Abstract
Simple Summary
The keratin-associated proteins (KAPs) are structural components of cashmere fibres. The human and sheep orthologous gene encoding high-sulphur (HS)-KAP24-1 is now described for the goat species. Our study identified the caprine KAP24-1 gene on goat chromosome 1, and we found that the gene was polymorphic and that variation in the gene affected cashmere fibre diameter.
Abstract
The keratin-associated proteins (KAPs) are structural components of cashmere fibres. The gene encoding the high-sulphur (HS)-KAP24-1 (KRTAP24-1) has been identified in humans and sheep, but it has not been described in goats. In this study, we report the identification of caprine KRTAP24-1, describe variation in this gene, and investigate the effect of this variation on cashmere traits. A search for sequences orthologous to the ovine gene in the goat genome revealed a 774 bp open reading frame on chromosome 1, which could encode an HS-KAP. Based on this goat genome sequence and comparison with ovine KRTAP24-1 sequences, polymerase chain reaction (PCR) primers were designed to amplify an 856 bp fragment that would contain the entire coding region of the putative caprine KRTAP24-1. Use of this PCR amplification with subsequent single-strand conformation polymorphism (SSCP) analysis of the amplicons identified four distinct patterns of DNA bands on gel electrophoresis, with these representing four different DNA sequences (A to D), in 340 Longdong cashmere goats reared in China. The variant sequences had the highest similarity to KRTAP24-1 sequences from sheep and humans, suggesting that they are variants of caprine KRTAP24-1. Nine single-nucleotide polymorphisms (SNPs) were detected in the gene, including four non-synonymous SNPs and an SNP in proximity to the ATG start codon. Of the three common genotypes (AA, AB, and BB) found in these Longdong cashmere goats, cashmere fibres from goats of genotype AA had lower mean fibre diameter (MFD) than did those of genotype AB, and cashmere fibres from goats of genotype AB had lower MFD than did those from goats of genotype BB.
Keywords: keratin associated protein 24-1 (KAP24-1) gene, variation, cashmere, fibre diameter, goat
1. Introduction
The fleece of cashmere goats comprises cashmere, which is the fibre produced by the secondary fibre follicles, and higher-fibre-diameter “guard hairs” that are produced by the primary fibre follicles. As cashmere is a valuable product, it is separated by combing from the guard hairs, which are discarded, and the value of goat fleece is determined by particular cashmere fibre traits. The weight fleece weight and the mean fibre diameter (MFD) are the most valuable of the cashmere traits that can be measured. They, in large part, control the financial return to cashmere producers [1]. It is known that both genetic and environmental factors affect variation in cashmere traits. Accordingly, the identification of genes that regulate cashmere quantity and quality offers some opportunity to improve cashmere production.
Structurally, cashmere fibres are composed of keratins, which are assembled into intermediate filaments (IFs), and keratin-associated proteins (KAPs), which form a semirigid matrix that cross-links with the keratins in the IFs. The KAPs are therefore believed to play an important role in determining the physico-mechanical properties of the fibre.
The KAPs characteristically possess a high content of either cysteine or both glycine and tyrosine, and they can be classified into three broad groups based on their amino acid composition: the high-sulphur (HS; ≤30 mol.% cysteine) KAPs, the ultra-high-sulphur (UHS; >30 mol.% cysteine) KAPs, and the high-glycine/tyrosine (HGT; 35−60 mol.% glycine and tyrosine) KAPs [2]. The KAPs can then be further subdivided into families based on sequence similarity, and, to date, 27 paralogous KAP families have been identified across mammalian species [3].
The KAPs are encoded by small intron-less genes called KRTAPs [3]. While there are over 80 functional KRTAPs from 25 families identified in humans, and 29 KRTAPs from 13 families reported in sheep [3,4,5], only 12 KRTAPs from 9 families have been described in goats, including the recently identified KRTAP20-2 [6,7]. Of these genes, variation in four (KRTAP13-1, KRTAP8-2, KRTAP20-1, and KRTAP20-2) has been described as being associated with cashmere fibre traits [7,8,9,10]. In sheep, where more studies have been carried out, additional KRTAPs have been found to be associated with wool traits, including KRTAP1-2 [11], KRTAP6-1 [12,13], KRTAP6-3 [14], KRTAP8-2 [15], KRTAP22-1 [5], and KRTAP26-1 [4]. This suggests that it would be worthwhile to identify new caprine KRTAPs and characterise variation in these genes, and then ascertain the effect of variation, should it exist, on cashmere fibre traits.
An HS-KAP gene named KRTAP24-1 has been identified in humans [16] and in sheep [17]. Despite being a member of the HS group, the KAP24-1 protein encoded by human and sheep KRTAP24-1 contains an unusually low level of cysteine when compared to other HS-KAPs [16,17]. Ovine KRTAP24-1 is clustered with other HGT-KRTAPs and some HS-KRTAPs in a chromosome region where a number of associations with a variety of wool fibre traits have been reported [4,5,11,12,13,14,15], and similar findings have been reported in goats [7,8,9,10]. To date, the ovine KRTAP24-1 orthologue has not been identified in goats, and its effect on fibre traits has not been reported in any other species. In this study, we report the identification of caprine KRTAP24-1, describe variation in this gene, and investigate the effect of this variation on cashmere traits.
