Abstract
Background: Gastric cancer (GC) is one of the most common diagnosed cancer with poor prognosis. Solute carrier (SLC) family 39 genes encode membrane transport proteins, which control the influx of zinc and may play important roles in human disease including cancer. However, the prognostic value of individual SLC family 39 gene in gastric cancer patients remain unclear. Methods: Genetic alteration frequency and mRNA expression level of SLC family 39 genes in GC were first assessed by using many online databases including cBioportal for Cancer Genomics, Oncomine, UCSC Xena browser and Ualcan database. The prognostic value of individual SLC family 39 gene in GC patients were further investigated via Kaplan-Meier plotter. Results: The analytic results of genetic alteration frequency showed that mRNA deregulation was one of the most important single factors for alteration in different kinds of gastric cancer. Compared with normal gastric tissues, 14 SLC family 39 genes were all significantly upregulated in GC tissue in Ualcan database, and SLC39A4, SLC39A5, SLC39A6, SLC39A10 mRNA expression were also higher in Oncomine database. The survival analysis indicated that most members of SLC family 39 genes were closely related with prognosis of GC patients, SLC39A7, SLC39A11, SLC39A14 were significantly associated with favorable overall survival (OS), the rest of SLC family 39 genes were importantly correlated with unfavorable OS except SLC39A10. Conclusion: Our analysis identified that 14 SLC family 39 genes are potential prognostic biomarkers of GC patients, and may offer effective and new strategies for GC therapy.
Keywords: Solute carrier (SLC) family 39 genes, gastric cancer, prognosis
Introduction
Gastric cancer (GC) is the second leading cause of cancer-related mortality worldwide [1], with about 90% of new cases of noncardiac gastric cancer are due to chronic infection of H. pylori bacteria [2]. As surgery can be a curative treatment in early stage, however, there are still a lot of patients being first diagnosed at advanced stage. Moreover, the median survival is only about 1 year in metastatic patients [3]. Therefore, it is extremely meaningful to successfully seek potential prognostic biomarkers for improving outcomes of GC patients [4].
The solute carrier (SLC) group of membrane transport proteins have 65 families, which participate in physiological processes ranging and may offer novel therapeutic targets for human disease [5,6]. Zinc transporters are one of membrane transport proteins of the solute carrier family including solute carrier family 30 (SLC30) and solute carrier family 39 (SLC39) [7,8]. The SLC family 39, or Zrt- and Irt-like protein (ZIP), which controls the influx of zinc, takes part in many signal pathways, plays a critical role in human diseases including tumor [9-11]. Besides, multiple studies have highlighted the key functions of SLC family 39 genes in different kinds of cancer, like lung cancer [12], esophageal carcinoma [13], prostate cancer [14], hepatocellular carcinoma [15], colorectal cancer [16]. Recent studies have demonstrated that part of SLC family 39 genes may be related with patient’s survival in some types of cancer and could serve as cancer diagnostic or prognostic biomarkers. For example, by comparing surgical and endoscopic ultrasound guided fine needle aspiration (EUS-FNA) specimens, Xu et al. have found that ZIP4 is a potential prognostic and diagnostic marker in pancreatic cancer [17]. In addition, Kagara, N et al. demonstrated that ZIP10 participates in migratory activity in metastatic breast cancer cells, indicating that ZIP10 can be utilized as a biomarker in metastatic phenotype of breast cancer and may offer a novel treatment strategy [18]. However, to our knowledge, studies regarding the expression pattern and prognostic values of the total SLC family 39 genes are still absent. In this article, we first focused on the expression pattern of individual SLC family 39 genes in GC, then explored the prognostic roles of these genes in GC. This study may provide a new insight into of SLC family 39 genes in GC and may be propitious for diagnosis, treatment and prognosis of patients with GC.
Materials and methods
cBioportal for cancer genomics
The relationship between SLC family 39 genes in GC patients and alteration frequency were analyzed through using data from cBioportal for Cancer Genomics (http://www.cbioportal.org/), choose “Stomach Adenocarcinoma (TCGA, Provisional)”, and results were shown in the Cancer Types Summary webpage.
