasmid 2, plasmid 3, or Cas9 + donor. Data points represent SEM of three independent experiments performed in triplicates. *p < 0.05, **p < 0.01, ***p < 0.001.
To analyze possible indel formation at the uncorrected Cas9 cleavage site where no HDR occurred in the integrated cFIX transgene and the endogenous hFIX sequence in Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells, we performed further T7E1 assays. As shown in Figure 4E and Figure S1, no indels could be detected at these target sites.
Correction of the Mutated cFIX Gene by Viral Delivery Using HCAdV5-Cas9 and ssAAV2-cFIX
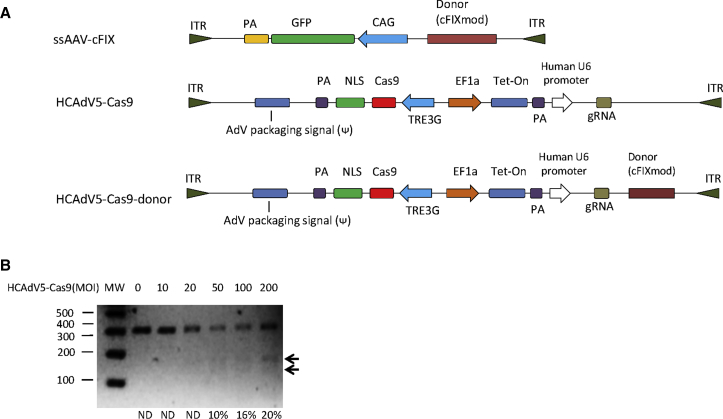
The rapid degradation of naked DNA impedes its clinical application, and, therefore, we moved on to investigating viral vectors for the delivery of CRISPR/Cas9 components and donor DNA. We intended to deliver the donor sequence to correct mutated cFIX using ssAAV2, which was shown to represent a potential candidate for in vivo gene delivery to treat hemophilia4, 31, 32 and as donor vector for HDR. Therefore, we inserted the modified cFIX as corrective donor template into ssAAV2, termed ssAAV2-cFIX (Figure 5A). Since the limited packaging capacity limits the usage of AAV vectors for delivering the Tet-on-inducible CRISPR/Cas9 system, we explored HCAdV for CRISPR/Cas9 delivery, which has the advantage of being able to carry large transgene cargos.33, 34
Figure 5.
Gene Correction of Mutated cFIX through the Viral Vectors HCAdV5-Cas9 and ssAAV2-cFIX
(A) Schematic outline of HCAdV5-Cas9 and ssAAV2-cFIX genomes. ITR, inverted terminal repeat; NLS, nuclear localization signal; PA, polyadenylation signal; TREG3, TRE3G promoter; EF1a, human elongation factor-1 alpha promoter; CAG, cytomegalovirus (CMV) enhancer fused to the chicken beta-actin promoter; gRNA, guide RNA; Tet-on, tetracycline-controlled transcriptional activation. (B) HCAdV5-Cas9 nuclease activity measured by T7E1 assay. MW, molecular-weight size marker; ND, no detection.
To identify the optimal adenoviral vector type for the transduction of different hepatocyte-derived cell lines, we first measured the transduction efficiencies of several different human adenovirus types from our recently described adenovirus library,35 in which the adenoviral vectors expressed luciferase. At 1 day post-infection with 3,000 viral particles (vps), luciferase expression levels were measured, and we found that adenovirus type 5 (Ad5) exhibits the highest transduction efficiencies and transgene expressions levels in analyzed Huh7 cells (Figure S2), which was in agreement with previous studies that Ad5 displayed high liver tropism.36, 37, 38 Therefore, we inserted the DOX-inducible CRISPR/Cas9 transgene and the gRNA transcription unit targeting the cFIX mutation site into a helper-dependent adenovirus vector based on type 5, termed HCAdV5-Cas9 (Figure 5A). Considering that P1 resulted in the most efficient correction efficiencies compared to P2 and P3 using non-viral gene transfer, we inserted the HDR donor elements of P1 into HCAdV5-Cas9, resulting in the single-vector HCAdV5-Cas9-donor (Figure 5A). Note that, for further gene correction studies, we used Huh7-cFIXmut cells, because these cells showed the highest correction efficiencies on the genome level in non-viral approaches (Figure 4B).
To optimize the co-transfection to conduct the desired modification with the two viral vector system, we investigated the co-transduction efficiencies of HCAdV5 and a single-stranded AAV2 vector. We transduced Huh7-cFIXmut with both HCAdV5-GFP and AAV2-mCherry concurrently, and we evaluated virus uptake using fluorescent microscopy. The results showed that the co-transduction was efficient when AAV2-mCherry was applied at an MOI 600 (Figure S3). We also determined transducing units for AAV and HCAdV5 vectors in the Huh7-cFIXmut target cell line to control incoming numbers of viral vector genomes (Figure S3). After determining the ratio of virus particle numbers to transducing units, we found that 8.6% of purified HCAdV5-Cas9 particles, 6.4% of HCAdV5-Cas9-donor particles, and 6.51% of ssAAV2-cFIX particles were active in transducing Huh7-cFIXmut cells (Figure S3).
