Abstract
Essential genes play an indispensable role in supporting the life of an organism. Identification of essential genes helps us to understand the underlying mechanism of cell life. The essential genes of bacteria are potential drug targets of some diseases genes. Recently, several computational methods have been proposed to detect essential genes based on the static protein–protein interactive (PPI) networks. However, these methods have ignored the fact that essential genes play essential roles under certain conditions. In this work, a novel method was proposed for the identification of essential proteins by fusing the dynamic PPI networks of different time points (called by FDP). Firstly, the active PPI networks of each time point were constructed and then they were fused into a final network according to the networks’ similarities. Finally, a novel centrality method was designed to assign each gene in the final network a ranking score, whilst considering its orthologous property and its global and local topological properties in the network. This model was applied on two different yeast data sets. The results showed that the FDP achieved a better performance in essential gene prediction as compared to other existing methods that are based on the static PPI network or that are based on dynamic networks.
Keywords: essential genes, dynamic network, network fusion, protein–protein interactive network
1. Introduction
Essential genes (and their encoded proteins) play an indispensable role in supporting the life of organisms, and without them, lethality or infertility is caused. Studying essential genes helps us to understand the basic requirements for cell viability and fertility [1]. Moreover, identifying the essential genes of bacteria contributes to finding potential drug targets for new antibiotics [2]. Recently, some researchers pointed out that essential genes have a close relationship with human diseases [3]. Studying essential genes also helps us to design novel strategies for disease therapy. However, the methods to experimentally discover essential genes in biology are time consuming and inefficient. Consequently, several recent computational methods have been proposed to identify essential genes [4,5]. Generally, these computational methods can be classified into three categories: sequence-based methods, network-based methods, and multi-biological information-based methods.
Sequence-based methods are based on the fact that essential genes evolve much slower than other genes, and that they usually conserve across different species [6,7,8]. This kind of method usually infers essential genes by comparing their sequences with the sequences of known essential genes in the same species or other species. However, with the exponential increase of sequencing gene data, few of them can find orthologous essential genes in the same species or in the species at a long evolution distance.
Network-based methods identify essential genes according to their topological properties in the protein–protein interactive (PPI) networks. Essential genes are supposed to be the center of the PPI network, because removing them from the networks would cause the lethality and break down of the network [9]. A series of genes’ centrality scores in the networks have been employed to identify the essential genes, including Degree Centrality (DC) [10], Betweenness Centrality (BC) [11], Closeness Centrality (CC) [12], Subgraph Centrality (SC) [13], Eigenvector Centrality (EC) [14], Information Centrality (IC) [15], and Edge Clustering Coefficient Centrality (NC) [16].
However, there are some limitations in the network-based methods, including incomplete and error-prone currently available PPI data, and the neglect of other intrinsic properties of the essential genes. To overcome these limitations, some methods integrate PPI networks with other biological information to improve the prediction accuracy of essential proteins. One of these methods refines the currently available PPI network by introducing other biological information, such as gene functional annotation data [17], gene expression data, a centrality measure by integrating protein-protein interaction data and gene expression data (PEC) [18], weighted degree centrality (WDC) [19]), both gene functional data and gene expression data [20], and subcellular information [21]. Another type of method predicts essential genes by comprehensively utilizing the inner biological properties and the topological properties of essential genes. For example, considering the conservative property of essential genes, Peng et al. [22] proposed a method for predicting essential genes by integrating the orthology with the PPI networks, namely ION. Considering the close relationship between genes’ essentiality and the protein domain of their coded proteins, the same research group [23] introduced protein domain information into the PPI network to predict essential genes. Since essential genes are highly likely to be rich in protein complexes, some methods use information on known protein complexes [24,25] to identify essential genes. Recently, researchers tried to improve the prediction performance of essential genes by combining more biological properties. Li et al. [20] combined subcellular localization information, orthology information, and PPI networks to predict essential genes. Zhang et al. [26] detected essential genes based on the network topology, gene expression data, and gene ontology information.
However, the aforementioned methods all ignore the fact that essential genes interact with each other and play essential roles under certain conditions. Most of these methods are based on the static PPI networks (called the S_PPI), which consist of interactions accumulated in different conditions and time points. In fact, the interactions between genes in cells change over time, environment, and different stages of the cell cycle [27]. Some researchers try to construct dynamic PPI networks by combining the S_PPI with gene expression profiles [28]. These methods firstly identify the active genes at each time point, according to their expression levels. After that, these active genes are mapped to the S_PPI and a serial of dynamic PPI networks (called the D_PPI) are constructed for each time point. Different methods use different strategies to select active genes at each time point. Tang et al. [29] select the active genes of each expression time point if their corresponding expression values are above a threshold. However, this method will filter out the active genes with low expression values. To overcome this shortcoming, Wang et al. [30] proposed a three-sigma principle-based method that sets a threshold for each gene by considering its own expression curve over different time points. Xiao et al. [4] divide the active genes into two categories: Time-dependent genes and time-independent genes. The active genes of each time point are identified using a k-sigma method, where the k-sigma method computes an active threshold for each gene according to the mean and standard deviation of its expression values. Li et al. [31] use both the subcellular information and the temporal information to construct a dynamical PPI network. Recently, some methods that detect essential genes based on the dynamical PPI networks have started to emerge. Xiao et al. [32] map all the active genes of each time point detected by their method in Reference [4] to a static PPI, and then predict the essential genes using some centrality methods, i.e., DC [10], NC [16], BC [11], CC [12], and SC [13]. Li et al. [33] refine the S_PPI according to whether or not the interactive genes are simultaneously active at a time point and locate in the same subcellular location. After that, more centrality methods besides the methods mentioned in Reference [32] are implemented on the refined networks to predict the essential genes. For all we know, these methods improve the prediction of essential proteins either by using active genes in the dynamic PPI network to refine the static PPI network [32,33,34], or by simply averaging the scores of the active genes at each time point to get final ranking scores. More complex strategies should be designed to predict the essential genes from dynamic PPI networks.
