Abstract
We hypothesized that identifying plasma glycoproteins that predict the development of heart failure following myocardial infarction (MI) could help to stratify subjects at risk. Plasma collected at visit 2 (2005–2008) from an MI subset of Jackson Heart Study participants underwent glycoproteomics and was grouped by the outcome: (1) heart failure hospitalization after visit 2 (n = 15) and (2) without hospitalization by 2012 (n = 45). Proteins were mapped for biological processes and functional pathways using Ingenuity Pathway Analysis and linked to clinical characteristics. A total of 198 glycopeptides corresponding to 88 proteins were identified (data available via ProteomeXchange with identifier PXD009870). Of these, 14 glycopeptides were significantly different between MI and MI + HF groups and corresponded to apolipoprotein (Apo) F, transthyretin, Apo C-IV, prostaglandin-D2 synthase, complement C9, and CD59 (p < 0.05 for all). All proteins were elevated in the MI + HF group, except CD59, which was lower. Four canonical pathways were upregulated in the MI + HF group (p < 0.05 for all): acute phase response, liver X receptor/retinoid X receptor, and macrophage reactive oxygen species generation. The coagulation pathway was significantly downregulated in the MI + HF group (p < 0.05). Even after adjustment for age and sex, Apo F was associated with the increased risk for heart failure (OR = 21.84; 95% CI 3.20–149.14). In conclusion, glycoproteomic profiling provided candidate early MI predictors of later progression to heart failure.
Introduction
Cardiovascular disease (CVD) is the leading cause of death in minorities in the United States.1−4 Among non-Hispanic African Americans aged 20 years and older, ∼45% have been diagnosed with CVD.1 Development of heart failure (HF) following myocardial infarction (MI) in African Americans carries higher morbidity.4 The Jackson Heart Study (JHS) is the largest single-site, prospective, epidemiologic CVD investigation in an African American population. JHS is the largest study in history to investigate inherited factors that affect high blood pressure, heart disease, strokes, and diabetes in the African American population.
The goal of the present study was to determine HF prediction markers, using plasma samples collected after MI and before the development of symptoms. A total of 60 JHS participants were included; all had been diagnosed with MI before visit 2 and none had progressed to HF by visit 2. We hypothesized that the evaluation of post-MI plasma samples, before HF was diagnosed, would reveal glycoprotein identifiers predictive of future HF. Because transporting drugs across the plasma membrane is technically challenging, enriching for extracellular proteins identifies targets most amenable to therapy.5
Results and Discussion
Glycoproteomic Analysis Identified 198 Peptides Belonging to 88 Unique Glycoproteins (Table S2)
Participant characteristics are shown in Table 1. Of the 60 participants, 15 (25%) experienced subsequent HF hospitalization after visit 2 (n = 15), which is consistent with past reports.6−9 The development of HF incidence rate was 19 per 100 person-years. A history of diabetes, hemoglobin A1C, and sex were different between groups (all p < 0.05).
Table 1. Clinical Characteristics in Groups Divided by Post-MI Congestive HF Statusa,b.
| MI (n = 45) | MI + HF (n = 15) | p value | |
|---|---|---|---|
| age (years) | 69 ± 9 | 70 ± 10 | 0.617 |
| sex, men/women (% women) | 24/21 (47%) | 3/12 (80%) | 0.036 |
| beta blocker (%) | 23 (51%) | 10 (64%) | 0.060 |
| body mass index (kg/m2) | 28.9 ± 4.3 | 34.8 ± 9.5 | 0.060 |
| calcium channel blocker (%) | 12 (27%) | 2 (13%) | 0.397 |
| diabetes history | 17 (38%) | 12 (80%) | 0.022 |
| diuretic (%) | 23 (51%) | 9 (60%) | 0.199 |
| diastolic blood pressure (mmHg) | 74 ± 1 | 71 ± 3 | 0.545 |
| fasting glucose (mg/dL) | 108 ± 7 | 112 ± 14 | 0.808 |
| fasting HDL (mg/dL) | 52 ± 3 | 64 ± 12 | 0.106 |
| fasting LDL (mg/dL) | 102 ± 7 | 103 ± 15 | 0.977 |
| fasting triglycerides (mg/dL) | 100 ± 9 | 189 ± 80 | 0.300 |
| hemoglobin A1C (%) | 6.1 ± 0.1 | 7.7 ± 0.5 | 0.008 |
| prevalent chronic kidney disease (%) | 0 (0%) | 1 (7%) | 0.083 |
| smokers (%) | 7 (16%) | 3 (20%) | 0.721 |
| statin use (%) | 15 (33%) | 2 (13%) | 0.683 |
| systolic blood pressure (mmHg) | 134 ± 3 | 138 ± 7 | 0.500 |
| time between MI and visit 2 (yrs) | 6 ± 5 | 5 ± 5 | 0.760 |
| QTc interval (ms; from visit 3) | 437 ± 32 | 437 ± 60 | 0.968 |
Values are taken from visit 2, except for QTc interval which was taken from visit 3.
Mean ± SD.
Although glycosylated albumin has been linked to diabetes prevalence, albumin does not contain the N–X–S/T motif for typical N-linked glycosylation.10 Using an N-linked glycoproteomic approach, therefore, removes the need to deplete albumin and enhances extracellular protein representation.11,12 A total of 198 glycopeptides corresponding to 88 unique proteins were identified (data available via ProteomeXchange with identifier PXD009870). Of the glycopeptides identified, 14 were significantly different between groups and corresponded to six unique glycoproteins (all p < 0.05). The six proteins significantly different by ANOVA were apolipoprotein F (Apo F), transthyretin, apolipoprotein C-IV (Apo C4), prostaglandin-H2 D-isomerase, complement C9, and CD59 (Tables 2 and S1, Figure S1).
Table 2. Proteins Differentially Expressed in MI + HF Subjects (all p < 0.05)a.
| accession # | protein name | MI + HF/MI ratio | p value |
|---|---|---|---|
| 4502165 | Apo F | 2.503 | 0.001 |
| 4507725 | transthyretin | 2.382 | 0.014 |
| 4502161 | Apo C4 | 1.667 | 0.014 |
| 32171249 | prostaglandin-H2 D-isomerase | 3.070 | 0.017 |
| 4502511 | complement C9 | 1.416 | 0.031 |
| 42761474 | CD59 glycoprotein | 0.521 | 0.043 |
MI + HF/MI ratio was obtained by dividing mean MI + HF intensity value by mean MI intensity value.
