Abstract
We previously identified 92 toxin-like peptides and proteins, including pilosulin-like peptides 1–6 from the predatory ant Odontomachus monticola, by transcriptome analysis. Here, to further characterize venom components, we analyzed the venom and venom sac extract by ESI-MS/MS with or without trypsin digestion and reducing agent. As the low-molecular-mass components, we found amino acids (leucine/isoleucine, phenylalanine, and tryptophan) and biogenic amines (histamine and tyramine) in the venom and venom sac extract. As the higher molecular mass components, we found peptides and proteins such as pilosulin-like peptides, phospholipase A2s, hyaluronidase, venom dipeptidyl peptidases, conotoxin-like peptide, and icarapin-like peptide. In addition to pilosulin-like peptides 1–6, we found three novel pilosulin-like peptides that were overlooked by transcriptome analysis. Moreover, pilosulin-like peptides 1–6 were chemically synthesized, and some of them displayed antimicrobial, hemolytic, and histamine-releasing activities.
Keywords: ant, venom, mass spectrometry analysis, pilosulin-like peptide
1. Introduction
Ants (Hymenoptera: Formicidae) have been believed to share the same ancestor with bees and wasps, and have many traits in common with them. Since most ant species have a sting with venoms including formic acid, hydrocarbons, amines, peptides, and proteins, for predatory purpose [1], the venom components have been attractive as potential lead compounds for drug development. One of the peptide components in the ant venom is the pilosulin-like peptide. Since Donovan et al. isolated pilosulin 1 cDNA from a Myrmecia pilosula cDNA library in 1993 [2], many pilosulin and pilosulin-like peptides have been identified from various ant species [3,4,5]. Some of the pilosulins and pilosulin-like peptides formed homo- or heterodimers by single or double disulfide bridges, and displayed various bioactivities [2,6,7].
Odontomachus monticola, a predatory ant species in the subfamily Ponerinae, is about 10 mm long, red-brown in color, and has long mandibles [8]. They prey on other insects using their venomous stings, which cause intense pain and prolonged itching in humans. In 2016, Aili et al. reported 528 molecular masses, including 27 disulfide-bonded, without information on their primary structures or biological activities, in the venom of O. hastatus by LC-MS analysis [9]. On the other hand, we identified 92 toxin-like peptides and proteins including pilosulin-like peptides 1–6 from O. monticola by transcriptome and peptidome analysis [10]. Despite the transcriptome and limited peptidome analysis, the venom components of the ant genus Odontomachus have not been fully understood.
In this study, we further analyzed the venom and venom sac extract by ESI-MS/MS with or without trypsin digestion and reducing agent. Our results uncovered 1244 molecular masses, including toxin-like peptides and toxin-like proteins in the venom and venom sac extract of O. monticola by ESI-MS/MS without trypsin digestion, and also confirmed the amino acid sequences, amidation, processing, and dimerization with a single disulfide bridge of pilosulin-like peptides. Furthermore, we characterized the physicochemical and biological properties of pilosulin-like peptides. Some of the pilosulin-like peptides have a cationic amphiphilic region and displayed antimicrobial, hemolytic, and histamine-releasing activities.
2. Results and Discussion
2.1. Low-Molecular-Mass Components in O. monticola Venom and Venom Sac Extract
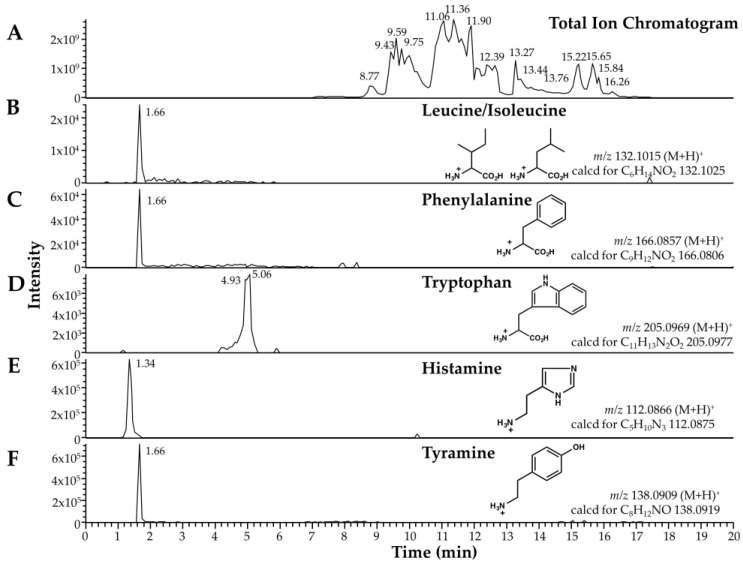
Among the low-mass components, we found two biogenic amines (histamine and tyramine) and three amino acids (leucine/isoleucine, phenylalanine, and tryptophan) in the venom and venom sac extract of O. monticola by retention time comparison and elemental analysis (Figure 1).
Figure 1.
Low-molecular-mass components of O. monticola venom and venom sac extract as analyzed by LC-ESI-MS. (A) Selected ion chromatograms of total ion chromatogram, (B) leucine/isoleucine, (C) phenylalanine, (D) tryptophan, (E) histamine, and (F) tyramine. The observed and calculated m/z values are shown with the corresponding structural formulae.
Histamine and tyramine interact with their specific receptors and activate specific neurons, being major neurotransmitters in insects, to regulate physiology and behavior. In addition, histamine is partly involved in pain-producing and itching reactions [11]. According to the early studies about ant venoms, histamine has been detected in several ant species [12]. In Myrmecia pyriformis, histamine accounted for approximately 2% of venom dry weight [13]. Histamine might be a major and common biogenic amine in ant venoms. Glutamic acid, a neurotransmitter and neurotoxin, was a major and common component in the venoms of five ant species, Myrmica ruginodis, Pogonomyrmex badins, Solenopsis saevissima, Tetramorium guineense, and T. caespitum L [12], but did not exist in the venom and venom sac extract of O. monticola. Most of the ant species might have biogenic amines and amino acids in various combinations. Although social and solitary wasps often have nucleosides (e.g., adenosine and guanosine) and nucleotides (e.g., AMP and ADP), this ant species lacks these compounds in its venom [14].
2.2. Overview of High-Molecular-Mass Components in Venom and Venom Sac Extract
We determined the amino acid sequences from ESI-MS/MS data and searched the amino acid sequences using PEAKS 8.5 against the major 192 components from O. monticola venom gland transcriptome data (Table S1). These components were selected from the transcripts which have high relative expression level in the transcriptome analysis or have some similarities with the transcripts of well-known toxins. MS/MS analysis under reducing and nonreducing conditions without trypsin digestion yielded 973 and 517 amino acid sequences (247 overwrapped sequences), respectively. Overall, 545 sequences from a total of 1244 sequences were derived from pilosulin-like peptides.
From the 1244 sequences, 193 were mapped to 50 high-molecular-mass components other than pilosulin-like peptides. The amino acid sequences from MS/MS data without trypsin digestion suggests that some of the proteins might be partially degraded before or after the extraction step. Due to the limited number of sequences in the reference database of the PEAKS 8.5 software, we considered that many of the de novo amino acid sequences were not assigned. Next, we searched the de novo amino acid sequences against 41,764 contig sequences of transcriptome analysis [10] with the tbalstn program to prevent oversight. In this way, 109 sequences were additionally mapped to 70 high-molecular-mass components other than pilosulin-like peptides (Table S2), and 397 sequences were unassigned. Including the MS/MS data with trypsin digestion with the data without trypsin digestion, the amino acid sequences were mapped to 104 components other than pilosulin-like peptides in the venom and venom sac extract by PEAKS 8.5 (Table 1). The total number of components identified from mass spectrometry analysis (183) is much smaller than that of contigs yielded from transcriptome analysis (41,764).