Before the conduction of HDR, we measured the activity of the HCAdV5-Cas9 in Huh7-cFIXmut by T7E1 assay, and the results revealed that HCAdV5-Cas9 showed high cutting activity at MOIs 100 and 200 of up to 20% (Figure 5B). The MOI of 100 was used for HCAdV5-Cas9 in the following HDR experiments.
Genotypic and Phenotypic Corrections of Mutated cFIX Using Co-infection with HCAdV5-Cas9 and ssAAV2-cFIX Vectors
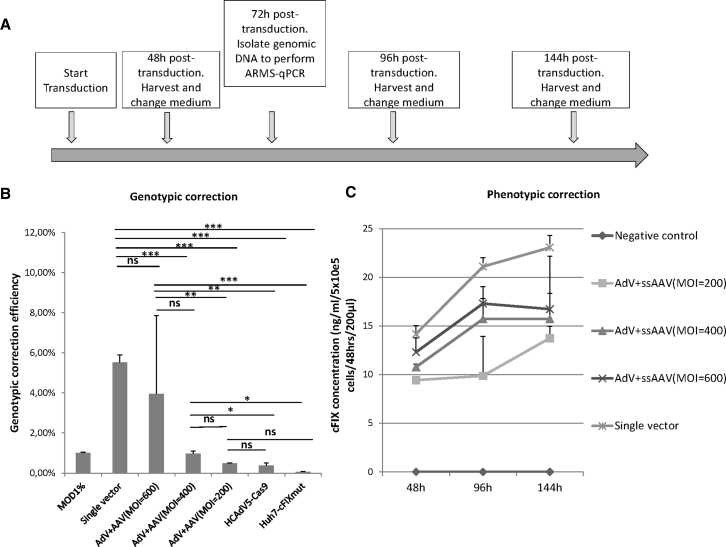
To conduct HDR in Huh7-cFIXmut cells, we co-transduced Huh7-cFIXmut cells with the two-vector system (HCAdV5-Cas9 at MOI 100 and a series of MOIs for ssAAV2-cFIX) and the single-vector system (HCAdV5-Cas9-donor, MOI 100). Cells were harvested 72 h post-infection, and genomic DNA was extracted to quantify gene correction efficiencies using ARMS-qPCR. The experimental setup is displayed in Figure 6A. The ARMS-qPCR results showed that the viral HDR efficiencies in Huh7-cFIXmut cells were 0.49%, 0.96%, and 3.95%, corresponding to the applied ssAAV2-cFIX MOIs of 200, 400, and 600, respectively, for the two-vector system, and 5.52% for the HCAdV5-Cas9-donor single-vector system (Figure 6B).
Figure 6.
Genotypic and Phenotypic Corrections in Huh7-cFIXmut Cells after Co-infection with the Gene Correction Vectors HCAdV5-Cas9-cFIX or HCAdV5-Cas9 and ssAAV2-cFIX
(A) Schematic outline of the experimental setup. Cellular genomic DNA was extracted at 72 h post-transduction for ARMS-qPCR assay, and cell media were collected at 48, 96, and 144 h post-transduction for the ELISA. (B) HCAdV5-Cas9-cFIX- or HCAdV5-Cas9- and ssAAV2-cFIX-mediated HDR events in mutated cFIX stable cells measured via ARMS-qPCR. Mod1%, controls containing 1% modified template; single vector, Huh7-cFIXmut cells transduced with HCAdV5-Cas9-cFIX; AdV + AAV (MOI 200), AdV + AAV (MOI 400),and AdV + AAV (MOI 600), Huh7-cFIXmut cells transduced with HCAdV5-Cas9 and ssAAV2-cFIX of different MOIs; Huh7-cFIXmut cells, mutated cFIX stable Huh7 cells; HCAdV5-Cas9, Huh7-cFIXmut cells transduced with HCAdV5-Cas9. (C) ELISA of cFIX concentration in the supernatant of Huh7-cFIXmut cells infected with HCAdV5-Cas9-cFIX or HCAdV-Cas9 and MOIs 200, 400, and 600 for ssAAV-cFIX. Data points represent SEM of three independent experiments performed in triplicates. *p < 0.05, **p < 0.01, ***p < 0.001.
To prove that this experimental setup also results in phenotypic correction on the protein level, we seeded Huh7-cFIXmut cells at 60%–70% confluency. The negative control cells received no treatment, while the experimental groups for HDR were treated with the single-vector system (HCAdV5-Cas9-donor) and the two-vector system (AdV and AAV) as described above. The cell supernatant was collected 48, 96, and 144 h post-transduction from both negative control and experimental groups. Levels of cFIX concentration secreted from both negative control and experimental groups were measured by ELISA (Figure 6C). Results revealed measurable cFIX levels in the supernatant of treated Huh7-cFIXmut cells, but not in the supernatant of untreated cells (Figure 6C). The highest cFIX levels in the supernatant were measured for the single-vector system.