In addition, there were three proteins that showed differential expression at the individual peptide level (complement C3, heparin cofactor 2, and polymeric immunoglobin receptor); when intensity values were added together for all glycopeptides identified in that protein, significance was lost (Table 3). One possible reason for individual peptides to show a difference that is not retained in the combined protein evaluation is that proteolytic processing of these proteins may be occurring such that differences in protein regions are observed. Proteolytic processing of extracellular matrix (ECM) proteins such as collagen and CD36 have been linked to adverse remodeling of the left ventricle following MI.13,14
Table 3. Peptides from Three Proteins were Differentially Expressed at the Individual Peptide but Not Whole Protein Level.
| accession # | protein | peptide sequence | MI + HF/MI ratio | p value |
|---|---|---|---|---|
| 115298678 | complement C3 | DAPDHQELNLDVSLQLPSR | 1.42 | 0.019 |
| VHQYFNVELIQPGAVK | 1.83 | 0.026 | ||
| VELLHNPAFCSLATTK | 1.68 | 0.045 | ||
| all peptides combined | 0.99 | 0.949 | ||
| 73858566 | heparin cofactor 2 | GGETAQSADPQWEQLNNKNLSMPLLPADFHK | 1.75 | 0.002 |
| GETHEQVHSILHFKDFVNASSK | 1.16 | 0.359 | ||
| DFVNASSK | 1.05 | 0.757 | ||
| NLSMPLLPADFHK | 1.00 | 0.974 | ||
| all peptides combined | 1.17 | 0.161 | ||
| 31377806 | polymeric immunoglobulin receptor | VPGNVTAVLGETLK | 1.79 | 0.006 |
| ANLTNFPENGTFVVNIAQLSQDDSGR | 1.49 | 0.254 | ||
| all peptides combined | 1.43 | 0.057 |
Immunoblotting for four of the six proteins and peptides matched direction and quantity with the glycoproteomic data (Tables S1 and S2). Polymeric immunoglobulin receptor, complement C9, Apo F, and CD59 showed good correlation between methods, whereas Apo C4 and transthyretin did not.
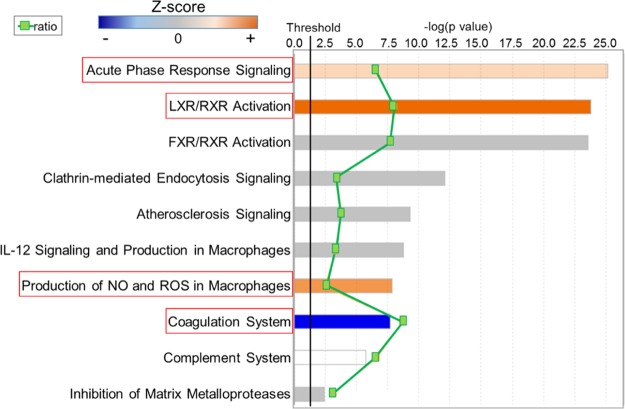
Canonical pathway analysis of the glycoproteomic data indicated that the 88 proteins identified belonged to 10 pathways (Figure 1). Of the 10 pathways, three were upregulated in participants who developed HF: acute phase response, liver X receptors/retinoid X receptor (LXR/RXR), and the activation of nitric oxide (NO) and reactive oxygen species (ROS) in macrophage pathways. The coagulation system was downregulated in participants who developed HF. The remaining six pathways showed no direction with HF, suggesting that these pathways were in response to MI or were due to a residual confounder and were not associated with HF. These six pathways included FXR/RXR, clathrin-mediated endocytosis, atherosclerosis signaling, interleukin (IL)-12 signaling in macrophages, complement, and inhibition of matrix metalloproteinase.
Figure 1.
Canonical pathway analysis identified pathways associated with HF. Canonical pathway analysis indicated acute phase response, LXR/RXR, and production of NO, and ROS pathways were upregulated, whereas the coagulation pathway was downregulated, with HF after MI. The top canonical pathways identified are listed, ranked by p value (columns), with the green squares showing the ratio of the number of pathway proteins identified in glycoproteomic data over the total number of proteins in that pathway. Multiple-testing-corrected p values were calculated using the Benjamini–Hochberg method. The threshold line corresponds to a p value of 0.05.
Partial least squares discriminate analysis also linked glycosylated Apo F, transthyretin, Apo C4, prostaglandin-H2 D-isomerase, complement C9, and CD59 to the post-MI development of HF because the important feature scores were high for all six (Figure 2A). Apo F, Apo C4, transthyretin, and prostaglandin-D2 synthase are linked to metabolic disease (specifically, to lipid metabolism), whereas C9 and CD59 are associated with inflammation. The increase in lipid metabolism is consistent with animal studies that have shown upregulation of cardiac lipid metabolism is an initiator stimulus for HF development.15 CD59 regulates complement activation by preventing C9 from polymerizing and forming the complement membrane attack complex.16
Figure 2.
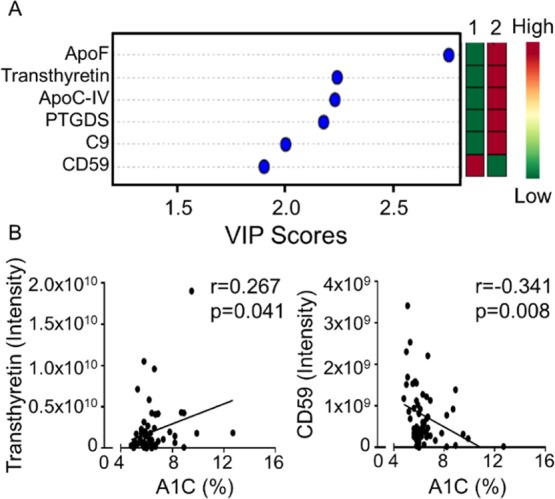
Bioinformatic analysis identified six candidates predictive of HF. (A) PLSDA linked upregulation of apolipoprotein (Apo) F, transthyretin, Apo C4, prostaglandin-H2 D-isomerase, and complement C9 and downregulation of CD59 to the development of post-MI HF. (B) Transthyretin and CD59 associated with elevated levels of hemoglobin A1C. VIP—important feature.
Of the six glycosylated proteins that were differentially expressed, transthyretin and CD59 were associated with elevated levels of hemoglobin A1C (Figure 2B). As CD59 inhibits complement activation, CD59 upregulation would protect cells from complement-mediated damage and stimulate inflammation resolution.16 The reduction of CD59 in the setting of diabetes provides a mechanism to dampen resolution and stimulate the development of HF. Optimal post-MI maintenance requires mechanisms that continually prevent inflammation by inhibiting proinflammatory cytokines, stimulating the clearance of inflammatory cells, and promoting ECM homeostasis to maintain a stable scar.17−22 In congestive HF, inflammation and ECM turnover is a major determinant of progressive cardiac remodeling,23 and metabolic alterations and immune-regulated ECM turnover are closely linked with HF progression.24 Our results suggest cross-talk between metabolism and immunity instigating the progression to HF, especially in participants with diabetes.
