Abstract
Breast cancer is a heterogeneous disease whose clinical management is very challenging. Although specific molecular features characterize breast cancer subtypes with different prognosis, the identification of specific markers predicting disease outcome within the single subtypes still lags behind. Both the non-canonical Wingless-type MMTV Integration site (WNT) and the Signal Transducer and Activator of Transcription (STAT)3 pathways are often constitutively activated in breast tumors, and both can induce the small GTPase Ras Homolog Family Member U RhoU. Here we show that RhoU transcription can be triggered by both canonical and non-canonical WNT ligands via the activation of c-JUN N-terminal kinase (JNK) and the recruitment of the Specificity Protein 1 (SP1) transcription factor to the RhoU promoter, identifying for the first time SP1 as a JNK-dependent mediator of WNT signaling. RhoU down-regulation by silencing or treatment with JNK, SP1 or STAT3 inhibitors leads to impaired migration and invasion in basal-like MDA-MB-231 and BT-549 cells, suggesting that STAT3 and SP1 can cooperate to induce high RhoU expression and enhance breast cancer cells migration. Moreover, in vivo concomitant binding of STAT3 and SP1 defines a subclass of genes belonging to the non-canonical WNT and the Interleukin (IL)-6/STAT3 pathways and contributing to breast cancer aggressiveness, suggesting the relevance of developing novel targeted therapies combining inhibitors of the STAT3 and WNT pathways or of their downstream mediators.
Keywords: RHOU, WNT, STAT3, IL-6, SP1, breast cancer, gene signatures
1. Introduction
Breast cancer (BC) is a heterogeneous disease that is routinely classified according to the immunohistochemical assessment of Estrogen and Progesterone Receptors (ER, PR), human epidermal growth factor receptor 2 (HER2) and Ki67 [1,2]. Alternatively, gene expression profiling according to the prediction analysis of microarray (PAM) 50 assay [3] can subdivide BC into intrinsic subtypes characterized by differential prognosis, i.e. luminal A, luminal B, HER2 positive, basal-like and normal-like. Gene signatures useful for informing therapeutic choices have been developed for early stage BC [3,4], and the dependence on ER, PR or HER2 activity is currently exploited for the application of targeted therapies [1], which have considerably improved patients’ survival. However, the identification of specific markers responsible for worse prognosis across all cell types, potentially amenable to targeted therapies, is still needful.
WNT signaling is mediated by the WNT family of ligands, which controls fundamental processes in embryonic development and adult tissue homeostasis [5,6]. According to the specific ligand involved, WNT signal transduction is broadly classified into canonical, β-catenin dependent, and non-canonical pathways, although the observation that specific co-receptors combinations in distinct cellular contexts can shape the pathway regardless of ligand identity challenges this rigid classification [7,8]. Aberrant WNT signaling has been amply implicated in malignant transformation, and WNT ligands are overexpressed in a variety of cancers correlating with tumor progression [9,10]. In particular, aberrant activation of the β-catenin-independent Planar Cell Polarity (PCP) pathway, which plays important roles in defining cell shape, adhesion and movement acting via the c-Jun N-terminal kinase (JNK) [6,10], is involved in the progression of many solid tumors, including those of the breast [11,12]. While transcriptional responses to the activation of beta-catenin-dependent WNT pathways are well characterized, those triggered by the PCP pathway are less well understood and have been mainly linked to the JNK-dependent activation of members of the AP1 family of transcription factors [13,14].
The small GTPase RhoU was identified as a WNT1 target gene, mediating tumor transformation and promoting cell migration/invasion by inhibiting the formation of focal adhesions [15,16]. Unlike most GTPases, RHOU displays high intrinsic guanine nucleotide exchange activity and is largely found in a GTP-loaded state [17,18]. Accordingly, its regulation mostly occurs at the transcriptional level [19]. Despite WNT1 being considered a canonical ligand, WNT1-induced RhoU transcription is β-catenin-independent and requires JNK activity instead, suggesting the involvement of the non-canonical PCP pathway [19]. However, the molecular mechanisms leading to WNT-mediated RhoU transcriptional activation are currently unknown. RhoU gene expression can also be controlled by Signal Transducer and Activator of Transcription (STAT) 3 [19], a pleiotropic transcription factor activated downstream of many cytokines and growth factor receptors and playing a multitude of different roles in regulating cell proliferation/apoptosis, differentiation, migration and metabolism [20,21,22]. STAT3 is considered as an oncogene, since its aberrant constitutive activity is required for the survival and proliferation of a wide variety of primary tumors and tumor cell lines [20,21,22,23,24].
Here, we identify the SP1 transcription factor as an essential mediator of RhoU transcriptional activation downstream of the WNT/PCP pathway, and we unveil a functional cooperation between WNT/PCP/JNK1, RHOU, SP1 and STAT3 to promote cell motility in basal-like human breast tumor cells. In vivo binding of both SP1 and STAT3 strikingly defines genes included in the non-canonical WNT and the IL-6 pathways. This novel signature is significantly correlated with low survival in breast cancer and is enriched in basal-like patients, suggesting that co-activation of the WNT-PCP and STAT3 pathways drives tumor aggressiveness.
2. Results
2.1. RhoU Expression Can Be Induced by both Canonical and Non-Canonical WNT Ligands
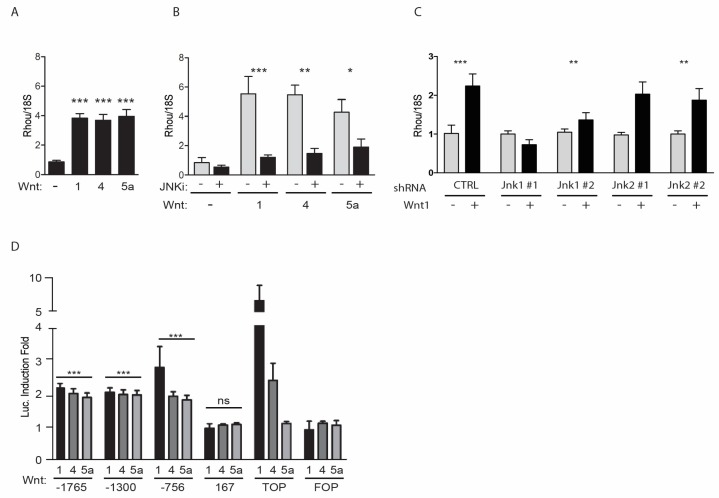
Our previous observation that the canonical WNT1 ligand triggers β-catenin-independent/JNK-dependent RhoU transcriptional induction in mouse embryonic fibroblasts (MEF) [19] prompted us to extend the analysis to other canonical or non-canonical WNT ligands. We thus measured RhoU mRNA in MEF cells stimulated with different WNT ligands, i.e. the canonical WNT1, the non-canonical WNT5a, and WNT4, which can activate both the canonical and the non-canonical pathway. Similar to WNT1, both WNT4 and WNT5a could trigger RhoU mRNA induction acting via the JNK-dependent PCP pathway, since the effect was abolished by the JNK inhibitor SP600125 (Figure 1A,B). Moreover, shRNA-mediated silencing of the two isoforms showed that JNK1, rather than JNK2, is involved (Figure 1C and Figure S2A). In order to identify the transcription factors (TFs) involved, deletion fragments of the TATA-less RhoU promoter fused to a luciferase reporter gene were transiently transfected in MEF cells, followed by co-culturing with WNT-expressing or wild-type HEK-293 cells. All three WNT ligands could induce transcription of the RhoU promoter at similar levels, involving the same promoter region between positions −756 and −167 (Figure 1D). As expected, the β-catenin-dependent control TOPflash was solely responsive to the canonical WNT1 and WNT4 ligands. Further dissection identified a minimal region required for both basal and WNT-inducible promoter activity between positions −366 and −234 (Figure S1A), conferring however weaker induction as compared to the sequence extending to position −756. Sequence analysis did not reveal binding sites for transcription factors known to be involved in WNT/PCP-mediated transcription such as those belonging to the AP1 family [19]. Therefore, in order to identify WNT-responsive elements we subjected the −756 construct Linker Scanning mutagenesis. Surprisingly, none of the mutations tested could affect promoter inducibility (Figure S1B), suggesting that WNT-mediated RhoU induction does not depend on a single responsive element but requires the cooperation of disseminated sites within the responsive region.
