Abstract
Streptococcus thermophilus is considered one of the most important species for the dairy industry. Due to their diffusion in dairy environments, bacteriophages can represent a threat to this widely used bacterial species. Despite the presence of a CRISPR-Cas system in the S. thermophilus genome, some lysogenic strains harbor cryptic prophages that can increase the phage-host resistance defense. This characteristic was identified in the dairy strain S. thermophilus M17PTZA496, which contains two integrated prophages 51.8 and 28.3 Kb long, respectively. In the present study, defense mechanisms, such as a lipoprotein-encoding gene and Siphovirus Gp157, the last associated to the presence of a noncoding viral DNA element, were identified in the prophage M17PTZA496 genome. The ability to overexpress genes involved in these defense mechanisms under specific stressful conditions, such as phage attack, has been demonstrated. Despite the addition of increasing amounts of Mitomycin C, M17PTZA496 was found to be non-inducible. However, the transcriptional activity of the phage terminase large subunit was detected in the presence of the antagonist phage vB_SthS-VA460 and of Mitomycin C. The discovery of an additional immune mechanism, associated with bacteriophage-insensitive strains, is of utmost importance, for technological applications and industrial processes. To our knowledge, this is the first study reporting the capability of a prophage integrated into the S. thermophilus genome expressing different phage defense mechanisms. Bacteriophages are widespread entities that constantly threaten starter cultures in the dairy industry. In cheese and yogurt manufacturing, the lysis of Streptococcus thermophilus cultures by viral attacks can lead to huge economic losses. Nowadays S. thermophilus is considered a well-stablished model organism for the study of natural adaptive immunity (CRISPR-Cas) against phage and plasmids, however, the identification of novel bacteriophage-resistance mechanisms, in this species, is strongly desirable. Here, we demonstrated that the presence of a non-inducible prophage confers phage-immunity to an S. thermophilus strain, by the presence of ltp and a viral noncoding region. S. thermophilus M17PTZA496 arises as an unconventional model to study phage resistance and potentially represents an alternative starter strain for dairy productions.
Keywords: Streptococcus thermophilus, lipoprotein (Ltp), noncoding region, bacteriophages, cryptic prophage
1. Introduction
The thermophilic Lactic Acid Bacterium (LAB) Streptococcus thermophilus is an extremely important starter culture, in the dairy industry, for production of cheeses and yogurts [1,2]. Its technological use is mostly linked to its ability to quickly acidify the substrate, a very important feature, since it is known that a pH decrease leads to modifications in bacterial [3,4] and also yeast [5,6] population composition. Its use in the dairy industry has an average market value of US$ 40 billion [7]. S. thermophilus starter cultures in the dairy environment are constantly threatened by bacterial viruses (bacteriophages or phages), which are the most abundant biological entities in the biosphere [8,9]. The lysis of starter culture cells leads to relevant economic losses, by lowering the quality of the end product, or even leading to a total process failure [10,11]. To overcome this problem, the dairy industry has adopted many different strategies to limit phage attacks against bacterial starter cultures [12]. During the past decades, molecular and genomic approaches have revealed diverse genetically defined resistance mechanisms directed against Streptococcus phages [13,14,15,16]. It was reported that S. thermophilus strains have natural adaptive immunity against phages, including CRISPR-Cas, Ltp lipoprotein, and noncoding viral DNA elements [15,16,17,18]. The last two mechanisms are part of the temperate bacteriophage driven immunity that protects the host (as well, as the prophage) from lysis, by blocking DNA injection and replication, respectively [15,17]. The phage-encoded lipoprotein Ltp is the prototype of a widely-distributed family of cell-surface-exposed lipoproteins, involved in superinfection exclusion (sie). The ltp gene belongs to the “moron” class, which is characterized by a common strategy to confer host benefits through phage-mediated horizontal gene transfer (HGT), by blocking invader DNA injection [15,19,20]. The ltp gene is usually located within the lysogeny module and is constitutively transcribed, while the virus is in the prophage state [15]. A different immunity system, encoded by prophages, is based on a mechanism determined by the increased copy number of a viral noncoding region, with characteristics of an origin of replication (ori) sequence. This region, that can block the accumulation of the invading phage DNA, was found for the first time in the DNA replication module of phage ΦSfi21 [17]. As previously demonstrated by Lamothe et al. (2005) [21], the transcription of cro-ori regions start 5 min after phage infection.
Due to the presence of these defense mechanisms in S. thermophilus, the identification of bacteriophage-insensitive starter cultures is extremely relevant and has significant economic importance. Additionally, the increasing amount of genomic data available on S. thermophilus phages can facilitate the identification of bacteriophage superinfection exclusion and the development of new resistance mechanisms. The discovery of new immune mechanisms against viral infection in S. thermophilus will help in the development of innovative strategies, to protect bacteria from phage attack. Here, we present the genomic analysis of two cryptic prophages, which naturally infected S. thermophilus M17PTZA496, a well-characterized strain [22,23,24] possessing interesting in vitro probiotic properties, along with anticancer activity and folic acid production [25]. The combined presence of two prophages features associated with phage immunity, i.e., ltp and a viral noncoding region in the same prophage, was investigated. Moreover, we evaluated the activity of these two immune mechanisms under mitomycin C (MmC) stress and phage attack conditions.
2. Materials and Methods
2.1. Strain and Growth Conditions
The bacterial strain S. thermophilus M17PTZA496 was isolated in the Valle d’Aosta Region (Italy) from Fontina, a protected designation of origin (PDO) cheese. This isolate was stored, as a frozen stock (−80 °C), in sterile reconstituted (10%, w/v) commercial nonfat skim milk, supplemented with 15% (v/v) glycerol. S. thermophilus M17PTZA496 was routinely grown at 44 °C, for 24 h, in modified M17 medium containing lactose 0.5% (w/v) (th-LM17) [26].
2.2. Bioinformatics Analysis
Following genomic analysis and strain comparison [27,28], a more detailed investigation was conducted by analyzing the prophages in the S. thermophilus M17PTZA496 chromosome. For this purpose, PHAge Search Tool Enhanced Release (PHASTER) [29] was used to identify and annotate all viral sequences. Functional information was obtained using UniProt and Pfam databases [30] and by manual refinement. Viral genome alignment visualization and manual inspection were performed using Progressive MAUVE [31]. The CGView Server [32] was used to generate a graphical map of the TP1-M17PTZA496 genome.
For comparative analysis, eighty-three S. thermophilus bacteriophages and five Lactococcus lactis phages whole genome sequences, downloaded from the National Center for Biotechnology Information (NCBI) database (Table S1), were used. A fragmented all-against-all comparison in the TBLASTX mode was performed with the Gegenees 2.0.0 software [33], setting the parameters to 50/25 (fragment-size/slide-size), as described by Barylski et al. (2014) [34]. A heat plot was generated by setting the maximum threshold (40%), in order to obtain the best phylogenomic overview. Unrooted phylogenetic tree was computed, using the SplitsTree4 following the neighbor joining method [35]. A whole genome comparison with pac-type phages from the genus Sfi11virus, TP1-M17PTZA496, and Streptococcus phage 20617, was performed, using the Easyfig comparison tool [36,37,38]. Finally, CRISPRdb [39] and HostPhinder [40] tools were used to predict the potential bacterial hosts of the identified prophages. The nucleotide sequence of S. thermophilus M17PTZA496 was taken from the GenBank database (accession No. NZ_CM002372.1).
2.3. S. thermophilus M17PTZA496 Prophage Induction Assay, DNA Extraction, Semi-Quantitative PCR, and Transmission Electron Microscopy
The prophage induction assay was conducted at a small scale (96-well microplates), as described by Oliveira et al. (2017) [41] and Arioli et al. 2018 [36]. Briefly, S. thermophilus M17PTZA496 was grown at 44 °C for 24 h, in th-LM17. The culture was then diluted with sterile th-LM17 medium to reach an optical density at 600 nm (OD600) of, approximately 0.1. A final volume of 200 µL was inserted into the wells of a 96-well microplate. In the chemically-treated groups, four phage-inducing agents (MmC −1, 2, 3, and 4 µg/mL; nalidixic acid −0.1, 0.2, and 0.4 µg/mL; NaCl −100, 200, and 400 mM, and H2O2 −100, 200, and 400 mM) were added to the early log-phase cultures. To evaluate the effect of low (0.1, 0.25, and 0.5% w/v) and high concentration (1.0% w/v) of lactose and sucrose on S. thermophilus M17PTZA496 growth curve and prophage induction, cells were prepared, as described previously, washed three times with the phosphate buffered saline (PBS: KH2PO4 144 mg/L, NaCl 9 mg/L, Na2HPO4·7H2O 795 mg/L, pH 7.4) and resuspended in the M17 medium, modified with lactose and fructose, at different concentrations, to reach an OD600 of 0.1. Then aliquots of 200 µL were transferred into the wells of a 96-well microplate. Measurements were automatically obtained by a Spark 10M (Tecan Trading AG, Männedorf, Switzerland) every 30 min. The entire assay was carried out in triplicates.