Oncomine
Some mRNA expression level data of SLC family 39 genes (SLC39A4, SLC39A5, SLC39A6, SLC39A10) in gastric cancer and in corresponding normal tissues were downloaded from Oncomine (http://www.oncomine.org/). Choose “Cancer VS. Normal analysis” and “Gene Summary View”, set p-value = 1E-4, fold change = 2, data type = mRNA and top 5% gene rank.
UCSC Xena browser
Different mRNA expression of SLC family 39 genes in primary STAD (TCGA Stomach Cancer, n = 591) were performed by UCSC Xena browser (http://xenabrowser.net/), analyzed the data from the Cancer Genome Atlas (TCGA).
Ualcan database
UALCAN is a web resource which provides a comprehensive cancer transcriptome data [19] (http://ualcan.path.uab.edu/). The whole solute carrier family 39 genes expression at mRNA level in gastric cancer tissues and in corresponding normal gastric tissues were assessed by using UALCAN database.
Kaplan-Meier plotter
In order to assess the potential prognostic values of SLC family 39 genes in GC, the relationship between expression level of SLC family 39 genes and survival analysis including OS, FP, PPS of GC patients were obtained by the Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p = service & cancer = gastric). Next, we further analyzed SLC family 39 genes prognostic values associated with relatively clinicopathological features also performed by this database.
Results
Relative mRNA expression and genetic alteration differences of SLC family 39 genes in GC
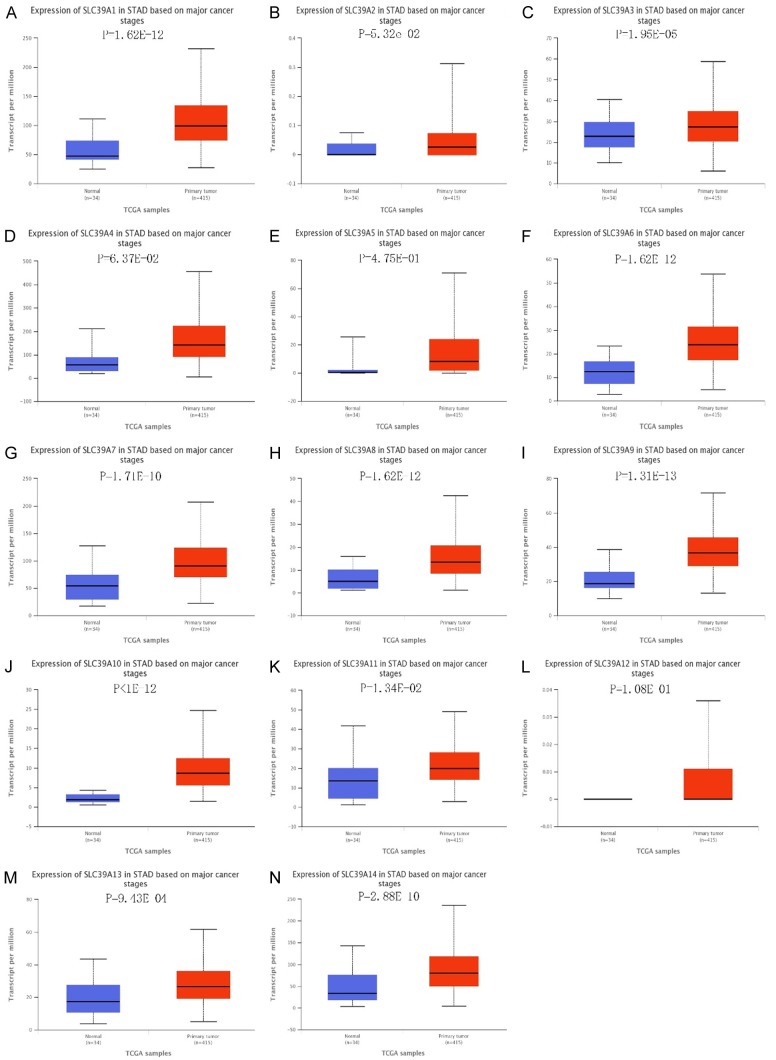
To explore the roles of SLC family 39 genes in GC, genetic alteration data of 14 SLC family 39 genes in GC patients from uBioPortal database were analyzed. Results showed that mRNA deregulation was one of the most important single factors for alteration in different kinds of gastric cancer (Figure 1A). We further compared the mRNA expression between different SLC family 39 genes in GC. In Oncomine database, only SLC39A4, SLC39A5, SLC39A6, SLC39A10 mRNA expression were measured in GC, all revealed significant upregulation in GC tissues than in corresponding normal tissues (Figure 1B). Besides, the mRNA level varied between 14 SLC family 39 members in stomach adenocarcinoma (STAD) by using UCSC Xena Browser, the mRNA expression of SLC39A12 was at lowest level, while the SLC39A14 mRNA level was the highest one (Figure 1C). As the result from Uaclan database, all the genes of solute carrier family 39 genes show high expression in gastric tumor tissues compared with in corresponding normal tissues (Figure 2).