Participants who developed HF had a higher prevalence of metabolic disease. Of the participants enrolled, 45% (27/60) were diagnosed with diabetes. Of the participants later hospitalized with HF, 80% (12/15) were diagnosed with diabetes compared to 38% (17/45) who did not progress to HF. MI subjects who developed HF did not have well-controlled glucose levels, as evidenced by higher hemoglobin A1C (Table 1).
Online Mendelian Inheritance in Man (OMIM) disease analysis indicated that complement deficiency and macular degeneration were linked with the proteins identified in the glycoproteomic screen.25 In particular, CD59, C3, and C9 were associated with complement deficiency, whereas C3 and Apo F were associated with macular degeneration. Interestingly, Cheng and colleagues have reported a link between macular degeneration and increased incidence of both MI and HF (Table 4).26 Complement component 3 has been shown to be necessary to preserve post-MI myocardium and myocardial physiology.27
Table 4. Disease Associationsa.
| name | p value | adjusted p value | Z-score | combined score | proteins |
|---|---|---|---|---|---|
| complement deficiency | 1.80 × 10–8 | 3.60 × 10–8 | –1.32 | 23.63 | CD59, C3, C9 |
| macular degeneration | 0.00807 | 0.00807 | –1.44 | 6.96 | C3, Apo F |
In the OMIM disease compendium, complement deficiency and macular degeneration were most linked with the proteins identified in the glycoproteomic screen.25
Apo F Predicted the Development of HF
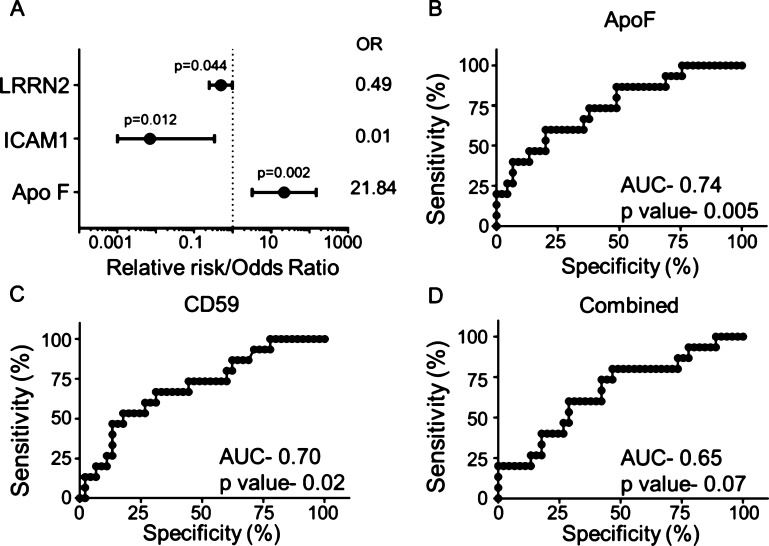
When stratified by age, sex, and time interval between MI and plasma collection, Apo F was strongly associated with an increased risk for HF (OR 21.84, 95% CI 3.20–149.14; Figure 3A). In addition, intercellular adhesion molecule 1 (ICAM1; OR 0.01, 95% CI 0.001–0.334) and leucine-rich repeat neuronal 2 (LRRN2) protein (OR 0.49, 95% CI 0.25–0.98) were associated with protection from the development of HF. After adjustment, transthyretin, ApoC4, prostaglandin-H2 D-isomerase, complement C9, and CD59 were no longer significant. Apo F was identified as a candidate biomarker for HF by both important feature analysis and by adjusted analysis. Apo F, an LXR/RXR pathway member, is also known as lipid transfer inhibitor protein for its ability to inhibit lipid transfer between lipoproteins.28 From our results, Apo F may also play a previously unidentified role in inflammation regulation.
Figure 3.
Apo F identified as the strongest biomarker for HF. (A) When stratified by age, sex, and time interval between MI and plasma collection, Apo F was associated with increased risk, and ICAM1 and LRRN2 were associated with decreased risk from the development of HF. (B) Apo F and (C) CD59 alone were strong predictors for HF compared to (D) all eight proteins combined (Apo F, transthyretin, Apo C4, prostaglandin-H2 D-isomerase, complement C9, CD59, ICAM1, and LRRN2).
Plasma glycoproteins identified were analyzed for predictability of the development of HF following MI. Receiver operating characteristic analysis identified that all eight markers detected by either important feature analysis (Apo F, transthyretin, ApoC4, prostaglandin-H2 D-isomerase, complement C9, and CD59) or adjusted analysis (ICAM1 and LRRN2) could predict the development of HF. Interestingly, Apo F and CD59 alone were stronger predictors for HF compared to all eight markers together, and Apo F was the strongest individual predictor for HF (Figure 3C–E). Neither IL-6 (AUC = 0.558; p = 0.516) or IL-6 receptor (AUC = 0.629; p = 0.147), which are known biomarkers for HF,29 were as powerful predictors as Apo F.
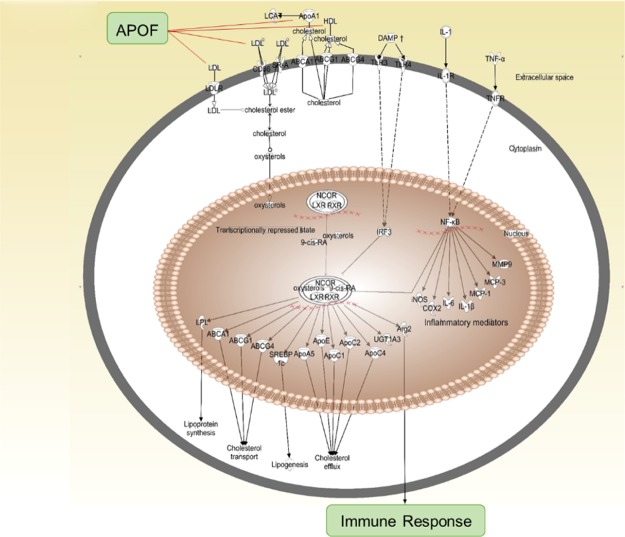
Apo F is a protein associated with both the acute phase response and with the LXR/RXR pathway and regulates macrophage physiology through mechanisms not fully understood. Apo F forms plasma membrane complexes with lipoproteins, including low-density lipoprotein (LDL) and high-density lipoprotein (HDL), to facilitate the uptake or esterification of cholesterol.30 Changes in these lipoprotein complexes promote inflammatory cell conversion from proinflammatory to anti-inflammatory phenotype or vice versa.31,32 This in turn could lead to prolonged inflammation to enable the development of congestive HF. Ingenuity pathway analysis (IPA) indicated that Apo F was an upstream regulator in the LXR/RXR pathway linked to changes in the immune response (Figure 4). The macrophage arm of the LXR/RXR pathway, therefore, may be vital for the post-MI progression to HF.