In this work, a novel method was developed for the identification of essential genes by fusing the dynamic PPI networks of different time points (FDP). Firstly, a serial of active PPI networks of each time point was constructed using Xiao’s method [4], and then these active PPI networks were fused into a final network using the method similar to that described in Reference [35]. In contrast to the method in Reference [35], FDP fuses the active PPI networks one by one according to their similarities. The nodes in the final network are active for at least one time point and their interactions are similar across all the time points. This idea relates to previous observations that the mRNA expression levels of essential genes tend to be high (active) [36] and vary, on average, within a narrow range, whereas the expression of non-essential genes fluctuates more widely [37]. Finally, a novel centrality method was designed to assign a ranking score to each node in the final network, whilst considering its orthologous property and both its global and local topological properties in the network. FDP, as well as eleven other existing methods were applied to predict yeast essential genes. Prediction results showed that FDP not only outperformed the existing methods that were based on the static PPI network, but it also outperformed the methods that were based on the dynamic PPI network.
2. Materials and Methods
2.1. Materials
FDP and other existing computational methods were applied to predict the essential genes of S. cerevisiae (Bakers’ Yeast). Two different PPI datasets of Saccharomyceas cerevisiae were adopted to evaluate our method. One dataset was the DIP_PPI, downloaded from the DIP database [38] published on 10 October 2010. There were a total of 5093 proteins and 24,743 interactions, excluding self-interactions and repeated interactions. The other dataset was the SC_net from Reference [39], which consisted of 4746 proteins and 15,166 distinct interactions.
The list of essential genes was integrated from the following databases: The Munich Information Center for Protein Sequences (MIPS) [40], Saccharomyces genome database (SGD) [41], Database of Essential Genes (DEG) [42], and Saccharomyces Genome Deletion Project (SGDP) [43]. There were 1285 essential genes, where only 1167 essential genes present in the DIP_PPI network and 1130 essential genes present in the SC_net network. The yeast’s gene expression data came from Reference [28], including 6777 gene products under 36 different time points of three life cycles. Therefore, there were 2759 genes or 2559 genes that appeared in the dynamic PPI networks constructed by combining the yeast’s gene expression data with the DIP_PPI network or the SC_net network, respectively. Moreover, 827 of the 2759 genes in the dynamic DIP_PPI network were essential genes and 785 of the 2559 genes in the dynamic SC_net network were essential genes. Table 1 lists the detailed information of the two yeast data sets.
Table 1.
Details of the two different yeast data sets. PPI: protein–protein interactive
| Network | Genes in S_PPI | Edges in S_PPI | Genes in D_PPI | Essential Genes in S_PPI | Essential Genes in D_PPI |
|---|---|---|---|---|---|
| DIP_PPI | 5093 | 24743 | 2759 | 1167 | 827 |
| SC_net | 4746 | 15166 | 2559 | 1130 | 785 |
Information on the orthologous proteins was taken from Version 7 of the InParanoid database. In our study, yeast proteins were mapped to another 99 species to find their orthologous proteins. Only the proteins in the seed orthologous sequence pairs of each cluster generated by InParaniod were chosen as the orthologous proteins.
2.2. Methods
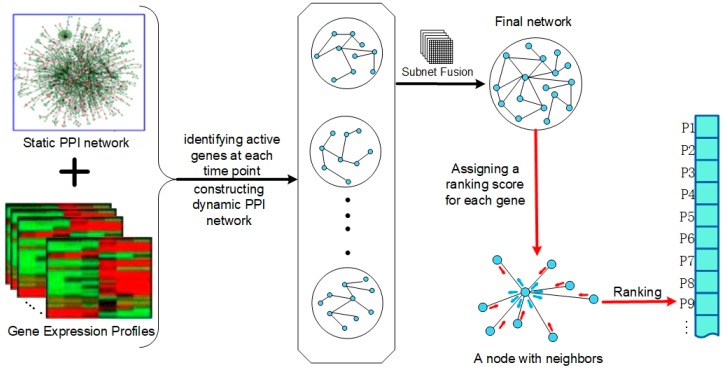
Figure 1 illustrates the workflow of the FDP. FDP takes three main steps to predict essential genes. Firstly, active PPI networks of each time point were constructed using Xiao’s method [4]. After that, a fusion method similar to Reference [35] was adopted to fuse the active PPI networks of different time points, and then a final network was constructed, in which the nodes were active for at least one time point and their interactions were similar across all the time points. Finally, a novel centrality method was designed to assign a ranking score for each node in the final network, and the nodes ranked on top were selected as the candidate essential genes.
Figure 1.
The workflow of our method.
2.2.1. Constructing Dynamic Protein–Protein Interactive Networks
Our dynamic PPI networks were constructed based on the gene expression profiles and the PPI network. The expression profiles consisted of periodically (time-dependent) and non-periodically (time-independent) expressed profiles and some inevitable noise. De Lichtenberg et al. [44] point out that periodically expressed genes are more likely to be dynamically deterministic than random. However, the non-periodically expressed genes are more likely to be random than dynamically deterministic. Therefore, the first step to construct the dynamic PPI networks was to detect the time-dependent genes and time-independent genes from the time-course gene expression profiles using an AR (autoregressive) model as in Reference [45].