Figure 1.
RhoU (Ras Homolog Family Member U) is a common Wingless-type MMTV Integration site (WNT) transcriptional target through the non-canonical c-JUN N-terminal kinase (JNK)-dependent pathway. (A–C) RhoU mRNA was measured by qRT-PCR in MEF cells co-cultured for 24 h with HEK-293 cells expressing or not the indicated WNTs (A), in the presence or absence of the JNK-inhibitor SP600125 (JNKi, B), or upon knockdown of JNK 1 or JNK 2 by means of 2 independent shRNAs/each (C). Data are mean ± SEM of the relative expression levels, normalized to the 18S rRNA. n = 3. (D) RhoU promoter activity in mouse embryonic fibroblasts (MEF) cells transiently co-transfected with the indicated reporter constructs and a Secreted Embryonic Alkaline Phosphatase (SEAP)-expressing vector for normalization. TOPflash (TOP) and FOPflash (FOP) were used as positive and negative controls, respectively, for the β-catenin-dependent pathway. Transfected cells were stimulated or not with WNT ligands as described above. Data are mean ± SEM of the luciferase induction fold, relative to unstimulated condition. All WNT ligands significantly induced the activity of the −1765, −1300 and −756 RhoU promoter constructs (p < 0.001), but not of the −167 unresponsive control (ns). * p < 0.05, ** p < 0.01, *** p < 0.001; n = 3.
2.2. WNT-Mediated Induction of the RhoU Promoter Rquires JNK-Dependent SP1 Recruitment
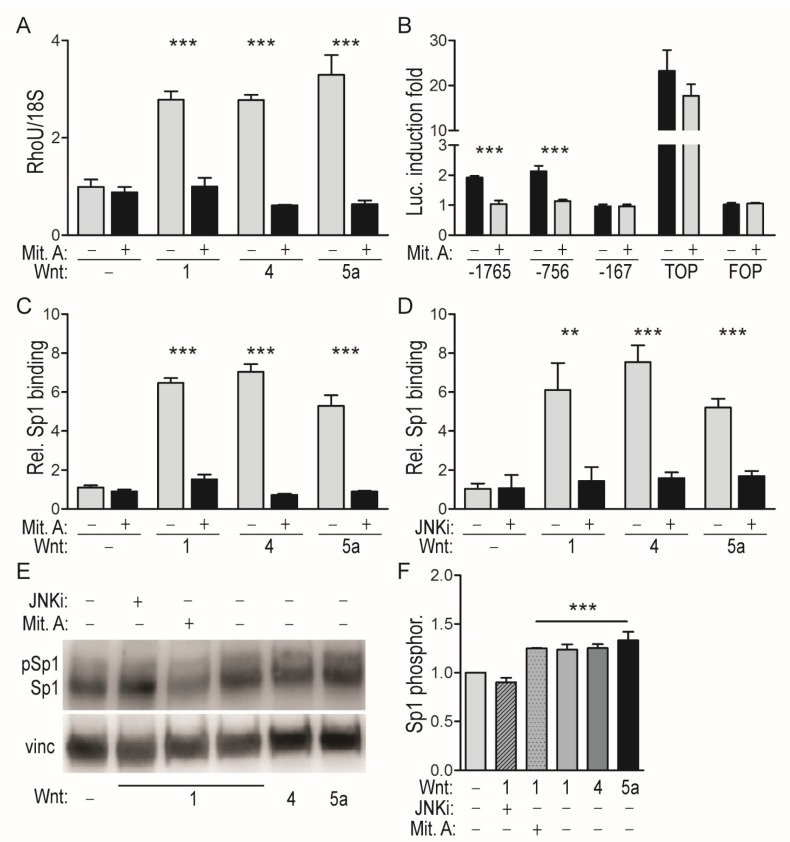
The failure to impair promoter inducibility with single mutations could be consistent with one or more TFs exhibiting diffused binding to the promoter due to high Total Binding Affinity (TBA), which considers the contribution of low-affinity binding sites to calculate the overall affinity of transcription factors for a chosen promoter region [25,26,27]. The TBA of the WNT-responsive RhoU promoter region for the TFs included in the JASPAR core vertebrata database was calculated (Table S1). Among high scoring TFs we focused our attention on SP1, known to play a major role in the transcription of GC-rich TATA-less promoters [28]. Although SP1 is mainly known as a basal activator of many housekeeping genes, it is also a target for JNK-mediated phosphorylation, which in turn can modulate its transcriptional and DNA binding activities at specific promoters [28,29,30,31]. We thus reasoned that WNT-activated JNK may mediate SP1 activation, enabling it to induce RhoU gene transcription via diffused binding to its promoter. SP1 involvement was confirmed by the observation that WNT-mediated RhoU mRNA induction was abolished by the SP1 inhibitor mithramycin A (Mit. A) (Figure 2A), which could also suppress the WNT1-mediated induction of the RhoU promoter but not of the TOPflash control (Figure 2B).
Figure 2.
JNK-mediated SP1 recruitment is responsible for RhoU promoter induction by WNT ligands. (A) RhoU mRNA levels measured by qRT-PCR in MEF cells stimulated for 24 h with the indicated WNT ligands as described in the legend to Figure 1, in the presence or absence of the SP1 inhibitor Mithramycin A (Mit. A). Mean ± SEM of the relative expression levels normalized to the 18S rRNA; n = 3. (B) The indicated RhoU promoter constructs were transfected in MEF cells as described in the legend to Figure 1C, followed by WNT1 stimulation in the presence or absence of Mit. A. Data are mean ± SEM of the luciferase induction fold, relative to unstimulated condition. n = 3. (C,D) SP1 binding to the RhoU promoter measured by ChIP assay in MEF cells stimulated for 24 h with the indicated WNTs, in the presence or absence of Mit. A (C) or of the JNK inhibitor JNKi (D). Immunoprecipitated chromatin was analyzed by qRT-PCR, and data (mean ± SEM) obtained upon normalization to total input (T.I.) and IgG. n = 3. (E) Western blot showing WNT-dependent increase of phosphorylated SP1 (upper band). MEF cells were stimulated for 24 h with the indicated WNTs, in the presence or absence of Mit. A or JNKi, and analyzed with anti-SP1 antibodies recognizing both the phosphorylated and unphosphorylated forms. Representative of one of four independent experiments. (F) Ratio between phosphorylated SP1 and total SP1 signals, expressed as fold induction relative to the unstimulated condition. Mean ± SEM of four independent experiments. ** p < 0.01, *** p < 0.001; n = 4.