Figure 4.
Macrophage arm of the LXR/RXR pathway is linked to the development of HF. Apo F regulates this process by binding to LDL and HDL, thus changing cell membrane lipoprotein complexes and facilitating inflammatory cell activation.
Conclusions
The goal of this study was to identify protein candidates in plasma samples from post-MI JHS participants that predicted future progression to HF. The three most salient findings were (1) Apo F was identified as a strong predictor for future HF development; (2) upregulation of the acute phase response and LXR/RXR signaling pathways and downregulation of the coagulation system associated with later HF; and (3) regulation of inflammation by metabolic pathways may be key in HF development. Participants who were diagnosed with HF before visit 2 were not included in this analysis to identify early markers of future HF. Our results indicate that participants with metabolic disease (e.g., diabetes) as a comorbidity had a higher risk of HF development, most likely because of enhanced and prolonged inflammation leading to adverse remodeling.
Apo F is a glycosylated protein highly expressed by the liver and secreted into the circulation where it attaches to lipoproteins.33 Apo F prefers binding to LDL over HDL, and binding to LDL inhibits its ability to transfer lipids.34 Whether glycosylation of Apo F affects lipid binding is not known. Future studies should be performed evaluating whether alterations to Apo F glycosylation status affect protein function and risk of developing HF. LDL also coordinates the activation of inflammatory and apoptotic responses in leukocytes.35 Overexpression of Apo F reduces HDL cholesterol (HDL-C) levels in mice by increasing the clearance of HDL-C esterase in macrophages.36 This is thought to play a role in polarizing inflammatory cells, specifically macrophages, from proinflammatory to anti-inflammatory phenotypes.31,32 Although elevated HDL-C associates with improved CVD outcomes,37 in patients with chronic disease such as diabetes, HDL undergoes structural modifications that can promote and prolong inflammation.32,38,39 Some of the same pathways, therefore, may be activated in opposite directions in the setting of comorbidities.
Nonenzymatic glycation of apolipoproteins is involved in the pathogenesis of vascular complications of metabolic diseases including diabetes. For example, glycosylation of ApoA-IV is associated with coronary artery disease severity in patients with type-2 diabetes by increasing the proinflammatory response through the nuclear receptor NR4A3.40 Similarly, IgG glycosylation associates with CVD risk possibly through direct interaction with Fcγ receptors and complement.41,42 In addition, Mayr and colleagues showed a strong association between very-LDL-associated apolipoproteins including C-II, C-III, and E with incident CVD similar to what we observed in our study.43 Although our study did not assess the glycosylation status of the identified proteins, future studies evaluating the effect of diabetes on protein glycosylation is required to understand their role in HF development. In the current guidelines for HF, natriuretic peptides are the recommended biomarker to aid with the diagnosis.1 Further comparison of the proteins identified in our study with more common biomarkers such as natriuretic peptides, C-reactive protein, and cardiac troponins are needed to comparatively weigh and rank different biomarkers that have been proposed. Combining biomarkers into composite scores will also provide further discriminatory capabilities.
Glycoproteomic evaluation of JHS samples served as a hypothesis-generating analysis to identify possible markers for HF. Additional studies that provide insight into the mechanisms behind Apo F regulation of lipoproteins and how glycosylation of Apo F affects inflammatory mechanisms are warranted. Studies delineating the connection between macular degeneration and HF development and mechanistic investigations exploring the role of Apo F in mediating macrophage proinflammatory signaling would also be informative.
Limitations to this study include small sample size and the glycoproteome was evaluated at a single time point, which varied in its temporal relation between MI and onset of HF across the cohort. Because of the small sample size, we had to limit the number of covariates assessed. Future studies with a larger sample size would be necessary to adjust for other important variables such as BMI, diabetes, and smoking status. Comparison of our findings with additional ethnicities/races, regions/countries, and ages will be needed to fully understand the efficacy of the proteins identified. In addition, future studies using serial samples are needed to understand the dynamic nature of the proteome. We have recently reported that LXR/RXR signaling and neutrophil phenotype following MI distinguish sex differences in remodeling, using this same cohort.44
In conclusion, glycosylated Apo F was the strongest predictor of future HF identified in post-MI plasma samples. Our data suggest that the increase in Apo F may facilitate HF development by extending the inflammatory response after MI.
Materials and Methods
Jackson Heart Study
JHS is a community-based cohort study that includes 5306 noninstitutionalized African American participants who were 21 to 94 years old at the time of enrollment and all residing in the Jackson, MS area. JHS sampled from urban and rural areas surrounding Jackson, MS, included representation from three metropolitan counties (Hinds, Madison, and Rankin). Participants were residents, volunteers, randomly selected individuals, and secondary family members of enrolled participants.45
The analysis included 60 participants diagnosed with MI before visit 2 (2005–2008), who had not been hospitalized with HF prior to the visit, and for whom informed consent was obtained for ancillary studies. JHS was approved by the institutional review board of the University of Mississippi Medical Center, and all participants provided written informed consent under IRB protocol #2014-0274. All participants that fit the criteria were included in this study. Later, HF events were formally adjudicated by JHS from 2005 to 2012. Participants were divided into two groups: (1) those who did not develop HF by 2013 (MI; n = 45) and (2) those who were hospitalized after 2009 with HF (MI + HF; n = 15). Classification of HF was based on the following: (1) Framingham criteria equal “HF present”, NHANES criteria equal “HF present”, and modified Boston criteria equal “definite or possible HF” and (2) Morbidity and Mortality Classification Committee (MMCC) HF classification is “chronic stable HF” or “no HF”. Two physician reviews were required for HF classification. If there was a disagreement in classification, a third adjudicator physician review was performed and that diagnosis was used. The time between MI and plasma collection was 6 ± 5 years (median = 4 years; time interval = 1–17 years). We previously evaluated this cohort for sex differences in remodeling.44
Electrocardiogram Analysis
Standard 12-lead electrocardiogram (ECG) measurements were recorded at visit 3 to evaluate if glycopeptides at visit 2 predicted ECG changes at visit 3. Average QT interval was corrected for the heart rate using the Bazett formula (QTc = QT/√RR).