A semiquantitative PCR test was used to estimate the amount of viral DNA in the bacterial cells and the integrity of the virus attachment sites. Sampling was performed after 0, 30, 60, and 90 min of bacterial growth and for each MmC concentration used. Bacterial DNA extraction was performed by adding 500 µL of bacterial culture to 50 µL of lysis buffer (NaOH 0.05 M; SDS 0.25%) in a 1.5 mL tube, mixed by vortexing for 2 min and incubated at 94 °C, for 15 min to achieve cell lysis. Cell debris was removed by centrifugation at 4,500× g for 10 min. DNA yield and purity were assessed by NanoDrop (NanoDrop 2000c, Thermo Scientific, Waltham, MA, USA).
Four primer couples were manually designed inside the coding region of the major capsid protein, in the attachment sites attL/attR and in a M17PTZA496 internal control region (Table 1), using the online tool NCBI/Primer-BLAST [42]. Primers were obtained from Invitrogen (Thermo Fisher Scientific, Rodano, MI, Italy). PCR reactions were performed using PureTaqTM Ready-To-GoTM PCR Bead kit (GE Healthcare, Munich, Germany), using 50 ng of bacterial DNA as the template.
Table 1.
Primers used for semiquantitative PCR and RT-qPCR assays.
| Primer | Sequence (5’ → 3’) | Reference | Genome position (bp) |
|---|---|---|---|
| PhageCntr_FW | CCAGCTCGCAAACAACTTGG | This study | 644,332–645,130 |
| PhageCntr_REV | CAGCGTTAACTGTGTTGTCAG | ||
| attL_FW | CACGCTGCTAACTCAATCCT | This study | 620,880–621,469 |
| attL_REV | GCTCTTTGGATATCCACACC | ||
| attR_FW | CTACGTAGTCAGAGGTCCG | This study | 664,158–664,626 |
| attR_REV | GATTAAAGGCCTATTCTAAGCC | ||
| M17ptza496_S71U_FW | GCAACCATTACACACATAAGGT | This study | 297,265–304,822 |
| M17ptza496_S71U_REV | CACAGCGACATCTATCATTGG | ||
| cas7_FW_1 | AGGAGCCTACCATACTTGATG | This study | 697,396–698,448 |
| cas7_REV_1 | GTAAGCGTGGGCAAGTGTTC | ||
| ltp_FW_4 | ACTAGCAAGACGTCAGAGGC | This study | 697,265–697,399 |
| ltp_REV_4 | CTGCTTAGCTTTCTCACCG | ||
| NC_FW_5 | CAACTTACAGACCAGACAAGG | This study | 626,422–626,895 |
| NC_REV_5 | CCTCAATATGCTTACCGGAC | ||
| Terminase large subunit_FW_2 | CATGGTGCTAAACGTGCTGG | This study | 639,113–639,784 |
| Terminase large subunit_REV_2 | GCAGGTACATCGTCAACGTC | ||
| gapdh_FW | CACCATCTTCCAGGAGCGAG | Conte et al. (2015) [43] | (not determined) |
| gapdh_REV | CACCATCTTCCAGGAGCGAG |
The presence of complete or defective viral particles in treated and untreated-MmC S. thermophilus cultures was evaluated by TEM, at different time points, namely 0, 30, 60, and 90 min. Bacterial cultures (1 mL) were harvested by centrifugation at 5,500× g for 15 min, at 4 °C, and the pellet was resuspended in 100 µL of SM buffer (5.8 g NaCl, 2.0 g MgSO4·7H2O, 50 mL Tris-HCl 1M, 5 mL gelatin 2%, pH 7.5 and H2O to 1 L). Chloroform (10% v/v) and NaCl 1M (final concentration) were added, in order to provoke cell lysis and release of viral particles. After 2 h of incubation at 4 °C, cellular debris were removed by centrifugation at 14,000× g for 20 min, at 4 °C, the supernatant was collected and transferred to the observation Formvar-coated grids and negatively stained with 2% (w/v) uranyl acetate. A transmission electron microscope (Tecnai 12, FEI Thermo Fisher Scientific, Eindhoven, The Netherlands), operating at an acceleration voltage of 80 kV, was used to examine the samples. Images were acquired at a resolution of 50 nm.
2.4. Phage-Susceptibility Screening, and Phage-Binding Assay
Six bacteriophages, namely, vB_SthS-VA214 [44], vB_SthS-VA353, vB_SthS-VA444, vB_SthS-VA460 [44], vB_SthS-VA698, and vB_SthS-VA720, were previously characterized, considering their variable region 2 (VR2) [45], and were selected and tested for their ability to infect S. thermophilus M17PTZA496. Bacteriophages were kindly provided by the Istituto per la Qualità e le Tecnologie Agroalimentari, Veneto Agricoltura (Thiene, Italy). Using a multiplicity of infection of 0.1, the six bacteriophages were independently added to a S. thermophilus M17PTZA496 growing culture (OD600 0.3), in th-LM17, supplemented with 1% CaCl2 1M. After incubation at 44 °C for 30 min, MmC (1 µg/mL) was added and samples were incubated for a further 30 min. Cells were then collected by centrifugation at 12,000× g for 4 min, at 4 °C, and the supernatant used for viral titration. Briefly, an aliquot of 100 µL was serially diluted (1:10) in SM buffer and each dilution was mixed with 0.2 mL of a host culture, previously grown overnight [46]. To this mix, 35 µL of CaCl2 1 M and 4 mL of molten th-LM17 soft agar (agar 0.75%) were added and poured onto a th-LM17 agar base plate (agar 1.5%), containing 1% CaCl2 [47]. Plates were incubated at 37 °C for 24 h. Viral titer was expressed in terms of plaques forming units per milliliter (PFU/mL). The percentage of phage particles bounded to S. thermophilus M17PTZA496 and the rate of attachment were calculated, as previously reported [48,49].
2.5. Transcriptional Activity Assay
An S. thermophilus M17PTZA496 overnight culture (OD600 1.4) was adjusted to an OD600 of 0.1 in 300 mL of th-LM17, containing 10 mM CaCl2 and 10 mM glycine, and incubated at 45 °C [45]. When the culture reached an OD600 of 0.3 (that was considered T0), 9 mL were transferred to two separate 15 mL polypropylene tubes, in which 1 mL of PBS (defined as Cntr1) and 1 mL of a vB_SthS-VA460 suspension (~107 PFU/mL) (defined as Pt1) were added, respectively. Tubes were incubated at 45 °C for 30 min. After this period, defined as T1, MmC (1 µg/mL) was added to both tubes (defined Cntr2 and Pt2, respectively) and a final incubation at 45 °C for 30 min, identified as T2, was performed. At each sampling time (T0, T1, and T2), a 100 µL aliquot was collected to determine the number of viable cells, using the drop plate methodology [50]. Viral titration was performed from Pt1 and Pt2 samples, as described above. All experiments were performed in triplicates. The experimental design is reported in Figure S1.
Samples obtained at T0, T1, and T2 were centrifuged at 4,500 g for 5 min, at 4 °C. The harvested cells were immediately frozen in liquid nitrogen and stored at −80 °C, to preserve RNA integrity. To perform cell lysis, 100 µL of lysozyme solution (10 mM Tris-HCl, 0.1 mM EDTA, 15 mg/mL lysozyme, pH 8.0) were added to the cell pellet and resuspended by vortexing. Afterwards, 0.5 μL of 10% (w/v) SDS were added, vortexed for 2 min and incubated for 5 min, at room temperature. After this, 350 μL of freshly prepared 1% (v/v) 2-mercaptoethanol lysis buffer and 50 mg of cold glass beads (diameter 0.6 mm, Sigma, St. Louis, MO, USA) were added and the samples were vortexed for approximately 2 min. Subsequently, 750 µL of TRIzol (Invitrogen, Rodano, Italy) were added, gently homogenized and incubated for 5 min, at room temperature. During this step, 1 µg of exogenous total RNA extracted from the skeletal muscle of Mus musculus was added to the sample, as an external housekeeping gene source, for real-time data normalization. Afterwards, 160 µL of chloroform were added, mixed, and incubated for 2 min, at room temperature. Following centrifugation at 12,000× g for 15 min at 4 °C, the colorless upper phase containing the RNA, was transferred to a fresh RNase-free tube containing 350 µL of isopropanol and incubated at −20 °C for 1 h. Tubes were then centrifuged at 12,000× g for 10 min at 4 °C, the pellet washed with 1 mL of 75% (v/v) ethanol and tubes centrifuged at 7,500× g for 5 min, at 4 °C, and left to dry at room temperature, for 5 min. After DNase I (Invitrogen, Rodano, Italy) treatment, the total RNA was purified using RNA Clean & Concentrator ™-5 (Zymo Research, Irvine, CA, USA), according to the manufacturer instructions.