2. Materials and Methods
2.1. Goats Investigated and Cashmere Data Collection
The animal experiments were carried out in accordance with the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval Number 2006-398), and the work was approved by Gansu Agricultural University, Lanzhou, China.
In total, 340 Longdong cashmere goats were investigated. These were the progeny of ten unrelated sires. The goats were farmed by the Yusheng Cashmere Goat Breeding Company, which is located in Huan County of the Gansu Province of China. At one year of age (i.e., at first combing), the weight of cashmere fibre retrieved by combing was recorded. Fibres collected from the mid-side region of the goats were measured to ascertain the crimped fibre length and the mean fibre diameter (MFD), which was assessed by the Inner Mongolia Agricultural University, Inner Mongolia, China. Blood samples were collected onto Munktell TFN paper (Munktell Filter AB, Falun, Sweden), and DNA for PCR amplification was purified using a washing procedure described by Zhou et al. (2006) [18].
2.2. Search for the Caprine KAP24-1 Gene
A previously reported sheep KRTAP24-1 sequence (GenBank accession no. JX112014) was used to BLAST search the Caprine Genome Assembly GCF_001704415.1 [19]. The sequence that shared the greatest similarity with JX112014 was assumed to be KRTAP24-1.
2.3. Polymerase Chain Reaction Single-Strand Conformation Polymorphism (PCR-SSCP) Analysis of Caprine KRTAP24-1
Based on the goat genome sequence identified above, and the comparison with sheep KRTAP24-1 sequences (GenBank accession no. JX112014-JX112017), two PCR primers (5′- AGCCACAGTCTCGCCATAC-3′ and 5′- AGGTGGCACCTGCACCTTG-3′) were designed to amplify an 856 bp fragment that would contain the entire coding region of the putative caprine KRTAP24-1. These primers were synthesised by the Takara Biotechnology Company Limited (Dalian, China). Amplifications were performed in a 20 μL reaction containing the genomic DNA purified from a 1.2 mm punch of dried blood, 0.25 μM of each primer, 150 μM of each dNTP (Takara, Dalian, China), 2.5 mM Mg2+, 0.5 U of Taq DNA polymerase (Takara, Dalian, China), and 1× the PCR buffer supplied with the enzyme. The thermal profile consisted of an initial denaturation for 2 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, 63 °C for 30 s, and 72 °C for 30 s, with a final extension of 5 min at 72 °C. Thermal cycling was undertaken in Bio-Rad S1000 thermal cyclers (Bio-Rad, Hercules, CA, USA).
For each amplicon, a separate 0.7 μL aliquot was added to 7 μL of loading dye (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). These samples were denatured at 95 °C for 5 min, then rapidly cooled on wet ice, prior to being loaded onto 16 × 18 cm, 12% acrylamide/bisacrylamide (37.5:1) (Bio-Rad, Hercules, CA, USA) gels. Electrophoresis was performed using Protean II xi cells (Bio-Rad, Hercules, CA, USA) for 22 h in 0.5 × TBE at 240 V and 17 °C, and the gels were stained using the method described by Byun et al. [20].
2.4. Sequencing of Allelic Variants and Sequence Analyses
For those amplicons that were deemed to be homozygous by PCR-SSCP analysis, DNA sequencing was performed directly and in both directions at the Beijing Genomics Institute, Beijing, China. For the variants that were typically rarer, and only found in a heterozygous form, DNA sequencing was undertaken using an approach described by Gong et al. [21]. In this approach, a band from the PCR-SSCP gel corresponding to the rare variant was excised, macerated, and then used to provide a DNA template for re-amplification with the original primers. After confirming that only this band had been amplified with a second round of SSCP analysis, this amplicon was sequenced as above. The BLAST algorithm was used to search the NCBI GenBank (http://www.ncbi.nlm.nih.gov/) databases for similar sequences.
DNAMAN version 5.2.10 (Lynnon BioSoft, Vaudreuil, QC, Canada) was used for the translation of open reading frames to amino acid sequences, the alignment of DNA sequences and amino acid sequences, and the creation of phylogenetic trees.
2.5. Statistical Analyses
The statistical analyses were performed using IBM SPSS Statistics version 24.0 (IBM, NY, USA). The General Linear Mixed-Effects Models (GLMMs) option was used to ascertain the effect of KRTAP24-1 genotype on the measured cashmere traits. Due to the multiple comparisons undertaken in these models, a Bonferroni correction was applied. Sire and gender were found to affect (p < 0.05) all the cashmere fibre traits, and, accordingly, they were included in the models (as random and fixed factors, respectively). Only main effects were tested, and associations were considered significant at the 5% level.
3. Results
3.1. Identification of Caprine KRTAP24-1
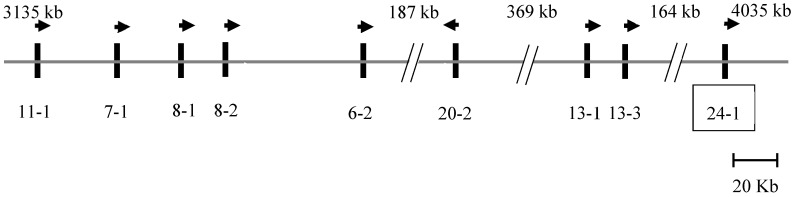
A BLAST search of the Caprine Genome Assembly GCF_001704415.1 using a sheep KRTAP24-1 sequence (JX112014) revealed a homologous region on goat chromosome 1, and an open reading frame of 774 bp was found at position 4878528_4879301 in the caprine genome sequence NC_022293.1. Eight previously described KRTAPs were identified upstream of this region [7], and in order from the centromere to the telomere, these were KRTAP11-1, KRTAP7-1, KRTAP8-1, KRTAP8-2, KRTAP6-2, KRTAP20-2, KRTAP13-1, and KRTAP13-3 (Figure 1).