Plasma Glycoproteomics
The glycopeptide capture method was used to enrich for extracellular proteins that had glycosylation sites; we did not evaluate the status of glycosylation. Plasma glycopeptides were isolated label-free and captured by the solid-phase extraction of glycopeptides method, as previously described.10,14,46,47 Plasma proteins were reduced, alkylated, and trypsin-digested into peptides.10,14,47 The trypsinized peptide mixture was oxidized and covalently conjugated to a solid support via hydrazide chemistry, and glycopeptides were N-linked to the column and released using PNGase F treatment. The released peptides were prepared using Sep-Pak Vac C18 cartridge (Waters, Milford, MA) and analyzed by liquid chromatography-tandem mass spectrometry using a Q Exactive (ThermoFisher, Waltham, MA) together with a 15 cm × 75 μm C18 column (5 μm particles with 100 Å pore size). A target value of 1 × 106 ions at a resolution setting of 70 000 was set and 1 × 105 ions at a resolution setting of 17 500 in MS2.
SEQUEST was searched for MS/MS spectra using Proteome Discoverer (version 1.4; Thermo Fisher) against the human RefSeq database (November 3, 2013 version) containing 53 918 sequences. To exclude low-probability protein identifications, the false discovery rate was set at 0.01. Spectral counting was used for peptide quantification, and total spectral counts for each sample were used for normalization. Missing values were replaced with 0.01 to calculate the ratio and p value. Samples were run in duplicate, and the results were combined for analysis. A total of 88 proteins were identified, of which 41% (36 proteins) were found in both runs. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009870 (http://www.ebi.ac.uk/pride/archive/).48
Immunoblotting
Immunoblotting was performed according to the guidelines for antibody use in cardiovascular research.49 Validation of glycoproteomic data was performed by regression analysis comparing mass spectroscopy peak intensity values to densitometry of protein measured by immunoblotting and normalized to the intensity of the total membrane stain (Table S1 and Figure S1). Samples were volume-loaded and separated on 4–12% Criterion XT Bis-Tris gels (Bio-Rad), transferred to a nitrocellulose membrane (Bio-Rad), and stained with MemCode Reversible Protein Stain Kit (Thermo Scientific) to verify the protein concentration and loading accuracy. Membranes were blocked with 5% nonfat milk (Bio-Rad), followed by overnight incubation with primary antibody [polymeric immunoglobulin receptor (Abcam ab96196; 1:1000), complement C9 (Abcam ab173302; 1:1000), Apo F (Raybiotech 102-10552; 1:250), and CD59 (Abcam ab124396; 1:1000)], followed by secondary antibody (anti-rabbit Vector PI-1000; 1:5000) and detection with ECL Prime Western Blotting Detection Substrate (Amersham).
IL6 and IL6R ELISA
IL6 and IL6R plasma concentrations were measured using human IL6 (Novus; NBP2-31051) and IL6R (Novus; NBP1-89870) ELISA kits, according to manufacturer instructions.
Statistics and Bioinformatics
Statistics are reported according to the guidelines, statistical considerations in reporting cardiovascular research.50 Data are presented as mean ± SD, and all data acquisition and statistical analyses were performed by investigators blinded to groups. The analysis of this study compared MI and MI + HF groups; as such, two group comparisons were analyzed by Students t-test. A statistical significance was set at p < 0.05 (two-sided).
A model was developed to estimate the odds ratio (OR) of HF associated with the identified glycoproteins. Logistic regression analyses were used to evaluate associations between plasma glycoprotein markers and HF development after MI in univariate and multivariable models, adjusting for age, sex, and time between MI and plasma collection. Receiver operating characteristic area under the curve (AUC) analysis was performed in GraphPad Prism 7, for the eight individual proteins identified as being significantly different between groups and for a composite assessment of all eight proteins combined.
For data visualization, heat maps were constructed using an online statistical tool provided in the Metaboanalyst 3.0 package (www.metaboanalyst.ca/).51,52 Signaling pathways were visualized using partial least squares discriminant analysis (PLSDA) and canonical analysis in IPA; (QIAGEN, Redwood City; www.qiagen.com/ingenuity). For IPA, MI + HF glycoproteomic data were normalized to MI-only datasets, to focus on pathways responsible for HF development and not in response to MI. Multiple-testing corrected p values were calculated using the Benjamini–Hochberg method.53
Acknowledgments
The JHS is supported and conducted in collaboration with the Jackson State University (HHSN268201300049C and HHSN268201300050C), Tougaloo College (HHSN268201300048C), and the University of Mississippi Medical Center (HHSN268201300046C and HHSN268201300047C) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. This work was also supported by the American Heart Association 15SDG22930009 to Y.M. and the National Institute of Health T32HL105324 to O.K.E.; HL075360, HL129823, and GM114833 to M.L.L.; and from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development Award 5I01BX000505 to M.L.L.; IK2BX003922 to K.Y.D.-P.; and by HL051971, GM104357, and GM115428.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b02207.
Regression analysis of mass spectrometry peak intensity values versus densitometry of protein measured by immunoblotting; protein quantification correlated with peak intensity of proteins identified by mass spectrometry and showed a positive relationship between the two methods; glycoproteomic results: values for individual samples are intensity units of the peptides; definition of terms: Xcorr—peptide score, m/z [Da]—mass/charge, and RT (min)—retention time; immunoblotting of top six proteins identified by glycoproteomics show strong correlation between two proteomic techniques; and n = 15 with no HF and 5 HF (PDF)
Glycoproteomic results (XLS)
Author Present Address
¶ Research Service, Ralph H Johnson Veterans Affairs Medical Center and Department of Medicine, Division of Cardiology, Medical University of South Carolina, Charleston, SC 29425.
Author Present Address
∇ Department of Molecular Pharmacology and Physiology, Morsani College of Medicine, University of South Florida, Tampa Bay, FL 33612.
Author Present Address
○ Merck Research Laboratories (MRL) South San Francisco, CA 94080.
Author Contributions
K.Y.D.-P. and M.L.L. conceived the hypothesis. K.Y.D.-P. analyzed all data; E.R.F. performed and analyzed mass spectroscopy experiments; K.Y.D.-P. and O.K.E. analyzed patient characteristics; I.E., S.K.M., R.S.V., M.E.H., and E.R.F. collected clinical samples, K.Y.D.-P., O.K.E., Y.M., R.P.I., E.F., I.E., S.K.M., R.S.V., M.E.H., E.R.F., and M.L.L. designed experimental procedures, interpreted data, and drafted manuscript. All authors read, edited, and approved the final manuscript.