Let x = {x1, …, xm, …, xM} be a time series of observation values at equally-spaced time points from a dynamic system. A gene is supposed to be time dependent if its gene expressions have linear relationships and can be modeled by an AR model of order p (see Equation (1)). A gene is regarded to be time independent if its gene expressions have nonlinear relationships and can be modeled by an AR model of order zero (see Equation (2)).
| (1) |
| (2) |
where βi (i = 0, 1, …, p) is the autoregressive coefficient, and εm (m = p + 1,…, M) denotes the random error, which follows a normal distribution with a mean of 0 and a variance of σ2. Since the order of the AR model in Equation (1) is unknown, similar to Reference [4], the p-values for all possible orders p (1 ≤ p ≤ (M − 1)/2) were calculated. A gene is regarded to be time dependent if one of these p-values calculated from its expression profile is smaller than a user-preset threshold value (threshold = 0.01). The expression profiles of a gene will be considered as noise if the gene is not only time-independent, but also if the mean of its expression values across all time points is very small (less than 0.5, according to the analysis in Reference [4]).
After identifying the time-dependent and the time-independent genes, and filtering out the noisy genes, the next step was to detect which of them were active at each time point. A gene was considered to be active when its expression value was above a given threshold (see Equation (3)). In this work, similar to Reference [34], we set the threshold for each gene using the following k-sigma principle, where k was set to 2.5, u and σ were the mean and standard deviation of their expression values.
| Active threshold = u + kσ × (1 − F) | (3) |
| (4) |
Therefore, a serial of active PPI networks was generated by mapping the active genes at each time point to the S_PPI and extracting the edges connecting them. Since the active genes were different at different time points, these active PPI networks dynamically changed over time. The details of the dynamic network construction algorithm are shown in Algorithm 1.
| Algorithm 1 Dynamic Network Construction |
| Input: A static PPI (S_PPI) network represented as Graph G = (V, E, W), a time series of the gene expression profile of each gene in G, parameter k. |
| Output: The active networks of each time point. |
| Step1: Identify two categories of genes, the time-dependent genes and the time-independent genes. using Equations (1) and (2), according to their expression profiles. |
| Step2: Filter out the noise genes in the time-independent genes. |
| Step3: Identify the active genes of each time point from the remaining two categories of genes by judging whether or not their expression values are above the threshold (calculated by Equation (3)). |
| Step4: Map the active genes of each time point to the S_PPI network and extract the active networks of each time point. |
2.2.2. Fusing the Active Protein–Protein Interactive Networks of Each Time Point
After constructing the active networks of each time point, the next step was to fuse them into a single network, which captured the shared and complementary network structure of all the active networks, offering insight into how the expression of proteins was similar across different time points from the view of the network structure. To formally define the process of fusing networks, the following variables were introduced.
A static PPI network (S_PPI) can be represented as an undirected graph G = (V, E, W), where a node v∈V represents a gene and an edge e(u,v) ∈E denotes an interaction between two genes v and u. w(u,v) denotes the weight of the edge e(u,v), which measures the similarity between genes v and u. A dynamic PPI can be represented as a serial of active networks of different time points G1, G2,.... Gi, … Gn, where Gi = (Vi, Ei, Wi) represents a subgraph of G at the ith time point. Vi∈V is the set of nodes that are active at the ith time point. Ei∈E is a subset of E that connects the active genes at the ith time point. Wi is an adjacency matrix of Gi, where its entry wi(ui,vi) measures the closeness of two nodes in the ith active network. The edges in the active network of each time point are weighted by Equation (5).
| (5) |
| (6) |
| (7) |
where Ni(ui) is the neighbor of ui in Gi. ECCi(ui,vi) is the edge-clustering coefficient of ei(ui,vi) in Gi, which is defined as the number of common neighbors of node ui and node vi in Gi divided by the number of common neighbors that might possibly exist between them. Since essential genes tend to form density clusters [22], their edge clustering coefficients can describe the degree to which two genes tend to cluster together. Similar to previous works [22,23,46,47], the edges in the active networks of each time are weighted by the edge clustering coefficients (see Equation (5)). Mean (Ecci(ui,N(ui))) is the average of the edge clustering coefficient values between ui and its neighbors in Gi. μ is a parameter that is empirically set to 0.5 according to the recommendation in Reference [35].
For the active network of the ith time point Gi, its adjacency matrix Wi has two derivatives, namely, matrix Pi and matrix Si. Matrix Pi carries the global information about the similarity of each gene to all the others obtained by performing normalization on Wi:
| (8) |
Matrix Si only encodes the similarity between each gene in Gi and its K nearest neighbors (K = 20 according to the recommendation in Reference [35]):
| (9) |
Given the number M of active networks at different time points, we could construct an adjacency matrix Wi of Gi using Equation (5) for the ith time point, i = 1, 2, 3, 4, … M. Pi and Si were obtained from Equations (8) and (9), respectively. The aim of the network fusion was to fuse the M active networks into a single network. The process was as follows.
Firstly, the similarities between any two networks were calculated based on the Euclidean distance of their adjacency matrixes Wi (i = 1, 2, 3 … M). Then the nearest two networks, i.e., i and j, were selected to fuse by the following iterative process.
| (10) |
| (11) |
Let and represent the initial two statuses at iteration step t = 0. and represent the status matrix of the active networks at the ith and the jth time point after t iteration steps, respectively. After t iteration steps, the fused network of the two networks was computed as
| (12) |
Then, R was the result of the fused active networks i and j. After that, the similarities between R and the remaining active networks were recomputed again. R and its closest active network were selected to fuse into one network by repeating the above process until all the active networks were fused into a single network. Algorithm 2 shows the algorithm for fusing active PPI networks.