Figure 1.
Location of KTRTAPs on caprine chromosome 1. The putative KRTAP24-1 (boxed) is clustered with eight previously identified KRTAPs [7]. The vertical bars represent the genes, and the arrows indicate the direction of transcription of these genes. The names of the genes are written below the bars (e.g., 11-1 represents KRTAP11-1). The spacing of the genes is only approximate and is based on the Caprine Genome Assembly. The nucleotide coordinates are given relative to GCF_001704415.1 [19] and are approximate. A pair of diagonal lines indicate that the spacing is not proportional to the length of the sequence, with the distance being shown above.
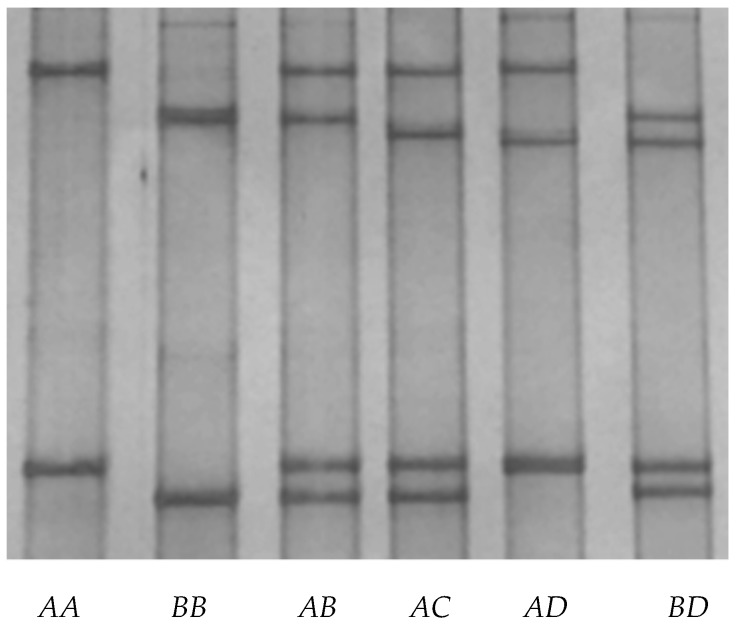
Four unique PCR-SSCP banding patterns (named A, B, C, and D) were detected for caprine KRTAP24-1. Either one or a combination of two different patterns was observed for each goat analyzed, these representing homozygous and heterozygous animals, respectively (Figure 2). DNA sequencing of the amplicons that produced these patterns revealed four unique nucleotide sequences, and while all of these sequences were different, they still had a similarity of over 99% with the putative KRTAP24-1 sequence (NC_022293.1) in the caprine genome assembly.
Figure 2.
Variation in the caprine KAP24-1 gene detected by PCR single-strand conformation polymorphism analysis. Four unique banding patterns representing four variants (A to D) were observed in either homozygous or heterozygous forms.
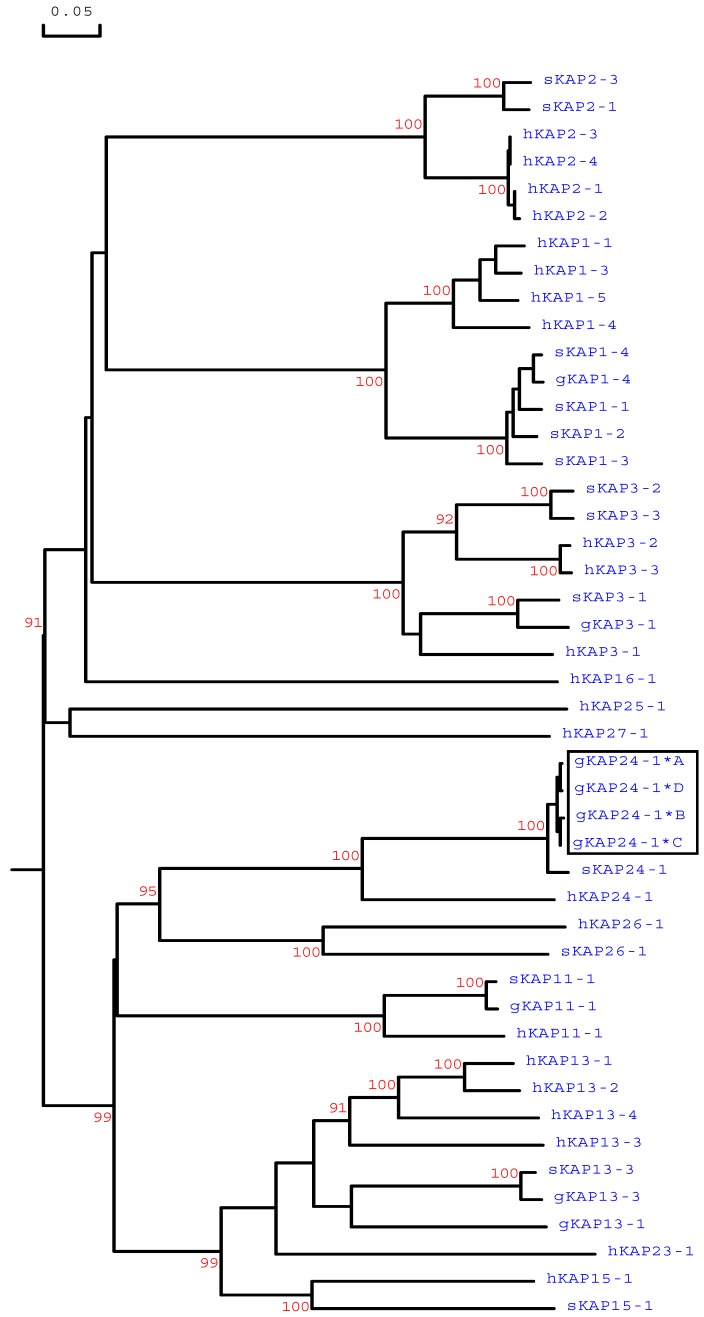
Phylogenetic analysis revealed that these caprine sequences were different to all of the caprine sequences identified to date but were most closely related to the KRTAP24-1 sequences from sheep and humans (Figure 3). This suggests that these newly identified sequences were caprine orthologous variants of KRTAP24-1. These variant sequences were deposited into GenBank with the accession numbers MG996011−MG996014.
