Abstract
MicroRNAs (miRNAs) act an important role in the progression of tumor. In this study, we showed that the serum expression of miR-365 was downregulated in the glioblastoma compared with in the healthy controls. We also demonstrated that miR-365 expression was downregulated in glioblastoma tissues compared with the adjacent normal tissues. Overexpression of miR-365 suppressed the glioblastoma cell proliferation and migration. Moreover, ectopic expression of miR-365 promoted the expression of Ecadherin while inhibited the expression of N-cadherin and Vimentin in U87 cell. Furthermore, we identified PAX6 as a direct target gene of miR-365 in U87 cell. Overexpression of miR-365 suppressed glioblastoma cell proliferation and migration and epithelial-to-mesenchymal transition through inhibiting PAX6 expression. These results suggested that miR-365 played a tumor suppressor in glioblastoma.
Keywords: Glioblastoma, microRNAs, miR-365, PAX6
Introduction
Gliomas are highly aggressive brain tumor with a high mortality and relapse rate [1-5]. Gliomas comprise of six subtypes including anaplastic astrocytoma, glioblastoma, anaplastic oligodendroglioma, astrocytoma, and malignant glioma [6-8]. Unfortunately, the prognosis of these tumors is not significantly improved during the last four decades because of no effective therapies [8-11]. Therefore, it is useful to search new early detection markers to improve diagnosis and treatment of gliomas.
MicroRNAs (miRNAs) are a class of non-coding, typically 22 nucleotides, and single-stranded and endogenous RNAs molecules [12-14]. MiRNAs play important role in the regulation of gene expression by binding to the 3’UTR (3’untranslated region) of mRNA and inhibiting mRNA expression or/and inducing mRNA degradation [15-17]. Increasing evidences have demonstrated that miRNAs are involved in many physiological processes including cell development, differentiation, migration and proliferation [18-20]. Recent research has indicated that deregulated miRNAs can be found in many tumors such as gastric, breast, ovarian, lung and bladder cancer and are involved in the development of these cancers [21-25]. In addition, cell-free, highly stable miRNAs have been identified in human serum [1,16,26]. Previous studies indicated that serum miRNAs might be a novel non-invasive diagnostic maker for cancers [26-28]. Previous studies suggested that miR-365 acted crucial roles in the development of tumors [29-32]. For instance, Nie et al. [33]. reported that miR-365 expression was downregulated in colon cancer samples compared to non-neoplastic mucosa tissues. Overexpression of miR-365 inhibited colon cell cycle progression, promoted cell apoptosis and suppressed tumorigenicity in the colon cancer cell via targeting Bcl-2 and Cyclin D1 expression. Sun et al. [34]. Showed that miR-365 expression was downregulated in non-small cell lung cancer (NSCLC) than that in normal tissue. Bai et al. [30]. also found that the expression of miR-365 was downregulated in malignant melanoma cell lines and tissues. Ectopic expression of miR-365 inhibited malignant melanoma cell proliferation and metastasis via regulating NRP1 expression. Recently, Liu et al. [35]. showed that the serum expression of miR-365 was downregulated in the NSCLC patients compared to that in healthy serum volunteers. However, the expression and role of miR-365 in the glioblastoma were still unknown.
In this study, we demonstrated that miR-365 expression was downregulated in the serum of glioblastoma compared to the healthy controls. Moreover, miR-365 expression was also lower in the in the glioblastoma tissue compared with the adjacent normal tissues. Overexpression of miR-365 suppressed the glioblastoma cell proliferation, migration and epithelial-to-mesenchymal transition (EMT).