Discussion
Recent research in a hemophilia B mouse model concluded that naked DNA surpassed human adenovirus type 5 in the delivery of HDR components for correction of mutated mouse FIX.21, 22 However, in contrast to our study, these latter studies explored early generation adenoviral vectors, which are known to induce stronger toxic side effects due to leaky expression of adenoviral genes remaining in the vector genome.39, 40 In fact there is strong evidence that the HCAdV vectors are superior and display an improved toxicity profile compared to early generation adenoviral vectors.39, 41, 42, 43, 44 Furthermore, the clinical systemic injection of naked DNA vectors is challenging, because high-pressure injection is not feasible at this time in larger mammals.
This research is the ongoing work of our former study for which the donor was designed for both TALEN- and CRISPR/Cas9-mediated gene correction. The HDR donor sequences flanking the gRNA-binding site were modified to prevent TALEN binding and cutting, as described in detail in our previous report.15 The cFIX HDR donor DNA used in this study comprised 208 bp and contained 15 modified bases, which represent silent mutations that do not result in amino acid changes.
AAV vectors are efficient vehicles for gene transfer because of their wide host-cell range, weak immunogenicity, and the ability to target liver cells.4 Furthermore, AAV2 is a nonpathogenic human parvovirus, which justifies its preferred choice to be used as cFIX donor sequence vehicle.45, 46, 47 The ssAAV2 expressing FIX was used in various canine studies48, 49, 50 and in the first liver-directed AAV clinical trial for hemophilia B.31 Here we successfully explored an AAV vector as donor template DNA, a strategy that was also used in previous studies.51, 52 One study explored two AAV vectors for the delivery of CRISPR/Cas9, gRNA, and donor DNA,51 and another study used AAV donor vectors in concert with CRISPR/Cas9 components, which were co-electroporated as non-viral vectors.52
Although single AAV vectors were used to deliver the CRISPR/Cas9 system based on Streptococcus pyogenes Cas9 (SpCas9) along with gRNA transcription units,53 the system would exceed the AAV-packaging size if an inducible promoter was used to control Cas9 expression, as chosen in the present study. Fortunately, human adenovirus vectors with high packaging capacities of up to 35 kb (HCAdVs) with reduced toxicity profiles were developed, which were shown to result in the efficient delivery of the CRISPR/Cas9 system in vitro.54 In future gene correction studies, the large packaging capacity of HCAdV may even allow researchers to combine the CRISPR/Cas9 machinery and the HDR donor sequence in a single vector.
The average genetic correction level of endogenous mutated cFIX alleles in Huh7-cFIXmut cells was 3.95%, and respective cFIX concentrations in the supernatants of modified Huh7 cells suggest that these correction levels are efficient to treat the disease. However, although genetic correction levels using viral vectors (Figure 6B) were higher compared to non-viral DNA-based approaches (Figure 4D), it remains to be analyzed whether these levels can be further improved by the molecular vector setup.
In the two-vector strategy, the genetic correction activity is not consistent with the phenotypic correction efficiencies. Especially when using an MOI 200 for the AAV donor on a molecular assay, the measured gene correction frequency was not above background, although it seemed to lead to significant cFIX protein production. One possible reason for this observation could be that expression of the cFIX gene is controlled by the strong hAAT promoter, which may lead to higher FIX levels compared to the endogenous gene. Another reason may be that the mutated cFIX could only be possibly corrected when both AdV-Cas9 and AAV-donor are present in the same Huh7-cFIXmut cell. However, in the process of transduction, the AdV-Cas9 and AAV-donor entered different Huh7-cFIXmut randomly, which may have induced obvious inconsistencies.
The highest cFIX concentrations were measured for the single-vector system (Figure 6). In Figure S4, we reanalyzed the data and calculated the amount of secreted cFIX protein per 1 × 10e6 cells. Treatments with the single-vector system led to secretion of up to 120 ng, possibly sufficient to reach therapeutic levels of FIX. To achieve therapeutic levels of hFIX in the plasma, 5% of normal levels (250 ng/mL) are required.55 The result in our approach showed that the corrected Huh7-cFIXmut produced up to 25 ng/mL cFIX per 5 × 10e5 cells in 48 h measured in 200 μL cell supernatant, which would equal up to 90 ng per 1 × 10e6 cells. We believe that, in total, this amount can be sufficient for the treatment of hemophilia B.
It remains to be mentioned that homologous recombination efficiencies in hepatocytes may not be equally efficient in vitro and in vivo and further experiments in primary cells or in vivo experiments should address this. However, one needs to consider that, in dividing cells, the donor DNA and the CRISPR/Cas9 cassette delivered by episomal vectors will get lost during cell division and that, in quiescent liver cells, in vivo or in vitro the episomal vector DNA is maintained longer, which in turn may increase correction efficiencies over time. Another option to increase coagulation factor levels would be sequential injections with different vector types, which may increase the correction efficiency and FIX serum levels.
In summary, we established an in vitro FIX mutation model for canine hemophilia B, and we detected restoration of the mutated FIX by CRISPR/Cas9-mediated genome editing upon adenoviral delivery. Moreover, we showed the potential of viral vector-delivered CRISPR/Cas9 and donor sequence to treat hemophilia B in dogs and patients. Based on our current knowledge, the one-vector system may be the most efficient gene correction tool, which we plan to analyze in further in vivo studies. In the future, enhancement of the HDR efficiency and in vivo experiments to evaluate possible side effects are interesting subjects for closer investigation.