Accordingly, ChIP analysis revealed strong in vivo binding of SP1 to the RHOU WNT-responsive promoter region following treatment with all three WNT ligands tested (Figure 2C,D). SP1 binding, undetectable in unstimulated cells, was abolished by both Mit.A treatment (Figure 2C) and JNK inhibition (Figure 2D). Thus, all tested WNT ligands can activate a non-canonical, JNK-mediated pathway that leads to SP1 recruitment to the RHOU promoter, driving its transcriptional activation. Accordingly, treatment with all three WNTs could increase the ratio of phosphorylated versus non-phosphorylated SP1 in a JNK-dependent manner, as shown by the relative increased intensity of the slower migrating band, corresponding to phosphorylated SP1 (Figure 2E,F). As expected Mit.A, which binds to GC-rich regions inhibiting SP1 DNA binding activity, did not affect WNT-mediated SP1 phosphorylation.
2.3. Migration of MDA-MB-231 Cells Downstream of Constitutively Active WNT5a Requires both JNK and SP1 Activities and RhoU Expression
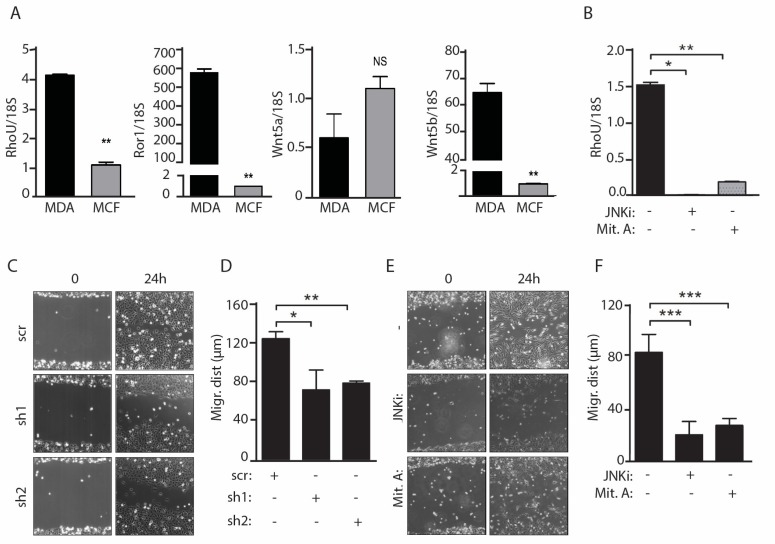
β-Catenin-independent WNT signaling is often aberrantly activated in both tumor and stromal cells [11,12,32,33], promoting JNK-dependent invasion [11,33]. To assess the functional role of RHOU in this context we analyzed the highly aggressive basal-like breast cancer cell line MDA-MB-231, whose motility and invasivity have been shown to require the expression of the non-canonical WNT5a and WNT5b ligands [11,33]. Indeed, as compared with the weakly motile MCF-7 cells, MDA-MB-231 cells express significantly higher levels of WNT5b and of the ROR1 co-receptor, implicated in the non-canonical WNT pathway [34] (Figure 3A). Accordingly, RHOU expression was significantly higher in MDA-MB-231 cells, and fully dependent on both JNK and SP1 activities (Figure 3A,B).
Figure 3.
Both RHOU expression and JNK and SP1 activities are required for migration of MDA-MB-231 cells. (A) qRT-PCR analysis of RHOU, Ror1, Wnt5a and Wnt5b mRNAs in MDA-MB-231 and MCF-7 cells. (B) RHOU mRNA downregulation in MDA-MB-231 cells treated with the JNK or SP1 inhibitors (JNKi, Mit. A). Data are mean ± SEM of the relative expression levels, normalized to 18S rRNA. (C–F) Wound healing assays. MDA-MB-231 cells were infected with lentiviral vectors expressing two distinct RHOU shRNAs or a scrambled control (sh1, sh2, scr) (C,D), or treated with the JNK or SP1 inhibitors (JNKi, Mit.A) (E,F), and migration was assessed by in vitro wound healing assays. The phase contrast pictures (C,E) are representative of three independent experiments (Field of View: 895 × 671 µm). Migrating distance (D,F) was calculated as described in the Materials and Methods, and shown as mean ± SEM. n = 3. * p < 0.05, ** p < 0.01, *** p < 0.001 between the indicated groups.
The motility of MDA-MB-231 cells in a wound healing assay could be significantly impaired both by shRNA-mediated silencing of RHOU (Figure 3C,D and Figure S3) and by treatment with the JNK or SP1 inhibitors (Figure 3E,F). Thus, SP1 activity is required to enhance cell migration downstream of the aberrant activation of the WNT/PCP/JNK pathway in basal-like cancer cells, at least partly via the induction of RHOU expression.
2.4. RhoU is also Involved in Mediating Cell Migration Downstream of STAT3 Signaling
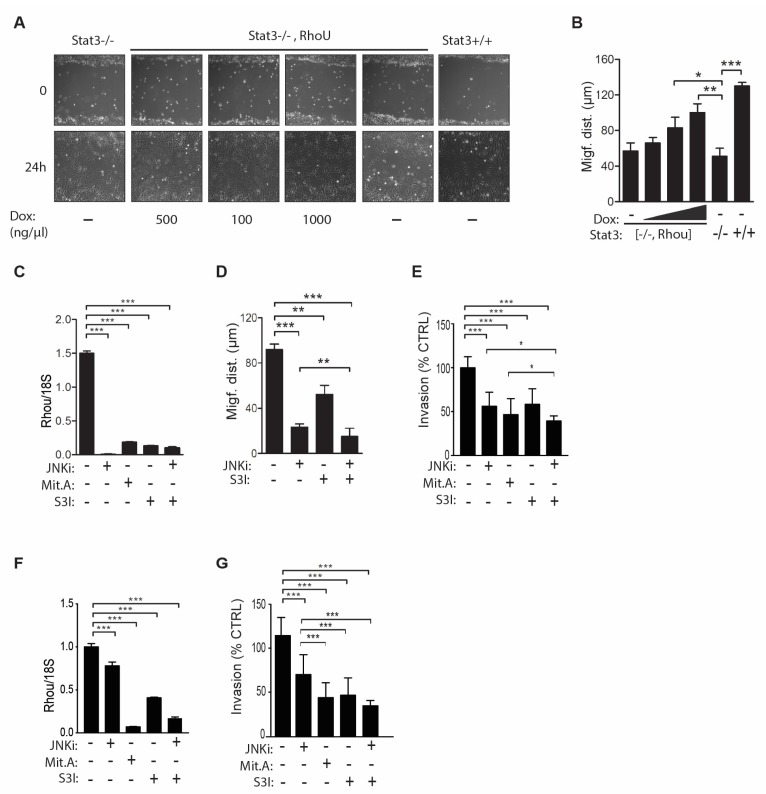
We recently identified RhoU as a STAT3 target gene downstream of gp130 cytokines, and reported a positive correlation between STAT3 activation and RHOU expression levels in a panel of human tumor cell lines, including MDA-MB-231 cells [19]. In order to investigate the role of RhoU in STAT3-induced migration, Stat3−/− MEFs, which express very low RhoU levels and display impaired migration compared to their wild-type counterparts [19,35], were stably transfected with a doxycycline-inducible RHOU-GFP expression vector and treated with increasing doxycycline concentrations, obtaining dose-dependent RHOU induction (Figure S2B). RhoU expression could significantly rescue the defective migration of Stat3−/− MEFs in a wound-healing assay (Figure 4A,B), demonstrating its involvement in STAT3-mediated migration.