Materials and methods
Sample and tissue collection
This research was approved by the ethics committee of The 2nd Affiliated Hospital, Harbin Medical University and all samples were contained from glioblastoma’s patients or control with written informed consents. Serum samples or tissues were collected from glioblastoma’s patients and healthy volunteers at our department between 2012 and 2014. None of the glioblastoma patients were received radiotherapy or chemotherapy before surgery.
qRT-PCR
Total RNA was isolated from serums, tissuse and cells using Trizol (Invitrogen, USA) following to instruction’s information. The expression of miR-365 was measured using qRT-PCR and qRT-PCR was performed on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, USA). The expression level of miR-365 was normalized to the endogenous snRNA U6. GAPDH was used to as the control for mRNA expression. The primers sequences were shown following: GAPDH forward, 5’-ACAACT TTGGTATCGTGGAAGG-3’, and reverse, 5’-GCCATCACGCCA CAGTTTC-3’; PAX6 forward, 5’-AACGATAACATACCAAGC GTGT-3’, and reverse, 5’-GGTCTGCCCGTTCAACATC-3’; Vimentin Forword: 5’-GAAGAGGTTAGTGGAGTGA-3’, Reverse: 5’-TGCTGTTCCTGAATCTGA-3’.
Cell lines cultured and tranfection
Four glioblastoma cell lines (U373, U87, A172 and U251) and one normal astrocyte line (NHAs) were collected from the American Tissue Culture Colection (ATCC) and was cultured in DMEM medium. miR-365 mimics and its scramble oligonucleotides were obtained from RiboBio (Guangzhou, China). Cells were transfected with the miR-365 mimic and scramble mimic, pcDNA-PAX6 and control using Lipofectamine 2000 (Invitrogen, USA) following to the manufacturer’s information.
Cell growth and migration assay
Cells were cultured in the 96-well plate and the cells proliferation ratio was measured with CCK-8 (Cell Counting Kit-8, Dojindo, Japan) following to the manufacturer’s information. Wound healing assay was performed to detect the cell migration. The wound was made using the Petri dishbottom. Cells were continued to culture for 48 hours and the migration rate was detected through measuring the wound distance.
Western blot
Total protein was extracted from cells in lysis buffer. Proteins were separated using electrophoresis on SDS-PAGE gels and transferred to nitrocellulose membranes (Pall Corp, NY, USA). The following antibodies were used as follows: PAX6, GAPDH, E-cadherin, N-cadherin and Vimentin (dilutions 1:1000, Abcam). Protein was visualized by an enhanced chemiluminescence (Amersham, UK).
Statistical analysis
Data was shown as mean ± SD (standard deviation). The differences between two groups were measured using Student’s t test and one-way ANOVA was performed to measure the differences between more than two groups. P < 0.05 was defined to be statistically significant.
Result
Serum expression level of miR-365 was downregulated in the glioblastoma
We firstly measure the serum expression level of miR-365 in glioblastoma and healthy controls. The serum expression of miR-365 was downregulated in glioblastoma compared with the healthy controls (Figure 1).
Figure 1.
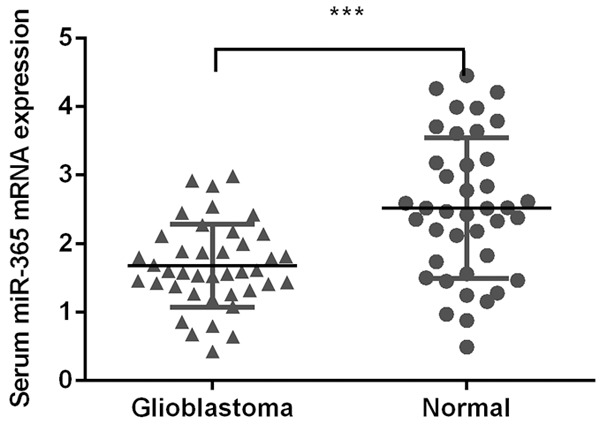
Serum expression level of miR-365 was downregulated in the glioblastoma. The serum expression of miR-365 in the glioblastoma and healthy controls was detected using qRT-PCR. ***P < 0.001. Statistical analysis was performed using Mann-Whitney U tests.
Expression level of miR-365 was decreased in glioblastoma tissue
The expression of miR-365 was downregulated in glioblastoma tissue compared to the adjacent normal tissues (Figure 2A). Among these patients, miR-365 expression was downregulated in 31 cases (31/40; 77%) compared with adjacent tissues (Figure 2B). In addition, the expression of miR-365 in glioblastoma tissue was positively associated with the expression of miR-365 in the serum of glioblastoma patients (Figure 2C).
Figure 2.
