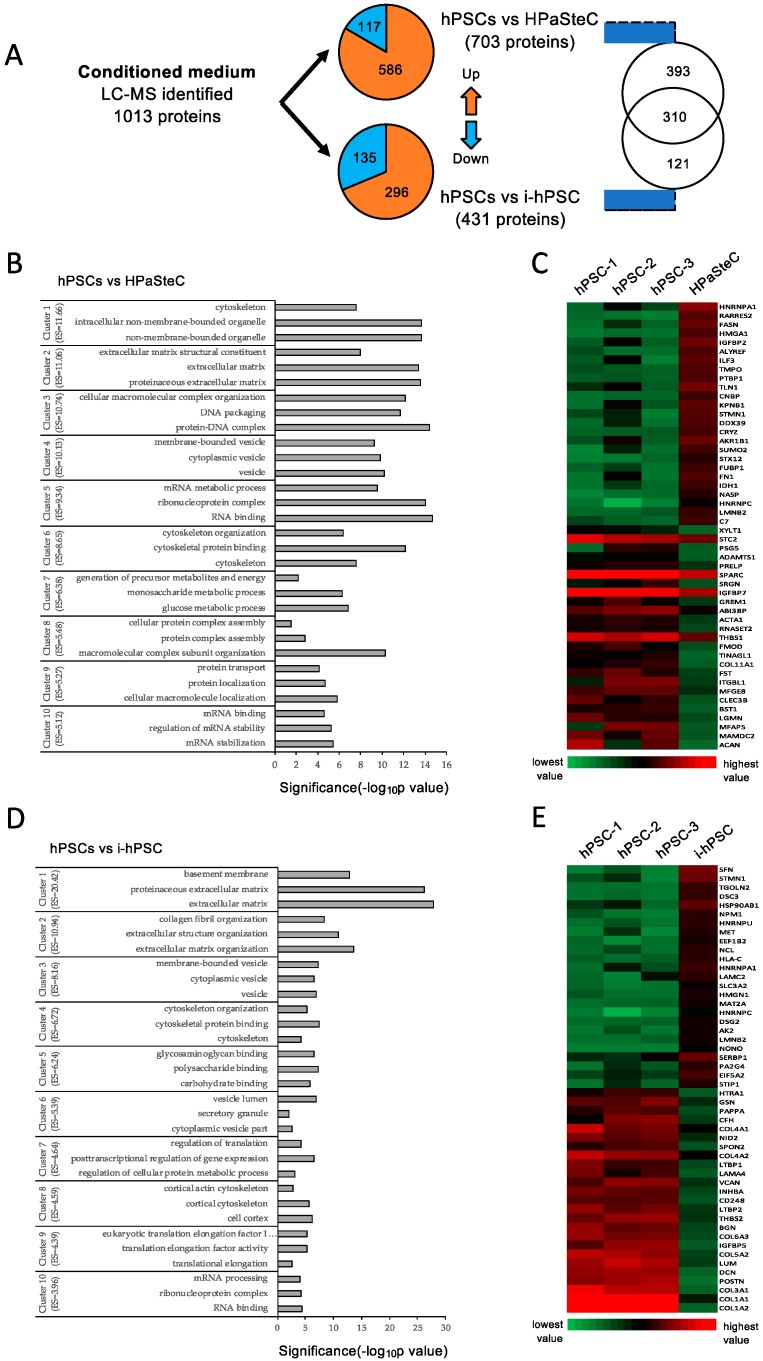
Figure 6.
Secretome analysis of PSC-CM. Conditioned medium from various PSC cultures were subjected to proteomics analysis using LC-MS/MS. (A) Protein identifications from three replicates for each PSC culture and number of differentially expressed genes. (B,D) Compared to hPSC, differentially secreted proteins by HPaSteC (B) and by i-hPSC (D) detected by LC-MS were interrogated in terms of functional annotation by the DAVID Bioinformatics Resource tool. The representative GO terms cluster groups with top 10 enrichment score are presented. The horizontal axis represents the significance (p-value) for each term, while the vertical axis represents the GO categories. (C,E) Heatmap of protein abundance pattern for the 50 most significantly downregulated and upregulated proteins (fold change). Red and green color indicates high and low expression, respectively. GO, Gene ontology; PSC, pancreatic stellate cell; hPSC, human primary PDAC-derived PSC culture; HPaSteC, PSCs from normal human pancreas; i-hPSC, immortalized human PSCs; PSC-CM, PSC-conditioned medium.