Figure 1.
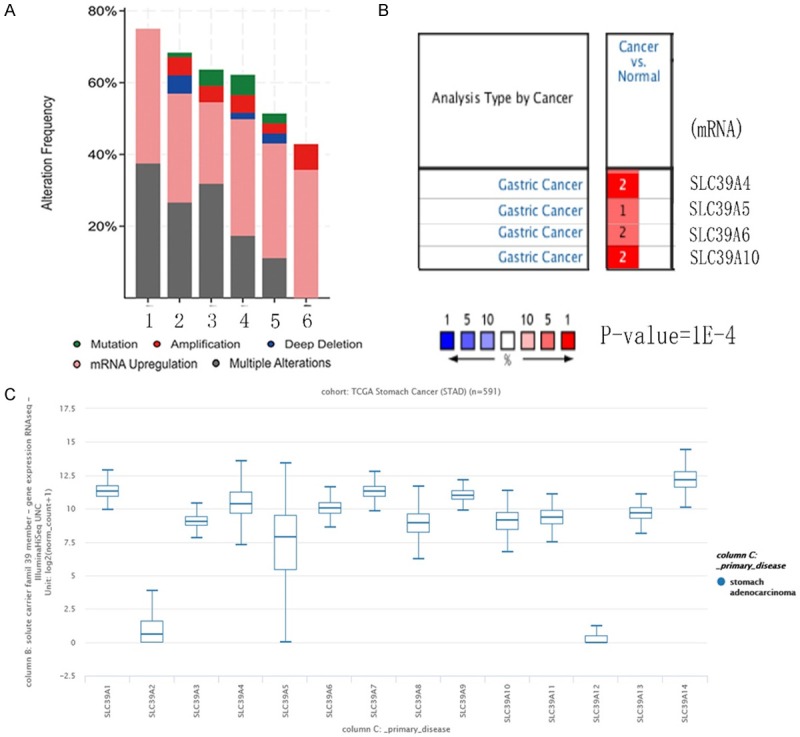
Alteration frequency and mRNA expression of SLC family 39 in GC. A. Alteration frequency analysis of SLC family 39 genes in GC. The analysis was performed using uBioPortal database. (1-6 means different types of gastric cancer, 1-Papillary Stomach Adencarcinoma, 2-Tubular Stomach Adencarcinoma, 3-Mucinous Stomach Adencarcinoma, 4-Stomach carcinoma, 5-Diffuse Type Stomach Adencarcinoma, 6-Signet Ring Cell Carcinoma of the Stomach). B. Partly mRNA expression of SLC family 39 genes (SLC39A4, SLC39A5, SLC39A6, SLC39A10) differences between cancer and normal tissues in GC using Oncomine, others are not measured. C. 14 SLC family 39 genes expression differences in GC using UCSC Xena browser.