Table 1.
High-molecular-mass components in the venom and venom sac extract detected by LC-ESI-MS.
| Peptide/Protein | Accession Number | Coverage (%) a | |||
|---|---|---|---|---|---|
| TR b−, DTT b− |
TR−, DTT+ |
TR+, DTT− |
TR+, DTT+ |
||
| acetylcholine esterase | FX985608 | 1 | |||
| apolipophorin 1 | FX985561 | 2 | 4 | 8 | 7 |
| apolipophorin 2 | FX985562 | 10 | 61 | 29 | |
| calcium-independent phospholipase A2 gamma | FX985510 | 12 | 7 | 7 | |
| carboxypeptidase D | FX985540 | 5 | 4 | 4 | |
| carboxypeptidase Q | FX985538 | 67 | 26 | ||
| CDV3 homolog | FX986049 | 8 | |||
| chymotrypsin inhibitor 1 | FX986026 | 11 | |||
| cytochrome P450 1 | FX985568 | 10 | 5 | 4 | |
| cytochrome P450 3 | FX985570 | 11 | |||
| cytochrome P450 4 | FX985571 | 4 | |||
| cytochrome P450 5 | FX985572 | 9 | 6 | 9 | |
| cytochrome P450 6 | FX985573 | 5 | 2 | ||
| cytosolic carboxypeptidase-like protein 5 | FX985541 | 4 | 7 | ||
| dishevelled homolog 3 | FX986050 | 3 | 4 | ||
| hyaluronidase | FX985505 | 5 | 68 | 47 | |
| lysosomal ProX carboxypeptidase | FX985539 | 3 | 7 | ||
| matrix metalloproteinase 14 | FX985528 | 20 | 18 | ||
| NADPH cytochrome P450 reductase | FX985574 | 5 | 6 | 5 | |
| neuroblastoma suppressor of tumorigenicity 1 | FX986056 | 7 | |||
| neuropeptide-like 1 | FX986015 | 7 | 7 | ||
| peptidyl-prolyl cis-trans isomerase 1 | FX986028 | 22 | 17 | ||
| peptidyl-prolyl cis-trans isomerase 2 | FX986029 | 4 | |||
| peptidyl-prolyl cis-trans isomerase 3 | FX986030 | 3 | 8 | ||
| peptidyl-prolyl cis-trans isomerase 4 | FX986031 | 7 | 8 | ||
| peptidyl-prolyl cis-trans isomerase 5 | FX986032 | 6 | 7 | ||
| peptidyl-prolyl cis-trans isomerase 7 | FX986034 | 1 | |||
| peptidyl-prolyl cis-trans isomerase 8 | FX986035 | 38 | |||
| pheromone binding protein 1 | FX985594 | 14 | |||
| pheromone binding protein 2 | FX985595 | 11 | |||
| pheromone binding protein 3 | FX985596 | 20 | |||
| pheromone binding protein 4 | FX985597 | 16 | |||
| pheromone binding protein 6 | FX985599 | 4 | |||
| phospholipase A2 isozyme 2 | FX985507 | 11 | 36 | 40 | |
| protein disulfide isomerase 1 | FX985548 | 8 | 14 | 8 | |
| protein disulfide isomerase 2 | FX985549 | 19 | 8 | ||
| royal jelly protein | FX986021 | 9 | |||
| UDP glucuronosyltransferase 2C1 | FX986048 | 14 | 4 | ||
| uncharacterized protein 1 | FX985636 | 4 | 5 | 3 | 6 |
| uncharacterized protein 3 | FX986042 | 19 | 64 | ||
| uncharacterized protein 4 | FX986043 | 54 | 64 | ||
| UPF0518 protein | FX986054 | 4 | 3 | 3 | 2 |
| VEGF C-like protein | FX985521 | 6 | |||
| venom allergen 1 | FX985511 | 7 | |||
| venom allergen 2 | FX985512 | 5 | |||
| venom allergen 3 | FX985513 | 21 | 5 | ||
| venom dipeptidyl peptidase 1 | FX985542 | 4 | 12 | ||
| venom dipeptidyl peptidase 2 | FX985543 | 4 | 4 | ||
| venom dipeptidyl peptidase 3 | FX985544 | 10 | 3 | 5 | |
| venom serine carboxypeptidase | FX985537 | 3 | |||
| venom serine protease 2 | FX985523 | 5 | |||
| venom serine protease 3 | FX985524 | 6 | |||
| very high density protein 1 | FX985566 | 3 | 3 | 3 | 4 |
| very high density protein 2 | FX985567 | 3 | 5 | ||
| vitellogenin 1 | FX985563 | 3 | 5 | 5 | 2 |
| vitellogenin 2 | FX985564 | 10 | |||
| waprin 1 | FX985515 | 22 | |||
| waprin 2 | FX985516 | 7 | 11 | ||
| 40S ribosomal protein SA * | FX985616 | 8 | 8 | ||
| 60S acidic ribosomal protein P0 * | FX985621 | 22 | 6 | 3 | |
| 60S ribosomal protein L10 * | FX985634 | 11 | |||
| 60S ribosomal protein L3 * | FX985633 | 10 | 7 | ||
| 60S ribosomal protein L34 * | FX985627 | 16 | |||
| 60S ribosomal protein L36 * | FX985629 | 15 | |||
| 60S ribosomal protein L4 * | FX985618 | 5 | 8 | ||
| 60S ribosomal protein L6 * | FX985625 | 5 | 11 | ||
| 60S ribosomal protein L7 * | FX985622 | 12 | |||
| 60S ribosomal protein L7a * | FX985617 | 13 | |||
| 60S ribosomal protein L9 * | FX985635 | 10 | |||
| actin, muscle * | FX985587 | 13 | 7 | 7 | |
| ATPase WRNIP1 * | FX986047 | 13 | |||
| elongation factor 1-alpha * | FX985554 | 10 | 6 | ||
| elongation factor 1-beta * | FX985557 | 5 | |||
| elongation factor 1-delta * | FX985559 | 8 | 29 | ||
| elongation factor 1-gamma * | FX985555 | 20 | 8 | 2 | |
| elongation factor 2 * | FX985553 | 4 | 4 | 2 | 8 |
| elongation factor G, mitochondrial * | FX985558 | 2 | 11 | ||
| elongation factor Tu, mitochondrial * | FX985556 | 10 | 3 | ||
| histone H2A * | FX985611 | 21 | |||
| histone H3 * | FX985610 | 15 | |||
| laminin subunit alpha 1 * | FX985614 | 4 | 6 | 5 | 8 |
| laminin subunit beta 1 * | FX985613 | 3 | 3 | 9 | |
| laminin subunit gamma 1 * | FX985612 | 3 | 3 | ||
| myosin heavy chain, muscle * | FX985575 | 4 | 3 | 11 | 8 |
| myosin heavy chain, nonmuscle * | FX985578 | 2 | 1 | 4 | |
| myosin IB * | FX985586 | 11 | 13 | ||
| myosin Ie * | FX985583 | 5 | 7 | 5 | 9 |
| myosin regulatory light chain * | FX985576 | 5 | |||
| myosin Va * | FX985582 | 2 | 5 | 4 | 8 |
| myosin VIIa * | FX985580 | 5 | 6 | 5 | |
| myosin XV * | FX985581 | 5 | 5 | 3 | 6 |
| myosin XVIIIa * | FX985584 | 2 | 5 | 4 | 3 |
| resistance to inhibitors of cholinesterase 3 * | FX986036 | 5 | |||
| transcription factor A, mitochondrial * | FX985588 | 12 | |||
| transmembrane protein 214A * | FX986055 | 10 | 4 | 4 | |
| TRPA channel d, * | FX985591 | 5 | |||
| TRPM channel d, * | FX985592 | 10 | 2 | 5 | 5 |
| TRPV channel d, * | FX985593 | 3 | |||
| voltage-gated potassium channel Shaker * | FX985601 | 9 | |||
| voltage-gated sodium channel beta subunit TipE * | FX985590 | 9 | 4 | 2 | |
| voltage-gated sodium channel Para * | FX985589 | 2 | 10 | ||
a The coverages were calculated by combining the peptide fragments with the same amino acid sequences and the same molecular masses by Peaks 8.5; b TR and DTT indicate MS/MS data with or without trypsin digestion and DTT treatment, respectively; c VEGF: Vascular Endothelial Growth Factor; d TRPA, TRPM, and TRPV channels are members of the transient receptor potential (TRP) channel superfamily; * The proteins might be derived from the venom sac.