Figure 4.
RhoU promotes cell migration also downstream of STAT3 in both MEF and tumor cells (A,B, D,E,G). (A,B) Wound healing assays with wild type (Stat3+/+) or Stat3-deficient MEF cells, either control (Stat3−/−), or expressing doxycycline (Dox)-inducible RhoU (Stat3−/−,RhoU), treated or not with the indicated amounts of Dox to induce increasing RhoU expression (A), quantified as described in the legend to Figure 3 (B). Field of View: 895 × 671 µm. (C–E) MDA-MB-231 cells were treated with the indicated combinations of Mit.A and of the JNK and STAT3 inhibitors (JNKi, S3I), followed by RhoU mRNA quantification by RT-PCR (C), wound healing assay (quantified in D), and transwell invasion assay (E). The same inhibitors were used to treat BT549 cells, followed by RNA quantification (F) and transwell invasion assay (G). (E,G) Invasion was calculated as percentage of control cells (vehicle treated). Mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 between the indicated groups.
We then explored potential synergic effects between the WNT and STAT3 pathways in MDA-MB-231 cells. Treatment with the STAT3 inhibitor S3I considerably reduced the levels of RHOU mRNA (Figure 4C), leading to impaired cell migration and invasion at 24 h, in synergy with the JNK inhibitor (p < 0.01) (Figure 4D,E and Figure S4A). In order to extend these observations, we performed the same invasion experiments on a second basal-like BC cell line, the BT-549 cells, obtaining similar results (Figure 4F,G and Figure S4B). These data are in line with the idea that RHOU indeed represents a common mediator of STAT3 and non-canonical WNT/SP1 functions, under both physiological and pathological conditions.
2.5. SP1 and STAT3 In Vivo Binding Defines a Subclass of Genes Belonging to Non-Canonical WNT or IL-6 Pathway
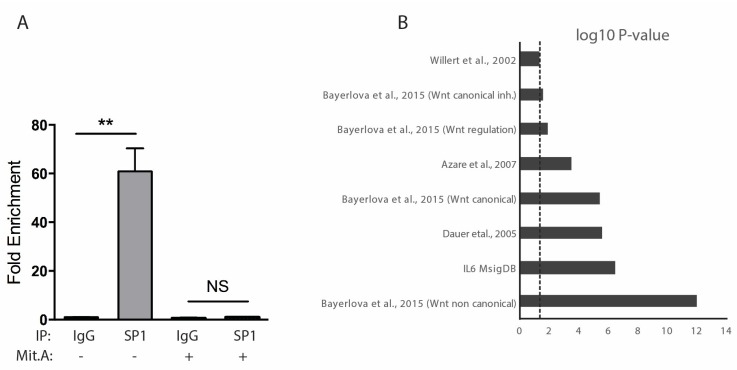
The TFs SP1 and STAT3, whose binding sites are often found close together in the regulatory regions of genes, are involved in several functions related to tumorigenesis including apoptosis, cell growth, angiogenesis, epithelial to mesenchymal transition (EMT) and invasion [36]. In order to investigate the STAT3 and SP1 regulatory network and to identify genes potentially regulated by both factors, we analyzed the Encyclopedia of DNA Elements (ENCODE) Transcription factor binding sites (TFBS) collapsed ChIP Sequencing track (wgEncodeRefTfbsClusteredV3) on H1-ESC, HeLa, MCF10A-Er-Src, K562, GM12878, HepG2 and HCT-116 cell lines, identifying ~3600 genes displaying ChIP-Sequencing peaks for both STAT3 and SP1 (see Section Materials and Methods). While STAT3 binding could be detected on the RHOU promoter in MCF10A-Er-Src cells, the only mammary cell line available, which display active STAT3 [37], SP1 binding could not be detected in any of the cell lines analyzed. We reasoned that STAT3 and SP1 may coordinately mediate RHOU transcription in a cell type-specific manner, depending on the activation status of both factors. To expand our analysis, we took advantage of the “Integrative ChIP-Seq analysis of regulatory regions (ReMap 2018) [38], which combines Public and ENCODE datasets, where we could detect in vivo SP1 binding to the RHOU promoter in HEK293T and A549 cells. The latter are lung adenocarcinoma cells displaying non-canonical WNT constitutive activity [39], and express high RHOU mRNA levels as detected by RNA sequencing. By ChIP-qPCR experiments we could demonstrate SP1 binding to the RHOU promoter in the MDA-MB-231 cells line, abolished in the presence of the SP1 inhibitor Mit. A (Figure 5A). Enrichment analysis of the genes presenting both STAT3 and SP1 binding in published gene sets involving the activation of either the STAT3 or the WNT pathways revealed a positive and significant enrichment (p < 0.05, Fisher exact test) for eight out of seventeen gene sets (Figure 5B). The ChIP-Seq traces for two representative genes are shown in Figure 5C. Of note, a high percentage of the peaks is located between positions −1000 and +1000 with respect to the TSS of the corresponding gene(s) (~74% for SP1, 57% for STAT3), compatibly with a regulatory role of the binding. Importantly, the two top-ranking gene sets according to significance were linked to IL-6 (Broad Institute, Boston, MA, USA) and to non-canonical WNT [40] signaling pathways, respectively (IL-6, ~37% overlap, 32 out of 87 genes; non-canonical WNT, ~23% overlap, 113 out of 489 genes).
Figure 5.
Concomitant activation of STAT3 and SP1 is associated with poor prognosis in breast cancer. (A) SP1 binding to the RHOU promoter measured by ChIP assay in MDA-MB-231 cells cultured in the presence or absence of Mit. A. Data are mean ± SEM of the fold enrichment obtained with anti-SP1 antibodies versus IgGs in three independent experiments. ** p < 0.005 (B) Bar plot showing the gene sets significantly enriched for STAT3 and SP1 bound genes. (C) IGV genome browser pictures showing binding of both SP1 and STAT3 in the indicated cell lines. Green: STAT3; Red: SP1. Two representative genes of the two top-ranking gene sets in (B), WNT5B and JUN, are shown. (D) Kaplan-Meier plot of overall survival for breast tumor patients, as a function of time in days. Patient samples from the METABRIC database (https://www.synapse.org/#!Synapse:syn1688369/) were subdivided according to low or high SP1-S3 score (median value). (E) Results of generalized linear models correlating grade, ER status, treatment, age at diagnosis and lymph node positivity to the SP1-STAT3 score. Coefficients (estimate) of the predictors and significance are indicated.