Figure 2.
mRNA expression of SLC family 39 genes varied in primary tumor and in corresponding normal tissues in GC patients using Uaclan database. A-N. All solute carrier family 39 genes show high expression in gastric tumor tissues compared with gastric normal tissues.
SLC family 39 genes have high prognostic values in GC patients using Kaplan-Meier plotter
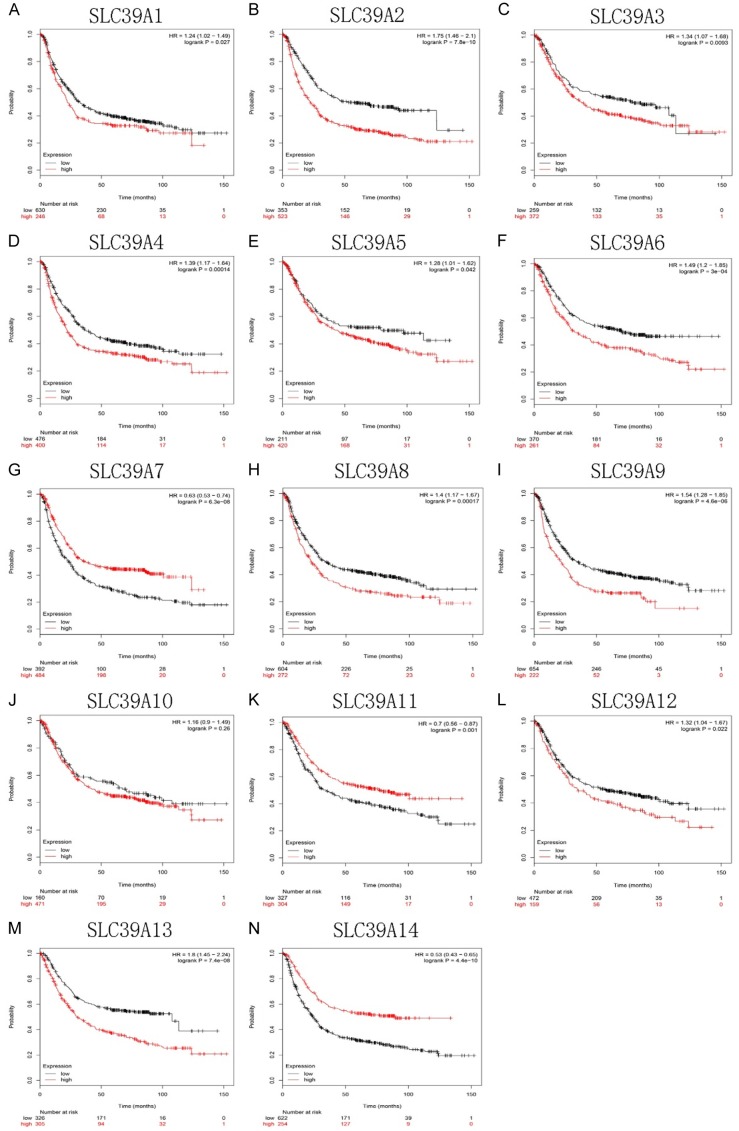
Then we using Kaplan-Meier plotter to investigate the potential prognostic values of SLC family 39 genes in GC. As shown in Figure 3, most of those genes showed a positively relation between high expression and significantly worse OS in GC patients, including SLC39A1, SLC39A2, SLC39A3, SLC39A4, SLC39A5, SLC39A6, SLC39A8, SLC39A9, SLC39A12, SLC39A13 (SLC39A1, HR 1.24 [95% CI: 1.02-1.49], P = 0.027; SLC39A2, HR 1.75 [95% CI: 1.46-2.1], P = 7.8e-10; SLC39A3, HR 1.34 [95% CI: 1.07-1.68], P = 0.0093; SLC39A4, HR 1.39 [95% CI: 1.17-1.64], P = 0.00014; SLC39A5, HR 1.28 [95% CI: 1.01-1.62], P = 0.042; SLC39A6, HR 1.49 [95% CI: 1.2-1.85], P = 3e-04; SLC39A8, HR 1.4 [95% CI: 1.17-1.67], P = 0.00017; SLC39A9, HR 1.54 [95% CI: 1.28-1.85], P = 4.6e-06; SLC39A12, HR 1.32 [95% CI: 1.04-1.67], P = 0.022; SLC39A13, HR 1.8 [95% CI: 1.45-2.24], P = 7.4e-08). In contrast, high mRNA levels of SLC39A7, SLC39A11 and SLC39A14 showed favorable OS (SLC39A7, HR 0.63 [95% CI: 0.53-0.74], P = 6.3e-08; SLC39A11, HR 0.7 [95% CI: 0.56-0.87], P = 0.001; SLC39A14, HR 0.53 [95% CI: 0.43-0.65], P = 4.4e-10). SLC39A10 was the only one which has no significance with OS in GC patients. (SLC39A10, HR 1.16 [95% CI: 0.9-1.49], P = 0.26).