Collecting the venom by electrostimulation has been successful in some large ant species [5]. Since most ant species are too small to apply electrostimulation for collecting venom, a method of dissecting and extracting the venom sac has been applied to collect venoms in many ant species. Because we collected the venom from dissected venom sacs, membrane proteins, cytoskeleton proteins, DNA-binding proteins, translation-related proteins, and replication-related proteins are not believed to represent venom components and are most likely contaminants from venom sac tissue.
The rest of the components were roughly classified into three groups: toxin-like components, non-toxin-like components, and uncharacterized components. Among the toxin-like components we found pilosulin-like peptides, phospholipase A2s, hyaluronidase, and venom dipeptidyl peptidases of which amino acid sequences have some similarities to those of Apis mellifera orthologues in our previous study [10]. These proteins might function in tissue damage, venom diffusion, and venom maturation. Nontoxin-like components included cytochrome P450s and pheromone-binding proteins.
According to the classification of the venom components in Solenopsis invicta [15], these proteins might be classified into the groups of self-venom protection and chemical communication for alarm. Among uncharacterized components, we found uncharacterized proteins 1, 3, and 4. Uncharacterized protein 3 has nine cysteine residues, like some of the well-characterized toxins which are rich in cysteine residues, allowing them form unique conformations [16,17].
2.3. Novel Pilosulin-Like Peptides
MS/MS analysis under reducing and nonreducing conditions without trypsin digestion showed that 545 sequences of 1244 total sequences were derived from pilosulin-like peptides; pilosulin-like peptides 1, 2, 3, 4, 5, and 6 have 47, 124, 98, 82, 34, and 101 derivatives, respectively. They included the precursors and degradation products (Table 2; Figure 2B; Figures S1–S6). Because pilosulin-like peptides 2 and 3 have closely related amino acid sequences, they share five common derivatives (Table 2; Figures S2 and S3). Furthermore, we found three novel pilosulin-like peptides in the de novo amino acid sequences, which we termed pilosulin-like peptides 7, 8, and 9 (Figure 2A). To isolate pilosulin-like peptides 7 and 8 cDNAs, we revised raw reads of the transcriptome data in our previous study and found several reads corresponding to pilosulin-like peptides 7 and 8. cDNA clones of pilosulin-like peptides 7 and 8 were isolated by RT-PCR (Figures S10 and S11), indicating that pilosulin-like peptides 4 and 7 and pilosulin-like peptides 5 and 8 each have related nucleotide sequences. Accordingly, four nucleotide sequences (pilosulin-like peptides 4, 5, 7, and 8) were integrated into two sequences (pilosulin-like peptides 4 and 5) by the cd-hit-est software during the assembly process of transcriptome analysis [10]. Pilosulin-like peptides 7 and 8 have 83 and 27 derivatives in the 1244 sequences, respectively. Pilosulin-like peptides 4 and 7 and pilosulin-like peptides 5 and 8 share 41 and 5 common derivatives, respectively (Table 2; Figure 2B; Figures S7 and S8).
Table 2.
Amino acid sequences of pilosulin-like peptide derivatives analyzed from MS/MS spectra.
| Toxin | Sequence | Molecular Mass | Length | Precursor Ion | RT | Intensity |
|---|---|---|---|---|---|---|
| PLP1 | GILDWGKKVMDWIKDKMGK | 2247.1907 | 19 | 750.0702 | 37.15 | 3.67 × 108 |
| GILDWGKKVMDWIKDKMG | 2119.0957 | 18 | 707.3729 | 38.28 | 9.58 × 109 | |
| GILDWGKKVMDWIKDKM-NH2 | 2061.0903 | 17 | 1031.5520 | 39.15 | 1.34 × 1010 | |
| GILDWGKKVMDWIKDKM | 2062.0742 | 17 | 516.5270 | 38.35 | 5.74 × 108 | |
| LDWGKKVMDWIKDKMGK | 2077.0852 | 17 | 520.2803 | 34.11 | 2.65 × 108 | |
| GILDWGKKVMDWIKDK | 1931.0338 | 16 | 483.7657 | 33.21 | 1.64 × 108 | |
| LDWGKKVMDWIKDKM-NH2 | 1890.9849 | 15 | 631.3347 | 38.44 | 1.79 × 108 | |
| PLP2 | EAGWGSIFKTVGKMIAKAAVKAAPEAISAMASQNE | 3561.8323 | 35 | 891.4648 | 36.4 | 6.36 × 108 |
| GWGSIFKTVGKMIAKAAVKAAPEAISAMASQNE | 3361.7527 | 33 | 1121.5916 | 34.85 | 6.41 × 109 | |
| SIFKTVGKMIAKAAVKAAPEAISAMASQNE | 3061.6304 | 30 | 766.4222 | 27.81 | 1.12 × 109 | |
| GWGSIFKTVGKMIAKAAVKAAPEAISAM | 2832.5393 | 28 | 709.1410 | 34.52 | 4.53 × 108 | |
| GWGSIFKTVGKMIAKAAVKAAPEAISA | 2701.4988 | 27 | 676.3826 | 33.09 | 6.43 × 108 | |
| KMIAKAAVKAAPEAISAMASQNE | 2329.2134 | 23 | 777.4113 | 17.04 | 4.98 × 108 | |
| KAAVKAAPEAISAMASQNE | 1885.9567 | 19 | 943.9856 | 15.44 | 9.06 × 108 | |
| PLP3 | KIKWGKIFKKGGKLIGKTALEAAANAAASEAISAMASQNE | 4101.2407 | 40 | 1026.3179 | 25.25 | 4.47 × 109 |