| Algorithm 2 Active PPI network fusion |
| Input: Active networks of each time point, parameter K. |
| Output: Final fused network. |
| Step1: Construct adjacency matrix Wi of the ith active network (I = 1, 2, 3, 4, …, M) using Equation (5). |
| Step2: Construct Pi and Si of the ith active network using Equations (8) and (9). |
| Step3: Calculate the similarities between any two networks based on the Euclidean distance of their adjacency matrixes. |
| Step4: Select the nearest two active networks Gi and Gj, , , t = 0. Step5: Compute and using Equations (10) and (11), let t = t + 1. Step6: Repeat step 5 until t = 20. Step7: Compute the fused network R of Gi and Gj using Equation (12). Step8: Let Wr = R, construct Pr and Sr of the fused network R using Equation (8) and (9). Step9: Find the nearest active network Gk to R from the remaining active networks, let = , t = 0. Step10: Compute and using Equations (10) and (11), let t = t + 1. Step11: Repeat step 10 until t = 20. Step12: Compute the fused network of R and Gk using Equation (12), the fused network is named as R. Step13: Remove Gk from active network list and repeat steps 8 to 12 until all the active networks are fused to a final network. Step14: Output the final fused network. |
2.2.3. Ranking Genes in the Fused Network
After fusing the active networks of different time points, an algorithm was designed to assign each gene in the fused network a ranking score. The ranking score measured the importance of the gene in the fused network from both the global and local perspectives.
A random walking process was implemented on the fused network to capture the global information of each gene. Let H be an F*F adjacency matrix of the final fused network. All its entries, i.e., h(i,j), were normalized by row. F is the number of genes in the network. In fact, F is the number of genes that are active at one of the time points. Let pr(i) be the ranking score of node i with respect to its global property in the fused network, which can be computed as follows.
| (13) |
| (14) |
where o(i) denotes the orthologous scores of node i, which is calculated by the number of times that the node has orthologs in the reference organisms. is the maximal orthologous score among all the nodes in the network. Similar to Reference [22], we adopted an iterative process to numerically solve Equation (13). Here, parameter а was set to 0.5 according to the recommendation in Reference [22].
The interaction frequency entropy (IFE) of a gene in the final fused network measured its local topological properties. For a gene i, since we only considered its local properties, the interactions connecting to its K closest neighbors were selected to calculate its IFE values (K = 20 according to the recommendation in Reference [35]).
| (15) |
| (16) |
where KNN(i,K) denotes the K closest neighbor set of node i, |KNN(i,K)| denotes the selected neighbor set size. Equation (16) was employed to perform the min-max normalization on the node’s IFE value.
Eventually, the ranking score of a node i in the final fused network, which was represented by FDP(i), equaled to the linear combination of its global topological score denoted as pr(i) and its local closest neighbors’ influence denoted as IFE(i). The parameter λ (0 ≤ λ ≤ 1) was used to adjust the weight of the two scores in the ranking score. Algorithm 3 shows the algorithm for computing the FDP values of genes.
| FDP(i) = λ*pr(i) + (1 − λ) IFE(i) | (17) |
| Algorithm 3 FDP |
| Input: A static PPI network represented as Graph G = (V, E, W), gene expression profile, orthologs data sets between Yeast and 99 other organisms (ranging from H.sapiens to E.coli), stopping error , parameter а, λ. |
| Output: FDP values of genes. |
| Step1: Construct active networks of each time point using the Dynamic Network Construction algorithm. |
| Step2: Fuse these active networks into a final fused network using the Active PPI network fusion algorithm. |
| Step3: Calculate the orthologous scores of each node in the final fused network using Equation (14). |
| Step4: Construct matrix H and normalize all its entries by row. |
| Step5: Initialize pr with pr0 = d, let t = 0. Step6: Compute prt+1 using Equation (13), let t = t + 1. Step7: Repeat step 6 until . |
| Step8: Calculate the IFE value of each gene in the final fused network using Equations (15) and (16). |
| Step9: Calculate the FDP value of each gene in the final fused network by linearly combining its pr value and IFE value (see Equation (17)). |
3. Results
In order to evaluate the performance of FDP in essential gene prediction, we compared the FDP with other existing methods (DC [10], BC [11], CC [12], SC [13], EC [14], IC [15], NC [16], PeC [18], ION [22], APPIN_DC [32], and APPIN_NC [32]). DC, BC, CC, SC, EC, IC, and NC are typical centrality-based methods that only consider the topological properties of genes in the S_PPI network. PeC and ION are two methods based on the S_PPI network that combine gene expression profiles or orthologous information with the S_PPI network. APPIN_DC and APPIN_NC are two methods based on the D_PPI network constructed using Xiao’s method [32]. The parameters in ION were selected according to the authors’ suggestion. All genes in the PPI network were ranked in descending order according to their ranking scores computed by the FDP, as well as other methods that were compared. After that, the top 100, 200, 300, and 400 of the ranked genes were selected as the candidates for essential genes. The performance of each method was judged according to how well the predicted genes matched the known genes. This evaluation method has been widely used in previous research procedures [16,18,22,48].
In this section, we first discuss the effect of parameter λ on the performance of the FDP. Then we compared the FDP with the other existing methods. After that, the results of the FDP and the other existing methods were analyzed in detail.
3.1. Effects of Parameter λ
In the FDP, parameter λ regulates the contribution of global network diffusion and local interaction frequency entropy when predicting essential genes based on the fused dynamic networks. This section focuses on the prediction accuracy analysis for parameter λ with different values, ranging from 0 to 1. When λ was set to 0, the ranking scores were calculated considering only the local topological properties of genes. When λ was set to 1, the ranking scores were calculated considering only the global topological properties of genes. The detailed results based on the DIP_PPI network and SC_net networks are listed in Table 2 and Table 3, respectively. Here, the parameter T was the number of selected candidate essential genes, ranging from 100 to 400. The prediction accuracy was measured in terms of the number of true essential genes in candidates.
Table 2.