Figure 3.
Phylogenetic tree of high-sulphur (HS)-KAPs identified in sheep, humans, and goats. Amino acid sequences or predicted amino acid sequences were used to construct the tree. In the tree, the numbers printed at the forks indicate the bootstrap confidence values. Only those values equal to or higher than 90% are shown. The sheep sequences are indicated with “s”, the human sequences are indicated with “h”, and the caprine KAPs are indicated with a prefix “g”. The four newly identified goat KAP24-1 sequences are shown in a box, and the GenBank/EMBL accession numbers for other HS-KAPs are JN012101.1 (gKAP1-4), NM_001285774 (gKAP3-1), NM_001285767.1 (gKAP11-1), AY510115 (gKAP13-1), JX426138 (gKAP13-3), X01610 (sKAP1-1 and sKAP1-4), HQ897973 (sKAP1-2), X02925 (sKAP1-3), P02443 (sKAP2-1), P02441 (sKAP2-3), P02446 (sKAP3-1), P02444 (sKAP3-2), P02445 (sKAP3-3), HQ595347 (sKAP11-1), JN377429 (sKAP13-3), KX817979 (sKAP15-1), JX112014 (sKAP24-1), KX644903 (sKAP26-1), NM_030967.2 (hKAP1-1), NM_030966.1 (hKAP1-3), NM_001257305.1 (hKAP1-4), NM_031957.1 (hKAP1-5), NM_001123387.1 (hKAP2-1), NM_033032.2 (hKAP2-2), NM_001165252.1 (hKAP2-3), NM_033184.3 (hKAP2-4), NM_031958.1 (hKAP3-1), NM_031959.2 (hKAP3-2), NM_033185.2 (hKAP3-3), NM_175858.2 (hKAP11-1), NM_181599.2 (hKAP13-1), NM_181621.3 (hKAP13-2), NM_181622.1 (hKAP13-3), NM_181600.1 (hKAP13-4), NM_181623.1 (hKAP15-1), NM_001146182.1 (hKAP16-1), NM_181624.1 (hKAP23-1), NM_001085455.2 (hKAP24-1), NM_001128598.1 (hKAP25-1), NM_203405.1 (hKAP26-1), and NM_001077711.1 (hKAP27-1).
The protein that might be encoded by caprine KRTAP24-1 would comprise 257 amino acids and would contain 8.6–9.0 mol.% of cysteine, 15.6 mol.% of serine, and 7.0 mol.% of tyrosine.