| KIFKKGGKLIGKTALEAAANAAASEAISAMASQNE | 3488.8660 | 35 | 873.2313 | 25.16 | 1.83 × 109 | |
| KIKWGKIFKKGGKLIGKTALEAAANAAASEAISAM | 3572.0276 | 35 | 894.0147 | 24.79 | 2.65 × 108 | |
| KKGGKLIGKTALEAAANAAASEAISAMASQNE | 3100.6187 | 32 | 776.1608 | 25.63 | 2.51 × 108 | |
| KGGKLIGKTALEAAANAAASEAISAMASQNE | 2972.5237 | 31 | 744.1398 | 27.22 | 2.09 × 108 | |
| GGKLIGKTALEAAANAAASEAISAMASQNE | 2844.4287 | 30 | 949.1498 | 28.73 | 5.85 × 108 | |
| KTALEAAANAAASEAISAMASQNE | 2319.1011 | 24 | 1160.5585 | 28.03 | 1.63 × 109 | |
| PLP4 | GVKELFGKAWGLVKKHLPKAC*GLLGYVKQ | 3223.8418 | 29 | 806.9683 | 28.76 | 1.14 × 1010 |
| GVKELFGKAWGLVKKHLPKAC*GLL | 2648.5352 | 24 | 663.1436 | 30.27 | 2.98 × 109 | |
| FGKAWGLVKKHLPKAC*GLLGYVKQ | 2697.5305 | 24 | 675.3940 | 20.53 | 5.56 × 108 | |
| GKAWGLVKKHLPKAC*GLLGYVKQ | 2550.4619 | 23 | 638.6231 | 18.8 | 4.34 × 109 | |
| AWGLVKKHLPKAC*GLLGYVKQ | 2365.3457 | 21 | 789.4556 | 19.93 | 6.44 × 108 | |
| GKAWGLVKKHLPKAC*GLL | 1975.1553 | 18 | 659.3927 | 17.99 | 6.76 × 108 | |
| KHLPKAC*GLLGYVKQ | 1710.9603 | 15 | 428.7475 | 16 | 1.13 × 109 | |
| PLP5 | IWGALLGTLIPAITSAIQG | 1894.0928 | 19 | 948.0541 | 43.31 | 3.50 × 108 |
| IWGALLGTLIPAITSAIQ-NH2 | 1836.0873 | 18 | 919.0532 | 44.83 | 7.46 × 109 | |
| IWGALLGTLIPAITSAIQ | 1837.0713 | 18 | 919.5428 | 42.58 | 3.75 × 107 | |
| ALLGTLIPAITSAIQ-NH2 | 1479.9025 | 15 | 740.9585 | 36.38 | 2.25 × 108 | |
| LLGTLIPAITSAIQ-NH2 | 1408.8654 | 14 | 705.4390 | 34.1 | 1.45 × 108 | |
| LLGTLIPAITSA | 1168.7067 | 12 | 585.3602 | 30.26 | 5.15 × 106 | |
| IWGALLGTLIP | 1152.6907 | 11 | 577.3530 | 38.86 | 1.34 × 107 | |
| PLP6 | IKGKKIMKNMGKAMKIAGKVAKAMAPIVVPLIVSAA-NH2 | 3704.2307 | 36 | 927.0673 | 28.61 | 1.58 × 109 |
| KIMKNMGKAMKIAGKVAKAMAPIVVPLIVSAA-NH2 | 3277.9353 | 32 | 820.4910 | 30.81 | 8.46 × 107 | |
| IKGKKIMKNMGKAMKIAGKVAKAMAPIVVPL | 3263.9561 | 31 | 816.9973 | 23.56 | 6.27 × 108 | |
| KNMGKAMKIAGKVAKAMAPIVVPLIVSAA-NH2 | 2905.7158 | 29 | 727.4368 | 30 | 2.21 × 108 | |
| GKAMKIAGKVAKAMAPIVVPLIVSAA-NH2 | 2532.5376 | 26 | 845.1871 | 29.7 | 1.46 × 108 | |
| AMKIAGKVAKAMAPIVVPLIVSAA-NH2 | 2347.4211 | 24 | 587.8628 | 31.32 | 1.60 × 108 | |
| KAMAPIVVPLIVSAA-NH2 | 1477.9054 | 15 | 739.9600 | 31.65 | 1.18 × 108 | |
| PLP7 | GVKELFGKAWGLVKKHLPKAC*GLMGYVKQ | 3241.7983 | 29 | 811.4578 | 28.54 | 4.63 × 109 |
| GVKELFGKAWGLVKKHLPKAC*GLMGY | 2886.5764 | 26 | 722.6530 | 29.82 | 5.00 × 108 | |
| FGKAWGLVKKHLPKAC*GLMGYVKQ | 2715.4868 | 24 | 679.8821 | 19.52 | 5.42 × 108 | |
| GVKELFGKAWGLVKKHLPKAC*GLM | 2666.4917 | 24 | 667.6287 | 29.1 | 2.48 × 109 | |
| GKAWGLVKKHLPKAC*GLMGYVKQ | 2568.4185 | 23 | 643.1163 | 17.44 | 4.67 × 109 | |
| LVKKHLPKAC*GLMGYVKQ | 2069.1641 | 18 | 518.2991 | 13.87 | 2.37 × 107 | |
| HLPKAC*GLMGYVKQ | 1600.8218 | 14 | 534.6163 | 16.8 | 9.41 × 108 | |
| PLP8 | FWGALLAAAIPAITSAIQG | 1870.0352 | 19 | 936.0242 | 42.69 | 2.01 × 108 |
| FWGALLAAAIPAITSAIQ-NH2 | 1812.0298 | 18 | 907.0258 | 44.13 | 2.77 × 109 | |
| GALLAAAIPAITSAIQ-NH2 | 1478.8820 | 16 | 740.4485 | 44.09 | 2.20 × 107 | |
| FWGALLAAAIPAITS | 1500.8340 | 15 | 751.4250 | 37.82 | 1.60 × 107 | |
| ALLAAAIPAITSAIQ-NH2 | 1421.8606 | 15 | 711.9372 | 32.08 | 7.32 × 107 | |
| LAAAIPAITSAIQ-NH2 | 1237.7394 | 13 | 619.8778 | 25.83 | 8.77 × 107 | |
| AAAIPAITSAIQ-NH2 | 1124.6553 | 12 | 563.3350 | 23.08 | 2.19 × 109 |
C* = S-(carbamoylmethyl)-L-cysteine. The amino acid sequences of the highest and second-highest intensities are highlighted by red and yellow, respectively.
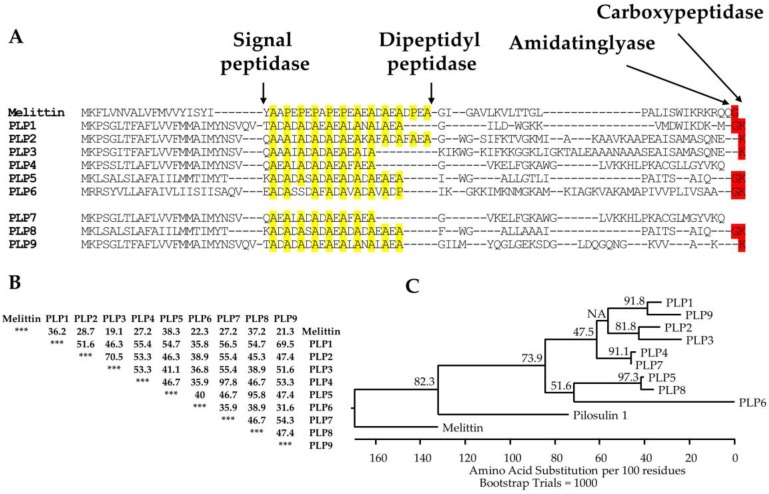
Figure 2.
Multiple alignment, identity matrix, and phylogenic analysis of pilosulin-like peptides. (A) The amino acid sequences of melittin and pilosulin-like peptides were aligned with ClustalW in Lasergene 12 (DNASTAR, Madison, WI, USA) and manually modified. Arrows indicate the putative processing and modification sites for signal peptidase, dipeptidyl peptidase, amidatinglyase, and carboxypeptidase. Proline and alanine residues in the spacer region between the signal and mature peptides of pilosulin-related peptides are highlighted in yellow. Nucleotide sequences for pilosulin-like peptides 7, 8, and 9 were assigned DDBJ/EMBL/GenBank Accession Numbers LC416796–LC416798, respectively. (B) Percentage amino acid sequence identities between melittin and pilosulin-like peptides are shown. (C) The alignment of pilosulin-like peptides, pilosulin 1, and melittin precursors by ClustalV in Lasergene 12 was used to construct a phylogenic tree using the neighbor-joining (NJ) method. The phylogenic tree rooted with the amino acid sequence of melittin. The numbers above the branches indicate the percentage of 1000 bootstrap replicates.