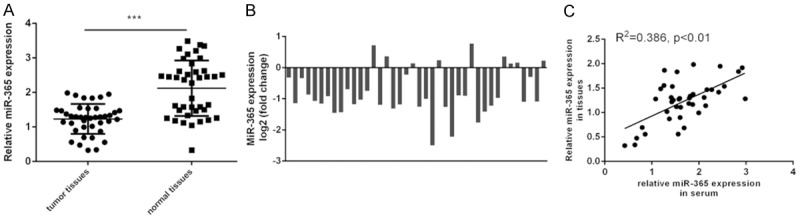
Expressionl level of miR-365 was decreased in the glioblastoma tissue. A. The expression of miR-365 was downregulated in the glioblastoma tissue compared to the adjacent normal tissues. Statistically significant difference was determined using Student’s t test. B. The miR-365 expression in 31 cases (31/40; 77%) compared to adjacent tissues. C. The expression of miR-365 in glioblastoma tissue was associated with those in glioblastoma patients’ serum. ***P < 0.001. Association between miR-365 expression in serum and matched glioma tissues was detected by Spearman correlation test.
miR-365 suppressed the glioblastoma cell proliferation and migration
miR-365 expression was downregulated in the glioblastoma cell lines (U373, U87, A172 and U251) compared with one normal astrocyte line (NHAs) (Figure 3A). The U87 cell was transfected with miR-365 mimic, qRT-PCR data showed that miR-365 mimic promoted the expression of miR-365 in the U87 cell (Figure 3B). Overexpression of miR-365 inhibited the U87 cell proliferation (Figure 3C). Moreover, miR-365 overexpression suppressed U87 cell migration (Figure 3D and 3E).
Figure 3.
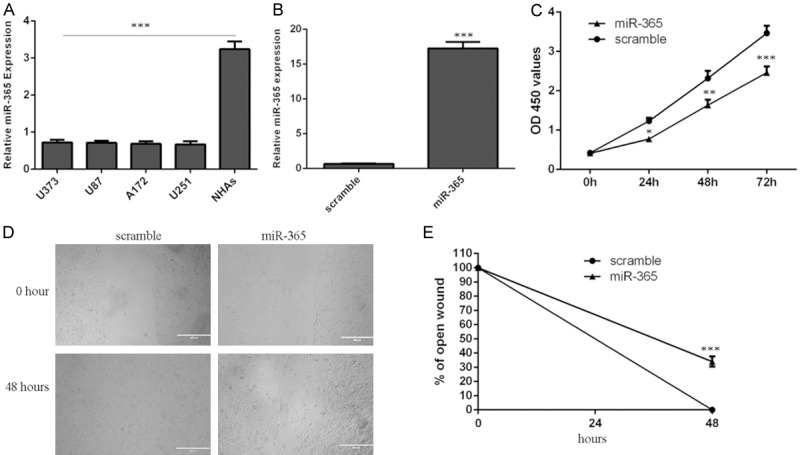
miR-365 suppressed the glioblastoma cell proliferation and migration. A. The expression of miR-365 in the glioblastoma cell lines (U373, U87, A172 and U251) and one normal astrocyte line (NHAs) was detecte using qRT-PCR. B. The expression of miR-365 was measured by qRT-PCR in the U87 cell. C. Overexpression of miR-365 suppressed the U87 cell proliferation. D. Overexpression of miR-365 suppressed the U87 cell migration. E. The relative open wound was shown. *P < 0.05, **P < 0.01 and ***P < 0.001. Statistically significant difference was determined using Student’s t test.
miR-365 inhibited the glioblastoma cell epithelial-to-mesenchymal transition
Ectopic expression of miR-365 promoted the mRNA expression of Ecadherin and suppressed the mRNA expression of N-cadherin and Vimentin mRNA in the U87 cell (Figure 4A). In line with this, overexpression of miR-365 increased the Ecadherin protein expression and suppressed the N-cadherin and Vimentin protein expression in the U87 cell (Figures 4B and S1).
Figure 4.

miR-365 inhibited the glioblastoma cell epithelial-to-mesenchymal transition. A. The mRNA expression of Ecadherin, N-cadherin and Vimentin was measured by qRT-PCR. B. The protein expression of Ecadherin, N-cadherin and Vimentin was detected by western blot.