Figure 3.
OS analysis of SLC family 39 genes in GC patients using Kaplan-Meier plotter.
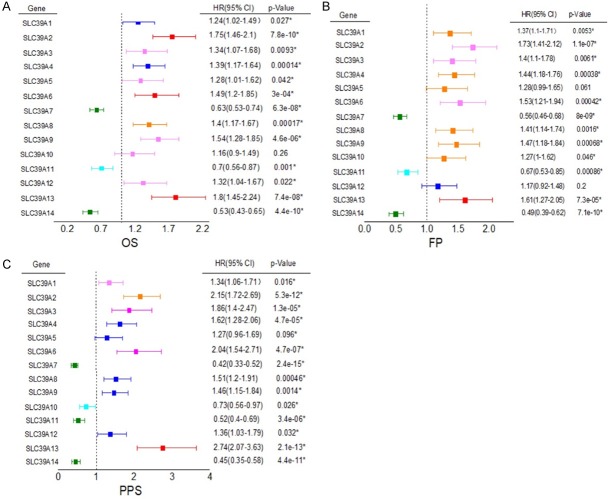
We further used Kaplan-Meier plotter to gather the correlation between GC patients OS, FP (First Progression), PPS (Post Progression Survival) and SLC39A family genes expression, showed by forest plot (Figure 4).
Figure 4.
The correlation between GC patients OS, FP (First Progression), PPS (Post Progression Survival) and SLC family 39 genes expression.
Association prognostic values of SLC family 39 genes in GC with different clinicopathological features
To further understand the significant roles of SLC family 39 genes in prognosis of patients with GC, we investigated the relationship between SLC family 39 genes and different kinds of clinicopathological features, such as different treatments, status of human epidermal growth factor receptor 2 (HER2), differentiation, Lauren classification and clinical stages. As shown in Table 1, SLC39A2, SLC39A3, SLC39A5, SLC39A6, SLC39A8, SLC39A13 high expression were significantly related with worse OS in surgery alone patients. SLC39A1, SLC39A4, SLC39A9, SLC39A10, SLC39A13 were associated with worse OS in 5-FU based adjuvant patients. And high mRNA expression of SLC39A10, SLC39A13 were also found to be correlated with worse OS in patients choose other adjuvant treatments. While SLC39A11, SLC39A14 high expression were related with better OS in both surgery alone and choose other adjuvant treatments patients. Besides, SLC39A3, SLC39A4, SLC39A7 were modestly correlated with better OS in patients choose other adjuvant treatments. HER2 infection is an important risk factor contributing the progress of GC [2]. From Table 2, SLC39A7, SLC39A11, SLC39A14 were found to be promising favorable prognosis factors in both HER2-negative and HER2-positive patients. However, SLC39A2, SLC39A4, SLC39A6, SLC39A8, SLC39A9, SLC39A12, SLC39A13 were significantly associated with unfavorable prognosis in HER2-negative patients and SLC39A2, SLC39A3, SLC39A6, SLC39A8, SLC39A13 were related with poor prognosis of HER2-positive patients. The following differentiation analysis indicated that high mRNA expression of SLC39A1, SLC39A4, SLC39A7, SLC39A10 were contributing to unfavorable OS in poorly differentiated GC patients. Meanwhile, SLC39A2, SLC39A3, SLC39A4, SLC39A10, SLC39A11 high expression were associated with shorter OS in moderately differentiated GC patients. In addition, SLC39A8, SLC39A9 were correlated with longer OS in poorly differentiated GC patients and