We found pilosulin-like peptide 9 in the annotation of de novo amino acid sequences. The unassigned de novo amino acid sequences of MS/MS analysis with trypsin digestion were compared against 41,764 contig sequences of transcriptome analysis [10] by the tblastn program. One of the amino acid sequences (Met-Tyr-Gln-Gly-Leu-Gly-Glu-Lys) (Figure S9) was matched to the contig Om11177_c0_g1_i2, which was annotated to aspartyl/glutamyl tRNA in our previous study [10]. Although tRNAs must be eliminated during the extraction step of total RNA and must not be transcribed by reverse transcriptase, the relative expression level of Om11177_c0_g1_i2 was high and accounted for 1.3% of all reads; so, we considered the contig Om11177_c0_g1_i2 as being a misassembled product of the Trinity software.
We selected the raw reads that encoded the amino acid sequence Met-Tyr-Gln-Gly-Leu-Gly-Glu-Lys and manually assembled the selected reads. The assembled nucleotide sequence encoded a similar leader sequence with pilosulin-like peptides 1–8, and we considered the manually assembled contig as encoding a novel member of the pilosulin-like peptide family. Using the contig sequence, we designed oligonucleotide primers and isolated a DNA fragment encoding the entire open reading frame (ORF) of pilosulin-like peptide 9 by RT-PCR (Figure S12).
Interestingly, the cDNA of pilosulin-like peptide 9 had a structure in which a nucleotide sequence (5′-ATGTACCAAG-3′) was inserted between the nucleotide positions 214 and 215 of the pilosulin-like peptide 1 cDNA. As the result of insertion and frame shifting, the downstream amino acid sequence of pilosulin-like peptide 9 is far different from that of pilosulin-like peptide 1. This may indicate the process of diversification of pilosulin-like peptides. Although we predicted an amino acid sequence of the mature peptide from the nucleotide sequence, only a partial amino acid sequence of pilosulin-like peptide 9 has been confirmed by MS/MS analysis.
2.4. Mature Forms of Pilosulin-Like Peptides
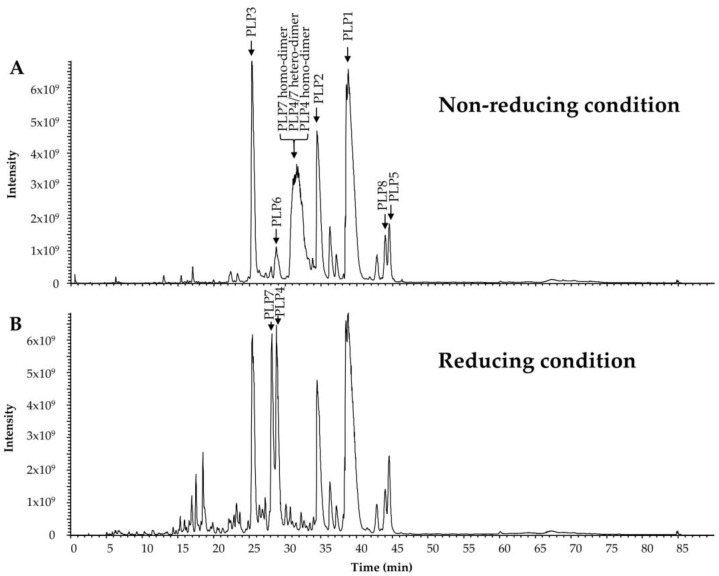
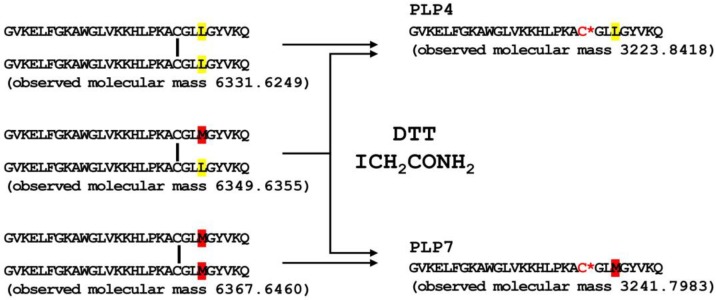
In our previous study, we predicted the N-termini of every mature pilosulin-like peptide, the elimination of Lys residues at the C-termini, and the amidation at the C-termini of some of the mature pilosulin-like peptides [10]. Signal peptidase, dipeptidyl peptidase, amidatinglyase, and carboxypeptidase, some of which are detected by ESI-MS/MS analysis (Table 1 and Table S2), might be involved in the processing and modification of pilosulin-like peptides (Figure 2A). The peptides of highest or second-highest signal intensity corresponded to the predicted mature peptides by nucleotide sequences. Since the signal intensity is affected by the abundance of the peptide in the venom and venom sac extract, the peptides of highest or second-highest signal intensity might be the major components in the venom. In addition to the mature peptides, we found the precursors of pilosulin-like peptides that have Gly or Gly–Lys residues at the C-termini and the degradation products of pilosulin-like peptides (Table 2). We compared the LC-ESI-MS profile of the crude venom and venom sac extract under reducing and nonreducing conditions. A broad peak (retention time around 31.58 min), which consisted of three masses (6331.6249, 6349.6355, and 6367.6460) under the nonreducing condition, was divided into two peaks (retention time 28.14 and 28.80 min), which consisted of two masses (3223.8418 and 3241.7983) under the reducing condition (Figure 3). These observed molecular masses were identical to the theoretical masses of mature pilosulin-like peptides 4 and 7 monomers, respectively. Consequently, we confirmed that pilosulin-like peptides 4 and 7 formed a homo- or heterodimer by a disulfide bridge (Figure 4).
Figure 3.
High-molecular-mass components of O. monticola venom and venom sac extract as analyzed by LC-ESI-MS. (A) The patterns of the total ion current of O. monticola venom and venom sac extract under nonreducing and (B) reducing conditions are shown. Peaks containing pilosulin-like peptides are labeled by arrows.
Figure 4.
Dimer formation of pilosulin-like peptides 4 and 7. Monomers of pilosulin-like peptides 4 and 7 were connected by a disulfide bridge at the amino acid position 21. Unique amino acid residues in pilosulin-like peptides 4 and 7 are highlighted in yellow and red, respectively. C* indicates S-(carbamoylmethyl)-L-cysteine.