PAX6 was a direct a target gene of miR-365 in the glioblastoma cell
Targetscan was used to found the potential target gene of miR-365. One putative binding sites of the miR-365 in the 3’UTR of PAX6 are shown in Figure 5A. PAX6 expression was upregulated in glioblastoma cell lines (U373, U87, A172 and U251) compared to one normal astrocyte line (NHAs) (Figure 5B). A luciferase reporter assay was conducted to confirm this prediction in U87 cell. The luciferase activity was decreased in the U87 cell co-transfected with miR-365 mimic and PAX6-3’UTR-WT luciferase reporter; however, this effect was abolished in the U87 cell co-transfected with PAX6-3’UTR-MUT luciferase reporter and miR-365 mimic (Figure 5C). Overexpression of miR-365 suppressed the PAX6 expression (Figures 5D, 5E and S2).
Figure 5.
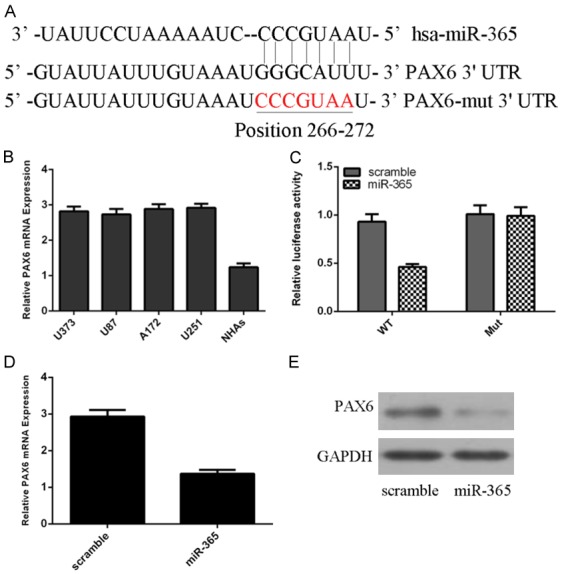
PAX6 was a direct a target gene of miR-365 in the glioblastoma cell. A. One putative binding sites of the miR-365 in the 3’UTR of PAX6 are shown. B. The expression of PAX6 in the glioblastoma cell lines (U373, U87, A172 and U251) and one normal astrocyte line (NHAs) was measured using qRT-PCR. C. The luciferase activity was decreased in the U87 cell co-transfected with miR-365 mimic and PAX6-3’UTR-WT luciferase reporter. D. Ecoptic expression of miR-365 suppressed the PAX6 mRNA expression. E. Overexpression of miR-365 inhibited the protein pression of PAX6.
miR-365 suppressed glioblastoma cell proliferation and migration by inhibiting PAX6
The mRNA expression of PAX6 was upregulated in the U87 cell after treated with pcDNA-PAX6 vector (Figure 6A). The protein expression of PAX6 was also increased in the U87 cell after treated with pcDNA-PAX6 vector (Figure 6B). We restored PAX6 expression through into the miR-365-overexpressing U87 cells by transfecting the pcDNA-PAX6 vector. We demonstrated that PAX6 overexpression promoted the miR-365-overexpressing U87 cell proliferation (Figure 6C). PAX6 Overexpression abrogated the reduction of migration ability caused by ectopic expression of miR-365 in U87 cells (Figure 6D). qRT-PCR and western blot assay demonstrated that epithelial marker (E-cadherin) was decreased, whereas mesenchymal markers (N-cadherin and vimentin) was increased accompanied with restoration of PAX6 (Figures 6E and 6F, S3).
Figure 6.