Materials and Methods
Generation of the Plasmid P7-cFIX for the Generation of Stable Cell Lines
Based on a wild-type cFIX transgene,56 we introduced the mutation at nucleotide 1,477 (G-A) to produce mutated cFIX through overlap PCR. The bridge primers were as follows: forward primer, 5′-GTT TCT TAA CTG AGA TTA TTA GCT GGG GTG-3′; and reverse primer, 5′-CAC CCC AGC TAA TAA TCT CAG TTA AGA AAC-3′. The end primers were as follows (SgrDI restriction site is underlined): forward primer, 5′-AAT CGT CGA CGA GTA GGC TC AGA GGC ACA CAG-3′; and reverse primer, 5′-AAT CGT CGA CGG CTG GTT CTT TCC GCC TCA GAA G-3′.
To amplify the necessary fragments separately, the extension PCR was performed with the bridge primers using an annealing temperature of 60°C. The PCR product was cleaned up by the my-Budget Double Pure Kit (Bio-Budget Technologies, Krefeld, Germany) and applied as template for the subsequent overlap PCR. The overlap PCR ran 15 PCR cycles using an annealing temperature of 60°C without primer, and it continued to cycle 15–20 rounds after end primers were added. The product was gel extracted using the my-Budget Double Pure Kit (Bio-Budget Technologies), and the complete mutated cFIX-coding sequence was obtained.
Then the complete mutated cFIX-coding sequence was inserted into the pJET1.2/blunt cloning vector for the construction of pJET1.2-cFIX. After validation by Sanger sequencing, the pJET1.2-cFIX was digested with SgrDI (Thermo Scientific, Waltham, MA, USA), and the digested mutated cFIX-coding sequence was gel extracted. The attB-bearing vector P7 containing neomycin resistance gene was also digested with SgrDI before being dephosphorylated by alkaline phosphatase (New England Biolabs, Frankfurt am Main, Germany). Afterward, the digested mutated cFIX-coding sequence and P7 vector were ligated using T4 DNA ligase (New England Biolabs) overnight, generating the phiC31 donor-cloning vector P7-cFIX.
Generation of the Tet-On-Inducible CRISPR/Cas9 Plasmid
The generation of the CRISPR/Cas9 plasmid was realized following the F. Zhang lab’s protocol.18 Briefly, the first gRNA sequence specific for the cFIX locus was predicted based on the website tool https://zlab.bio/guide-design-resources. The first and complementary oligonucleotides were synthesized, phosphorylated, and annealed. Subsequently, the oligonucleotides were inserted into the cloning vector pX330-U6-chimeric_BB-CBh-hSpCas9 (Addgene, Cambridge, MA, USA) by performing a digestion-ligation reaction. The ligation mix was transformed into E. coli strain DH10B, and positive clones were confirmed by restriction enzyme digestion and the Sanger sequencing. The final vector was constructed by PCR amplifying the gRNA expression cassette and cloning into the pShV-TRE3G-Cas9-EF1α-Tet-ON3G plasmid via the restriction enzyme NheI-HF (New England Biolabs). To construct the single-vector system, the donor sequence was inserted into the latter plasmid using restriction enzyme I-CeuI.
Generation of cFIXmut Stable Cell Lines
Huh7 cells were cultured in DMEM (PAN Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS) (PAN Biotech) and 1% penicillin-streptomycin (PAN Biotech). PLC/PRF/5 and Hep3B cells were cultured in modified Eagle’s medium (MEM; PAN Biotech) supplemented with 10% FBS and 1% penicillin-streptomycin (PAN Biotech). Cells were transfected with phiC31 integrase vector pPhiC31 and the phiC31 donor-cloning vector P7-cFIX using FuGENE 6 transfection reagent (Promega, Madison, WI, USA). 3 days later, the cells were split into a serial dilution and transferred into 10-cm dishes. 24 h later, the media were replaced with selective media containing G418 (Carl Roth, Karlsruhe, Germany), using the following concentrations: Huh7-cFIXmut, 550 μg/mL; PLC/PRF/5-cFIXmut, 500 μg/mL; and Hep3B-cFIXmut, 300 μg/mL. The selection was maintained for 3 weeks with media changes performed every 3 days to eliminate dead cells. The mutated cFIX stable cell genome was extracted, and the mutated cFIX fragment was PCR amplified and sequenced.
Correction of Mutated cFIX via HDR with Naked DNA
For HDR experiments, Huh7-cFIXmut, PLC/PRF/5-cFIXmut, and Hep3B-cFIXmut cells were seeded in 24-well plates and transfected at approximately 75% confluency using the FuGENE 6 transfection reagent. Briefly, 800 ng Tet-on-inducible CRISPR/Cas9 plasmid and/or optimized donor sequence (PCR amplicon) was transfected per well, according to the manufacturer’s protocol. For HDR approaches, the molar ratio for Tet-on-inducible CRISPR/Cas9 plasmid to the donor sequence was 1:21. For the single-vector system, 800 ng P1, P2, and P3 were transfected. The cells transfected with the Tet-on-inducible CRISPR/Cas9 plasmid were also treated with 100 ng/mL DOX. Cells were harvested 72 h post-transfection, and gDNA was isolated and purified for the measurement of genetic correction efficiencies with the ARMS-qPCR approach.