2.5. Structural and Physicochemical Properties of Pilosulin-Like Peptides
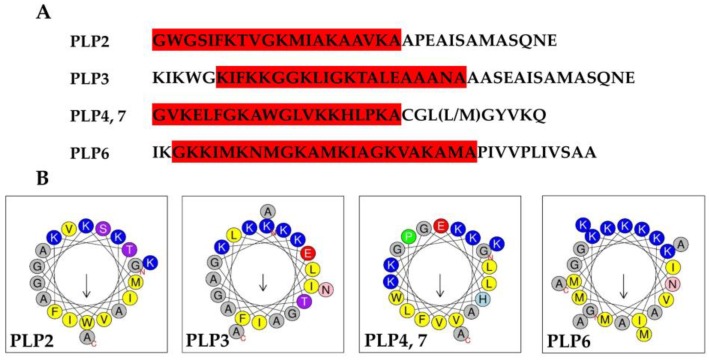
Pilosulins are α-helical cationic antimicrobial peptides [2,6,7]. We examined the secondary structure of pilosulin-like peptides using Proteus [18]. Proteus showed that mature regions of pilosulin-like peptides 1–8 are α-helical structures, but pilosulin-like peptide 9 is a coiled-coil structure. The pI of the mature pilosulin-like peptides, without consideration of amidation at C-termini, was calculated with IPC [19]. IPC demonstrated that the mature pilosulin-like peptides 5 and 8 are acidic peptides (calculated pI = 5.98) and the others are basic peptides. The calculated pIs of the mature pilosulin-like peptides 1, 2, 3, 4, 6, 7, and 9 are 8.46, 8.89, 9.06, 9.18, 9.96, 9.18, and 8.46, respectively. Taken together, pilosulin-like peptides 1–4, 6, and 7 may function as α-helical cationic antimicrobial peptides. To test whether pilosulin-like peptides have a cationic, amphiphilic helical conformation, the net charge and hydrophobic indexes of pilosulin-like peptides were examined using HeliQuest [20]. Parameters to search cationic amphipathic α-helix regions of pilosulin-like peptides were determined based on a well-known cationic amphipathic antimicrobial peptide, cecropin A (GenBank accession No. AAA29185): hydrophobicity 0–0.6, hydrophobic moment 0.1–1.0, and net charge 3–10. HeliQuest found cationic amphipathic α-helix sequences in pilosulin-like peptides 2–4, 6, and 7 (Figure 5A). Furthermore, helical wheel projections demonstrated that they are typical cationic α-helical amphiphilic peptides, in which hydrophobic amino acids are located on one side and basic amino acids are on the other side (Figure 5B). However, HeliQuest could not find an α-helical region that showed a typical cationic amphiphilic peptide in pilosulin-like peptide 1, since the mature form of pilosulin-like peptide 1 is shorter than the default setting (18 amino acids) of HeliQuest and the net charge is less than two.
Figure 5.
Amino acid sequences and helical wheel projection of pilosulin-like peptides 2, 3, 4, 6, and 7. (A) Amino acid sequences of mature pilosulin-like peptides. The cationic amphipathic helix regions in pilosulin-like peptides (PLP) 2, 3, 4, 6, and 7 predicted by HeliQuest are highlighted in red. (B) Helical wheel projections of pilosulin-like peptides 2, 3, 4, 6, and 7 drawn by HeliQuest. Nonpolar amino acids (F, I, L, M, V, and W), basic amino acids (K), acidic amino acids (D), small polar amino acids (A and G), aromatic polar amino acids (H), hydroxyl-containing polar amino acids (S and T), amide-containing polar amino acids (N), and proline (P) are highlighted by yellow, blue, red, gray, sky-blue, purple, pink, and green color, respectively.
2.6. Biological Activities of Pilosulin-Like Peptides
Pilosulin-like peptides 1–6 were chemically synthesized by the 9-fluorenylmethyloxycarbonyl (Fmoc) method, and we examined their biological activities (Table 3). We synthesized homodimeric pilosulin-like peptide 4, which was linked by a single disulfide bond. Pilosulin-like peptides 1–4 and 6 with a cationic α-helix displayed antimicrobial activities against Escherichia coli and Staphylococcus aureus. These activities were higher than those of magainin, a well-known Xenopus laevis antimicrobial peptide [21]. Interestingly, pilosulin-like peptide 4 also had high antimicrobial activities against Saccharomyces cerevisiae. In support of these experimental results, the antifungal peptide prediction server Antifp predicted that pilosulin-like peptide 4, but not other pilosulin-like peptides, was an antifungal peptide [22]. Virtual alanine scanning of pilosulin-like peptide 4 by Antifp revealed that the replacement of a cysteine residue remarkably reduced the index of antifungal activity. Cysteine residues are known to be abundant in antifungal peptides in general [22], and the replacement of cysteine with serine in brevinin-1B Ya, a frog antimicrobial peptide, reduced the antifungal activity [23]. The cysteine residue of pilosulin-like peptide 4 may be important for antifungal activity.
Table 3.
Biological properties of O. monticola pilosulin-like peptides.
| Peptide | MIC a (μM) | Hemolytic Activity (%) | Histamine-Releasing Activity at 10 μM (%) |
|||
|---|---|---|---|---|---|---|
|
E. coli (NBRC 14237) |
S. aureus (NBRC 12732) |
S. cerevisiae (NBRC 10217) |
at 10 μM | at 50 μM | ||
| PLP1 | <3.1 | <3.1 | <50 | Negative | 32.9 | |
| PLP2 | <6.2 | <6.2 | <50 | Negative | 10.4 | 30.1 |
| PLP3 | <3.1 | <25 | <50 | Negative | 37.5 | |
| PLP4 | <3.1 | <3.1 | <3.1 | Negative | 10.5 | 66.4 |
| PLP5 | <50 | Negative | Negative | 6.9 | 94.8 | 28.3 |
| PLP6 | <3.1 | <3.1 | Negative | Negative | 33.6 | |
| Magainin | <12.5 | <25 | Negative | - | - | |
| Mastoparan | - | - | - | 13.5 b | - | 31.1 b |
| Melittin | - | - | - | 100.0 b | - | 64.3 b |
a MIC: minimum inhibitory concentration; b The activities indicate from Shigeri et al. [24]; -: not determined.
Pilosulin-like peptide 5 with an acidic α-helix had no or low antimicrobial activities against all the microbes tested in this study, but had the highest hemolytic activity among pilosulin-like peptides 1–6. This hemolytic activity may depend on higher hydrophobicity. Previous studies suggested a correlation between peptide hydrophobicity and hemolytic activity [25]. For example, melittin, a honey bee hemolytic peptide, shows a higher hydrophobicity (0.45–0.88). Hydrophobicity of pilosulin-like peptide 5 is 0.914, which is the highest among pilosulin-like peptides.
Pilosulin-like peptides are major components of O. monticola venom sac extract. The first step in the antimicrobial mechanism has been suggested to be these peptides binding to the anionic phospholipids that are abundant in bacterial membranes, which is then followed by pore formation by α-helical cationic peptides as the second step [26]. The variations of antimicrobial activities might reflect on the difference of the physicochemical properties among pilosulin-like peptides.
Pilosulin-like peptide 4 had histamine-releasing activity against rat mast cells comparable to that of melittin, which displays histamine-releasing activity.
3. Conclusions
In this study, we have analyzed the venom and venom sac extract of O. monticola by mass spectrometry analysis. Because of the high sensitivity of the analysis, the extract accounting for just one fifth of a venom sac was enough for a single MS/MS analysis. We determined 1244 amino acid sequences by ESI-MS/MS analysis without trypsin digestion. In total, 545 of them were derived from pilosulin-like peptides 1–8, and 302 amino acid sequences corresponded to the high molecular mass components that were identified from our previous transcriptome analysis. This MS/MS analysis suggests that the majority of the venom components might be 2–6.5-kDa peptides.
Synthetic pilosulin-like peptides 1–4 and 6 displayed antimicrobial and histamine-releasing activities. Moreover, pilosulin-like peptide 5 showed the highest hemolytic activity among pilosulin-like peptides. Some of the ant toxins show various biological activities, such as neurotoxic [27,28] and enzymatic activity [29]. In future studies, we will further explore the multiple activities of pilosulin-like peptides.
4. Materials and Methods
4.1. Ants
One O. monticola colony was collected in Musashimurayama, Tokyo, Japan, on 1 July 2016. The species was morphologically identified.