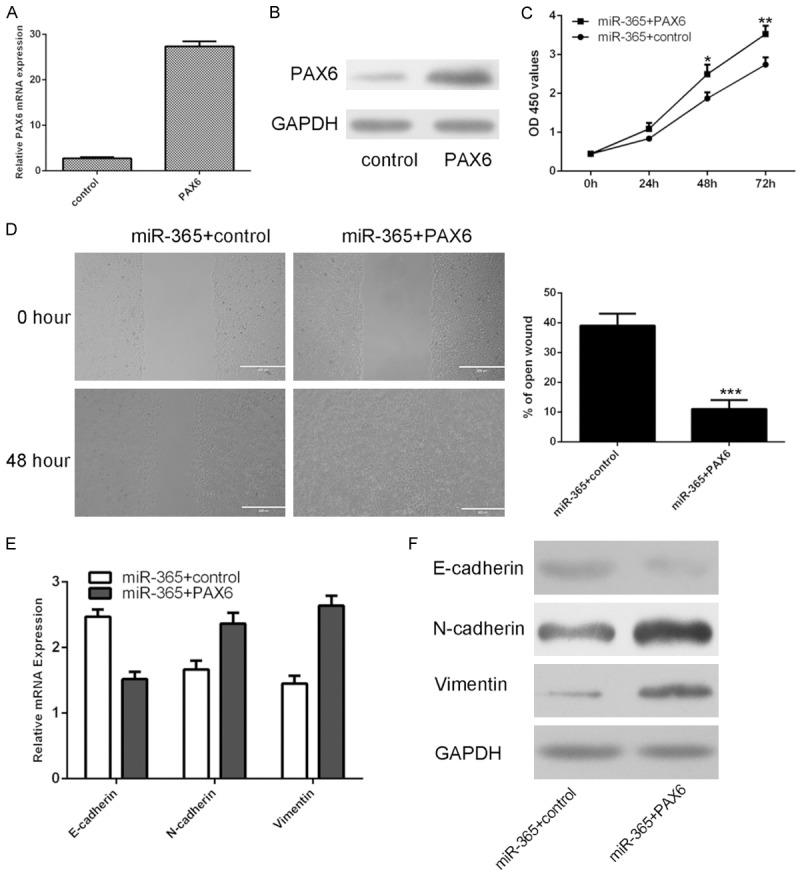
miR-365 suppressed glioblastoma cell proliferation and migration by inhibiting PAX6. A. The mRNA expression of PAX6 was determined by using qRT-PCR. B. The protein expression of PAX6 was detected by using western blot. C. PAX6 overexpression promoted the miR-365-overexpressing U87 cell proliferation. D. PAX6 Overexpression abrogated the reduction of migration ability caused by ectopic expression of miR-365 in U87 cells. The relative migrative wound was shown. E. The mRNA expression of E-cadherin, N-cadherin and vimentin was determined by qRT-PCR. F. The protein expression of E-cadherin, N-cadherin and vimentin was determined by western blot. *P < 0.05, **P < 0.01 and ***P < 0.001. Statistically significant difference was determined using Student’s t test.
Discussion
In this study, we showed that the serum expression of miR-365 was downregulated in the glioblastoma compared with healthy controls. We further measured the expression of miR-365 in glioblastoma tissues. We demonstrated that miR-365 expression was downregulated in the glioblastoma tissue compared with the adjacent normal tissues. Interestingly, we also found that miR-365 expression in glioblastoma tissue was associated with those in glioblastoma patients’ serum. Overexpression of miR-365 inhibited the U87 cell proliferation and migration. Moreover, ectopic expression of miR-365 promoted the expression of Ecadherin and suppressed the expression of N-cadherin and Vimentin in the U87 cell. These results suggested that overexpression of miR-365 inhibited glioblastoma cell epithelial-to-mesenchymal transition. Furthermore, we identified PAX6 as direct target gene of miR-365 in the U87 cell. Overexpression of miR-365 suppressed glioblastoma cell proliferation and migration and epithelial-to-mesenchymal transition by inhibiting PAX6 expression. These results suggested that miR-365 played a tumor suppressor role through inhibiting PAX6 expression in glioblastoma.