Effects of parameter λ on the performance of the FDP based on the DIP_PPI network.
| T | 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 47 | 70 | 79 | 82 | 85 | 87 | 90 | 90 | 89 | 90 | 92 |
| 200 | 108 | 111 | 130 | 147 | 151 | 154 | 159 | 163 | 164 | 165 | 168 |
| 300 | 156 | 166 | 176 | 193 | 201 | 211 | 215 | 215 | 223 | 229 | 226 |
| 400 | 201 | 212 | 225 | 230 | 242 | 252 | 249 | 273 | 280 | 285 | 277 |
Table 3.
Effects of parameter λ on the performance of the FDP based on the SC_net network.
| T | 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 37 | 58 | 77 | 82 | 88 | 88 | 90 | 91 | 90 | 91 | 90 |
| 200 | 87 | 100 | 134 | 147 | 152 | 154 | 158 | 163 | 162 | 164 | 167 |
| 300 | 131 | 156 | 182 | 199 | 209 | 217 | 222 | 222 | 226 | 232 | 221 |
| 400 | 176 | 199 | 224 | 248 | 255 | 266 | 269 | 275 | 280 | 277 | 272 |
Table 2 and Table 3 showed that the performance of the FDP based only on the local topological properties (λ = 0) is very poor. It was because the final fused network was a fully connected graph, which would introduce many false positive connections. However, the performance of the FDP considering the global topological properties rose sharply, because the orthologous properties of genes in the global topological property scores made a great contribution to ranking real essential genes. The performance of the FDP where it only combined the genes’ orthologous property with the genes’ global topological property (λ = 1) achieved the best performance when predicting a small number of essential genes. However, it was slightly poorer than the performance that considered both the local and global topological properties (λ ranging from 0.8 to 0.9), with an increase in the number of candidate genes selected. The reason may have been that the essential genes with high orthologous scores tended to rank in the top place, whilst the essential genes with high local centrality scores tended to rank at a slightly lower place. Consequently, we set λ to 0.8 in this work to make the FDP achieve good performance when predicting both a small and large number of essential genes.
3.2. Comparing with Other Methods
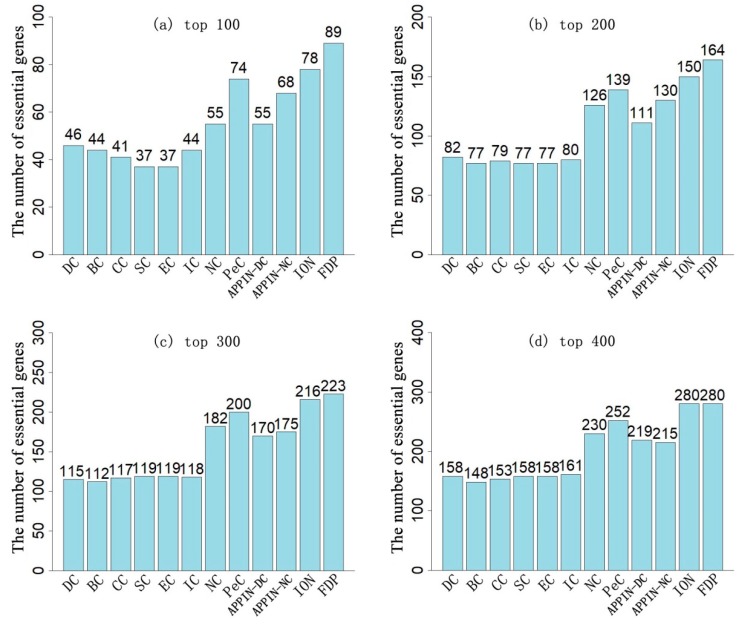
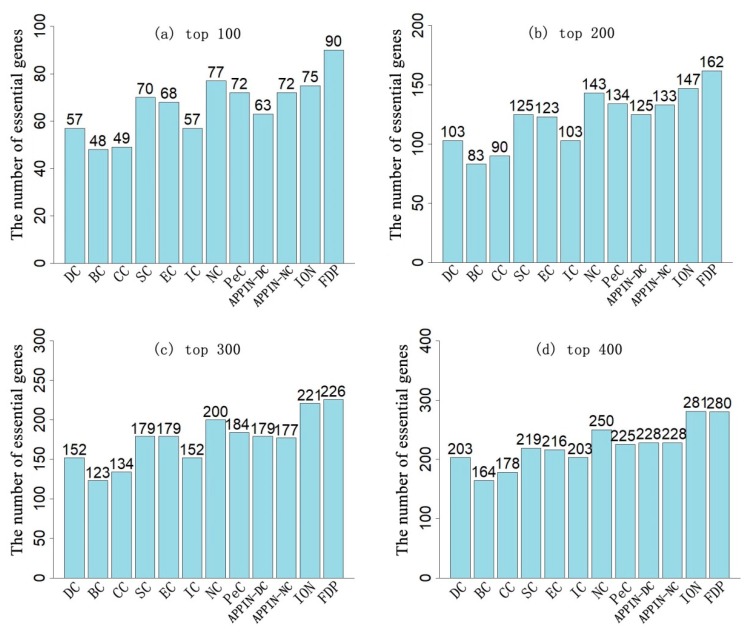
To assess the prediction performance of the FDP, the number of real essential genes identified by the FDP and other existing methods were compared, when the various top numbers of ranked genes were selected as candidates. Figure 2 and Figure 3 illustrate the results based on the DIP_PPI network and the SC_net network, respectively.
Figure 2.
Comparison of the number of essential genes identified by the FDP and other existing methods based on the DIP_PPI network. (a–d) respectively show the results of these methods when selecting the top 100, 200, 300, and 400 of the ranked genes as candidate essential proteins. The data labels above the bars are the number of true essential proteins identified by the corresponding methods in each top number of ranked genes.
Figure 3.