4.2. Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis for Low-Molecular-Weight Components
Twenty O. monticola venom sacs were collected and extracted with 50% acetonitrile containing 0.1% (v/v) trifluoroacetic acid (50 μL) for 2 h at 4 °C. A single venom sac contains 50–150 nL of venom. The extract was passed through a 0.45-μm filter and successively diluted with the extraction solvent to a final concentration of 0.04 sacs/μL. This dilution was used for the LC-MS analysis. The LC conditions were: solvent A, 0.1% (v/v) aqueous formic acid; solvent B, 0.1% (v/v) formic acid in acetonitrile; 5–65% linear gradient of solvent B in solvent A at a flow rate of 200 μL/min; column, Capcell Pak C18 UG 120 (1.5 × 150 mm, Shiseido, Tokyo, Japan); column temperature, 25 °C. The molecular weights of the ant peptides were verified by LTQ Orbitrap XL-ETD (Thermo Fisher Scientific, Waltham, MA, USA). The MS conditions were: ionization, electrospray in positive mode; ion spray voltage, 4.6 kV; capillary temperature, 350 °C; capillary and tube lens voltages, 19 V and 35 V, respectively; detector, an Orbitrap at a resolution of 60,000 at m/z 400. MS scan range was m/z 100–2000. The mass spectrometer was calibrated with polytyrosine, and the resolution was usually 1–3 ppm after measurement.
4.3. Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis for Peptide and Protein Components
After filtration of the venom sac extract, the extract was diluted 10 times with 50 mM ammonium bicarbonate, pH 8.0, to improve separation and prevent peak tailing in HPLC. The diluted extract was reduced with DTT at a concentration of 10 mM (Thermo Fisher Scientific), alkylated with iodoacetamide at a concentration of 20 mM (Thermo Fisher Scientific), or digested by trypsin at a concentration of ca. 30 μg/mL (Promega, Madison, WI, USA) overnight at 37 °C. Separation was achieved by Zaplous α pep C18 analytical column (3 μm 120A, 1.5 × 150 mm, AMR, Tokyo, Japan) with L-Trap column (5 μm, 0.3 × 5 mm, AMR, Tokyo, Japan) at a flow rate of 500 μL/min using two mobile phases, 0.1% (v/v) aqueous formic acid (solvent A) and 0.1% (v/v) formic acid in acetonitrile (solvent B). The following gradient was used: 0–60 min, 5–65% solvent B; 60–70 min, 65–95% solvent B; 70–80 min, 95% solvent B.
The molecular weights of the ant peptides were verified by Q Exactive (Thermo Fisher Scientific, Waltham, MA, USA). The MS conditions were: ionization, nanoelectrospray (CaptiveSpray Ionization: CSI) in positive mode; ion spray voltage, 1.4 kV; capillary temperature, 250 °C; S-lens level, 50; detector, an Orbitrap at a resolution of 70,000 from m/z 350–2000. The mass spectrometer was calibrated with a calibrant of LTQ Velos ESI Positive Ion Calibration Solution (Thermo Fisher Scientific), and the mass accuracy was usually 1–3 ppm after measurement. The raw mass spectrum was processed by using Xcalibur (Thermo Fisher Scientific). Peptide sequences were determined from MS/MS spectra by PEAKS 8.5 (Bioinformatics Solutions, Waterloo, Canada; parent mass error tolerance 10.0 ppm, fragment mass error tolerance 0.02 Da, score threshold 15.0) and manually checked. We selected 192 amino acid sequences derived from major transcripts in the venom gland transcriptome analysis and the 116 amino acid sequences of common external contaminants from cRAP (Global Proteome Machine Organization). After construction of a FASTA format file including 308 amino acid sequences, the file was incorporated into PEAKS 8.5 software as the reference database.
To assign the de novo amino acid sequences, tblastn (parameters: E-value 0.1, matrix PAM40, word size 3) was performed using 41,764 contig sequences of transcriptome analysis.
4.4. Pilosulin-Like Peptides 7–9 cDNA Cloning and Sequencing
Total RNA isolated from the ant venom glands with sacs was reverse-transcribed to cDNA and amplified by PCR with KOD-Plus-Neo (Toyobo, Osaka, Japan). The oligonucleotide primers used for pilosulin-like peptide 7 were Pilo U1 (5′-ATGAAACCGTCGGGTATCAC-3′), corresponding to nucleotides (nt) 9–28 of pilosulin-like peptide 2; 41CPas (5′-TTGCTTTACGTATCCCAT-3′); 41CP (5′-CCAAAGCGTGTGGACTGA-3′); and Oligo-dT (5′-GAGTCGACTCGAGAA(T)17-3′). 41CP and 41CPas primers were designed from transcriptome analysis data and the amino acid sequences: Met-Gly-Tyr-Val-Lys-Gln; Lys-Ala-Cys-Gly-Leu-Met. The oligonucleotide primers used for pilosulin-like peptide 8 were 51S (5′-TATGTGTGAAAGCTCTTC-3′) and 51AS (5′-CCAATGTAATGCCAATCG-3′), designed based on the 5′ and 3′ untranslated regions predicted by transcriptome analysis. The oligonucleotide primers used for pilosulin-like peptide 9 were Pilo U1, 9CPas (5′-CCCCAGTCCTTGGTACAT-3′), 9CP (5′-ATGTACCAAGGACTGGGG-3′), and Oligo-dT. 9CP and 9CPas primers were designed from the transcriptome analysis data and the amino acid sequence: Met-Tyr-Gln-Gly-Leu-Gly-Glu-Lys. The amplified products of the cDNAs were cloned into the EcoRI site of pBluescript II SK(-) (Agilent Technologies, La Jolla, CA, USA). All inserts were sequenced using a Model 3500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
4.5. Synthesis and Biological Activities of Pilosulin-Like Peptides
Pilosulin-like peptides 1–6 were prepared using Fmoc chemistry by GenScript (Nanjing, Jiangsu, China). The peptides were purified by RP-HPLC with a preparative C18 column. The purity and molecular weight of the final peptides were verified by HPLC and MS. Antimicrobial, hemolytic, and histamine-releasing activities were measured as described previously [6].
4.6. WEB Server Used to Analyze the Physiochemical Properties
Secondary structure prediction was performed by Proteus (http://www.proteus2.ca/proteus/). The pI of peptides was calculated with IPC (http://isoelectric.org/index.html). The physicochemical properties were examined using HeliQuest (http://heliquest.ipmc.cnrs.fr/). The antifungal peptide prediction was carried out using the Antifp server (http://webs.iiitd.edu.in/raghava/antifp/index.html).
Acknowledgments
We thank Toshiyuki Sato (Tokyo University of Agriculture and Technology), Mamoru Terayama (University of Tokyo), Katsuyuki Eguchi (Tokyo Metropolitan University), Rijal Satria (Tokyo Metropolitan University) for their assistance in the field survey, and Masaki Okano for the arrangement of ESI-MS/MS usage. We are also grateful to Leslie Sargent Jones (Appalachian State University, Retired) for her careful reading of our manuscript.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/11/1/50/s1, Figures S1–S9 (MS spectra of pilosulin-like peptides 1–9), Figures S9–S12 (Nucleotide and deduced amino acid sequences of pilosulin-like peptides 7–9), Table S1 (Major 192 components of O. monticola venom), and Table S2 (Protein assignment of the de novo amino acid sequences).
Author Contributions
N.T., K.K. (Kazuma), and K.K. (Konno) performed the ESI-MS; K.M. corrected and classified the ant species; H.I. examined the biological activities; Y.O. checked physicochemical properties; Y.S. and K.K. (Konno) prepared pilosulin-like peptides; Y.O., Y.S., and H.I. wrote the paper.
Funding
This work was supported by JSPS KAKENHI (Grant Nos. 15K07805 and 15K01814) and the program of the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
Overview the venom compositions and bioactivities of the predatory ant Odontomachus monticola.