RNA quality and quantity were verified by NanoDrop (ThermoFisher Scientific, Waltham, MA, USA), while RNA integrity was evaluated on denaturing formaldehyde agarose gel (0.5% w/v). The RNA quality obtained was good, according to the standard parameters for real-time expression analysis (Figure S2).
Primer pairs for the cas7 (csn2), ltp, viral non-coding region, and terminase large subunit (Table 1) were designed using the on-line tool NCBI/Primer-BLAST [42] and manually checked on the PCR templates. Primers were obtained from Invitrogen (Thermo Fisher Scientific, Rodano, MI, Italy). The glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene from Mus musculus was used as reference control [43].
One µg of RNA was reverse transcribed, using the RevertAid Reverse Transcriptase (Thermo Scientific) and random hexamer primers, according to the manufacturer recommendations. qRT-PCR was performed using Power SYBR® Green PCR master mix (Life Technologies).
2.6. Statistical Analysis
Statistical analysis was performed with the GraphPad Instat 3 software (GraphPad, La Jolla, CA, USA), using the one-way analysis of variance (ANOVA), to evaluate the phage-binding capability and S. thermophilus M17PTZA496 viable cell counts, under different conditions. Assays were set up in triplicates and Tukey’s test was used as the post hoc test.
The relative expression level of each gene was normalized, taking into consideration the gene gapdh of Mus musculus as reference. Transcript levels from the RNA samples were evaluated using three technical replicates and the difference in the transcription of each specific gene was calculated with the 2−ΔΔCT method [51]. Paired Student t-test was used to calculate the significance of the difference between the relative expression of the cas7, ltp, Siphovirus Gp157, and terminase large subunit, under different conditions. Statistically significant values were defined for p < 0.05.
3. Results
3.1. Genomic Analysis of the S. thermophilus M17PTZA496 Prophages
In the present study, a global genomic investigation was performed to describe the prophages identified in the S. thermophiles, from a taxonomic and functional perspective. The presence of two prophages in the S. thermophilus M17PTZA496 chromosome was predicted (Figure S3). According to their position in the bacterial genome they were named TP1-M17PTZA496 (bacterial genome position: 617,680–669,542 bp; 51.8 Kb in length) and TP2-M17PTZA496 (bacterial genome position: 890,763–919,156 bp). The estimated length of TP2-M17PTZA496 is 28.3 Kb, with a GC content of 34.74% and twenty-eight predicted protein-encoding genes. The TP2-M17PTZA496 gene annotation revealed some features with a relevant technological role, such as a methionyl-tRNA synthetase, a carotenoid biosynthetic protein, an exopolysaccharide associated protein (Eps4Q), a bacteriocin and a glucose transporter (Table S2). As described elsewhere, a temperate prophage can be considered non-inducible when the lysis module is not identified [52,53]. The same applies to TP2-M17PTZA496, even though it was classified as “intact” by the PHASTER software. For this reason, only TP1-M17PTZA496 was considered for further analysis.
The genomic region representative of the TP1-M17PTZA496 has an estimated length of 51.8 Kb, a GC content of 39.13% and a total of sixty-one protein-encoding genes predicted between the attL and attR attachment sites (Figure 1). Results obtained from the similarity searches were manually curated, improving the overall annotation quality, and leading to the clarification of the functional role of integrated genes. Downstream to the left host-phage junction (attL), bacterial genes encoding enolase, glycogen synthase, and glycerate kinase were found, while upstream to the right junction (attR), bacterial genes encoding isoleucyl-tRNA synthetase, lipoteichoic acid synthase, methyltransferase, and 3-dehydroquinate dehydratase were identified. These integration sites have high similarity with those identified in the temperate bacteriophage Φ20617 [37], as revealed by the nucleotide BLAST analysis that showed 84% of identity between both viruses and a query cover of 76%. Presence of the tRNA synthetase genes in the att sites is a common feature, since tRNA coding regions have been recognized as integration site “hotspots” for viruses [54]. In the present study, isoleucyl-tRNA and methyonil-tRNA synthetase close to the att sites, showed this “preference rule” in the S. thermophilus.
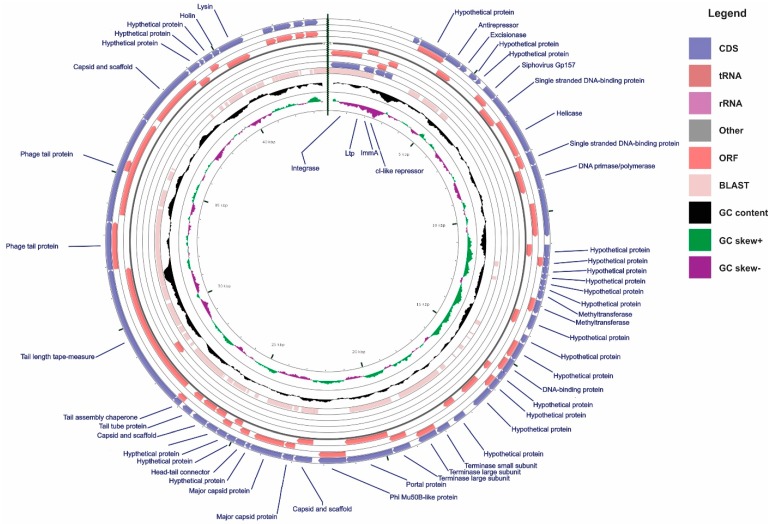
Figure 1.
Genome map of the TP1-M17PTZA496. The linear genome was circularized to improve its visualization. CDS, ORF, BLAST against the Streptococcus phage 20617, GC content, GC skew+, and GC skew−, are reported in circles from the outside inwards. Streptococcus phage 20617 whole genome sequence was used as a reference for the BLAST analysis. Only the ORFs between the attL and attR are displayed for the TP1-M17PTZA496.
In TP1-M17PTZA496, fifty-seven ORFs are transcribed from the positive strand. Most of the genes were assigned to putative functions and only seventeen ORFs remained as sequences coding for hypothetical proteins (Table S3). Some interesting features related to gene content and phage structure were identified; for example, ORF 2 (Ltp) and ORF 11, annotated as a gene with high similarity to the Gp157 of the S. thermophilus bacteriophages members of the genus Sfi11virus, commonly located in a cro-ori region, was found to be related to phage immunity. Regarding the lysogeny prophage module, ORF4 was annotated as CI-like repressor, while a cro-like coding sequence was missing.