Comparison of the number of essential genes identified by the FDP and other existing methods based on the SC_net network. (a–d) respectively show the results of these methods when selecting top 100, 200, 300, and 400 of the ranked genes as candidate essential proteins. The data labels above the bars are the number of true essential proteins identified by corresponding methods in each top number of ranked genes.
By selecting the top 100 of genes, the FDP achieved an 89% and 90% prediction accuracy on the DIP_PPI and SC_net networks, respectively. This was a 14% and 20% improvement compared to the ION, which had the best performance amongst all the other methods being compared on the two corresponding networks. When the top 200 genes were selected, the prediction accuracy of the FDP achieved about 82% accuracy on the two networks, which was nearly 10% higher than the ION. When the top 300 of genes were selected as candidates, the FDP still had a nearly 75% prediction accuracy on the two networks, which was 3% and 2% higher than the ION on the DIP_PPI and SC_net networks, respectively. When selecting the top 400 of genes, the FDP had a comparable prediction performance to the ION.
PeC predicts essential genes by integrating gene expression profiles with the static PPI network. Compared with PeC, when selecting the top 100, top 200, top 300, and top 400 of proteins as candidates, the accuracies of the FDP improved by 20.3%, 18%, 11.5%, and 11.1%, respectively, on the DIP_PPI network, and the accuracies improved by 23.3%, 20.9%, 22.8%, and 22.4%, respectively on the SC_net network. As for APPIN_NC, which predicts essential genes based on a dynamic PPI network and edge-clustering coefficient, in each top number of selected genes, the performance of the FDP was 30.9%, 26.2%, 27.4%, and 30.2% higher than that of the APPIN_NC on the DIP_PPI network, and it was 25%, 21.8%, 27.7%, and 22.8% higher than that of the APPIN_NC on the SC_net network. NC had the best performance among the seven centrality methods based on the static PPI network (DC, BC, CC, SC, EC, IC, and NC). Compared to the NC, in each top number (top 100, top 200, top 300, and top 400), the prediction accuracy of the FDP improved by 61.82%, 30.16%, 22.53%, and 21.74% on the DIP_PPI network, respectively, and it improved by 16.88%, 13.29%, 13%, and 12% on the SC_net network, respectively. Hence, overall, the FDP outperformed all the other comparative methods in the prediction of essential genes. Especially, with the small number of candidate genes selected, the advantage of the FDP becomes increasingly obvious.
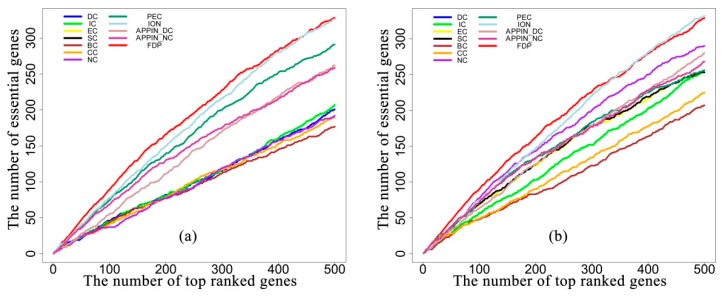
3.3. Evaluation in Terms of Jackknife Curves
To investigate the performance of all the testing methods when selecting the different number of genes ranked at the top as candidates, jackknife curves were employed to show the results, where the x-axis represents the number of genes ranked at the top in descending order, according to their ranking scores computed by the corresponding methods. The y-axis is the cumulative count of the real essential genes within the ranked genes. Figure 4a,b illustrate the jackknife curves of all the methods based on the DIP_PPI network and the SC_net network, respectively. The two figures show that the FDP dramatically outperformed the methods based on the centrality of the static PPI network, such as the DC, IC, EC, SC, BC, NC, and CC. The FDP also outperformed the methods based on the centrality of the dynamic PPI network, such as the APPIN_DC and APPIN_NC. The FDP consistently exceeds the PeC which identifies essential genes by integrating gene expression data with the static PPI data. Compared with the ION that identifies the essential genes by integrating orthologous information with the static PPI data, the FDP also achieved better prediction performance when selecting less than 400 candidate genes. With more candidates selected, the curves of the two methods were very close.
Figure 4.
Jackknife curves of the FDP and other existing methods based on the DIP_PPI network (a) and the SC_net network (b).
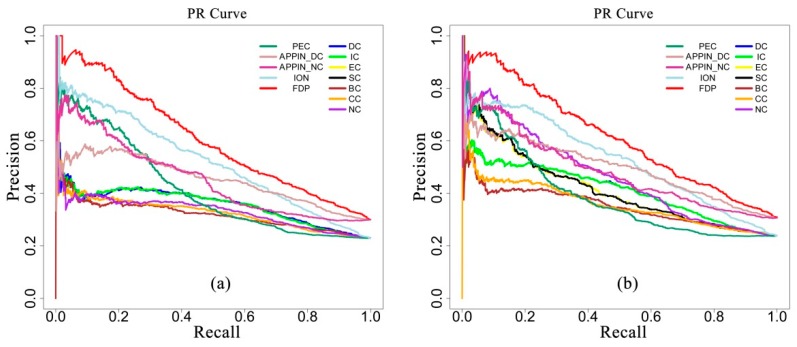
3.4. Evaluation in Terms of Precision-Recall Curve
Precision-recall (PR) curves were also plotted to further show the overall performance of the comparative methods. Precision measures the percentage with which the predicted essential genes match the known genes in all the predicted genes. Recall measures the percentage that known essential genes matched the predicted ones over all the known essential genes. Figure 5a,b illustrate the PR curves of all the methods based on the DIP_PPI network and SC_net network, respectively. The figures show that the PR curves of the FDP are clearly above the curves of all the other methods on both the DIP_PPI network and SC_net network.