Previous studies demonstrated that miR-365 played crucial roles in the initiation and development of cancers [29-32]. For example, Nie et al. [33] Showed that miR-365 expression was downregulated in colon cancer tissues compared with non-neoplastic mucosa tissues. Restored expression of miR-365 suppressed colon cell cycle progression, increased cell apoptosis and inhibited tumorigenicity in the colon cancer cell through targeting Bcl-2 and Cyclin D1 expression. Sun et al. [34]. demonstrated that miR-365 expression was lower in non-small cell lung cancer (NSCLC) than that in normal tissue. Bai et al. [30] also found that miR-365 expression was decreased in malignant melanoma cell lines and tissues. Ectopic expression of miR-365 suppressed malignant melanoma cell proliferation and metastasis through regulating NRP1 expression. However, Zhou et al. [36] demonstrated that miR-365 expression was upregulated in cutaneous squamous cell carcinoma (CSCC) tissues and miR-365 promoted CSCC development through regulating the expression of Nuclear Factor I/B (NFIB). Recently, Liu et al. [35] showed that the serum expression of miR-365 was downregulated in the NSCLC patients compared with that in healthy serum volunteers. However, the expression and role of miR-365 in the glioblastoma were still unknown. In our study, we showed that the serum expression of miR-365 was downregulated in the glioblastoma compared with healthy controls. We further measured the expression of miR-365 in the glioblastoma tissues. We demonstrated that miR-365 expression was downregulated in glioblastoma tissue compared with the adjacent normal tissues. Among these patients, the miR-365 expression was downregulated in 31 cases (31/40; 77%) compared with adjacent tissues. Moreover, the expression of miR-365 in glioblastoma tissue was associated with the expression of miR-365 in glioblastoma patients’ serum. Overexpression of miR-365 inhibited the U87 cell proliferation and migration. Moreover, ectopic expression of miR-365 promoted the expression of Ecadherin and suppressed the expression of N-cadherin and Vimentin in the U87 cell. These results suggested that miR-365 acted as a potential tumor suppressor gene in the development of glioblastoma.
PAX6 is one member of the paired box (PAX) families, which is located on the chromosome 11p13 and plays a crucial role in the development of neuroectodermal epithelial tissues [37-39]. Recent studies showed that PAX6 existed in tumor tissues and acted an important role in the development of cancer [40,41]. For example, Xia et al. [40] demonstrated that PAX6 expression was upregualted in invasive ductal breast cancer and higher expression of PAX6 was associated with poor prognosis in breast cancer patients. Meng et al. [42] showed that miR-335 suppressed cell proliferation, colony formation, cell-cycle progression, and invasion through inhibiting PAX6 expression in breast cancer cells. Luo et al. [43] demonstrated that miR-7 suppressed the cell proliferation and invasion in non-small cell lung cancer cell through targeting PAX6 expression. In line with these data, we showed that PAX6 was a direct a target gene of miR-365 in glioblastoma cell.
In conclusion, we demonstrated that miR-365 expression was downregulated in the glioblastoma compared with healthy controls. The expression of miR-365 was also lower in the in the glioblastoma tissue compared with the adjacent normal tissues. Overexpression of miR-365 suppressed the glioblastoma cell proliferation, migration and EMT partly through targeting PAX6 expression. These results suggested that miR-365 played a tumor suppressor role in glioblastoma.
Acknowledgements
First prize of Heilongjiang Postdoctoral Science Foundation (BS142785). Second prize of China Postdoctoral Science Foundation (2014M561373). Foundation for Returness of Ministry of Education of China. Doctoral Fund of The second affiliated to Harbin Medical University (BS2012-18). The Since Returning Foundation of Heilongjiang Province (LC2013C40).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wei X, Chen D, Lv T, Li G, Qu S. Serum MicroRNA-125b as a potential biomarker for glioma diagnosis. Mol Neurobiol. 2016;53:163–170. doi: 10.1007/s12035-014-8993-1. [DOI] [PubMed] [Google Scholar]