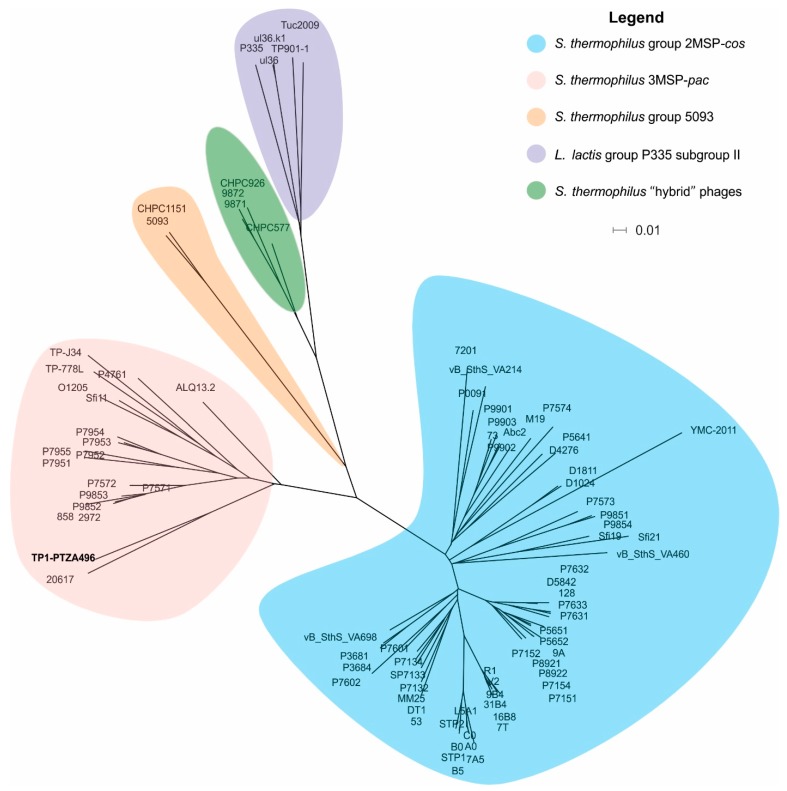
A phylogenomic tree was constructed, based on a “fragmented all-against-all comparison” with the aim of determining the taxonomic relationship that exist between prophage TP1-M17PTZA496 and other phages infecting the S. thermophilus and Lactococcus lactis. The phylogenetic tree evidenced that these viruses were clustered into five groups (Figure 2). According to this classification, the TP1-M17PTZA496 was associated with members of the S. thermophilus group 3MSP-pac and it is closely related to the temperate bacteriophage Φ20617 [14,37,54,55,56,57,58].
Figure 2.
Phylogenomic tree constructed using the whole genome sequence of the TP1-M17PTZA496, 83 S. thermophilus and 5 L. lactis bacteriophages. GenBank accession numbers are reported in Table S1. The scale bar represents a 1% difference on the average tBLASTx score.
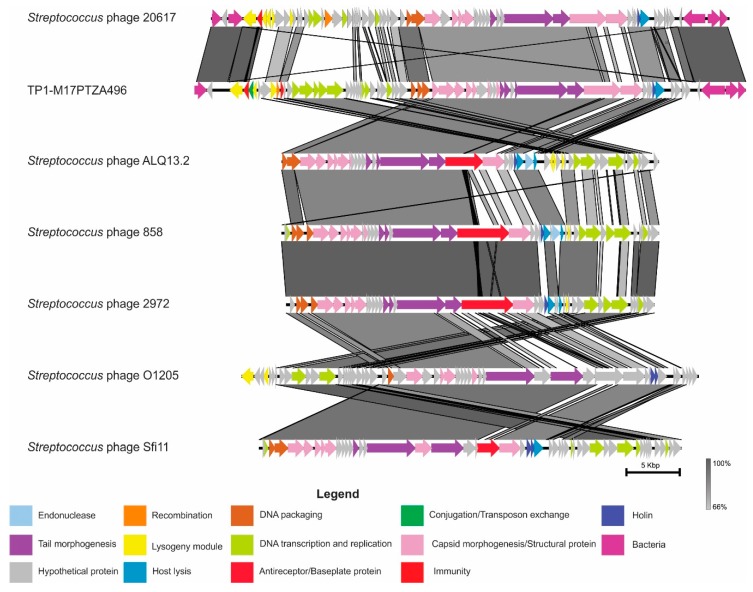
According to the TP1-M17PTZA496 phylogenomic tree, the genome homology among the viruses grouped as 3MSP-pac was investigated, based on their translated nucleotide sequence (Figure 3). Despite this, the analysis revealed that the virus TP1-M17PTZA496 has a modular organization related to the virus φ20617, major differences were identified on the lysogeny module. The main changes are related to the absence of the coding sequence for cro and the presence of immA, which encodes an anti-immunity system involved in triggering, between the lytic and the lysogenic cycle [59]. The modules coding for structural proteins, tail morphogenesis, and host lysis (including holin) displayed a high similarity with other members of the Sfi11virus genus, mainly with the Streptococcus phage ALQ13.2.
Figure 3.
Sequence alignment among the Streptococcus phage 20617, TP1-M17PTZA496, and the members of the genus Sfi11virus. Gray shading corresponds to the percentage of identity of the nucleotide sequences.
Phage-host interactions were analyzed, considering the spacer acquisition in the CRISPR arrays. This finding demonstrated that twenty-seven spacers associated with the TP1-M17PTZA496, were predicted in three species of the genus Streptococcus, indicating mainly S. thermophilus (24 spacers) and, to a lesser extent, S. salivarius (2 spacers) and S. macedonicus (1 spacer) as the main potential hosts (Table S4).
3.2. Prophage Induction Evaluation
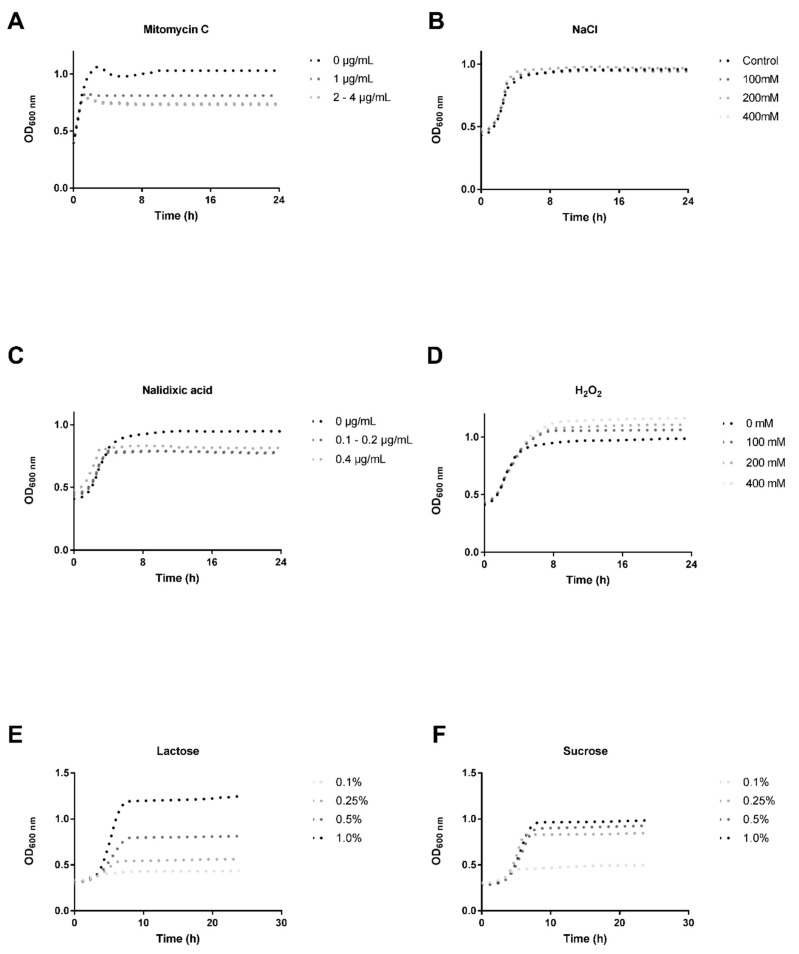
The ability of the S. thermophilus M17PTZA496 to release inducible prophages was evaluated by treatment with four different phage-inducing agents (MmC, nalidixic acid, H2O2 and NaCl) added at the beginning of the log-phase (corresponding to OD600 ~ 0.3). It is worth mentioning that low concentrations of MmC (0.1, 02 and 0.5 µg/mL) were evaluated but no positive results were obtained [60]. Moreover, we used low (0.1, 0.25, and 0.5%) and high (1.0% w/v) concentrations of lactose and sucrose, in order to evaluate the effect of energy starvation on the bacteriophage induction. Comparison between the growth curves (24 h, 44 °C) of the treated and the untreated cultures, revealed that S. thermophilus M17PTZA496 possesses cryptic prophages. The presence of the prophages was clearly evidenced by the fact that the OD600 value in the treated samples, did not present significant decreases, which is typically represented by a growth inhibition, phage production, and host death (Figure 4). Based on these observations, we proceeded with further investigations, using only Mitomycin C.
Figure 4.
Phage-induction evaluation performed using four different chemicals: (A) Mitomycin C (1–4 µg/mL); (B) NaCl (100, 200, and 400 mM); (C) Nalidixic acid (0.1, 0.2, and 0.4 µg/mL); (D) H2O2 (100, 200, and 400 mM); (E) Lactose (0.1, 0.25, 0.5, and 1.0% w/v); (F) Sucrose (0.1, 0.25, 0.5, and 1.0% w/v). The same legend was used for different concentrations of the MmC (2–4 µg/mL) and the Nalidixic acid (0.1–0.2 µg/mL), since these growth curves overlapped each other.