2.6. High Expression of SP1 and STAT3 Dual Targets is Associated with Low Survival in Breast Cancer
We then assessed the functional relevance of combined STAT3 and SP1 activity in breast cancer. To this end, we examined the METABRIC breast cancer dataset [41] for the expression of a signature composed of the genes belonging to the IL-6 or the WNT non-canonical pathways characterized by in vivo binding of both STAT3 and SP1, as identified above, denominated the SP1-S3 signature (Tables S2 and S3). All profiled tumors (n = 1982) where classified according to the cumulative expression of the SP1-S3 signature (SP1-S3 score, high and low). This score was significantly higher in basal-like tumors compared to other subtypes, in keeping with the known frequent activation of both STAT3 and WNT pathways in this subset (Figure S5A). Patients with a high SP1-S3 score displayed significantly shorter overall survival as compared to patients scored as low (Figure 5D). To exclude that this correlation was due to the contribution of basal-like tumors, which can predict shorter survival when considered as a whole versus all other subtypes (p < 0.005), we repeated the survival analysis excluding basal-like tumors. Even under these conditions, a high SP1-S3 score was still significantly predictive of poor prognosis, supporting the idea that SP1 and STAT3 can functionally cooperate to enhance tumor aggressiveness independently of the tumor subtype (Figure S5B). A high SP1-S3 score was also significantly positively associated with tumor grade, higher degree of lymph node positivity and chemotherapy treatment, and negatively correlated with age at diagnosis and ER positivity (Figure 5E).
3. Discussion
Molecular classification of tumors based on gene signatures correlated with clinical history may guide prognosis, therapeutic choices and, finally, the development of novel specific therapies [4]. Both the WNT and the STAT3 pathways are often altered in solid tumors including breast cancer, correlating with enhanced tumor cells migration, invasion and metastasis [10,42]. In an attempt to assess potential synergistic mechanisms between the two pathways, we focused on their common transcriptional target, the atypical small GTPase RhoU [19]. Our observation that RhoU induction via the JNK-dependent PCP pathway can be equally mediated by canonical and non-canonical WNT ligands suggests that this small GTPase may act as a common effector of both, expanding the scope for its potential relevance in mediating the pathogenic effects of disrupted WNT signaling. Moreover, we could determine that JNK1, rather than JNK2, is involved in WNT-mediated RhoU induction.
Small GTPases belonging to the Rho family are well known to mediate downstream effects of the JNK/PCP pathway [43] that, although best characterized for its transcription-independent functions, is also known to lead to the transcriptional modulation of several genes, mainly via JNK-dependent activation of members of the AP1 family of transcription factors [13,14]. The finding that RhoU is one of these transcriptional targets expands the repertoire of Rho GTPases downstream of the pathway. Moreover, our data identify SP1 as a novel transcriptional mediator of the JNK/PCP non canonical pathway, since its JNK-dependent recruitment to the RhoU promoter is essential for transcriptional induction downstream of both canonical and non-canonical WNT ligands. Although we cannot discriminate between direct or indirect JNK-mediated SP1 activation, several pieces of evidence implicate this factor in inducible transcription upon both JNK and MAPK-mediated phosphorylation [29,30,31].
Our data establish a clear functional correlation between constitutive activation of the PCP pathway, JNK activity, SP1-mediated RhoU transcription and tumor cell motility. Indeed, RhoU downregulation dramatically impairs migration and invasion of the basal-like MDA-MB-231 cells, which display constitutively high expression of the WNT5b ligand and of the ROR1 co-receptor, both implicated in non-canonical WNT pathways. Importantly, we show that also STAT3 converges on RhoU to support cell migration, since RhoU ectopic expression can partially rescue the defective migration of STAT3 null MEFs. Coordinated inhibition of both STAT3 and JNK cooperatively suppresses MDA-MB-231 cells migration and invasion, supporting the idea that the WNT/PCP/SP1 and STAT3 pathways can functionally synergize. The similar results achieved with BT-549 cells further strengthen our observations.
Strikingly, the analysis of genes displaying in vivo binding for both SP1 and STAT3 detects a significant proportion of genes listed as non-canonical WNT or IL-6/STAT3 pathways, suggesting that the molecular mechanisms regulating RhoU transcription here identified may indeed extend to a subclass of WNT and IL-6/STAT3 responsive genes. The significant correlation of this gene signature with lower survival, high tumor grade, lymph node positivity, early age at diagnosis and chemotherapy treatment suggests that indeed the functional convergence of WNT/JNK/SP1 and STAT3 on downstream genes can confer aggressive features to breast tumors, independent of tumor subtype. Analysis of the genes of the signature reveals several interesting groups of genes, also exemplified by the results of the gene ontology analysis. Among the most enriched GO categories are “Chromatin Assembly”, “Chromatin Organization” and “DNA Metabolic Process”, including the genes encoding histone variants H2AX, essential for DNA repair and linked to chemoresistance in breast cancer [44] and H2AZ, whose high expression correlates with BC progression, lymph node metastasis and survival [45]. Other enriched categories are “Signal Transduction”, “Cell Communication” and “Response to Stimulus”, and include cytokine receptors involved in inflammation and immune response, integrins, important for migration and invasion, and several MAP Kinases. Indeed, the category “MAPK Cascade” is also significantly enriched. MAP kinase genes showing dual SP1 and STAT3 binding are, for example, those encoding for MEK1, the proto-oncogenic MAP3K8 and p38, all well-known players in Growth Factor and stress signaling linked to tumoral growth. Notably also JNK, the SP1 activator downstream of WNT, is a dual SP1-STAT3 target. Another significantly enriched category is “proteolysis”, and indeed it includes a striking number of proteasomal subunits, whose levels and activity are increased in a high percentage of primary breast cancers [46]. Among the “Regulation of Gene Expression, Epigenetic” and the “Transcription, DNA-dependent” categories are included many genes encoding subunits of the Mediator complex, often observed to contribute to the progression of several types of cancer [47], and several well-known oncogenic transcription factors such as JUN and YAP. Finally, suggesting positive feed-forward mechanisms, SP1 itself as well as the WNT ligands WNT5b and WNT2b, the latter involved in both canonical and non-canonical signaling, are also dual SP1 and STAT3 targets.
Our data, shedding light on the molecular mechanisms regulating WNT/PCP-mediated transcriptional induction and pointing towards a cooperative co-regulation of many genes and gene families involved in tumor progression and aggressiveness by WNT/SP1 and STAT3, provide the rationale for efforts towards obtaining a coordinated inhibition of both pathways.