HCAdV5-Cas9 Production
The procedure was described in detail before.33, 34 Briefly, 116 cells were cultured in MEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 40 μg/mL hygromycin B (PAN Biotech, Aidenbach, Germany). The 116-cell media contained no hygromycin B during HCAdV5-Cas9 and HCAdV5-Cas9-donor vector amplification. Before transfection, the HCAdV5-Cas9 and HCAdV5-Cas9-donor plasmids were linearized by the restriction enzyme NotI-HF (New England Biolabs) digestion. Then the linearized HCAdV5-Cas9 and HCAdV5-Cas9-donor constructs were transfected into 116 producer cells. At 16–18 h post-transfection, the 116 cells were infected with helper virus. After viral amplification for 18–24 days until passage 6, the viruses were purified using cesium chloride (CsCl) density gradient centrifugation. The total particles, helper virus contamination, and infectious units of the purified HCAdV5-Cas9 and HCAdV5-Cas9-donor viruses were measured by qPCR.
ssAAV2-cFIX Production
HEK293 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. The pAAV-RC, pHelper, and pZAC-T2B constructs were co-transfected into HEK293 cells using calcium phosphate transfection. The media were changed the second day. After 72 h, all media and cells were harvested, and ssAAV2-cFIX vectors were purified using ViraBind AAV Purification Kit (Cell Biolabs, San Diego, CA, USA). The total particles and infectious units of the purified ssAAV2-cFIX vectors were measured by qPCR.
Co-transduction of HCAdV5-GFP and ssAAV2-Cherry into Huh7-cFIXmut
To analyze the co-transduction of HCAdV5 and ssAAV2 into Huh7-cFIXmut in the two viral vector strategy, we seeded Huh7-cFIXmut in 24-well plates with medium containing 10% FBS. The cells were transduced with HCAdV5-GFP (MOI 100) and/or ssAAV2-mCherry (MOIs 200, 400, and 600) on the second day. Ar 3 days after co-transduction, the fluorescence in transduced Huh7-cFIXmut cells was measured using fluorescence microscopy and analyzed with ImageJ software.
T7E1 Assay
The T7E1 assay was employed to determine the CRISPR/Cas9 nuclease efficacy and off-target effects. Briefly, a 25-μL PCR reaction using Phusion High-Fidelity DNA Polymerase (New England Biolabs) was set up to amplify the target locus with primers cFIX long fwd6 (5′-CGA TCG GCT TCA ATT CTT CA-3′) and cFIX long rev4 (5′-CCT AAA CGT GTC AAC CTT GGA-3′). To measure off-target effects, the following primer pairs were used: hFIX: cFIX-OFF-F9F, 5′-TGGATCTGGCTATGTAAGTGGCTG-3′ and cFIX-OFF-F9R, 5′-GGAAGATGGAATGCTGCTAAGGAG-3′; HDAC7: cFIX-OFF-2F, 5′- TTCCTCCCATTCCTCTCCCTC-3′ and cFIX-OFF-2R, 5′-TACAATGGTGGCGCCTCACG-3′; H3K27AC: cFIX-OFF-3F, 5′-CTTGCTGTGATATACCCATCACCC-3′ and cFIX-OFF-3R, 5′-TGCACGTCTGCTGTCTCAGG-3′; CEP192: cFIX-OFF-4F, 5′-AGATTGTCTTCCCCTGCTAAACCC-3′ and cFIX-OFF-4R, 5′-TTATACACAGCTGCCTGCCCTC-3′; ZNRF2: cFIX-OFF-5F, 5′-AGCCATGTGAGCTGTACCCAAG-3′ and cFIX-OFF-5R, 5′-CACAGCTTCTTACCCGACACTAAC-3′.
The PCR program was as follows: 98°C, 30 s; 30 cycles × (98°C 10 s, 60°C 30 s, and 72°C 7 s); and 72°C, 10 min. For heteroduplex formation, the PCR amplification was purified via ethanol precipitation and dissolved in 17.5 μL double-distilled water. The purified PCR product was mixed with 2 μL NEB2 buffer (New England Biolabs), and the mixture was transferred into a PCR machine using the following protocol: 95°C, 5 min; 95°C decrease to 85°C (−2°C/s); and 85°C decrease to 25°C (−0.1°C/s). Then 0.5 μL T7E1 (New England Biolabs) was added to the annealed PCR products, and the reaction was incubated for 20 min at 37°C. The reaction was stopped by adding loading dye (New England Biolabs) and subsequently loaded on a 2% agarose gel. The band strengths were analyzed with the Bio-Rad Gel Doc EZ Imager and the Image Lab 5.2 software (Bio-Rad, Munich, Germany). The nuclease efficacy was estimated using the following formula: fractional modification = (1 − (1 − (fraction of cleaved bands))1/2).57
Genetic Correction of Mutated cFIX via HDR Using the Viral Vectors HCAdV5-Cas9 and ssAAV2-cFIX
To analyze HDR mediated by virus-delivered nucleases and donor in the context of hemophilia B, Huh7-cFIXmut cells were seeded in 24-well plates with medium containing 10% FBS. On the second day, the cells were transduced with HCAdV5-Cas9 (MOI 100) and/or ssAAV2-cFIX (MOIs 200, 400, and 600). The cells transduced with HCAdV5-Cas9 were also treated with 100 ng/mL DOX. For the ARMS-qPCR assay, cells were harvested and total cellular genomic DNA was isolated at 72 h post-transduction. For the ELISA, cell media were collected at 48, 72, and 96 h post-transduction. New medium was added at 48 and 72 h post-transduction.