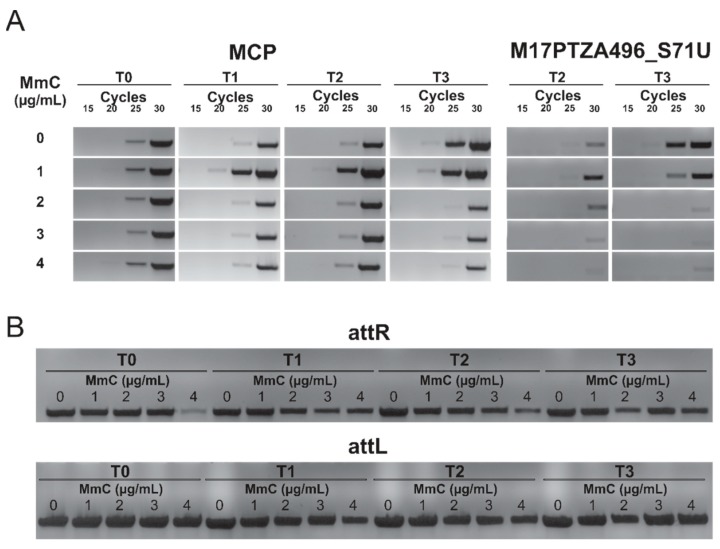
Additionally, in order to determine the copy number of the viral DNA in the lysogenic S. thermophilus M17PTZA496, semi-quantitative PCR analyses were performed, using the total genomic DNA as a template, and two different primer couples. Specific primers targeting the MCP gene were used to determine the prophage TP1-M17PTZA496 copy number, while primers targeting a genomic region of the bacterium (Scaffold 71; 297,265–304,822 bp) were used to determine the number of copies of the bacterial chromosome. Analyses were performed on bacteria cultures grown at different MmC concentrations and at multiple time points, after treatment (Figure 5A). Results showed that the MCP copy-number increased after 30 (T1) and 60 min (T2), following the MmC addition (1 µg/mL), as revealed by the presence of a detectable signal, after twenty amplification cycles. In fact, Arioli et al. (2018) [37] described the heterogeneity of S. thermophilus DSM20617T determined by the phage excision events, even in the non-cured cells. On the other hand, when higher amounts of MmC were used (2, 3 and 4 µg/mL), a reduction in the MCP content was observed and a detectable signal was identified at the 25th cycle. As described by Oliveira et al. (2017) [41], high concentrations of MmC might have a toxic effect on the Lactococcus lactis culture harboring prophage and we observed the same phenomenon in the S. thermophilus M17PTZA496. In all the conditions tested, the complete sequences of the attL and the attR sites were identified (Figure 5B), confirming that prophage TP1-M17PTZA496 is not able to be completely excised from the host chromosome.
Figure 5.
(A) Semi-quantitative PCR analysis showing the MCP levels at 30, 60, and 90 min, after addition of different amounts of MmC. M17PTZA496_S71U was used as the bacterial chromosome control region. (B) attL and attR sites amplification to check the absence of phage excision. T0, T1, T2, and T3 are the sampling times, namely 0, 30, 60, and 90 min, respectively under diverse MmC concentrations (1–4 µg/mL).
Finally, TEM analysis of the samples obtained from the treated and the untreated cultures was performed, considering four time points after exposure to MmC. Results were in agreement with the semi-quantitative PCR and revealed that no complete or defective phage particles were present in the lysogenic S. thermophilus M17PTZA496 strain.
3.3. S. thermophilus M17PTZA496 Phage-Susceptibility Assay
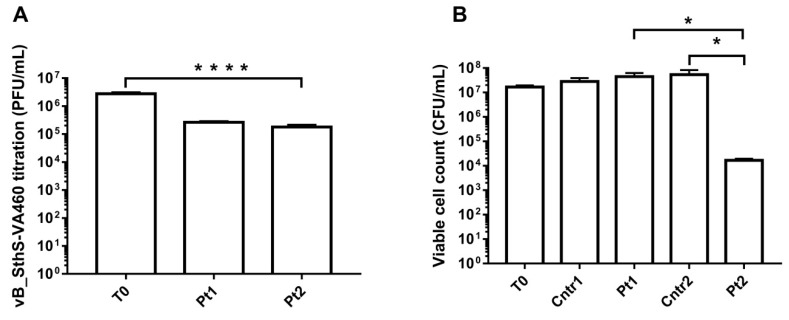
Six different cos-type bacteriophages, previously characterized in terms of their variable region (VR2), were used to perform phage-susceptibility screenings. S. thermophilus M17PTZA496 was only found to be susceptible but not permissive, to phage vB_SthS-VA460 (VA460), thus, suggesting the presence of bacterial mechanisms blocking it from starting the lytic cycle. After viral titration was performed on VA460, a 1-log reduction of the viral content was observed (90.4% of phages bound, Figure 6A). Measurement of the phage VA460 attachment rate was performed using the S. thermophilus M17PTZA496 cells as target, and it was calculated to be 7.79 × 10−9 mL/min.
Figure 6.
(A) Phage titration of the S. thermophilus cultures. Samples were taken at three different time points, namely at 0 (T0), 30 (Pt1), and 60 min (Pt2). (A) After addition of the VA460 plus MmC. (B) S. thermophilus M17PTZA496 viable cell counts, after PBS (Cntr1), VA460 (Pt1), Mitomycin C (Cntr2 and Pt2), and VA460 plus MmC (Pt2) addition. Asterisks indicate different levels of statistical significance (*: p ≤ 0.05; ****: p < 0.0001).
To evaluate the effectiveness of S. thermophilus M17PTZA496 phage defense mechanisms, the expression level of cas7, ltp, and the ORF11 (Siphovirus Gp157 located in a viral cro-ori region), was investigated [61,62,63]. Additionally, to determine the capability of the prophage TP1-M17PTZA496 to package its genomic DNA, the expression level of the terminase large subunit gene was analyzed. RT-PCR analysis showed that MmC provided at a concentration of 1 µg/mL, led to a small but significant reduction in the expression level of both cas7 and terminase large subunits (0.4 and 0.15-fold, respectively, Table 2). A 0.4-fold reduction was also observed for the ltp, Siphovirus Gp157, and high subunit expression levels, when only phage VA460 was added. Conversely, when MmC and VA460 were provided together, the ltp expression level increased 2.2-fold. A very similar result was obtained for the terminase large subunit gene (1.8 fold).
Table 2.
Relative expression of the cas7, ltp, Siphovirus Gp157, and terminase large subunit. Only statistically significant values (p < 0.05) were considered. (Cntr1—PBS; Pt1—VA460; Cntr2—MmC; Pt2—VA460 plus MmC).
| cas7 | ltp | Siphovirus Gp157 | Terminase Large Subunit | |||||
|---|---|---|---|---|---|---|---|---|
| Conditions | p-value | X-fold | p-value | X-fold | p-value | X-fold | p-value | X-fold |
| Cntr1 vs Pt1 | 0.093 | 0.50 | 0.012 | 0.40 | 0.016 | 0.40 | 0.012 | 0.40 |
| Pt2 vs Cntr2 | 0.154 | 1.80 | 0.022 | 2.20 | 0.756 | 1.10 | 0.026 | 1.75 |
| Cntr1 vs Cntr2 | 0.025 | 0.40 | 0.320 | 0.70 | 0.250 | 0.80 | 0.000 | 0.15 |
In accordance with the gene expression results, no reduction of the S. thermophilus M17PTZA496 viable cells was observed when the MmC or VA460 were independently added to the bacterial culture. Instead, a significant 3-log reduction (Figure 6B) was observed when S. thermophilus M17PTZA496 was exposed to a combination of the VA460 and the MmC.
4. Discussion
According to Hols et al. (2005) [64], HGT among bacteria contributes to the development of important industrially-relevant phenotypic characteristics. However, phage-mediated HGT is favored by the virus capability to infect different hosts [65,66], a quite uncommon feature for S. thermophilus bacteriophages, which are normally characterized by a narrow host spectrum [45,47]. An in silico host range prediction revealed that prophage TP1-M17PTZA496 has the potential capability to infect multiple Streptococcus species, including S. thermophilus, S. macedonicus, and S. salivarius. This interesting characteristic suggests that TP1-M17PTZA496 could have a potential role in the HGT.