3.2. Variation in Caprine KRTAP24-1
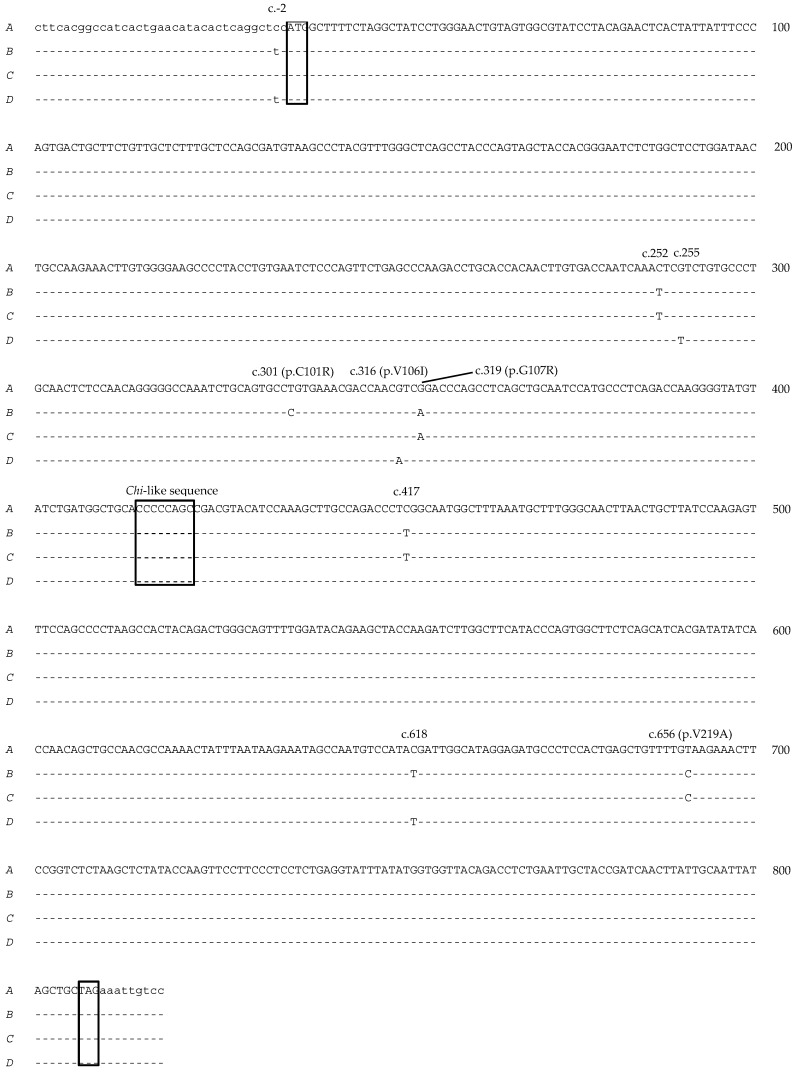
Nine single-nucleotide polymorphisms (SNPs) were detected across the caprine variant sequences. Of these SNPs, eight were within the coding region, and four of them would result in amino acid changes (Figure 4). It is notable that there was a C/T SNP located two nucleotides upstream from the predicted ATG codon. In the regions around this position and at c.618, variants A and C were identical, while B and D were identical. However, in the regions around c.252 and c.656 and the region spanning c.319 to c.417, A and D were identical, and B and C were identical (Figure 4).
Figure 4.
Nucleotide sequences of caprine KRTAP24-1 variants A–D. The sequences exclude the PCR primer binding regions. Nucleotides in the coding region are shown in uppercase, while those outside the coding region are in lowercase. A putative Chi-like sequence (c.380 to c.387) and the predicted ATG start codon and TAG stop codon are boxed. Dashes signify homology with the KRTAP24-1 variant A sequence. The single-nucleotide polymorphism (SNP) positions are shown above the sequences, and those that would result in amino acid changes are indicated. The numbering of nucleotides and amino acids follows the HGVS nomenclature guidelines [22].
3.3. Association between Variation in KRTAP24-1 and Cashmere Traits
Six genotypes (AA, AB, BB, AC, AD, and BD) were found in the cashmere goats investigated. Their frequencies were 32.06%, 47.06%, 17.65%, 1.18%, 0.88%, and 1.18% for AA, AB, BB, AC, AD, and BD, respectively. AC, AD, and BD were each detected at a frequency of less than 5%; hence, the association of these genotypes with cashmere traits was not investigated given this low frequency and potential for bias. Associations were accordingly only tested for the three common genotypes: AA, AB, and BB.
Genotype was found to have an effect on cashmere MFD. Cashmere produced by AA goats had the lowest MFD, while cashmere produced by BB goats had the highest MFD. Cashmere produced by AB heterozygote goats had an intermediate MFD, but it was significantly different to the MFDs of goats with the AA and BB genotypes (Table 1). No associations were found between the genotypes and cashmere weight or crimped fibre length.
Table 1.
The effect of KRTAP24-1 genotype on various cashmere fibre traits (mean ± SE) 1 in Longdong cashmere goats.
| Cashmere Trait (Unit) | Mean ± SE | p Value | ||
|---|---|---|---|---|
| AA (n = 109) | AB (n = 160) | BB (n = 60) | ||
| Cashmere weight (g) | 414 ± 4.6 | 418 ± 3.8 | 411 ± 5.4 | 0.457 |
| Mean fibre diameter (μm) | 13.4 ± 0.04 C | 13.6 ± 0.04 B | 13.8 ± 0.05 A | <0.001 |
| Crimped fibre length (cm) | 4.2 ± 0.05 | 4.3 ± 0.04 | 4.2 ± 0.06 | 0.301 |
1 Estimated marginal means and standard errors (SEs) derived from General Linear Models. In these models, “sire” was fitted as a random factor and “gender” as a fixed factor. A Bonferroni correction was used to correct for multiple comparisons made in the models. Means within rows that do not share a superscript letter (e.g., A, B, or C) are different at p < 0.01 and shown in bold.
4. Discussion
This study describes the identification of a new caprine KAP gene called KRTAP24-1 that encodes an HS-KAP protein. The gene was clustered with eight previously described KAP genes including three HS- and five HGT-KAP genes on goat chromosome 1. It exhibited the highest similarity with the KRTAP24-1 sequence from sheep and human when compared to any previously described HS-KAP sequences from goat, sheep, and human. It was concluded that the newly identified gene represents the orthologous caprine KRTAP24-1.