Figure 5.
Precision-recall (PR) curves of the FDP and other existing methods based on the DIP_PPI network (a) and SC_net network (b).
4. Conclusions
Essential genes play important roles in cell life under certain conditions and their mRNA expression levels tend to change within a narrow range. Under these observations, in this work, a novel method was proposed to identify essential genes by fusing the dynamic PPI networks of different time points. Compared with previous methods, our method hierarchically fuses the active networks of different time points into a single one. Moreover, it comprehensively utilizes the genes’ orthologous property and both their global and local topological properties to select the candidate essential genes from the fused network. The prediction results on two yeast PPI network datasets, show that our method improves essential gene prediction significantly, compared to the methods based on the static PPI network, including the methods considering the topological properties, i.e., DC, NC, and also the methods combining the PPI network with other biological properties, i.e., PeC and ION. Moreover, our method also outperformed the methods based on Xiao’s dynamic PPI network [4], i.e., APPIN_DC and APPIN_NC. All the results indicated that fusing the dynamic PPI networks and combining proteins’ orthologous properties with the PPI network improved the performance in the prediction of essential genes.
Compared with the existing methods, the FDP shows outstanding performance when selecting a small number of genes as the candidate essential genes. It may benefit from the construction of a dynamic network, which filters out the non-active genes of each time point. However, some real essential genes that consistently express low values across different time points have also been regarded as noise and have been ignored. It causes the decrease of the FDP’s prediction performance when selecting a large number of candidates. Hence, our future work is to construct a high quality dynamic network from the expression profiles that are full of mRNA isoforms and inevitable background noise. The prediction of essential genes also has great relations with the biological properties of known essential genes. New potential correlations between biological events and essential genes will be mined, such as alternative splicing. Moreover, the fused network is fully connected, which introduces some false interactions between the genes and causes poor performance when only considering the topological properties in the network. Therefore, another future work for us is to develop a more efficient strategy to fuse the active networks of different time points.
Author Contributions
F.Z. collected the datasets and developed the program. W.P. designed the FDP method and drafted the manuscript. F.Z., W.P., Y.Y. and W.D. analyzed the results and extensively discussed about the manuscript. J.S., W.P., F.Z. participated in revisiting and modifying the draft. All authors have read and approved the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China under grant No. 31560317, No. 61502214, No. 61472133, No. 61502166 and No. 81560221. It was also supported in part by Natural Science Foundation of Yunnan Province of China under grant No. 2016FB107.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wang J., Peng W., Wu F X. Computational approaches to predicting essential proteins: A survey. J. Proteom. 2013;7:181–192. doi: 10.1002/prca.201200068. [DOI] [PubMed] [Google Scholar]
- 2.Clatworthy A.E., Pierson E., Hung D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 3.Furney S., Alba M.M., Lopez-Bigas N. Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC Genom. 2006;7:165. doi: 10.1186/1471-2164-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Q., Wang J., Peng X., Wu F.-X. Detecting protein complexes from active protein interaction networks constructed with dynamic gene expression profiles. Proteome Sci. 2013;11:S20. doi: 10.1186/1477-5956-11-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Acencio M.L., Lemke N. Predicting essential genes and proteins based on machine learning and network topological features: A comprehensive review. Front. Physiol. 2016;7:75. doi: 10.3389/fphys.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser H.B., Hirsh A.E., Steinmetz L.M., Scharfe C., Feldman M.J. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 7.Jordan I.K., Rogozin I.B., Wolf Y.I., Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batada N.N., Hurst L.D., Tyers M. Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2006;2:e88. doi: 10.1371/journal.pcbi.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong H., Mason S.P., Barabási A.-L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 10.Hahn M.W., Kern A.D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- 11.Joy M.P., Brock A., Ingber D.E., Huang S. High-betweenness proteins in the yeast protein interaction network. J. Biomed. Biotechnol. 2016;2005:96–103. doi: 10.1155/JBB.2005.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuchty S., Stadler P.F. Centers of complex networks. J. Theor. Biol. 2003;223:45–53. doi: 10.1016/S0022-5193(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 13.Estrada E., Rodriguez-Velazquez J.A. Subgraph centrality in complex networks. Phys. Rev. E. 2005;71:056103. doi: 10.1103/PhysRevE.71.056103. [DOI] [PubMed] [Google Scholar]
- 14.Bonacich P. Power and centrality: A family of measures. Am. J. Sociol. 1987;92:1170–1182. doi: 10.1086/228631. [DOI] [Google Scholar]
- 15.Stephenson K., Zelen M. Rethinking centrality: Methods and examples. Soc. Netw. 2002;11:1–37. doi: 10.1016/0378-8733(89)90016-6. [DOI] [Google Scholar]
- 16.Wang J., Li M., Wang H., Pan Y. Identification of essential proteins based on edge clustering coefficient. IEEE/ACM Trans. Comput. Biol. Bioinform./IEEEACM. 2012;9:1070–1080. doi: 10.1109/TCBB.2011.147. [DOI] [PubMed] [Google Scholar]
- 17.Li M., Wang J.X., Wang H.A., Pan Y. Essential proteins discovery from weighted protein interaction networks. Lect. Note Bioinform. 2010;6053:89–100. [Google Scholar]
- 18.Li M., Zhang H., Wang J., Pan Y. A new essential protein discovery method based on the integration of protein-protein interaction and gene expression data. BMC Syst. Biol. 2012;6:15. doi: 10.1186/1752-0509-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X., Wang J., Zhong J., Pan Y. Predicting essential proteins based on weighted degree centrality. IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB) 2014;11:407–418. doi: 10.1109/TCBB.2013.2295318. [DOI] [PubMed] [Google Scholar]