ARMS-qPCR
For the detection of HDR efficiency, a modified version of established ARMS-qPCR was employed as previously described.30 The outer PCR primer pair sequences amplifying the identical region of all samples were as follows: forward primer, 5′-GTT CCA CTT GTT GAC CGA GC-3′; and reverse primer, 5′-CCT AAA CGT GTC AAC CTT GGA-3′. The detection PCR primer pair specific for cFIX donor sequences was as follows (the underlined bases match donor, but not mutated cFIX): forward primer, 5′-CGA TCG GCT TCA ATT CTT CA-3′; and reverse primer, 5′-CAG GAA ATA ATC CCA GTC AAG-3′. Plasmid mixtures of mutated cFIX and cFIX donor sequence contain 1% and 100% of the cFIX donor sequence. As described in our previous report,15 MOD1% represents the adjusted sample containing 1% of modified cFIX plasmid mixed with mutated cFIX plasmid to a final molar ratio of 1:99, respectively.
Both detection PCR and outer PCR were performed separately in a total volume of 10 μL containing 5 μL DNA sample, 2 μL 5× EvaGreen-qPCR-Mix-II (Bio-Budget, Krefeld, Germany), 200 nmol/L of each primer, and 0.5 μL DMSO. qPCR was performed as follows: initial incubation 95°C for 15 min and 40 cycles × (95°C for 15 s, 60°C for 20 s, and 72°C for 20 s). The Pfaffl method was used to calculate the HDR efficiency (percentage of modification) and the formula was as follows: Percentage of modification = (EdetPCR)ΔCT, detPCR (calibrator-test)/(EoutPCR)ΔCT, outPCR (calibrator-test) (EdetPCR and EoutPCR are amplification efficiencies of detPCR and outPCR, respectively). Furthermore, the following samples were analyzed: calibrator = 100% of the cFIX donor sequence; test = analyzed samples; ΔCT detPCR (calibrator-test) = CT of the detPCR in the calibrator minus the CT of the detPCR in the analyzed sample; ΔCT outPCR (calibrator-test) = CT of the outPCR in the calibrator minus the CT of the outPCR in the analyzed sample; and CT = Cycle number.
ELISA
The capture monoclonal antibody (Affinity Biologicals, Ancaster, ON, Canada) specific for cFIX was coated onto polystyrene microtiter ELISA plates (Greiner Bio-One, Kremsmünster, Austria) at 4°C overnight, and the remaining protein-binding sites in the coated wells were blocked by Tris-buffered saline with Tween 20 (TBST) with 5% BSA. Then the samples were incubated with horseradish peroxidase (HRP)-conjugated second antibody (Affinity Biologicals, Ancaster, ON, Canada). Afterward the addition of substrate solution resulted in a color reaction, and the absorbance was measured at 490 nm using BioPhotometer Plus (Eppendorf, Hamburg, Germany).
Statistical Analyses
All experiments were performed using at least triplicates. Statistical significance was determined by unpaired Student’s t test. All data are reported as mean ± SEM. Statistical comparison was performed using the two-tailed Student’s t test, and a value of p < 0.05 was considered to be relevant compared to the respective control group.
Author Contributions
J.G. designed the research, analyzed the data, and wrote the paper. T.B. developed and tested the CRISPR/Cas9 cassette. W.Z. constructed ssAAV2-cFIX plasmid and provided adenoviruses expressing luciferase. E.E.-S. performed research involving ARMS-qPCR. E.E.-S., M.S., and W.Z. revised the paper. A.E. designed the research, revised the paper, and approved the final submitted version.
Conflicts of Interest
All the authors declare no competing financial interests in relation to this work.
Acknowledgments
This project was supported by the China Scholarship Council to J.G.; internal research funding of the Witten/Herdecke University to E.E.-S., M.S., and W.Z.; and the Else Kröner-Fresenius Foundation to A.E.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.12.008.