Lysogeny is an infrequent process in S. thermophilus [67,68]. In the present study, the analysis of the prophage TP1-M17PTZA496 lysogeny module identified a putative cI-like repressor, while a gene coding for a cro-like repressor was absent. This is relevant to increase our knowledge of phage DNA replication properties and gene expression of the S. thermophilus phages, for which little information is currently available [21]. Cro and CI proteins mutually repress each other and are involved in the lytic/lysogenic switch [69,70]. As discussed below, TP1-M17PTZA496 is considered a non-inducible prophage and this behavior can be associated with the lack of a cro-like repressor [71]. Moreover, the terminase large subunit was flanked by genes involved in the lysis/morphogenesis and a group I intron was also identified in the CDS. The homing of group I introns are horizontally transferred, via mixed infections, as described for pac-type Streptococcus phage 2972, and removed through phage mRNA splicing [72]. Similar to the TP1-M17PTZA496, the phage 2972 possessed two group I intron sequences located in the genes coding for the terminase large subunit and for the endolysin. An alignment performed between the prophage M17PTZA496 and Streptococcus phage 2972 sequences, revealed that the same intron was present in the terminase large subunit of both viruses. This result corroborated our hypothesis that the prophage TP1-M17PTZA496 originated via mixed infection between the cos- and the pac-S. thermophilus phages. Despite the process that led to the classification of the TP1-M17PTZA496, as a mix between the cos- and the pac- S. thermophilus bacteriophages, phylogeny assigned it to the S. thermophilus group 3MSP-pac. This group includes other pac-type Streptococcus phages, such as O1205, 858, TP-J34, and Sfi11 [15,73,74,75,76].
Bioinformatics analysis revealed that the TP1-M17PTZA496 has interesting features associated with phage immunity, along with the absence of cro, in its lysogeny module. To verify this finding, the ability of the prophage to excise was checked, both using four different chemical compounds, and also by testing lactose- and sucrose-limiting conditions. The growth profiles of the treated cultures indicated that the TP1-M17PTZA496 can be considered non-inducible. Additional PCR-based and TEM analyses were in agreement in asserting that the TP1-M17PTZA496 can be considered a cryptic prophage with a very low excision rate, unable to form the phage particles. This result is consistent with previous findings indicating that different prophages in the E. coli K-12 can have very low excision rates, with values that may reach less than 1 prophage per 100,000 cells, or adopt the pseudo-lysogeny life cycle [37,73,77,78]. To further support results related to the excision ability, gene expression analyses were used. Since the small and large terminase subunits are components of the packaging machinery [79], their expression is representative of the tailed phages ability to pack their DNA. The large terminase relative expression analysis confirmed that TP1-M17PTZA496 has different activity levels, which slightly decreased after the MmC and VA460 treatments, but markedly increased, after the combined application of both stimuli. Interestingly, after the combined application, a marked reduction of the S. thermophilus M17PTZA496 viable cells was observed, although no cellular lysis was detected. This observation might be due to SOS system response activation and the phage-attack response, which justifies the absence of viable cells after plating.
Six cos-type S. thermophilus bacteriophages were screened for their capability to infect the S. thermophilus M17PTZA496. After viral titration, it was demonstrated that VA460 was able to adsorb S. thermophilus M17PTZA496, without provoking host lysis (rate of attachment 7.79 × 10−9 mL/min). Our result was in agreement with those of Bull et al. (2014) [80], who reported similar constant adsorption rate values (from 10−8 to 10−9 mL/min), as a result of the collision between phage and bacteria that culminated in viral infection, a relevant step to evaluate the bacterium self-defense mechanisms against phages. For this reason, VA460 was chosen for the phage immunity evaluation of S. thermophilus M17PTZA496. In particular, sie and host protection were tested by means of the Ltp type protein and the Siphovirus Gp157 expression.
A significant increase in the ltp expression (2.2 fold) was observed when the S. thermophilus M17PTZA496 was treated with a combination of VA460 and MmC. However, when only VA460 was added, a 0.4-fold decrease in expression (p < 0.05) was noticed. It was previously reported that ltp is transcribed only in the prophage state [15]; accordingly, we suggest that TP1-M17PTZA496 is a non-inducible prophage that actively expresses ltp. However, its low expression level could be attributed to the prophage transient excision activity level and, consequently, to the ltp expression reduction. It is worth mentioning that the relative expression level of each gene was normalized, using the gapdh gene of the Mus musculus as an external reference control. This normalization was needed considering that the addition of 1 µg/mL of MmC impaired the stable expression of the housekeeping genes gapdh, recA, and the rpoD of the S. thermophilus M17PTZA496 [60]. As described elsewhere [81,82], the use of an external reference gene is a valuable solution when the expression levels of the internal reference genes are not stable under determined conditions.
With regard to the short viral noncoding region, its function has been predicted to be similar to a 302-bp noncoding viral element (system PER—phage-encoded resistance) found in the S. thermophilus bacteriophage ΦSfi21 genome [83]. This region acts as an independent replication origin activated by viral infection and its action is copy-number dependent. In the present study, a 0.4-fold reduction in the number of copies of the Siphovirus Gp157 was observed in the presence of the phage VA460. This finding can be explained by the activity of this element, which is able to bind to the infected viral proteins and to decrease their free copy-number. As reported by Foley et al. (1998) [17], this region protected the S. thermophilus Sf1 from infection by seventeen out of the twenty-five evaluated phages. Noncoding oris regions were also previously reported in other bacteriophages, such as lactococcal phage P335 [84], S. thermophilus bacteriophages DT1, Sfi19, Sfi21, O1205, 7201, and κ3 [21].
Finally, the unexpected presence of two prophages in the S. thermophilus M17PTZA496 genome was previously hypothesized to be related to the low number of CRISPR-Cas modules found in this strain [28]. Relative expression of the cas7, a CRISPR protein involved in spacer acquisition [61], was examined with the aim of evaluating the influence of the bacterial adaptive immune system, under phage and antibiotic treatments. It was observed that only MmC influenced the cas7 expression leading to a 0.4-fold reduction. The generally low activity of the CRISPR-Cas defense mechanism highlights the importance of the previously described mechanism in host superinfection exclusion. Moreover, using the S. thermophilus and the virulent phage 2972 as models, Vale et al. (2015) [85] suggested that the maintenance of the CRISPR-Cas defense system requires a great load of energy for the cell, that can result in decreased bacterial fitness. Since the MmC determines DNA damage and induces the SOS response, a process that requires a conspicuous amount of energy [86], it could be hypothesized that the MmC suppresses the bacterial adaptive immune system, in order to save energy.
5. Conclusions
Although the CRISPR-Cas system plays a relevant role in the phage-host defense, the presence of cryptic prophages can enhance bacterial defenses. Here, we reported the occurrence of Ltp and a noncoding viral DNA element in the non-inducible prophage TP1-M17PTZA496. Thanks to these additional immune mechanisms, S. thermophilus M17PTZA496 can be proposed as an alternative model to study bacterial resistance to phages. In conclusion, considering the enormous importance of this bacterial species for the industrial production of fermented foods, S. thermophilus M17PTZA496 could be considered as a promising alternative to the phage-sensitive starter cultures for the manufacturing of dairy products.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4915/11/1/7/s1, Figure S1: RT-qPCR experimental design, Figure S2: RNA integrity was evaluated on denaturing formaldehyde agarose gel (0.5% w/v), Figure S3, PHASTER analysis indicating the two prophages (positions 617,680–669,542 and 890,763–919,156) integrated in the S. thermophilus M17PTZA496 chromosome, named (A) TP1-M17PTZA496 and (B) TP2-M17PTZA496, Table S1: Genbank accession number of the bacteriophages sequences (or genome sequences?) used to construct the phylogenomic tree, Table S2: TP2-M17PTZA496 annotation considering three different databases. Asterisks indicate lack of information according to each database, Table S3: 61 ORFs were predicted among attL and attR attachment sites, Table S4: 27 different spacers were predicted for the prophage M17PTZA496 in 3 diverse species of the genus Streptococcus (24 S. thermophilus, 2 S. salivarius and 1 S. macedonicus) using the CRISPRdb database.
Author Contributions
Conceptualization, V.d.S.D.; S.G.; S.C., and L.T.; Data curation, V.d.S.D.; S.C.; L.T. and A.A.; Funding acquisition, S.O.d.P.; A.G., and V.C.; Investigation, V.d.S.D.; S.G.; A.A., and A.T.; Resources, A.A. and S.O.d.P.; Supervision, L.T. and A.G.; Writing—original draft, V.d.S.D.; S.C., and L.T.; Writing—review & editing, A.G.