- 2.Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH, Van Haastert RJ, Yousefi A, Mastrobattista E, Storm G, Berezikov E, Cuppen E, Woodle M, Schaapveld RQ, Prevost GP, Griffioen AW, Van Noort PI, Schiffelers RM. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget. 2014;5:6687–6700. doi: 10.18632/oncotarget.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Yang BB. Stress response of glioblastoma cells mediated by miR-17-5p targeting PTEN and the passenger strand miR-17-3p targeting MDM2. Oncotarget. 2012;3:1653–1668. doi: 10.18632/oncotarget.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang CH, Yue J, Pfeffer SR, Fan M, Paulus E, Hosni-Ahmed A, Sims M, Qayyum S, Davidoff AM, Handorf CR, Pfeffer LM. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3) J Biol Chem. 2014;289:25079–25087. doi: 10.1074/jbc.M114.593863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Luo H, Wang S, Chen W, Chen Z, Wang HW, Chen Y, Yang J, Zhang X, Wu W, Zhang SY, Shen S, Dong Q, Zhang Y, Jiang T, Lu D, Zhao S, You Y, Liu N, Wang H. MicroRNA-377 inhibited proliferation and invasion of human glioblastoma cells by directly targeting specificity protein 1. Neuro Oncol. 2014;16:1510–1522. doi: 10.1093/neuonc/nou111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, Jiang T, Chen CC. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang F, Nam S, Brown CE, Zhao R, Starr R, Ma Y, Xie J, Horne DA, Malkas LH, Jove R, Hickey RJ. A novel berbamine derivative inhibits cell viability and induces apoptosis in cancer stem-like cells of human glioblastoma, via up-regulation of miRNA-4284 and JNK/AP-1 signaling. PLoS One. 2014;9:e94443. doi: 10.1371/journal.pone.0094443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Gong X, Purow B, Zhao Z. Uncovering MicroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol. 2012;8:e1002488. doi: 10.1371/journal.pcbi.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setty M, Helmy K, Khan AA, Silber J, Arvey A, Neezen F, Agius P, Huse JT, Holland EC, Leslie CS. Inferring transcriptional and microRNA-mediated regulatory programs in glioblastoma. Mol Syst Biol. 2012;8:605. doi: 10.1038/msb.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao SA, Arimappamagan A, Pandey P, Santosh V, Hegde AS, Chandramouli BA, Somasundaram K. miR-219-5p inhibits receptor tyrosine kinase pathway by targeting EGFR in glioblastoma. PLoS One. 2013;8:e63164. doi: 10.1371/journal.pone.0063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Xie Q, Yan Y, Huang Z, Zhong X, Huang L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology. 2014;34:455–464. doi: 10.1111/neup.12129. [DOI] [PubMed] [Google Scholar]
- 15.Yan W, Zhang W, Sun L, Liu Y, You G, Wang Y, Kang C, You Y, Jiang T. Identification of MMP-9 specific microRNA expression profile as potential targets of anti-invasion therapy in glioblastoma multiforme. Brain Res. 2011;1411:108–115. doi: 10.1016/j.brainres.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Li L, Jiang C. Identification and Evaluation of Serum MicroRNA-29 Family for Glioma Screening. Mol Neurobiol. 2015;52:1540–1546. doi: 10.1007/s12035-014-8937-9. [DOI] [PubMed] [Google Scholar]
- 17.Shang C, Guo Y, Hong Y, Liu YH, Xue YX. MiR-21 up-regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting FASLG. Mol Biol Rep. 2015;42:721–7. doi: 10.1007/s11033-014-3820-3. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Li Z, Chen G, Wu WK. MicroRNA-10b induces vascular muscle cell proliferation through Akt pathway by targeting TIP30. Curr Vasc Pharmacol. 2015;13:679–686. doi: 10.2174/1570161113666150123112751. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J Cell Mol Med. 2016;20:10–6. doi: 10.1111/jcmm.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Yu X, Shen J, Liu Y, Chan MT, Wu WK. MicroRNA dysregulation in rhabdomyosarcoma: a new player enters the game. Cell Prolif. 2015;48:511–516. doi: 10.1111/cpr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, Huang WL, Zeng YX, Shao JY. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–3562. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 23.McMillen BD, Aponte MM, Liu Z, Helenowski IB, Scholtens DM, Buttin BM, Wei JJ. Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod Pathol. 2012;25:1644–1653. doi: 10.1038/modpathol.2012.118. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Chi YL, Wang PY, Wang YQ, Zhang YX, Deng J, Lv CJ, Xie SY. miR-511 and miR-1297 inhibit human lung adenocarcinoma cell proliferation by targeting oncogene TRIB2. PLoS One. 2012;7:e46090. doi: 10.1371/journal.pone.0046090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Zhu Y, Li S, Zheng X, Xie L. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36:62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY, Zhang C. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132:116–127. doi: 10.1002/ijc.27657. [DOI] [PubMed] [Google Scholar]