Supplemental Information
References
- 1.Nathwani A.C., Tuddenham E.G. Epidemiology of coagulation disorders. Baillieres Clin. Haematol. 1992;5:383–439. doi: 10.1016/s0950-3536(11)80025-9. [DOI] [PubMed] [Google Scholar]
- 2.Sun J., Hua B., Livingston E.W., Taves S., Johansen P.B., Hoffman M., Ezban M., Monroe D.M., Bateman T.A., Monahan P.E. Abnormal joint and bone wound healing in hemophilia mice is improved by extending factor IX activity after hemarthrosis. Blood. 2017;129:2161–2171. doi: 10.1182/blood-2016-08-734053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kay M.A., Rothenberg S., Landen C.N., Bellinger D.A., Leland F., Toman C., Finegold M., Thompson A.R., Read M.S., Brinkhous K.M. In vivo gene therapy of hemophilia B: sustained partial correction in factor IX-deficient dogs. Science. 1993;262:117–119. doi: 10.1126/science.8211118. [DOI] [PubMed] [Google Scholar]
- 4.Kay M.A., Manno C.S., Ragni M.V., Larson P.J., Couto L.B., McClelland A., Glader B., Chew A.J., Tai S.J., Herzog R.W. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 2000;24:257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 5.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma A., Easow Mathew M., Sriganesh V., Reiss U.M. Gene therapy for haemophilia. Cochrane Database Syst. Rev. 2016;12:CD010822. doi: 10.1002/14651858.CD010822.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crudele J.M., Finn J.D., Siner J.I., Martin N.B., Niemeyer G.P., Zhou S., Mingozzi F., Lothrop C.D., Jr., Arruda V.R. AAV liver expression of FIX-Padua prevents and eradicates FIX inhibitor without increasing thrombogenicity in hemophilia B dogs and mice. Blood. 2015;125:1553–1561. doi: 10.1182/blood-2014-07-588194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arruda V.R., Doshi B.S., Samelson-Jones B.J. Novel approaches to hemophilia therapy: successes and challenges. Blood. 2017;130:2251–2256. doi: 10.1182/blood-2017-08-742312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair N., Rincon M.Y., Evens H., Sarcar S., Dastidar S., Samara-Kuko E., Ghandeharian O., Man Viecelli H., Thöny B., De Bleser P. Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood. 2014;123:3195–3199. doi: 10.1182/blood-2013-10-534032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J., Cuker A., Sullivan L.M., Majumdar S., Teitel J. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N. Engl. J. Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F., Valdmanis P.N., Spector L.P., Porteus M.H., Gaensler K.M., Kay M.A. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anguela X.M., Sharma R., Doyon Y., Miller J.C., Li H., Haurigot V., Rohde M.E., Wong S.Y., Davidson R.J., Zhou S. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122:3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Haurigot V., Doyon Y., Li T., Wong S.Y., Bhagwat A.S., Malani N., Anguela X.M., Sharma R., Ivanciu L. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergmann T., Ehrke-Schulz E., Gao J., Schiwon M., Schildgen V., David S., Schildgen O., Ehrhardt A. Designer nuclease-mediated gene correction via homology-directed repair in an in vitro model of canine hemophilia B. J. Gene Med. 2018;20:e3020. doi: 10.1002/jgm.3020. [DOI] [PubMed] [Google Scholar]
- 16.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 17.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan Y., Ma Y., Li Q., Sun Z., Ma L., Wu L., Wang L., Zeng L., Shao Y., Chen Y. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huai C., Jia C., Sun R., Xu P., Min T., Wang Q., Zheng C., Chen H., Lu D. CRISPR/Cas9-mediated somatic and germline gene correction to restore hemostasis in hemophilia B mice. Hum. Genet. 2017;136:875–883. doi: 10.1007/s00439-017-1801-z. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T.H., Anegon I. Successful correction of hemophilia by CRISPR/Cas9 genome editing in vivo: delivery vector and immune responses are the key to success. EMBO Mol. Med. 2016;8:439–441. doi: 10.15252/emmm.201606325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans J.P., Brinkhous K.M., Brayer G.D., Reisner H.M., High K.A. Canine hemophilia B resulting from a point mutation with unusual consequences. Proc. Natl. Acad. Sci. USA. 1989;86:10095–10099. doi: 10.1073/pnas.86.24.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groth A.C., Olivares E.C., Thyagarajan B., Calos M.P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrhardt A., Xu H., Dillow A.M., Bellinger D.A., Nichols T.C., Kay M.A. A gene-deleted adenoviral vector results in phenotypic correction of canine hemophilia B without liver toxicity or thrombocytopenia. Blood. 2003;102:2403–2411. doi: 10.1182/blood-2003-01-0314. [DOI] [PubMed] [Google Scholar]