Funding
We are grateful to the following Brazilian agencies for their financial support (scholarship during the doctoral period, exchange modality, at the University of Padova): Fundação de Amparo à Pesquisa do Estado de Minas Gerais (Fapemig), Coordenacão de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Financiadora de Estudos e Projetos (Finep), Sistema Nacional de Laboratórios em Nanotecnologias (SisNANO)/Ministério da ciência, tecnologia e Informação (MCTI).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Calasso M., Ercolini D., Mancini L., Stellato G., Minervini F., Di Cagno R., De Angelis M., Gobbetti M. Relationships among house, rind and core microbiotas during manufacture of traditional Italian cheeses at the same dairy plant. Food Microbiol. 2016;54:115–126. doi: 10.1016/j.fm.2015.10.008. [DOI] [Google Scholar]
- 2.Marco M.L., Heeney D., Binda S., Cifelli C.J., Cotter P.D., Foligné B., Gänzle M., Kort R., Pasin G., Pihlanto A., et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017;44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Maragkoudakis P.A., Nardi T., Bovo B., D’Andrea M., Howell K.S., Giacomini A., Corich V. Biodiversity, dynamics and ecology of bacterial community during grape marc storage for the production of grappa. Int. J. Food Microbiol. 2013;162:143–151. doi: 10.1016/j.ijfoodmicro.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Coton E., Leguerinel I. Encyclopedia of Food Microbiology. Elsevier; Amsterdam, The Netherlands: 2014. Ecology of Bacteria and Fungi in Foods | Effects of pH; pp. 577–586. [Google Scholar]
- 5.Bovo B., Giacomini A., Corich V. Effects of grape marcs acidification treatment on the evolution of indigenous yeast populations during the production of grappa. J. Appl. Microbiol. 2011;111:382–388. doi: 10.1111/j.1365-2672.2011.05060.x. [DOI] [PubMed] [Google Scholar]
- 6.Dantigny P., Burgain A., Deniel F., Bensoussan M. A model for the effect of pH on the growth of chalk yeasts. Int. J. Food Microbiol. 2014;186:49–54. doi: 10.1016/j.ijfoodmicro.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Hutkins R.W. Microbiology and Technology of Fermented Foods. Wiley-Blackwell; Hoboken, NJ, USA: 2007. [Google Scholar]
- 8.Clokie M.R., Millard A.D., Letarov A.V., Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcó M.B., Moineau S., Quiberoni A. Bacteriophages and dairy fermentations. Bacteriophage. 2012;2:149–158. doi: 10.4161/bact.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruttin A., Desiere F., D’Amico N., Guérin J.P., Sidoti J., Huni B., Lucchini S., Brüssow H. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 1997;63:3144–3150. doi: 10.1128/aem.63.8.3144-3150.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moineau S., Lévesque C. Bacteriophages. CRC Press; Boca Raton, FL, USA: 2004. Control of Bacteriophages in Industrial Fermentations; pp. 285–296. [Google Scholar]
- 12.Garneau J.E., Moineau S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 2011;10:S20. doi: 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonnell B., Mahony J., Hanemaaijer L., Kouwen T.R.H.M., van Sinderen D. Generation of bacteriophage-insensitive mutants of Streptococcus thermophilus via an antisense RNA CRISPR-Cas silencing approach. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.01733-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szymczak P., Janzen T., Neves A.R., Kot W., Hansen L.H., Lametsch R., Neve H., Franz C.M.A.P., Vogensen F.K. Novel Variants of Streptococcus thermophilus Bacteriophages Are Indicative of Genetic Recombination among Phages from Different Bacterial Species. Appl. Environ. Microbiol. 2017;83:e02748-16. doi: 10.1128/AEM.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali Y., Koberg S., Heßner S., Sun X., Rabe B., Back A., Neve H., Heller K.J. Temperate Streptococcus thermophilus phages expressing superinfection exclusion proteins of the Ltp type. Front. Microbiol. 2014;5:98. doi: 10.3389/fmicb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The Biology of CRISPR-Cas: Backward and Forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 17.Foley S., Lucchini S., Zwahlen M.C., Brüssow H. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology. 1998;250:377–387. doi: 10.1006/viro.1998.9387. [DOI] [PubMed] [Google Scholar]
- 18.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 19.Cumby N., Davidson A.R., Maxwell K.L. The moron comes of age. Bacteriophage. 2012;2:225–228. doi: 10.4161/bact.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X., Göhler A., Heller K.J., Neve H. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology. 2006;350:146–157. doi: 10.1016/j.virol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Lamothe G., Lévesque C., Bissonnette F., Cochu A., Vadeboncoeur C., Frenette M., Duplessis M., Tremblay D., Moineau S. Characterization of the cro-ori region of the Streptococcus thermophilus virulent bacteriophage DT1. Appl. Environ. Microbiol. 2005;71:1237–1246. doi: 10.1128/AEM.71.3.1237-1246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarrah A., Noal V., Treu L., Giaretta S., da Silva Duarte V., Corich V., Giacomini A. Short communication: Comparison of growth kinetics at different temperatures of Streptococcus macedonicus and Streptococcus thermophilus strains of dairy origin. J. Dairy Sci. 2018 doi: 10.3168/jds.2018-14731. [DOI] [PubMed] [Google Scholar]
- 23.Tarrah A., Noal V., Giaretta S., Treu L., da Silva Duarte V., Corich V., Giacomini A. Effect of different initial pH on the growth of Streptococcus macedonicus and Streptococcus thermophilus strains. Int. Dairy J. 2018;86:65–68. doi: 10.1016/j.idairyj.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Tarrah A., Treu L., Giaretta S., Duarte V., Corich V., Giacomini A. Differences in Carbohydrates Utilization and Antibiotic Resistance Between Streptococcus macedonicus and Streptococcus thermophilus Strains Isolated from Dairy Products in Italy. Curr. Microbiol. 2018 doi: 10.1007/s00284-018-1528-7. [DOI] [PubMed] [Google Scholar]
- 25.Tarrah A., de Castilhos J., Rossi R.C., da Duarte V.S., Ziegler D.R., Corich V., Giacomini A. In vitro Probiotic Potential and Anti-cancer Activity of Newly Isolated Folate-Producing Streptococcus thermophilus Strains. Front. Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krusch U., Neve H., Luschei B., Teuber M. Characterization of virulent bacteriophages of Streptococcus salivarius subsp. thermophilus by host specificity and electron microscopy. Kieler Milchwirtschaftliche Forschungsberichte. 1987;39:155–167. [Google Scholar]
- 27.Treu L., Vendramin V., Bovo B., Campanaro S., Corich V., Giacomini A. Genome sequences of Streptococcus thermophilus strains MTH17CL396 and M17PTZA496 from fontina, an Italian PDO cheese. Genome Announc. 2014;2 doi: 10.1128/genomeA.00067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vendramin V., Treu L., Campanaro S., Lombardi A., Corich V., Giacomini A. Genome comparison and physiological characterization of eight Streptococcus thermophilus strains isolated from Italian dairy products. Food Microbiol. 2017;63:47–57. doi: 10.1016/j.fm.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:1–6. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M., et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2016:gkw1107. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant J.R., Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ågren J., Sundström A., Håfström T., Segerman B. Gegenees: Fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS ONE. 2012 doi: 10.1371/journal.pone.0039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barylski J., Nowicki G., Goździcka-Józefiak A. The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae. PLoS ONE. 2014 doi: 10.1371/journal.pone.0086632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006 doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 36.Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018 doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arioli S., Eraclio G., Della Scala G., Neri E., Colombo S., Scaloni A., Fortina M.G., Mora D. Role of Temperate Bacteriophage ϕ20617 on Streptococcus thermophilus DSM 20617T Autolysis and Biology. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villarroel J., Kleinheinz K.A., Jurtz V.I., Zschach H., Lund O., Nielsen M., Larsen M.V. HostPhinder: A phage host prediction tool. Viruses. 2016;8:116. doi: 10.3390/v8050116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira J., Mahony J., Hanemaaijer L., Kouwen T.R.H.M., Neve H., MacSharry J., van Sinderen D. Detecting Lactococcus lactis Prophages by Mitomycin C-Mediated Induction Coupled to Flow Cytometry Analysis. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NCBI/Primer-BLAST. [(accessed on 25 September 2018)]; Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast.