References
- 1.Aili S.R., Touchard A., Escoubas P., Padula M.P., Orivel J., Dejean A., Nicholson G.M. Diversity of peptide toxins from stinging ant venoms. Toxicon. 2014;92:166–178. doi: 10.1016/j.toxicon.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Donovan G.R., Baldo B.A., Sutherland S. Molecular cloning and characterization of a major allergen (Myr p I) from the venom of the Australian jumper ant, Myrmecia pilosula. Biochim. Biophys. Acta. 1993;1171:272–280. doi: 10.1016/0167-4781(93)90065-L. [DOI] [PubMed] [Google Scholar]
- 3.Torres A.F., Huang C., Chong C.M., Leung S.W., Prieto-da-Silva A.R., Havt A., Quinet Y.P., Martins A.M., Lee S.M., Radis-Baptista G. Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: Insights into the polypeptide toxin arsenal of hymenopterans. PLoS ONE. 2014;9:e87556. doi: 10.1371/journal.pone.0087556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touchard A., Tene N., Song P.C.T., Lefranc B., Leprince J., Treilhou M., Bonnafe E. Deciphering the molecular diversity of an ant venom peptidome through a venomics approach. J. Proteome Res. 2018;17:3503–3516. doi: 10.1021/acs.jproteome.8b00452. [DOI] [PubMed] [Google Scholar]
- 5.Robinson S.D., Mueller A., Clayton D., Starobova H., Hamilton B.R., Payne R.J., Vetter I., King G.F., Undheim E.A.B. A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci. Adv. 2018;4:eaau4640. doi: 10.1126/sciadv.aau4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inagaki H., Akagi M., Imai H.T., Taylor R.W., Kubo T. Molecular cloning and biological characterization of novel antimicrobial peptides, pilosulin 3 and pilosulin 4, from a species of the Australian ant genus Myrmecia. Arch. Biochem. Biophys. 2004;428:170–178. doi: 10.1016/j.abb.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Inagaki H., Akagi M., Imai H.T., Taylor R.W., Wiese M.D., Davies N.W., Kubo T. Pilosulin 5, a novel histamine-releasing peptide of the Australian ant, Myrmecia pilosula (Jack Jumper Ant) Arch. Biochem. Biophys. 2008;477:411–416. doi: 10.1016/j.abb.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Kubota M., Imai H.T., Kondo M., Onoyama K., Ogata K., Terayama M., Yoshimura M. Japanese Ant Image Database. [(accessed on 21 November 2018)]; Available online: http://ant.miyakyo-u.ac.jp/
- 9.Aili S.R., Touchard A., Koh J.M., Dejean A., Orivel J., Padula M.P., Escoubas P., Nicholson G.M. Comparisons of protein and peptide complexity in poneroid and formicoid ant venoms. J. Proteome Res. 2016;15:3039–3054. doi: 10.1021/acs.jproteome.6b00182. [DOI] [PubMed] [Google Scholar]
- 10.Kazuma K., Masuko K., Konno K., Inagaki H. Combined venom gland transcriptomic and venom peptidomic analysis of the predatory ant Odontomachus monticola. Toxins. 2017;9:323. doi: 10.3390/toxins9100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson S., Giesler G.J. The multiple pathways for itch and their interactions with pain. Trends Neurosci. 2010;33:550–558. doi: 10.1016/j.tins.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vonsicard N.A.E., Candy D.J., Anderson M. The biochemical-composition of venom from the pavement ant (Tetramorium-Caespitum L.) Toxicon. 1989;27:1127–1133. doi: 10.1016/0041-0101(89)90006-8. [DOI] [PubMed] [Google Scholar]
- 13.De la Lande I.S., Lewis J.C. Constituents of the venom of the Australlian bull ant, Myrmecia pyriformis. Memorias do Instituto Butantan. 1966;33:951–955. [PubMed] [Google Scholar]
- 14.Hisada M., Satake H., Masuda K., Aoyama M., Murata K., Shinada T., Iwashita T., Ohfune Y., Nakajima T. Molecular components and toxicity of the venom of the solitary wasp, Anoplius samariensis. Biochem. Biophys. Res. Commun. 2005;330:1048–1054. doi: 10.1016/j.bbrc.2005.03.087. [DOI] [PubMed] [Google Scholar]
- 15.dos Santos Pinto J.R., Fox E.G., Saidemberg D.M., Santos L.D., da Silva Menegasso A.R., Costa-Manso E., Machado E.A., Bueno O.C., Palma M.S. Proteomic view of the venom from the fire ant Solenopsis invicta Buren. J. Proteome Res. 2012;11:4643–4653. doi: 10.1021/pr300451g. [DOI] [PubMed] [Google Scholar]
- 16.Escoubas P., Diochot S., Corzo G. Structure and pharmacology of spider venom neurotoxins. Biochimie. 2000;82:893–907. doi: 10.1016/S0300-9084(00)01166-4. [DOI] [PubMed] [Google Scholar]
- 17.Goudet C., Chi C.W., Tytgat J. An overview of toxins and genes from the venom of the Asian scorpion Buthus martensi Karsch. Toxicon. 2002;40:1239–1258. doi: 10.1016/S0041-0101(02)00142-3. [DOI] [PubMed] [Google Scholar]
- 18.Montgomerie S., Cruz J.A., Shrivastava S., Arndt D., Berjanskii M., Wishart D.S. PROTEUS2: A web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res. 2008;36:W202–W209. doi: 10.1093/nar/gkn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlowski L.P. IPC—Isoelectric point calculator. Biol. Direct. 2016;11:55. doi: 10.1186/s13062-016-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier R., Douguet D., Antonny B., Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 21.Zasloff M., Martin B., Chen H.C. Antimicrobial activity of synthetic magainin peptides and several analogues. Proc. Natl. Acad. Sci. USA. 1988;85:910–913. doi: 10.1073/pnas.85.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal P., Bhalla S., Chaudhary K., Kumar R., Sharma M., Raghava G.P.S. In silico approach for prediction of antifungal peptides. Front. Microbiol. 2018;9:323. doi: 10.3389/fmicb.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal T., Abraham B., Sonnevend A., Jumaa P., Conlon J.M. Brevinin-1BYa: A naturally occurring peptide from frog skin with broad-spectrum antibacterial and antifungal properties. Int. J. Antimicrob. Agents. 2006;27:525–529. doi: 10.1016/j.ijantimicag.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Shigeri Y., Horie M., Yoshida T., Hagihara Y., Imura T., Inagaki H., Haramoto Y., Ito Y., Asashima M. Physicochemical and biological characterizations of Pxt peptides from amphibian (Xenopus tropicalis) skin. J. Biochem. 2016;159:619–629. doi: 10.1093/jb/mvw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Guarnieri M.T., Vasil A.I., Vasil M.L., Mant C.T., Hodges R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007;51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers J.P., Hancock R.E. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 27.Touchard A., Brust A., Cardoso F.C., Chin Y.K., Herzig V., Jin A.H., Dejean A., Alewood P.F., King G.F., Orivel J., et al. Isolation and characterization of a structurally unique beta-hairpin venom peptide from the predatory ant Anochetus emarginatus. Biochim. Biophys. Acta. 2016;1860:2553–2562. doi: 10.1016/j.bbagen.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 28.Johnson S.R., Rikli H.G., Schmidt J.O., Evans M.S. A reexamination of poneratoxin from the venom of the bullet ant Paraponera clavata. Peptides. 2017;98:51–62. doi: 10.1016/j.peptides.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt J.O., Blum M.S., Overal W.L. Comparative enzymology of venoms from stinging Hymenoptera. Toxicon. 1986;24:907–921. doi: 10.1016/0041-0101(86)90091-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.