SLC39A4, SLC39A14 were related with longer OS in well differentiated GC patients (Table 3). In terms of Lauren classification, GC patients can be divided into three subtypes: intestinal, diffuse, or mixed type. From Table 4, GC patients with intestinal type, high SLC39A2, SLC39A3, SLC39A4, SLC39A6, SLC39A8, SLC39A9, SLC39A13 expression were strongly associated with worse OS while SLC39A7, SLC39A11, SLC39A14 were related with better OS. High mRNA expression level of SLC39A2, SLC39A3, SLC39A6, SLC39A10, SLC39A12, SLC39A13 were found to be unfavorable prognosis factors in diffuse type GC patients. Nevertheless, SLC39A7, SLC39A9, SLC39A11, SLC39A14 were correlated with better OS. Moreover, high expression of SLC39A2, SLC39A5 were contributing to poor OS while SLC39A6, SLC39A10 were contributing to favorable OS in mixed type patients. As the results in Table 5, increased the mRNA expression of SLC39A3, SLC39A6, SLC39A12, SLC39A13 were indicated poor OS in stage III/IV GC while SLC39A11, SLC39A14 were associated with better OS in stage III/IV GC. Besides, high SLC39A5, SLC39A8 and SLC39A2, SLC39A10 expression were related with worse OS in stage I/II GC, respectively. SLC39A1, SLC39A2, SLC39A4, SLC39A8, SLC39A9 were only associated with poor prognosis in stage III GC patients. Furthermore, the better OS for stage IV GC patients, the higher expression of SLC39A1, SLC39A7, SLC39A10. In addition, SLC39A11 was a good prognostic factor for stage II GC patients. Besides, SLC39A13 was also significantly with poor OS in stage I GC.
Table 1.
The association between SLC family 39 genes expression and treatments of GC patients
| Gene | Treatments | Cases | HR (95% CI) | p-value |
|---|---|---|---|---|
| SLC39A1 | Surgery alone | 380 | 0.8 (0.59-1.08) | 0.15 |
| 5-FU based adjuvant | 153 | 1.36 (0.96-1.93) | 0.085* | |
| Other adjuvant | 76 | 1.59 (0.53-4.77) | 0.4008 | |
| SLC39A2 | Surgery alone | 380 | 1.51 (1.1-2.07) | 0.01* |
| 5-FU based adjuvant | 153 | 1.4 (0.98-2) | 0.066 | |
| Other adjuvant | 76 | 0.74 (0.3-1.8) | 0.5 | |
| SLC39A3 | Surgery alone | 380 | 1.4 (1.01-1.92) | 0.041* |
| 5-FU based adjuvant | 34 | 2.41 (0.67-8.61) | 0.16 | |
| Other adjuvant | 76 | 0.4 (0.17-0.98) | 0.037* | |
| SLC39A4 | Surgery alone | 380 | 1.26 (0.91-1.73) | 0.16 |
| 5-FU based adjuvant | 153 | 1.64 (1.14-2.35) | 0.0073* | |
| Other adjuvant | 76 | 0.41 (0.17-1.01) | 0.044* | |
| SLC39A5 | Surgery alone | 380 | 1.41 (1.03-1.94) | 0.032* |
| 5-FU based adjuvant | 34 | 1.44 (0.58-3.61) | 0.43 | |
| Other adjuvant | 76 | 0.35 (0.1-1.2) | 0.082 | |