4. Materials and Methods
4.1. Cell lines and Treatments
Stat3+/+ and Stat3−/− immortalized MEF [48] and HEK-293 cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium, Thermo Fisher Scientific, Waltham, MA, USA); MDA-MB-231 and BT-549 cells were purchased from ATCC (Manassas, VA, USA) and cultured according to the manufacturer’s instructions. Both media were supplemented with heat-inactivated 10% fetal bovine serum, 100 units/mL penicillin and 100 µg/mL streptomycin. WNT1/WNT4/WNT5a-expressing HEK-293 cells were obtained by stable transfection with vectors expressing the indicated WNT ligands. Cells were treated for 24 h with the following inhibitors (Sigma-Aldrich, Saint Louis, MO, USA): 50 µM SP600125 (JNK inhibitor), 50 mM S3I-201 (STAT3 inhibitor) and 500 nM mithramycin A (SP1 inhibitor).
4.2. Cell Transfection and Transduction, Co-Culture and Luciferase Assays
Mouse RhoU promoter fragments were generated by PCR using a genomic BAC vector as a template. The primers used are reported in Table S4. PCR products were cloned into the promoterless luciferase reporter vector pGL3 basic (Promega, Madison, WI, USA). Linker Scanning mutagenesis constructs, carrying 10 bp mutations starting from the indicated position (Figure S1), were generated from the RhoU −756 promoter fragment. The primers used are shown in Table S5. The Super8X-TOPflash (TOP) and Super8X-FOPflash (FOP) constructs [6] were used to monitor WNT pathway activation.
Transient transfections, co-cultures with WNT-expressing cells (24 h), luciferase and SEAP assays were performed essentially as previously described [19]. Briefly, transfections were performed with LipofectamineTM (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. Each construct was co-transfected at a 10:1 ratio with a vector expressing secreted embryonic alkaline phosphatase (SEAP) as an internal control for transfection efficiency.
MDA-MB-231 cells were stably infected with pLKO lentiviral vectors expressing two different shRNA sequences targeting RHOU (assays No. 77507 and 48657, Thermo Fisher Scientific, Waltham, MA, USA) or a scrambled sequence as a control, followed by puromycin selection (Sigma-Aldrich, 1 µg/mL). MEF cells were stably infected with pLKO lentiviral vectors expressing shRNA against Jnk1 and Jnk2 (TRCN0000301243, TRCN0000304297, TRCN0000348049, TRCN0000333881, Sigma-Aldrich).
4.3. Chromatin Immunoprecipitation Assays
ChIP experiments were performed as previously described [19] on three independent biological samples, using the following antibodies: anti-SP1 sc-59X (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-histone H3 06-755 (Merck Millipore, Darmstadt, Germany); IgG (ChIP-IT Control Kit-Mouse; Active Motif, La Hulpe, Belgium). Non-immunoprecipitated chromatin was used as Total Input control (T.I.). Immunoprecipitated chromatin was analyzed by quantitative real-time PCR using Platinum SYBR Green PCR SuperMix-UDG with ROX (Life Technologies, Carlsbad, CA, USA). Values were normalized to T.I. and to IgG signals. The β-globin promoter region was used as a negative control (Table 1).
Table 1.
Primers used for ChIP-qPCR.
| RhoU Primers |
| mRhoU-forward 5′-AGGGCAGGAGGAACTGGAGAGC-3′ |
| mRhoU-reverse, 5′-TACCCCTGGCCCCTGCTGTG-3′ |
| hRHOU-forward, 5′-ATAAAGGTTCACGGCATGCC-3′ |
| hRHOU-reverse, 5′-TAACTGCAGCTGATCGTGTG-3′ |
| β-globin Primers |
| β-glo-forward, 5′-CTCCCCCTCACTCTGTTCTG-3′ |
| β-glo-reverse, 5′-AGGAGGAGGGGAAGCTGATA-3′ |
4.4. Wound Healing Migration Assay
In vitro wound healing assays were performed essentially as described by Liang et al. [49], with some modifications. In brief, confluent cells in 60-mm dish were serum-starved O/N, followed by mitomycin A treatment (10 μg/mL) for 2 h. The monolayer was then scraped with a sterile pipette tip, washed with PBS and incubated in fresh medium containing 3% FBS. Phase-contrast images were taken at the indicated times (Axiovert 200M microscope, Carl Zeiss, Wetzlar, Germany) (10×). Wound’s width was measured with the software ImageJ as the distance between the edges (National Institute of Health, Bethesda, MD, USA), and the migrating distance calculated as the difference between widths with respect to time 0, in three individual fields per experiment. As the edges were often irregular, the distance in each field was integrated from measures taken at three different position. Data are shown as mean ± SEM of three independent experiments.
4.5. Matrigel Invasion Assay
Tumor cell invasion assays were performed with 8-μm pores Transwell chambers, and membranes pre-coated with Matrigel (VWR International, Radnor, PA, USA) from Corning (New York, NY, USA). Briefly, cells were harvested and re-suspended in DMEM (MDA-MB-231 cells) or RPMI (BT-459 cells) medium without FBS, and 7.5 × 104 cells were seeded into the upper chamber of a 24-well plate. The lower chambers were filled with 10% FBS medium. After 24 h, invading cells were fixed with 4% Paraformaldehyde, stained with Crystal Violet and visualized under an inverted light microscope (Olympus, Tokyo, Japan). Invasion was quantified by Image J, and calculated as percentage of the control cells.
4.6. Protein Extracts and Immunoblotting
Whole protein extractions and immunoblotting were performed as described [19] using the following antibodies: anti-SP1 sc-59 (Santa Cruz Biotechnology); mouse monoclonal anti-GFP and mouse monoclonal anti-vinculin (home-made production).
4.7. RNA Extraction, Retrotranscription and Quantitative Real-Time PCR (qRT-PCR)
These procedures were previously described [19]. qRT-PCR reactions were performed with the ABI Prism 7300 real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) using Platinum Quantitative or SYBR Green PCR SuperMix-UDG with ROX (Life Technologies). Primers and probes used are reported in Supplementary Table S6. The 18S rRNA pre-developed TaqMan assay (Applied Biosystems) was used as an internal control.
4.8. Statistical Analysis
Results were assessed for statistical significance by a standard two-tailed Student’s t test, using the GraphPad Prism v.5 software (GraphPad Software Inc., San Diego, CA, USA). Statistically significant p values: p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), with the exception of experiments of Figure 4E,G, where a two-way ANOVA test was performed.
4.9. Bioinformatics Analysis
4.9.1. Total Binding Affinity (TBA)
TBA was computed as described in [26,27] for all TFs included in the JASPAR core vertebrata database (Version 4) with the promoter regions around the transcription start site (positions −756 to +235 bp, the same experimentally determined for the RhoU promoter) of all RefSeq genes (USCS genome browser version mm9, Boston, MA, USA).
4.9.2. Source Data from Database
mRNA normalized expression data and clinical feature were obtained by permission from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC, Syn1688369), downloaded at probe-level. Data from a total of 1981 breast invasive cancer tissue (BRCA) were obtained.
4.9.3. ChIP-Seq Data Analysis
ChIP-Seq data were downloaded from the UCSC browser [50]. Peaks were filtered according to the absolute distance (5 Kb) from the closest gene. Genes displaying binding sites for both TFs were selected.
4.9.4. Gene set Data
Genes related to WNT and STAT3 signaling were obtained either from published manually curated gene sets [39,51,52,53], or from the “Molecular Signatures Database” (IL6, MsigDB C2) [54] (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The SP1-S3 signature was defined as follows: genes defined by the ChIP-Seq data analysis were assessed for their enrichment with the above gene sets (Fisher exact test, p < 0.05). A combined gene signature was obtained from the intersection between the two top-ranking signatures and the ChIP-Seq derived signature, obtaining 144 genes, listed in Table S2).