Views expressed are those of the authors and do not necessarily represent views of the NHLBI, National Institutes of Health, U.S. Department of Health and Human Services, American Heart Association, or the Veterans Administration. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Benjamin E. J.; Virani S. S.; Callaway C. W.; Chamberlain A. M.; Chang A. R.; Cheng S.; Chiuve S. E.; Cushman M.; Delling F. N.; Deo R.; de Ferranti S. D.; Ferguson J. F.; Fornage M.; Gillespie C.; Isasi C. R.; Jiménez M. C.; Jordan L. C.; Judd S. E.; Lackland D.; Lichtman J. H.; Lisabeth L.; Liu S.; Longenecker C. T.; Lutsey P. L.; Mackey J. S.; Matchar D. B.; Matsushita K.; Mussolino M. E.; Nasir K.; O’Flaherty M.; Palaniappan L. P.; Pandey A.; Pandey D. K.; Reeves M. J.; Ritchey M. D.; Rodriguez C. J.; Roth G. A.; Rosamond W. D.; Sampson U. K. A.; Satou G. M.; Shah S. H.; Spartano N. L.; Tirschwell D. L.; Tsao C. W.; Voeks J. H.; Willey J. Z.; Wilkins J. T.; Wu J. H.; Alger H. M.; Wong S. S.; Muntner P. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 2018, 137, e67–e492. 10.1161/cir.0000000000000558. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D.; Benjamin E. J.; Go A. S.; Arnett D. K.; Blaha M. J.; Cushman M.; de Ferranti S.; Després J.-P.; Fullerton H. J.; Howard V. J.; Huffman M. D.; Judd S. E.; Kissela B. M.; Lackland D. T.; Lichtman J. H.; Lisabeth L. D.; Liu S.; Mackey R. H.; Matchar D. B.; McGuire D. K.; Mohler E. R.; Moy C. S.; Muntner P.; Mussolino M. E.; Nasir K.; Neumar R. W.; Nichol G.; Palaniappan L.; Pandey D. K.; Reeves M. J.; Rodriguez C. J.; Sorlie P. D.; Stein J.; Towfighi A.; Turan T. N.; Virani S. S.; Willey J. Z.; Woo D.; Yeh R. W.; Turner M. B. Heart Disease and Stroke Statistics-2015 Update. Circulation 2015, 131, e29–322. 10.1161/cir.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Go A. S.; Mozaffarian D.; Roger V. L.; Benjamin E. J.; Berry J. D.; Borden W. B.; Bravata D. M.; Dai S.; Ford E. S.; Fox C. S.; Franco S.; Fullerton H. J.; Gillespie C.; Hailpern S. M.; Heit J. A.; Howard V. J.; Huffman M. D.; Kissela B. M.; Kittner S. J.; Lackland D. T.; Lichtman J. H.; Lisabeth L. D.; Magid D.; Marcus G. M.; Marelli A.; Matchar D. B.; McGuire D. K.; Mohler E. R.; Moy C. S.; Mussolino M. E.; Nichol G.; Paynter N. P.; Schreiner P. J.; Sorlie P. D.; Stein J.; Turan T. N.; Virani S. S.; Wong N. D.; Woo D.; Turner M. B. Heart Disease and Stroke Statistics-2013 Update. Circulation 2013, 127, e6–e245. 10.1161/cir.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisant L. M.; Thomas K. L.; Lewis E. F.; Huang Z.; Francis G. S.; Weaver W. D.; Pfeffer M. A.; McMurray J. J. V.; Califf R. M.; Velazquez E. J. Racial analysis of patients with myocardial infarction complicated by heart failure and/or left ventricular dysfunction treated with valsartan, captopril, or both. J. Am. Coll. Cardiol. 2008, 51, 1865–1871. 10.1016/j.jacc.2007.12.050. [DOI] [PubMed] [Google Scholar]
- Wang X.; Wang R.; Zhang Y.; Zhang H. Evolutionary survey of druggable protein targets with respect to their subcellular localizations. Genome Biol. Evol. 2013, 5, 1291–1297. 10.1093/gbe/evt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin E. J.; Blaha M. J.; Chiuve S. E.; Cushman M.; Das S. R.; Deo R.; de Ferranti S. D.; Floyd J.; Fornage M.; Gillespie C.; Isasi C. R.; Jiménez M. C.; Jordan L. C.; Judd S. E.; Lackland D.; Lichtman J. H.; Lisabeth L.; Liu S.; Longenecker C. T.; Mackey R. H.; Matsushita K.; Mozaffarian D.; Mussolino M. E.; Nasir K.; Neumar R. W.; Palaniappan L.; Pandey D. K.; Thiagarajan R. R.; Reeves M. J.; Ritchey M.; Rodriguez C. J.; Roth G. A.; Rosamond W. D.; Sasson C.; Towfighi A.; Tsao C. W.; Turner M. B.; Virani S. S.; Voeks J. H.; Willey J. Z.; Wilkins J. T.; Wu J. H.; Alger H. M.; Wong S. S.; Muntner P. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation 2017, 135, e146–e603. 10.1161/cir.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami H.; Kronmal R.; Bluemke D. A.; Olson J.; Shea S.; Liu K.; Burke G. L.; Lima J. A. C. Differences in the incidence of congestive heart failure by ethnicity: the multi-ethnic study of atherosclerosis. Arch. Intern. Med. 2008, 168, 2138–2145. 10.1001/archinte.168.19.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker W. H.; Mullooly J. P.; Getchell W. Changing incidence and survival for heart failure in a well-defined older population, 1970-1974 and 1990-1994. Circulation 2006, 113, 799–805. 10.1161/circulationaha.104.492033. [DOI] [PubMed] [Google Scholar]
- Yeung D. F.; Boom N. K.; Guo H.; Lee D. S.; Schultz S. E.; Tu J. V. Trends in the incidence and outcomes of heart failure in Ontario, Canada: 1997 to 2007. Can. Med. Assoc. J. 2012, 184, E765–E773. 10.1503/cmaj.111958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoux A.; Tian Y.; DeLeon-Pennell K. Y.; Nguyen N. T.; de Castro Brás L. E.; Flynn E. R.; Cannon P. L.; Griswold M. E.; Jin Y.-F.; Puskarich M. A.; Jones A. E.; Lindsey M. L. Plasma Glycoproteomics Reveals Sepsis Outcomes Linked to Distinct Proteins in Common Pathways. Crit. Care Med. 2015, 43, 2049–2058. 10.1097/ccm.0000000000001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey M. L.; Hall M. E.; Harmancey R.; Ma Y. Adapting extracellular matrix proteomics for clinical studies on cardiac remodeling post-myocardial infarction. Clin. Proteomics 2016, 13, 19. 10.1186/s12014-016-9120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey M. L.; Zamilpa R. Temporal and spatial expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases following myocardial infarction. Cardiovasc. Ther. 2010, 30, 31–41. 10.1111/j.1755-5922.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey M. L.; Iyer R. P.; Zamilpa R.; Yabluchanskiy A.; DeLeon-Pennell K. Y.; Hall M. E.; Kaplan A.; Zouein F. A.; Bratton D.; Flynn E. R.; Cannon P. L.; Tian Y.; Jin Y.