While the putative protein encoded by caprine KRTAP24-1 could be assigned into the HS-KAP group, the caprine KAP24-1 protein would contain a much lower content of cysteine compared to any other HS-KAP. In contrast, this protein would have a relatively high content of serine and tyrosine. Cysteine residues are thought to form disulfide bonds that cross-link with the IFs [23], whereas tyrosine residues in HGT-KAPs may regulate the arrangement of IFs via cation–π interactions [24]. It has also been suggested that with ovine KAP11-1 and KAP13-3, the serine residues may be phosphorylated [25,26] and that phosphorylation might therefore affect the assembly and organisation of the keratins [27]. Having a lower content of cysteine and a higher content of serine and tyrosine is not common in HS-KAPs. This phenomenon has been described previously for KAP24-1 in other species (human and sheep) [16,17] and for KAP11-1 and KAP13-3 in sheep [25,26]. The functional significance of this unusual amino acid composition is unknown.
The amount of variation found in the caprine KRTAP24-1 sequence is comparable to that reported for ovine KRTAP24-1 [17]. It is also consistent with what has been observed with other KRTAPs in goats [7,8,28] and sheep [29,30]. There is some evidence that gene conversion events or non-reciprocal genetic exchange may have occurred with these KRTAP24-1 sequences, as similarities are observed with what has been described to underpin the nucleotide variation observed in KRTAP1-n and KRTAP15-1, the suggestion being that it has been driven by the activity of Chi-like sequences [31,32]. Chi (crossover hotspot instigator, χ) is an octamer sequence (5′-GCTGGTGG-3′) associated with recombination hotspots in E. coli and is where recombination is initiated by double-strand DNA breaks [33]. Variants of this Chi-motif are suggested to have partial recombinogenic activity, and analysis of the KRTAP24-1 sequences reveals a Chi-like sequence from c.380 to c.387 (5′-CCCCCAGC-3′, reverse complementary to 5-GCTGGGGG-3′). Together, this suggests that further research is needed into the evolutionary origins of the KAP genes.
The observation that there were more AA and AB genotypes and fewer BB genotypes in Longdong cashmere goats appears to be consistent with the association results obtained here. Genotypes AA and AB were found to be associated with finer cashmere fibre than genotype BB. This is consistent with this trait been selected for in Longdong cashmere goats, leading to AA and AB becoming more common in the population.
The KAP genes are clustered together by chromosomal region, and research in sheep has revealed that all of the KAP genes identified to date are polymorphic [2,4,5]. This raises the possibility that the associations described here for KRTAP24-1 may be due to linkage with other KRTAPs clustered in the same chromosome region. Eight other KRTAPs have been found to be located upstream from KRTAP24-1 on goat chromosome 1, and these are KRTAP13-3, KRTAP13-1, KAP20-2, KRTAP6-2, KRTAP8-2, KRTAP8-1, and KRTAP11-1 [7]. The association detected for KRTAP24-1 is similar to that reported for KRTAP8-2 [9] but is different to that described for KRTAP20-2 [7] and KRTAP13-1 [10]. Given that KRTAP24-1 is physically closer to KRTAP20-2 and KRTAP13-1 than KRTAP8-2 on the chromosome (Figure 1), it would suggest that the association obtained here is not because of the effect of the other nearby KRTAPs but instead represents the independent effect of KRTAP24-1.
Variation in caprine KRTAP24-1 may affect cashmere traits in different ways. Firstly, a number of the SNPs that were found were non-synonymous and would result in amino acid changes in the putative KAP24-1 protein. These amino acid changes may affect the protein structure, its property, or its interaction with IFs, and consequently affect fibre traits. There were three non-synonymous SNPs (c.301T/C, c.319G/A, and c.656T/C) when comparing variants A and B for which a difference in MFD was detected here. Two of these SNPs are worthy of attention. SNP c.301T/C would result in gain or loss of cysteine, whereas SNP c.319G/A would result in gain or loss of glycine. Cysteine is usually the first-limiting amino acid for wool or cashmere fibre synthesis [34] and is essential for the formation of disulphide bonds between KAPs and IFs, whereas glycine is a small residue that may make the KAP protein more flexible and thus better able to form a compact structure with IFs. It is logical that the gain of cysteine and glycine for variant A compared to B may favour KAP24-1 forming a more compact structure with the IFs, and thus lead to a finer fibre, this being consistent with the association results. However, as described below, these SNPs may simply be linked to other genetic variation that might affect the expression of KRTAP24-1.
Synonymous SNPs can affect gene expression or protein structure. It has been suggested that “silent” variation can affect in vivo protein folding and, consequently, protein function by creating less common codons in mRNA, which slow the rate of translation and protein formation, leading to structural changes [35,36].
Finally, SNPs near to ATG start codons may influence the efficacy of translation and, thus, gene expression. One SNP was located two base pairs upstream of the start codon, with the nucleotide being a cytosine (C) in variants A and C but being substituted to a thymine (T) in variants B and D (Figure 4). In eukaryotes, the ribosome requires a specific sequence on a mRNA molecule as the translational start site, with gcc(A/G)ccAUGG from c.−6 to c.4 being proposed as the optimal sequence for initiation of translation [37], where a lowercase letter denotes the most common base at a position where the base can, nevertheless, vary. Uppercase letters indicate highly conserved bases, and the typical AUG initiator codon is underlined. While a cytosine at c.−2 is not absolutely conserved, it can contribute to the overall strength of ribosome binding [38]. The substitution of cytosine (C) to thymine (T) seen in variants B and D may therefore affect the translation of the KRTAP24-1 mRNA, and, hence, less KAP24-1 protein is produced to cross-link the IFs. This may lead to a less compact fibre being produced and, hence, an association with high MFD, as was described for variant B when compared to variant A.