4.9.5. Survival Analysis
Time-to-event analyses were performed using the Kaplan-Meier method (R, survival package). Patients were subdivided in two groups according to the median value of the SP1-S3 score, defined as the sum of SP1-S3 gene expression for each patient. Unstratified log-rank statistics were used to evaluate the effect of the change in SP1-S3 score on overall survival.
4.9.6. Univariate Analysis
All univariate analyses were performed using the glm R function to assess the relationship between the SP1-S3 score and clinical features using the univariate Gaussian distribution. Logistic regression was applied for binary variables (https://www.R-project.org).
4.9.7. Plots
All plots were made using ggplot2 [55], survminer (https://CRAN.R-project.org/package=survminer) and R base graphics.
5. Conclusions
In conclusion, we demonstrated that JNK1 can mediate the functions of the WNT/PCP pathways by regulating transcription of the RhoU gene and possibly of other targets via the transcription factor SP1. Importantly, our data also show the cooperative regulation of RhoU, and of RhoU-mediated triple negative breast cancer cells migration and invasion, by WNT/JNK1/SP1 and STAT3. These data suggest that a family of SP1/STAT3 target genes could be responsible of mediating WNT/PCP-STAT3 convergent functions. Identifying them and assessing their relevance in different cancer subtypes where the two pathways are known to play a key pathogenic role could in turn support the pre-clinical experimentation of combinations of STAT3 and JNK1 inhibitors in triple-negative breast cancer. Several STAT3 and JNK inhibitors are under clinical trial, and could be tested with this purpose.
Acknowledgments
The authors wish to thank S. Oliviero and his group and G. Merlo (University of Torino) for helpful suggestions and generous gift of reagents, P. P. Pandolfi (University of Torino and Harvard) for helpful discussion, and S. Cabodi, P. Defilippi, F. Di Cunto, A. Camporeale and A. Camperi (University of Torino) for critically reading the manuscript.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/11/1/101/s1, Figure S1: RhoU promoter analysis in MEF cells, Figure S2: Downregulation of Jnk1 or Jnk2 mRNAs by the indicated shRNAs as measured by qRT-PCR in MEF cells, Figure S3: Manipulation of RhoU levels in MEF and MDA-MB-231 cells, Figure S4: Migration and invasion assays upon treatment with various inhibitors, Figure S5: SP1 and STAT3 can functionally cooperate to enhance tumor aggressiveness independently of the tumor subtype. Table S1: Total Binding Affinity of JASPAR Transcription Factors (TF) with the mouse RhoU promoter, Table S2: List of genes included in the SP1-STAT3 signature, Table S3: GO of SP1-S3 signature, Table S4: Primers used to generate the RhoU promoter fusions, Table S5: Primers used to generate the Linker Scanning constructs, Table S6: Primers used for qRT-PCR.
Author Contributions
Conceptualization, V.P., P.P., E.M. (Emanuele Monteleone), V.O.; Validation, P.C. and L.A.; Investigation, E.M. (Emanuele Monteleone), V.O, D.S., L.A.; Formal Analysis, E.M. (Emanuele Monteleone), E.M. (Enrico Moiso), A.S., I.M.; Supervision, V.P. and P.P.; Visualization, V.O., E.M. (Emanuele Monteleone); Writing-Original Draft Preparation, V.P. and V.O.; Writing-Review & Editing, V.P. and E.M. (Emanuele Monteleone); Project Administration, V.P.; Funding Acquisition, V.P., P.P.
Funding
This work was supported by the Italian Cancer Research Association (AIRC IG13009 and IG16930 to V.P.); the Italian Ministry of University and Research (MIUR PRIN 2012 9JLHSY to V.P., MIUR PRIN 2015 YYLBTR to P.P.); the Ateneo/Compagnia di San Paolo (02_2012_0062 to V.P. and 02_2012_0094 to P.P.), and the Truus and Gerrit van Riemsdijk Foundation, Liechtenstein, donation to V.P. Em.M. was the recipient of a PhD fellowship from Fondazione CRT.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Goldhirsch A., Ingle J.N., Gelber R.D., Coates A.S., Thürlimann B., Senn H.J. Thresholds for therapies: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2009. Ann. Oncol. 2009;20:1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viale G. The current state of breast cancer classification. Ann. Oncol. 2012;23(Suppl. 10) doi: 10.1093/annonc/mds326. [DOI] [PubMed] [Google Scholar]
- 3.Perou C.M., Sørile T., Eisen M.B., van de Rijn M., Jeffrey S.S., Ress C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Prat A., Ellis M.J., Perou C.M. Practical implications of gene-expression-based assays for breast oncologists. Nat. Rev. Clin. Oncol. 2012;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan C.Y., Nusse R. the Wnt Signaling Pathway in Development and Disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 6.Veeman M.T., Axelrod J.D., Moon R.T. A second canon: Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell. 2003;5:367–377. doi: 10.1016/S1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 7.Peradziryi H., Kaplan N.A., Podleschny M., Liu X., Wehner P., Borchers A., Tolwinski N.S. PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling. EMBO J. 2011;30:3729–3740. doi: 10.1038/emboj.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent J.P., Beckett K. Off-track takes Frizzled off the canonical path. EMBO J. 2011;30:3665–3666. doi: 10.1038/emboj.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilyas M. Wnt signalling and the mechanistic basis of tumour development. J. Pathol. 2005;205:130–144. doi: 10.1002/path.1692. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y. Wnt/Planar cell polarity signaling: A new paradigm for cancer therapy. Mol. Cancer Ther. 2009;8:2103–2109. doi: 10.1158/1535-7163.MCT-09-0282. [DOI] [PubMed] [Google Scholar]
- 11.Klemm F., Bleckmann L., Siam H.N., Chuang E., Rietktter D., Behme M., Schulz M., Schaffrinski S., Schindler L., Trümper F., et al. β-catenin-independent WNT signaling in basal-like breast cancer and brain metastasis. Carcinogenesis. 2011;32:434–442. doi: 10.1093/carcin/bgq269. [DOI] [PubMed] [Google Scholar]
- 12.MacMillan C.D., Leong H.S., Dales D.W., Robertson A.E., Lewis J.D., Chambers A.F., Tuck A.B. Stage of Breast Cancer Progression Influences Cellular Response to Activation of the WNT/Planar Cell Polarity Pathway. Sci. Rep. 2014;3:6315. doi: 10.1038/srep06315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lien W.H., Fuchs E. Wnt some lose some: Transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schambony A., Wedlich D. Wnt-5A/Ror2 Regulate Expression of XPAPC through an Alternative Noncanonical Signaling Pathway. Dev. Cell. 2007;12:779–792. doi: 10.1016/j.devcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Ory S., Brazier H., Blangy A. Identification of a bipartite focal adhesion localization signal in RhoU/Wrch-1, a Rho family GTPase that regulates cell adhesion and migration. Biol. Cell Auspices Eur. Cell Biol. Organ. 2007;99:701–716. doi: 10.1042/BC20070058. [DOI] [PubMed] [Google Scholar]
- 16.Tao W., Pennica D., Xu L., Kalejta R.F., Levine A.J. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saras J., Wollberg P., Aspenström P. Wrch1 is a GTPase-deficient Cdc42-like protein with unusual binding characteristics and cellular effects. Exp. Cell Res. 2004;299:356–369. doi: 10.1016/j.yexcr.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Shutes A., Berzat A.C., Chenette E.J., Cox A.D., Der C.J. Biochemical analyses of the Wrch atypical Rho family GTPases. Methods Enzymol. 2006;406:11–26. doi: 10.1016/S0076-6879(06)06002-2. [DOI] [PubMed] [Google Scholar]