- 43.Conte M., Vasuri F., Bertaggia E., Armani A., Santoro A., Bellavista E., Degiovanni A., D’Errico-Grigioni A., Trisolino G., Capri M., et al. Differential expression of perilipin 2 and 5 in human skeletal muscle during aging and their association with atrophy-related genes. Biogerontology. 2015;16:329–340. doi: 10.1007/s10522-014-9549-5. [DOI] [PubMed] [Google Scholar]
- 44.da Silva Duarte V., Giaretta S., Treu L., Campanaro S., Pereira Vidigal P.M., Tarrah A., Giacomini A., Corich V. Draft Genome Sequences of Three Virulent Streptococcus thermophilus Bacteriophages Isolated from the Dairy Environment in the Veneto Region of Italy. Genome Announc. 2018;6 doi: 10.1128/genomeA.00045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binetti A.G., Del Rio B., Martin M.C., Alvarez M.A. Detection and Characterization of Streptococcus thermophilus Bacteriophages by Use of the Antireceptor Gene Sequence. Appl. Environ. Microbiol. 2005;71:6096–6103. doi: 10.1128/AEM.71.10.6096-6103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svensson U., Christiansson A. Methods for phage monitoring [easy to use in dairy laboratories] Bull. Int. Dairy Fed. 1991;263:29–39. [Google Scholar]
- 47.Quiberoni A., Tremblay D., Ackermann H.-W., Moineau S., Reinheimer J.A. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J. Dairy Sci. 2006;89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- 48.Valyasevi R., Sandine W.E., Geller B.L. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 1990;56:1882–1889. doi: 10.1128/aem.56.6.1882-1889.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kropinski A.M. Measurement of the Rate of Attachment of Bacteriophage to Cells. Methods Mol. Biol. 2009;501:151–155. doi: 10.1007/978-1-60327-164-6_15. [DOI] [PubMed] [Google Scholar]
- 50.Naghili H., Tajik H., Mardani K., Razavi Rouhani S.M., Ehsani A., Zare P. Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. Vet. Res. Forum Int. Q. J. 2013;4:179–183. [PMC free article] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Chopin A., Bolotin A., Sorokin A., Ehrlich S.D., Chopin M. Analysis of six prophages in Lactococcus lactis IL1403: Different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 2001;29:644–651. doi: 10.1093/nar/29.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catalão M.J., Gil F., Moniz-Pereira J., São-José C., Pimentel M. Diversity in bacterial lysis systems: Bacteriophages Show the way. FEMS Microbiol. Rev. 2013;37:554–571. doi: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 54.Fouts D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brüssow H., Probst A., Frémont M., Sidoti J. Distinct Streptococcus thermophilus Bacteriophages Share an Extremely Conserved DNA Fragment. Virology. 1994 doi: 10.1006/viro.1994.1256. [DOI] [PubMed] [Google Scholar]
- 56.Le Marrec C., Van Sinderen D., Walsh L., Stanley E., Vlegels E., Moineau S., Heinze P., Fitzgerald G., Fayard B. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 1997 doi: 10.1128/aem.71.10.6096-6103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonnell B., Mahony J., Neve H., Hanemaaijer L., Noben J.P., Kouwen T., van Sinderen D. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 2016 doi: 10.1128/AEM.00835-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahony J., Martel B., Tremblay D.M., Neve H., Heller K.J., Moineau S., Van Sinderen D. Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13. Appl. Environ. Microbiol. 2013 doi: 10.1128/AEM.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravin N.V., Svarchevsky A.N., Dehò G. The anti-immunity system of phage-plasmid N15: Identification of the antirepressor gene and its control by a small processed RNA. Mol. Microbiol. 1999 doi: 10.1046/j.1365-2958.1999.01658.x. [DOI] [PubMed] [Google Scholar]
- 60.Tarrah A. (Department of Agronomy Food Natural Resources Animals and Environment, University of Padova, 35020 Legnaro, Italy). Page induction tests using low concentrations of Mitomycin C. 2018.
- 61.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seed K.D. Battling Phages: How Bacteria Defend against Viral Attack. PLoS Pathog. 2015;11:e1004847. doi: 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pare K.R. Bacterial toxin-antitoxin systems. Mob. Genet. Elements. 2011;1:283–290. doi: 10.4161/mge.18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hols P., Hancy F., Fontaine L., Grossiord B., Prozzi D., Leblond-Bourget N., Decaris B., Bolotin A., Delorme C., Dusko Ehrlich S., et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 2005;29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 65.Beumer A., Robinson J.B. A Broad-Host-Range, Generalized Transducing Phage (SN-T) Acquires 16S rRNA Genes from Different Genera of Bacteria. Appl. Environ. Microbiol. 2005;71:8301–8304. doi: 10.1128/AEM.71.12.8301-8304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watson B.N.J., Staals R.H.J., Fineran P.C. CRISPR-Cas-Mediated Phage Resistance Enhances Horizontal Gene Transfer by Transduction. mBio. 2018;9:e02406-17. doi: 10.1128/mBio.02406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brussow H., Fremont M., Bruttin A., Sidoti J., Constable A., Fryder V. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl. Environ. Microbiol. 1994;60:4537–4543. doi: 10.1128/aem.60.12.4537-4543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canchaya C., Proux C., Fournous G., Bruttin A., Brüssow H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Achigar R., Magadan A.H., Tremblay D.M., Julia Pianzzola M., Moineau S. Phage-host interactions in Streptococcus thermophilus: Genome analysis of phages isolated in Uruguay and ectopic spacer acquisition in CRISPR array. Sci. Rep. 2017;7:43438. doi: 10.1038/srep43438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vohradsky J. Lambda phage genetic switch as a system with critical behaviour. J. Theor. Biol. 2017;431:32–38. doi: 10.1016/j.jtbi.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 71.Schubert R.A., Dodd I.B., Egan J.B., Shearwin K.E. Cro’s role in the CI Cro bistable switch is critical for {lambda}’s transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–2472. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lévesque C., Duplessis M., Labonté J., Labrie S., Fremaux C., Tremblay D., Moineau S. Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 2005;71:4057–4068. doi: 10.1128/AEM.71.7.4057-4068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanley E., Fitzgerald G.F., Marrec C.L., Fayard B., van Sinderen D. Sequence analysis and characterization of ∅O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology. 1997 doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 74.Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lucchini S., Desiere F., Ssow H.B. Comparative Genomics of Streptococcus thermophilus Phage Species Supports a Modular Evolution Theory. J. Virol. 1999 doi: 10.1111/j.1751-7915.2010.00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guglielmotti D.M., Deveau H., Binetti A.G., Reinheimer J.A., Moineau S., Quiberoni A. Genome analysis of two virulent Streptococcus thermophilus phages isolated in Argentina. Int. J. Food Microbiol. 2009;136:101–109. doi: 10.1016/j.ijfoodmicro.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 77.Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:147–149. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chou W.C., Huang S.C., Chiu C.H., Chen Y.Y.M. YMC-2011, a temperate phage of Streptococcus salivarius 57.I. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.03186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun S., Gao S., Kondabagil K., Xiang Y., Rossmann M.G., Rao V.B. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc. Natl. Acad. Sci. USA. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bull J.J., Vegge C.S., Schmerer M., Chaudhry W.N., Levin B.R. Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0094690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Płachetka-Bożek A., Augustyniak M. Evaluation of Candidate Reference Genes for Quantitative Gene Expression Analysis in Spodoptera exigu a after Long-time Exposure to Cadmium. Sci. Rep. 2017;7:8338. doi: 10.1038/s41598-017-08630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young N.J., Thomas C.J., Collins M.E., Brownlie J. Real-time RT-PCR detection of Bovine Viral Diarrhoea virus in whole blood using an external RNA reference. J. Virol. Methods. 2006 doi: 10.1016/j.jviromet.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hill C., Miller L.A., Klaenhammer T.R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 1990;172:6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madsen S.M., Mills D., Djordjevic G., Israelsen H., Klaenhammer T.R. Analysis of the Genetic Switch and Replication Region of a P335-Type Bacteriophage with an Obligate Lytic Lifestyle on Lactococcus lactis. Appl. Environ. Microbiol. 2001;67:1128–1139. doi: 10.1128/AEM.67.3.1128-1139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vale P.F., Lafforgue G., Gatchitch F., Gardan R., Moineau S., Gandon S. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proceedings. Biol. Sci. 2015;282:20151270. doi: 10.1098/rspb.2015.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Breivik J., Gaudernack G. Resolving the evolutionary paradox of genetic instability: A cost-benefit analysis of DNA repair in changing environments. FEBS Lett. 2004;563:7–12. doi: 10.1016/S0014-5793(04)00282-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.