- 20.Li G., Li M., Wang J., Wu J., Wu F.-X., Pan Y. Predicting essential proteins based on subcellular localization, orthology and ppi networks. BMC Bioinform. 2016;17:279. doi: 10.1186/s12859-016-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng X., Wang J., Wang J., Wu F.-X., Pan Y. Rechecking the centrality-lethality rule in the scope of protein subcellular localization interaction networks. PLoS ONE. 2015;10:e0130743. doi: 10.1371/journal.pone.0130743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng W., Wang J., Wang W., Liu Q., Wu F.-X., Pan Y. Iteration method for predicting essential proteins based on orthology and protein-protein interaction networks. BMC Syst. Biol. 2012;6:87. doi: 10.1186/1752-0509-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng W., Wang J., Cheng Y., Lu Y., Wu F., Pan Y. Udonc: An algorithm for identifying essential proteins based on protein domains and protein-protein interaction networks. IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB) 2015;12:276–288. doi: 10.1109/TCBB.2014.2338317. [DOI] [PubMed] [Google Scholar]
- 24.Ren J., Wang J.X., Li M., Wang H., Liu B.B. Prediction of essential proteins by integration of PPI network topology and protein complexes information. Bioinform. Res. Appl. 2011;6674:12–24. [Google Scholar]
- 25.Luo J., Qi Y. Identification of essential proteins based on a new combination of local interaction density and protein complexes. PLoS ONE. 2015;10:e0131418. doi: 10.1371/journal.pone.0131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W., Xu J., Li Y., Zou X. Detecting essential proteins based on network topology, gene expression data and gene ontology information. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016;15:109–116. doi: 10.1109/TCBB.2016.2615931. [DOI] [PubMed] [Google Scholar]
- 27.Przytycka T.M., Singh M., Slonim D.K. Toward the dynamic interactome: It’s about time. Brief. Bioinform. 2010;11:15–29. doi: 10.1093/bib/bbp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Peng X., Peng W., Wu F.X. Dynamic protein interaction network construction and applications. Proteomics. 2014;14:338–352. doi: 10.1002/pmic.201300257. [DOI] [PubMed] [Google Scholar]
- 29.Tang X., Wang J., Liu B., Li M., Chen G., Pan Y. A comparison of the functional modules identified from time course and static PPI network data. BMC Bioinform. 2011;12:339. doi: 10.1186/1471-2105-12-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Peng X., Li M., Pan Y. Construction and application of dynamic protein interaction network based on time course gene expression data. Proteomics. 2013;13:301–312. doi: 10.1002/pmic.201200277. [DOI] [PubMed] [Google Scholar]
- 31.Li M., Meng X., Zheng R., Wu F.-X., Li Y., Pan Y., Wang J. Identification of protein complexes by using a spatial and temporal active protein interaction network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017 doi: 10.1109/TCBB.2017.2749571. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Q., Wang J., Peng X., Wu F.-x., Pan Y. Identifying essential proteins from active PPI networks constructed with dynamic gene expression. BMC Genom. 2015;16(Suppl. 3):S1. doi: 10.1186/1471-2164-16-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M., Ni P., Chen X., Wang J., Wu F., Pan Y. Construction of refined protein interaction network for predicting essential proteins. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017 doi: 10.1109/TCBB.2017.2665482. [DOI] [PubMed] [Google Scholar]
- 34.Shang X., Wang Y., Chen B. Identifying essential proteins based on dynamic protein-protein interaction networks and RNA-seq datasets. Sci. China Inf. Sci. 2016;59:070106. doi: 10.1007/s11432-016-5583-z. [DOI] [Google Scholar]
- 35.Wang B., Mezlini A.M., Demir F., Fiume M., Tu Z., Brudno M., Haibe-Kains B., Goldenberg A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods. 2014;11:333. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 36.Krylov D.M., Wolf Y.I., Rogozin I.B., Koonin E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawoong Jeong Z.N.O. Albert-László Barabási. Prediction of protein essentiality based on genomic data. ComPlexUs. 2002;2003:10. [Google Scholar]
- 38.Xenarios I., Salwinski L., Duan X.Q.J., Higney P., Kim S.M., Eisenberg D. DIP, the database of interacting proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharan R., Suthram S., Kelley R.M., Kuhn T., McCuine S., Uetz P., Sittler T., Karp R.M., Ideker T. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mewes H.W., Frishman D., Mayer K.F.X., Münsterkötter M., Noubibou O., Pagel P., Rattei T., Oesterheld M., Ruepp A., Stümpflen V. MIPS: Analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34:169–172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherry J.M. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26:9. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R., Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37:D455–D458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Chu A.M. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 44.De Lichtenberg U., Jensen L.J., Fausbøll A., Jensen T.S., Bork P., Brunak S. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2004;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 45.Wu F.-X., Xia Z., Mu L. Finding significantly expressed genes from time-course expression profiles. Int. J. Bioinform. Res. Appl. 2009;5:50–63. doi: 10.1504/IJBRA.2009.022463. [DOI] [PubMed] [Google Scholar]
- 46.Peng W., Wang J., Zhao B., Wang L. Identification of protein complexes using weighted pagerank-nibble algorithm and core-attachment structure. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12:179–192. doi: 10.1109/TCBB.2014.2343954. [DOI] [PubMed] [Google Scholar]
- 47.Peng W., Li M., Chen L., Wang L. Predicting protein functions by using unbalanced random walk algorithm on three biological networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017;14:360–369. doi: 10.1109/TCBB.2015.2394314. [DOI] [PubMed] [Google Scholar]
- 48.Estrada E.J.P. Virtual identification of essential proteins within the protein interaction network of yeast. Proteomics. 2010;6:35–40. doi: 10.1002/pmic.200500209. [DOI] [PubMed] [Google Scholar]