Further investigation is needed to reveal how variation in KRTAP24-1 may affect cashmere traits. Nevertheless, the association found in the study suggests that KRTAP24-1 could be a gene marker for fine cashmere fibre.
5. Conclusions
This study identified the caprine KAP24-1 gene and revealed variation in the gene. Variation in the caprine KAP24-1 gene was found to affect cashmere fibre diameter. These results may be useful in the future development of breeding programs based on improving cashmere fibre diameter.
Author Contributions
Conceptualisation, J.W., H.Z., Y.L. and J.G.H.H.; validation, M.Z. and Z.H.; formal analysis, J.W., H.Z. and H.G.; investigation, J.W., M.Z. and Z.H.; resources, J.W. and Y.L.; data curation, J.W., H.Z. and H.G.; writing—original draft preparation, J.W., H.Z., Y.L., J.H. and J.G.H.H.; writing—review and editing, J.W. and J.G.H.H.; visualisation, J.W.; supervision, J.H.; project administration, J.W. and J.H.; funding acquisition, J.W. and J.H.
Funding
This research was funded by the fund of Discipline construction fund projects of Gansu Agricultural University (GSAU-XKJS-2018-020).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Franck R.R. Silk, Mohair, Cashmere and Other Luxury Fibres. Woodhead Publishing Limited; Cambridge, UK: CRC Press; Boca Raton, FL, USA: Textile Institute; Cambridge, UK: 2001. Appendix 10: Quality Assessment of Goat Hair for Textile Use; pp. 227–233. [Google Scholar]
- 2.Gong H., Zhou H., Forrest R.H., Li S., Wang J., Dyer J.M., Luo Y., Hickford J.G. Wool keratin-associated protein genes in Sheep-A Review. Genes. 2016;7:24. doi: 10.3390/genes7060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong H., Zhou H., McKenzie G.W., Yu Z., Clerens S., Dyer J.M., Plowman J.E., Wright M.W., Arora R., Bawden C.S., et al. An updated nomenclature for keratin-associated proteins (KAPs) Int. J. Biol. Sci. 2012;8:258–264. doi: 10.7150/ijbs.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S., Zhou H., Gong H., Zhao F., Hu J., Luo Y., Hickford J.G.H. Identification of the ovine keratin-associated protein 26-1 gene and its association with variation in wool traits. Genes. 2017;8:225. doi: 10.3390/genes8090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Luo Y., Hickford J.G. Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits. Genes. 2017;8:27. doi: 10.3390/genes8010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parris D., Swart L.S. Studies on the high-sulphur proteins of reduced mohair. The isolation and amino acid sequence of protein scmkb-m1.2. Biochem. J. 1975;145:459–467. doi: 10.1042/bj1450459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Che L., Hickford J.G., Zhou H., Hao Z., Luo Y., Hu J., Liu X., Li S. Identification of the caprine keratin-associated protein 20-2 (KAP20-2) gene and its effect on cashmere traits. Genes. 2017;8:328. doi: 10.3390/genes8110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Hao Z., Zhou H., Luo Y., Hu J., Liu X., Li S., Hickford J.G.H. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small. Ruminant. Res. 2018;167:104–109. doi: 10.1016/j.smallrumres.2018.08.014. [DOI] [Google Scholar]
- 9.Liu H., Li N., Jia C., Zhu X., Jia Z. Effect of the polymorphisms of keratin associated protein 8.2 gene on fibre traits in Inner Mongolia cashmere goats. Asian Australas. J. Anim. Sci. 2007;20:821–826. doi: 10.5713/ajas.2007.821. [DOI] [Google Scholar]
- 10.Fang Y., Liu W.J., Zhang F.Q., Shao Y.G., Yu S.G. The polymorphism of a novel mutation of KAP13.1 gene and its associations with cashmere traits on Xinjiang local goat breed in China. Asian. J. Anim. Vet. Adv. 2010;5:34–42. [Google Scholar]
- 11.Gong H., Zhou H., Hodge S., Dyer J.M., Hickford J.G.H. Association of wool traits with variation in the ovine KAP1-2 gene in Merino cross lambs. Small. Ruminant. Res. 2015;124:24–29. doi: 10.1016/j.smallrumres.2015.01.009. [DOI] [Google Scholar]
- 12.Zhou H., Gong H., Li S., Luo Y., Hickford J.G.H. A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter. J. Anim. Breed. Genet. 2015;132:301–307. doi: 10.1111/jbg.12138. [DOI] [PubMed] [Google Scholar]
- 13.Tao J., Zhou H., Gong H., Yang Z., Ma Q., Cheng L., Ding W., Li Y., Hickford J.G. Variation in the KAP6-1 gene in Chinese Tan sheep and associations with variation in wool traits. Small. Ruminant. Res. 2015;154:129–132. doi: 10.1016/j.smallrumres.2017.08.001. [DOI] [Google Scholar]
- 14.Li S., Zhou H., Gong H., Zhao F., Wang J., Luo Y., Hickford J.G. Variation in the ovine KAP6-3 gene (KRTAP6-3) is associated with variation in mean fibre diameter-associated wool traits. Genes. 2017;8:204. doi: 10.3390/genes8080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao J., Zhou H., Yang Z., Gong H., Ma Q., Ding W., Li Y., Hickford J.G.H. Variation in the KAP8-2 gene affects crimping and grow rate of wool in Chinese Tan sheep. Small. Ruminant. Res. 2017;149:77–80. doi: 10.1016/j.smallrumres.2017.01.001. [DOI] [Google Scholar]
- 16.Rogers M.A., Winter H., Langbein L., Wollschläger A., Praetzel-Wunder S., Jave-Suarez L.F., Schweizer J. Characterization of human KAP24.1, a cuticular hair keratin-associated protein with unusual amino-acid composition and repeat structure. J. Invest. Dermatol. 2007;127:1197–1204. doi: 10.1038/sj.jid.5700702. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H., Gong H., Yan W., Luo Y., Hickford J.G.H. Identification and sequence analysis of the keratin-associated protein 24-1 (KAP24-1) gene homologue in sheep. Gene. 2012;511:62–65. doi: 10.1016/j.gene.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H., Hickford J.G., Fang Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006;354:159–161. doi: 10.1016/j.ab.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 19.GCF_001704415.1. [(accessed on 12 October 2017)]; Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_001704415.1.