-F.; Lange R. A.; Tokmina-Roszyk D.; Fields G. B.; de Castro Brás L. E. A Novel Collagen Matricryptin Reduces Left Ventricular Dilation Post-Myocardial Infarction by Promoting Scar Formation and Angiogenesis. J. Am. Coll. Cardiol. 2015, 66, 1364–1374. 10.1016/j.jacc.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon-Pennell K. Y.; Tian Y.; Zhang B.; Cates C. A.; Iyer R. P.; Cannon P.; Shah P.; Aiyetan P.; Halade G. V.; Ma Y.; Flynn E.; Zhang Z.; Jin Y.-F.; Zhang H.; Lindsey M. L. CD36 Is a Matrix Metalloproteinase-9 Substrate That Stimulates Neutrophil Apoptosis and Removal During Cardiac Remodeling. Circ.: Cardiovasc. Genet. 2016, 9, 14–25. 10.1161/circgenetics.115.001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg I. J.; Trent C. M.; Schulze P. C. Lipid metabolism and toxicity in the heart. Cell Metab. 2012, 15, 805–812. 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Qiao F.; Abagyan R.; Hazard S.; Tomlinson S. Defining the CD59-C9 binding interaction. J. Biol. Chem. 2006, 281, 27398–27404. 10.1074/jbc.m603690200. [DOI] [PubMed] [Google Scholar]
- DeLeon-Pennell K. Y.; Meschiari C. A.; Jung M.; Lindsey M. L.. Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Progress in Molecular Biology and Translational Science; Elsevier, 2017; Vol. 147, pp 75–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R. P.; Jung M.; Lindsey M. L. MMP-9 signaling in the left ventricle following myocardial infarction. Am. J. Physiol.: Heart Circ. Physiol. 2016, 311, H190–H198. 10.1152/ajpheart.00243.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey M. L.; Saucerman J. J.; DeLeon-Pennell K. Y. Knowledge gaps to understanding cardiac macrophage polarization following myocardial infarction. Biochim. Biophys. Acta, Gen. Subj. 2016, 1862, 2288–2292. 10.1016/j.bbadis.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Iyer R. P.; Jung M.; Czubryt M. P.; Lindsey M. L. Cardiac Fibroblast Activation Post-Myocardial Infarction: Current Knowledge Gaps. Trends Pharmacol. Sci. 2017, 38, 448–458. 10.1016/j.tips.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschiari C. A.; Ero O. K.; Pan H.; Finkel T.; Lindsey M. L. The impact of aging on cardiac extracellular matrix. GeroScience 2017, 39, 7–18. 10.1007/s11357-017-9959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinale F. G.; Frangogiannis N. G.; Hinz B.; Holmes J. W.; Kassiri Z.; Lindsey M. L. Crossing Into the Next Frontier of Cardiac Extracellular Matrix Research. Circ. Res. 2016, 119, 1040–1045. 10.1161/circresaha.116.309916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannad F.; Alla F.; Dousset B.; Perez A.; Pitt B. Limitation of excessive extracellular matrix turnover may contribute to survival benefit of spironolactone therapy in patients with congestive heart failure: insights from the randomized aldactone evaluation study (RALES). Rales Investigators. Circulation 2000, 102, 2700–2706. 10.1161/01.cir.102.22.2700. [DOI] [PubMed] [Google Scholar]
- Shapiro H.; Lutaty A.; Ariel A. Macrophages, meta-inflammation, and immuno-metabolism. Sci. World J. 2011, 11, 2509–2529. 10.1100/2011/397971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis A. D.Searching Online Mendelian Inheritance in Man (OMIM) for Information on Genetic loci involved in human disease. Current Protocols in Human Genetic; Wiley, 2012; Chapter 9, Unit 9.13.1-10. [DOI] [PubMed] [Google Scholar]
- Cheng Q.; Saaddine J. B.; Klein R.; Rothenberg R.; Chou C.-F.; Il’yasova D. Early Age-related Macular Degeneration with Cardiovascular and Renal Comorbidities: An Analysis of the National Health and Nutrition Examination Survey, 2005-2008. Ophthalmic Epidemiol 2017, 24, 413–419. 10.1080/09286586.2017.1337911. [DOI] [PubMed] [Google Scholar]
- Wysoczynski M.; Solanki M.; Borkowska S.; van Hoose P.; Brittian K. R.; Prabhu S. D.; Ratajczak M. Z.; Rokosh G. Complement component 3 is necessary to preserve myocardium and myocardial function in chronic myocardial infarction. Stem Cells 2014, 32, 2502–2515. 10.1002/stem.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagor W. R.; Fields D. W.; Khetarpal S. A.; Kumaravel A.; Lin W.; Weintraub N.; Wu K.; Hamm-Alvarez S. F.; Drazul-Schrader D.; de la Llera-Moya M.; Rothblat G. H.; Rader D. J. The effects of apolipoprotein F deficiency on high density lipoprotein cholesterol metabolism in mice. PLoS One 2012, 7, e31616. 10.1371/journal.pone.0031616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held C.; White H. D.; Stewart R. A. H.; Budaj A.; Cannon C. P.; Hochman J. S.; Koenig W.; Siegbahn A.; Steg P. G.; Soffer J.; Weaver W. D.; Östlund O.; Wallentin L. Inflammatory Biomarkers Interleukin-6 and C-Reactive Protein and Outcomes in Stable Coronary Heart Disease: Experiences From the STABILITY (Stabilization of Atherosclerotic Plaque by Initiation of Darapladib Therapy) Trial. J. Am. Heart Assoc. 2017, 6, e005077. 10.1161/jaha.116.005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren E.; McConathy W. J.; Alaupovic P. Isolation and characterization of simple and complex lipoproteins containing apolipoprotein F from human plasma. Biochemistry 1982, 21, 5347–5351. 10.1021/bi00264a035. [DOI] [PubMed] [Google Scholar]