- 27.Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112(Suppl):1241–1246. doi: 10.1038/bjc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao X, Sun Y, Tang J. Serum miR-21 is a diagnostic and prognostic marker of primary central nervous system lymphoma. Neurol Sci. 2014;35:233–238. doi: 10.1007/s10072-013-1491-9. [DOI] [PubMed] [Google Scholar]
- 29.Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Cancer Biomark. 2015;15:599–608. doi: 10.3233/CBM-150500. [DOI] [PubMed] [Google Scholar]
- 30.Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Int J Clin Exp Pathol. 2015;8:4913–4922. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L, Wang Y, Ou C, Lin Z, Wang J, Liu H, Zhou M, Ding Z. microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int J Biochem Cell Biol. 2015;65:182–191. doi: 10.1016/j.biocel.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Huang Z, Ye Q, Ming Y, Zhang S, Zhao Y, Liu L, Wang Q, Cheng K. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- 33.Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y, Du X, Han W. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2012;33:220–225. doi: 10.1093/carcin/bgr245. [DOI] [PubMed] [Google Scholar]
- 34.Sun R, Liu Z, Ma G, Lv W, Zhao X, Lei G, Xu C. Associations of deregulation of mir-365 and its target mRNA TTF-1 and survival in patients with NSCLC. Int J Clin Exp Pathol. 2015;8:2392–2399. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Zhang G, Li H, Han L, Fu A, Zhang N, Zheng Y. Serum microRNA-365 in combination with its target gene TTF-1 as a non-invasive prognostic marker for non-small cell lung cancer. Biomed Pharmacother. 2015;75:185–190. doi: 10.1016/j.biopha.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Zhou M, Zhou L, Zheng L, Guo L, Wang Y, Liu H, Ou C, Ding Z. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB) PLoS One. 2014;9:e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curto GG, Nieto-Estevez V, Hurtado-Chong A, Valero J, Gomez C, Alonso JR, Weruaga E, Vicario-Abejon C. Pax6 is essential for the maintenance and multi-lineage differentiation of neural stem cells, and for neuronal incorporation into the adult olfactory bulb. Stem Cells Dev. 2014;23:2813–2830. doi: 10.1089/scd.2014.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasamoto Y, Hayashi R, Park SJ, Saito-Adachi M, Suzuki Y, Kawasaki S, Quantock AJ, Nakai K, Tsujikawa M, Nishida K. PAX6 Isoforms, along with reprogramming factors, differentially regulate the induction of cornea-specific genes. Sci Rep. 2016;6:20807. doi: 10.1038/srep20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas MG, Welch C, Stone L, Allan P, Barker RA, White RB. PAX6 expression may be protective against dopaminergic cell loss in Parkinson’s disease. CNS Neurol Disord Drug Targets. 2016;15:73–79. doi: 10.2174/1871527314666150821101757. [DOI] [PubMed] [Google Scholar]
- 40.Xia X, Yin W, Zhang X, Yu X, Wang C, Xu S, Feng W, Yang H. PAX6 overexpression is associated with the poor prognosis of invasive ductal breast cancer. Oncol Lett. 2015;10:1501–1506. doi: 10.3892/ol.2015.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Yang X, Wang J, Liang T, Gu Y, Yang D. Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int J Clin Exp Pathol. 2015;8:11452–11457. [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Y, Zou Q, Liu T, Cai X, Huang Y, Pan J. microRNA-335 inhibits proliferation, cell-cycle progression, colony formation, and invasion via targeting PAX6 in breast cancer cells. Mol Med Rep. 2015;11:379–385. doi: 10.3892/mmr.2014.2684. [DOI] [PubMed] [Google Scholar]
- 43.Luo J, Li H, Zhang C. MicroRNA-7 inhibits the malignant phenotypes of nonsmall cell lung cancer in vitro by targeting Pax6. Mol Med Rep. 2015;12:5443–5448. doi: 10.3892/mmr.2015.4032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