- 20.Byun S.O., Fang Q., Zhou H., Hickford J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009;385:174–175. doi: 10.1016/j.ab.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Gong H., Zhou H., Hickford J.G. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011;38:31–35. doi: 10.1007/s11033-010-0074-6. [DOI] [PubMed] [Google Scholar]
- 22.Sequence Variant Nomenclature. [(accessed on 12 October 2017)]; Available online: http://varnomen.hgvs.org/
- 23.Powell B.C., Rogers G.E. The Role of Keratin Proteins and Their Genes in the Growth, Structure and Properties of Hair. In: Jollès P., Zahn H., Höcker H., editors. Formation and Structure of Human Hair. Birkhäuser Verlag; Basel, Switzerland: 1997. pp. 59–148. [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga R., Abe R., Ishii D., Watanabe S., Kiyoshi M., Nöcker B., Tsuchiya M., Tsumoto K. Bidirectional binding property of high glycine-tyrosine keratin-associated protein contributes to the mechanical strength and shape of hair. J. Struct. Biol. 2013;183:484–494. doi: 10.1016/j.jsb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Gong H., Zhou H., Dyer J.M., Hickford J.G. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011;38:5429–5433. doi: 10.1007/s11033-011-0697-2. [DOI] [PubMed] [Google Scholar]
- 26.Gong H., Zhou H., Dyer J.M., Plowman J.E., Hickford J.G.H. Identification of the keratin-associated protein 13-3 (KAP13-3) gene in sheep. Open J. Genet. 2011;1:60–64. doi: 10.4236/ojgen.2011.13011. [DOI] [Google Scholar]
- 27.Ku N.O., Liao J., Chou C.F., Omary M.B. Implications of intermediate filament protein phosphorylation. Cancer. Metastasis. Rev. 1996;15:429–444. doi: 10.1007/BF00054011. [DOI] [PubMed] [Google Scholar]
- 28.Li M., Liu X., Wang J., Li S., Luo Y. Molecular characterization of caprine KRTAP13-3 in Liaoning cashmere goat in China. J. Appl. Anim. Res. 2014;42:140–144. doi: 10.1080/09712119.2013.822813. [DOI] [Google Scholar]
- 29.Gong H., Zhou H., McKenzie G.W., Hickford J.G., Yu Z., Clerens S., Dyer J.M., Plowman J.E. Emerging issues with the current keratin-associated protein nomenclature. Int. J. Trichol. 2010;2:104–105. doi: 10.4103/0974-7753.77519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Gong H., Wang J., Dyer J.M., Luo Y., Hickford J.G. Identification of four new gene members of the KAP6 gene family in sheep. Sci. Rep. 2016;6:24074. doi: 10.1038/srep24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers G.R., Hickford J.G.H., Bickerstaffe R. Polymorphisms in two genes for B2 high sulfur proteins of wool. Anim. Genet. 1994;25:407–415. doi: 10.1111/j.1365-2052.1994.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Zhou H., Zhu J., Hu J., Liu X., Li S., Luo Y., Hickford J.G. Identification of the ovine keratin-associated protein 15-1 gene (KRTAP15-1) and genetic variation in its coding sequence. Small. Ruminant. Res. 2017;153:131–136. doi: 10.1016/j.smallrumres.2017.06.007. [DOI] [Google Scholar]
- 33.Smith G.R. Homologous recombination near and far from DNA breaks: Alternative roles and contrasting views. Annu. Rev. Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 34.Reis P.J., Tunks D.A., Munro S.G. Effects of the infusion of amino acids into the abomasum of sheep, with emphasis on the relative value of methionine, cysteine and homocysteine for wool growth. J. Agr. Sci. 1990;114:59–68. doi: 10.1017/S0021859600071008. [DOI] [Google Scholar]
- 35.Duan J., Wainwright M.S., Comeron J.M., Saitou N., Sanders A.R., Gelernter J., Gejman P.V. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 36.Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 38.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]