- Pajkrt D.; Doran J. E.; Koster F.; Lerch P. G.; Arnet B.; van der Poll T.; ten Cate J. W.; van Deventer S. J. Antiinflammatory effects of reconstituted high-density lipoprotein during human endotoxemia. J. Exp. Med. 1996, 184, 1601–1608. 10.1084/jem.184.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lenten B. J.; Hama S. Y.; de Beer F. C.; Stafforini D. M.; McIntyre T. M.; Prescott S. M.; La Du B. N.; Fogelman A. M.; Navab M. Anti-inflammatory HDL becomes pro-inflammatory during the acute phase response. Loss of protective effect of HDL against LDL oxidation in aortic wall cell cocultures. J. Clin. Invest. 1995, 96, 2758–2767. 10.1172/jci118345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagor W. R.; Fields D. W.; Bauer R. C.; Crawford A.; Abt M. C.; Artis D.; Wherry E. J.; Rader D. J. Genetic manipulation of the Apo F/Stat2 locus supports an important role for type I interferon signaling in atherosclerosis. Atherosclerosis 2014, 233, 234–241. 10.1016/j.atherosclerosis.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Driscoll D. M.; Morton R. E. Molecular cloning and expression of lipid transfer inhibitor protein reveals its identity with apolipoprotein F. J. Biol. Chem. 1999, 274, 1814–1820. 10.1074/jbc.274.3.1814. [DOI] [PubMed] [Google Scholar]
- Galkina E.; Ley K. Immune and inflammatory mechanisms of atherosclerosis (*). Annu. Rev. Immunol. 2009, 27, 165–197. 10.1146/annurev.immunol.021908.132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagor W. R.; Brown R. J.; Toh S.-A.; Millar J. S.; Fuki I. V.; de la Llera-Moya M.; Yuen T.; Rothblat G.; Billheimer J. T.; Rader D. J. Overexpression of apolipoprotein F reduces HDL cholesterol levels in vivo. Arterioscler., Thromb., Vasc. Biol. 2009, 29, 40–46. 10.1161/atvbaha.108.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckert E.; Hansel B. HDL-c is a powerful lipid predictor of cardiovascular diseases. Int. J. Clin. Pract. 2007, 61, 1905–1913. 10.1111/j.1742-1241.2007.01509.x. [DOI] [PubMed] [Google Scholar]
- Navab M.; Anantharamaiah G. M.; Reddy S. T.; Van Lenten B. J.; Fogelman A. M. HDL as a biomarker, potential therapeutic target, and therapy. Diabetes 2009, 58, 2711–2717. 10.2337/db09-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbstein D.; Levy A. P. HDL dysfunction in diabetes: causes and possible treatments. Expert Rev. Cardiovasc. Ther. 2012, 10, 353–361. 10.1586/erc.11.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.; Shen Y.; Li Q. R.; Ding F. H.; Wang X. Q.; Liu H. J.; Yan X. X.; Wang L. J.; Yang K.; Wang H. B.; Chen Q. J.; Shen W. F.; Zhang R. Y.; Lu L. Glycated Apolipoprotein A-IV Induces Atherogenesis in Patients With CAD in Type 2 Diabetes. J. Am. Coll. Cardiol. 2017, 70, 2006–2019. 10.1016/j.jacc.2017.08.053. [DOI] [PubMed] [Google Scholar]
- Menni C.; Gudelj I.; Macdonald-Dunlop E.; Mangino M.; Zierer J.; Bešić E.; Joshi P. K.; Trbojević-Akmačić I.; Chowienczyk P. J.; Spector T. D.; Wilson J. F.; Lauc G.; Valdes A. M. Glycosylation Profile of Immunoglobulin G Is Cross-Sectionally Associated With Cardiovascular Disease Risk Score and Subclinical Atherosclerosis in Two Independent Cohorts. Circ. Res. 2018, 122, 1555–1564. 10.1161/circresaha.117.312174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomp R.; Ruhaak L. R.; Uh H. W.; Reiding K. R.; Selman M.; Houwing-Duistermaat J. J.; Slagboom P. E.; Beekman M.; Wuhrer M. Subclass-specific IgG glycosylation is associated with markers of inflammation and metabolic health. Sci. Rep. 2017, 7, 12325. 10.1038/s41598-017-12495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlaner R.; Tsimikas S.; Yin X.; Willeit P.; Baig F.; Santer P.; Oberhollenzer F.; Egger G.; Witztum J. L.; Alexander V. J.; Willeit J.; Kiechl S.; Mayr M. Very-Low-Density Lipoprotein-Associated Apolipoproteins Predict Cardiovascular Events and Are Lowered by Inhibition of APOC-III. J. Am. Coll. Cardiol. 2017, 69, 789–800. 10.1016/j.jacc.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon-Pennell K. Y.; Mouton A. J.; Ero O. K.; Ma Y.; Padmanabhan Iyer R.; Flynn E. R.; Espinoza I.; Musani S. K.; Vasan R. S.; Hall M. E.; Fox E. R.; Lindsey M. L. LXR/RXR signaling and neutrophil phenotype following myocardial infarction classify sex differences in remodeling. Basic Res. Cardiol. 2018, 113, 40. 10.1007/s00395-018-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. G.; Rotimi C. N.; Ekunwe L.; Royal C. D.; Crump M. E.; Wyatt S. B.; Steffes M. W.; Adeyemo A.; Zhou J.; Taylor Jr H. A.; Jaquish C. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis 2005, 15, 30–37. [PubMed] [Google Scholar]
- Tian Y.; Kelly-Spratt K. S.; Kemp C. J.; Zhang H. Mapping tissue-specific expression of extracellular proteins using systematic glycoproteomic analysis of different mouse tissues. J. Proteome Res. 2010, 9, 5837–5847. 10.1021/pr1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Koganti T.; Yao Z.; Cannon P.; Shah P.; Pietrovito L.; Modesti A.; Aiyetan P.; DeLeon-Pennell K.; Ma Y.; Halade G. V.; Hicks C.; Zhang H.; Lindsey M. L. Cardiac extracellular proteome profiling and membrane topology analysis using glycoproteomics. Proteomics: Clin. Appl. 2014, 8, 595–602. 10.1002/prca.201400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A.; Csordas A.; del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T.; Xu Q.-W.; Wang R.; Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, 11033. 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks H. L.; Lindsey M. L. Guidelines for authors and reviewers on antibody use in physiology studies. Am. J. Physiol.: Heart Circ. Physiol. 2018, 314, H724–h732. 10.1152/ajpheart.00512.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey M. L.; Gray G. A.; Wood S. K.; Curran-Everett D. Statistical considerations in reporting cardiovascular research. Am. J. Physiol.: Heart Circ. Physiol. 2018, 315, H303–H313. 10.1152/ajpheart.00309.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T. H.; Shipley J. M.; Bergers G.; Berger J. E.; Helms J. A.; Hanahan D.; Shapiro S. D.; Senior R. M.; Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 1998, 93, 411–422. 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Sinelnikov I. V.; Han B.; Wishart D. S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J. A. The Benjamini–Hochberg method in the case of discrete test statistics. Int. J. Biostat. 2007, 3, 11. 10.2202/1557-4679.1065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.