| SLC39A6 | Surgery alone | 380 | 1.34 (1.01-1.8) | 0.044* |
| 5-FU based adjuvant | 34 | 0.5 (0.18-1.41) | 0.18 | |
| Other adjuvant | 76 | 0.26 (0.06-1.13) | 0.053 | |
| SLC39A7 | Surgery alone | 380 | 0.72 (0.51-1) | 0.052 |
| 5-FU based adjuvant | 153 | 0.68 (0.46-1.02) | 0.062 | |
| Other adjuvant | 76 | 0.34 (0.14-0.81) | 0.011* | |
| SLC39A8 | Surgery alone | 380 | 1.38 (1.03-1.85) | 0.032* |
| 5-FU based adjuvant | 153 | 1.42 (0.93-2.18) | 0.1 | |
| Other adjuvant | 76 | 0.56 (0.2-1.55) | 0.26 | |
| SLC39A9 | Surgery alone | 380 | 0.78 (0.56-1.06) | 0.11 |
| 5-FU based adjuvant | 153 | 1.48 (1.02-2.15) | 0.038* | |
| Other adjuvant | 76 | 2.38 (0.91-6.21) | 0.067 | |
| SLC39A10 | Surgery alone | 380 | 1.2 (0.9-1.6) | 0.21 |
| 5-FU based adjuvant | 34 | 6.83 (2.3-20.31) | 9.6e-05* | |
| Other adjuvant | 76 | 5.23 (1.21-22.54) | 0.013* | |
| SLC39A11 | Surgery alone | 380 | 0.74 (0.55-0.99) | 0.044* |
| 5-FU based adjuvant | 34 | 0.74 (0.3-1.84) | 0.51 | |
| Other adjuvant | 76 | 0.24 (0.09-0.66) | 0.0027* | |
| SLC39A12 | Surgery alone | 380 | 1.24 (0.9-1.7) | 0.18 |
| 5-FU based adjuvant | 34 | 0.42 (0.14-1.28) | 0.12 | |
| Other adjuvant | 76 | 1.79 (0.74-4.29) | 0.19 | |
| SLC39A13 | Surgery alone | 380 | 1.39 (1.04-1.86) | 0.026* |
| 5-FU based adjuvant | 34 | 3.53 (1.26-9.87) | 0.011* | |
| Other adjuvant | 76 | 3.76 (1.53-9.22) | 0.0019* | |
| SLC39A14 | Surgery alone | 380 | 0.6 (0.44-0.82) | 0.0011* |
| 5-FU based adjuvant | 153 | 1.39 (0.98-1.97) | 0.067 | |
| Other adjuvant | 76 | 0.23 (0.1-0.57) | 0.00044* |
The * is mean significant p value.
Table 2.
The association between SLC family 39 genes expression and HER2 status of GC patients
| Gene | HER2 status | Cases | HR (95% CI) | p-value |
|---|---|---|---|---|
| SLC39A1 | Negative | 532 | 1.21 (0.96-1.52) | 0.1 |
| Positive | 344 | 1.23 (0.94-1.62) | 0.14 | |
| SLC39A2 | Negative | 532 | 1.76 (1.39-2.39) | 2.6e-06* |
| Positive | 344 | 1.78 (1.33-2.39) | 8.2e-05* | |
| SLC39A3 | Negative | 429 | 1.32 (1-1.75) | 0.053 |
| Positive | 202 | 1.64 (1.07-2.53) | 0.022* | |
| SLC39A4 | Negative | 532 | 1.56 (1.23-1.97) | 0.00021* |
| Positive | 344 | 0.84 (0.64-1.09) | 0.19 | |
| SLC39A5 | Negative | 429 | 1.29 (0.98-1.7) | 0.064 |
| Positive | 202 | 0.68 (0.45-1.01) | 0.055 | |
| SLC39A6 | Negative | 429 | 1.55 (1.19-2.03) | 0.0012* |
| Positive | 202 | 1.71 (1.08-2.71) | 0.021* | |
| SLC39A7 | Negative | 532 | 0.57 (0.46-0.72) | 7.8e-07* |
| Positive | 344 | 0.72 (0.54-0.94) | 0.017* | |
| SLC39A8 | Negative | 532 | 1.44 (1.13-1.82) | 0.0028* |