- 19.Schiavone D., Dewilde S., Vallania F., Turkson J., di Cunto F., Poli V. The RhoU/Wrch1 Rho GTPase gene is a common transcriptional target of both the gp130/STAT3 and Wnt-1 pathways. Biochem. J. 2009;421:283–292. doi: 10.1042/BJ20090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demaria M., Giorgi C., Lebiedzinska M., Esposito G., D’Angeli L., Bartoli A., Gough D.J., Turkson J., Levy D.E., Watson C.J., et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging. 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D.E., Lee C.K. What does Stat3 do? J. Clin. Investig. 2002;109:1143–1148. doi: 10.1172/JCI0215650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H., Jove R. The stats of cancer-New molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 23.Avalle L., Pensa S., Regis G., Novelli F., Poli V. STAT1 and STAT3 in tumorigenesis. Jak-Stat. 2012;1:65–72. doi: 10.4161/jkst.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiquee K., Zhang S., Guida W.C., Blaskovich M.A., Greedy B., Lawrence H.R., Yip M.L.R., Jove R., McLaughlin M.M., Lawrence N.J., et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. USA. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foat B.C., Morozov A.V., Bussemaker H.J. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–e149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- 26.Grassi E., Zapparoli E., Molineris I., Provero P. Total binding affinity profiles of regulatory regions predict transcription factor binding and gene expression in human cells. PLoS ONE. 2015 doi: 10.1371/journal.pone.0143627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molineris I., Grassi E., Ala U., di Cunto F., Provero P. Evolution of promoter affinity for transcription factors in the human lineage. Mol. Biol. Evol. 2011;28:2173–2183. doi: 10.1093/molbev/msr027. [DOI] [PubMed] [Google Scholar]
- 28.Beishline K., Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 29.Benasciutti E., Pagès G., Kenzior O., Folk W., Blasi F., Crippa M.P. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood. 2004;104:256–262. doi: 10.1182/blood-2003-08-2661. [DOI] [PubMed] [Google Scholar]
- 30.Hammoud L., Burger D.E., Lu X., Feng Q. Tissue inhibitor of metalloproteinase-3 inhibits neonatal mouse cardiomyocyte proliferation via EGFR/JNK/SP-1 signaling. Am. J. Physiol. Cell Physiol. 2009;296:C735–C745. doi: 10.1152/ajpcell.00246.2008. [DOI] [PubMed] [Google Scholar]
- 31.Reddy V.S., Prabhu S.D., Mummidi S., Valente A.J., Venkatesan B., Shanmugam P., Delafontaine P., Chandrasekar B. Interleukin-18 induces EMMPRIN expression in primary cardiomyocytes via JNK/Sp1 signaling and MMP-9 in part via EMMPRIN and through AP-1 and NF- B activation. AJP Heart Circ. Physiol. 2010;299:H1242–H1254. doi: 10.1152/ajpheart.00451.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurayoshi M., Oue N., Yamamoto H., Kishida M., Inoue A., Asahara T., Yasui W., Kikuchi A. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 33.Pukrop T., Klemm F., Hagemann T., Gradl D., Schulz M., Siemes S., Trümper L., Binder C. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc. Natl. Acad. Sci. USA. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minami Y., Oishi I., Endo M., Nishita M. Ror-family receptor tyrosine kinases in noncanonical Wnt signaling: Their implications in developmental morphogenesis and human diseases. Dev. Dyn. 2010;239:1–15. doi: 10.1002/dvdy.21991. [DOI] [PubMed] [Google Scholar]
- 35.Ng D.C.H., Bao H.L., Cheh P.L., Huang G., Zhang T., Poli V., Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 2006;172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C., Xie K. Crosstalk of Sp1 and Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth Factor Rev. 2012;23:25–35. doi: 10.1016/j.cytogfr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iliopoulos D., Hirsch H.A., Struhl K. An Epigenetic Switch Involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B. ReMap 2018: An updated regulatory regions atlas from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic. Acids Res. 2017;46:267–275. doi: 10.1093/nar/gkx1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartis D., Csongei V., Weich A., Kiss E., Barko S., Kovacs T., Avdicevic M., D’Souza V.K., Rapp J., Kvell K., et al. Down-Regulation of Canonical and Up-Regulation of Non-Canonical Wnt Signalling in the Carcinogenic Process of Squamous Cell Lung Carcinoma. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0057393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayerlová M., Klemm F., Kramer F., Pukrop T., Beißbarth T., Bleckmann A. Newly constructed network models of different WNT signaling cascades applied to breast cancer expression data. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0144014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 43.Schlessinger K., Hall A., Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 2009;23:265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- 44.Gruosso T., Mieulet V., Cardon M., Bourachot B., Kieffer Y., Devun F., Dubois T., Dutreix M., Vincent-Salomon A., Miller K.M., et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol. Med. 2016;8:527–549. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangasamy D. Histone variant H2A.Z can serve as a new target for breast cancer therapy. Curr. Med. Chem. 2010;17:3155–3161. doi: 10.2174/092986710792231941. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65:5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 47.Syring I., Klümper N., Offermann A., Braun M., Deng M., Boehm D., Queisser A., von Mässenhausen A., Brägelmann J., Vogel W., et al. Comprehensive analysis of the transcriptional profile of the Mediator complex across human cancer types. Oncotarget. 2016;7:23043–23055. doi: 10.18632/oncotarget.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa-Pereira A.P., Tininini S., Strobl B., Alonzi T., Schlaak J.F., Is’harc H., Gesualdo I., Newman S.J., Kerr I.M., Poli V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. USA. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 50.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler A.D. The Human Genome Browser at UCSC. Genom. Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azare J., Leslie K., Al-Ahmadie H., Gerald W., Weinreb P.H., Violette S.M., Bromberg J. Constitutively Activated Stat3 Induces Tumorigenesis and Enhances Cell Motility of Prostate Epithelial Cells through Integrin 6. Mol. Cell. Biol. 2007;27:4444–4453. doi: 10.1128/MCB.02404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dauer D.J., Ferraro B., Song L., Yu B., Mora L., Buettner R., Enkemann S., Jove R., Haura E.B. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 53.Willert J., Epping M., Pollack J.R., Brown P.O., Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2002;2 doi: 10.1186/1471-213X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson L. ggplot2: Elegant Graphics for Data Analysis by WICKHAM, H. Biometrics. 2011 doi: 10.1111/j.1541-0420.2011.01616.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.