- 27.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pliatsika V., Rigoutsos I. “Off-Spotter”: very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biol. Direct. 2015;10:4. doi: 10.1186/s13062-015-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrke-Schulz E., Bergmann T., Schiwon M., Doerner J., Saydaminova K., Lieber A., Ehrhardt A. Quantification of designer nuclease induced mutation rates: a direct comparison of different methods. Mol. Ther. Methods Clin. Dev. 2016;3:16047. doi: 10.1038/mtm.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 32.High K.A., Anguela X.M. Adeno-associated viral vectors for the treatment of hemophilia. Hum. Mol. Genet. 2016;25(R1):R36–R41. doi: 10.1093/hmg/ddv475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer D., Ng P. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Jager L., Hausl M.A., Rauschhuber C., Wolf N.M., Kay M.A., Ehrhardt A. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat. Protoc. 2009;4:547–564. doi: 10.1038/nprot.2009.4. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Fu J., Liu J., Wang H., Schiwon M., Janz S., Schaffarczyk L., von der Goltz L., Ehrke-Schulz E., Dörner J. An Engineered Virus Library as a Resource for the Spectrum-wide Exploration of Virus and Vector Diversity. Cell Rep. 2017;19:1698–1709. doi: 10.1016/j.celrep.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Alba R., Bradshaw A.C., Parker A.L., Bhella D., Waddington S.N., Nicklin S.A., van Rooijen N., Custers J., Goudsmit J., Barouch D.H. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood. 2009;114:965–971. doi: 10.1182/blood-2009-03-208835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waddington S.N., McVey J.H., Bhella D., Parker A.L., Barker K., Atoda H., Pink R., Buckley S.M., Greig J.A., Denby L. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Kalyuzhniy O., Di Paolo N.C., Silvestry M., Hofherr S.E., Barry M.A., Stewart P.L., Shayakhmetov D.M. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:5483–5488. doi: 10.1073/pnas.0711757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mian A., Guenther M., Finegold M., Ng P., Rodgers J., Lee B. Toxicity and adaptive immune response to intracellular transgenes delivered by helper-dependent vs. first generation adenoviral vectors. Mol. Genet. Metab. 2005;84:278–288. doi: 10.1016/j.ymgme.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Bangari D.S., Mittal S.K. Current strategies and future directions for eluding adenoviral vector immunity. Curr. Gene Ther. 2006;6:215–226. doi: 10.2174/156652306776359478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morral N., O’Neal W., Rice K., Leland M., Kaplan J., Piedra P.A., Zhou H., Parks R.J., Velji R., Aguilar-Córdova E. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. USA. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehrhardt A., Kay M.A. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood. 2002;99:3923–3930. doi: 10.1182/blood.v99.11.3923. [DOI] [PubMed] [Google Scholar]
- 43.Flynn R., Buckler J.M., Tang C., Kim F., Dichek D.A. Helper-dependent adenoviral vectors are superior in vitro to first-generation vectors for endothelial cell-targeted gene therapy. Mol. Ther. 2010;18:2121–2129. doi: 10.1038/mt.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muruve D.A., Cotter M.J., Zaiss A.K., White L.R., Liu Q., Chan T., Clark S.A., Ross P.J., Meulenbroek R.A., Maelandsmo G.M., Parks R.J. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 2004;78:5966–5972. doi: 10.1128/JVI.78.11.5966-5972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blacklow N.R., Hoggan M.D., Sereno M.S., Brandt C.D., Kim H.W., Parrott R.H., Chanock R.M. A seroepidemiologic study of adenovirus-associated virus infection in infants and children. Am. J. Epidemiol. 1971;94:359–366. doi: 10.1093/oxfordjournals.aje.a121331. [DOI] [PubMed] [Google Scholar]
- 46.Mayor H.D., Houlditch G.S., Mumford D.M. Influence of adeno-associated satellite virus on adenovirus-induced tumours in hamsters. Nat. New Biol. 1973;241:44–46. doi: 10.1038/newbio241044b0. [DOI] [PubMed] [Google Scholar]
- 47.Hermonat P.L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 48.Niemeyer G.P., Herzog R.W., Mount J., Arruda V.R., Tillson D.M., Hathcock J., van Ginkel F.W., High K.A., Lothrop C.D., Jr. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood. 2009;113:797–806. doi: 10.1182/blood-2008-10-181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arruda V.R., Stedman H.H., Haurigot V., Buchlis G., Baila S., Favaro P., Chen Y., Franck H.G., Zhou S., Wright J.F. Peripheral transvenular delivery of adeno-associated viral vectors to skeletal muscle as a novel therapy for hemophilia B. Blood. 2010;115:4678–4688. doi: 10.1182/blood-2009-12-261156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Nichols T.C., Read M.S., Bellinger D.A., Verma I.M. Sustained expression of therapeutic level of factor IX in hemophilia B dogs by AAV-mediated gene therapy in liver. Mol. Ther. 2000;1:154–158. doi: 10.1006/mthe.2000.0031. [DOI] [PubMed] [Google Scholar]
- 51.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bak R.O., Porteus M.H. CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep. 2017;20:750–756. doi: 10.1016/j.celrep.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrke-Schulz E., Schiwon M., Leitner T., Dávid S., Bergmann T., Liu J., Ehrhardt A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 2017;7:17113. doi: 10.1038/s41598-017-17180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herzog R.W., Hagstrom J.N., Kung S.H., Tai S.J., Wilson J.M., Fisher K.J., High K.A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc. Natl. Acad. Sci. USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hausl M.A., Zhang W., Müther N., Rauschhuber C., Franck H.G., Merricks E.P., Nichols T.C., Kay M.A., Ehrhardt A. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol. Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.