| Positive | 344 | 1.45 (1.11-1.88) | 0.0053* | |
| SLC39A9 | Negative | 532 | 1.56 (1.22-1.99) | 0.00031* |
| Positive | 344 | 1.26 (0.96-1.65) | 0.091 | |
| SLC39A10 | Negative | 429 | 0.85 (0.62-1.16) | 0.3 |
| Positive | 202 | 1.5 (0.98-2.31) | 0.061 | |
| SLC39A11 | Negative | 429 | 0.66 (0.51-0.86) | 0.0021* |
| Positive | 202 | 0.64 (0.44-0.93) | 0.019* | |
| SLC39A12 | Negative | 429 | 1.56 (1.18-2.05) | 0.0015* |
| Positive | 202 | 0.7 (0.48-1.03) | 0.068 | |
| SLC39A13 | Negative | 429 | 1.72 (1.32-2.24) | 4.8e-05* |
| Positive | 202 | 1.99 (1.32-2.99) | 0.00078* | |
| SLC39A14 | Negative | 532 | 0.5 (0.39-0.64) | 9.6e-09* |
| Positive | 344 | 0.68 (0.51-0.93) | 0.013* |
The * is mean significant p value.
Table 3.
The association between SLC family 39 genes expression and differentiation of GC patients
| Gene | Differentiation | Cases | HR (95% CI) | p-value |
|---|---|---|---|---|
| SLC39A1 | Poorly differentiated | 165 | 1.67 (1.01-2.76) | 0.042* |
| Moderately differentiated | 67 | 1.72 (0.89-3.32) | 0.1 | |
| Well differentiated | 32 | 0.68 (0.26-1.75) | 0.42 | |
| SLC39A2 | Poorly differentiated | 165 | 1.23 (0.74-2.06) | 0.42 |
| Moderately differentiated | 67 | 2.02 (1-4.09) | 0.046* | |
| Well differentiated | 32 | 2.44 (0.71-8.34) | 0.14 | |
| SLC39A3 | Poorly differentiated | 121 | 0.73 (0.43-1.23) | 0.24 |
| Moderately differentiated | 67 | 2.28 (1.17-4.43) | 0.013* | |
| SLC39A4 | Poorly differentiated | 165 | 2.48 (1.46-4.21) | 5e-04* |
| Moderately differentiated | 67 | 1.97 (1-3.89) | 0.047* | |
| Well differentiated | 32 | 0.16 (0.04-0.53) | 0.00079* | |
| SLC39A5 | Poorly differentiated | 121 | 1.3 (0.8-2.13) | 0.29 |
| Moderately differentiated | 67 | 0.57 (0.29-1.11) | 0.096 | |
| SLC39A6 | Poorly differentiated | 121 | 0.78 (0.47-1.29) | 0.33 |
| Moderately differentiated | 67 | 0.56 (0.29-1.1) | 0.091 | |
| SLC39A7 | Poorly differentiated | 165 | 1.54 (1.03-2.31) | 0.036* |
| Moderately differentiated | 67 | 0.63 (0.32-1.22) | 0.17 | |
| Well differentiated | 32 | 0.6 (0.25-1.46) | 0.26 | |
| SLC39A8 | Poorly differentiated | 165 | 0.52 (0.31-0.87) | 0.011* |
| Moderately differentiated | 67 | 1.54 (0.81-2.95) | 0.19 | |
| Well differentiated | 32 | 1.65 (0.7-3.92) | 0.25 | |
| SLC39A9 | Poorly differentiated | 165 | 0.68 (0.45-1.03) | 0.069 |
| Moderately differentiated | 67 | 0.61 (0.3-1.24) | 0.17 | |
| Well differentiated | 32 | 1.49 (0.54-4.08) | 0.44 | |
| SLC39A10 | Poorly differentiated | 121 | 1.7 (1.05-2.75) | 0.03* |
| Moderately differentiated | 67 | 2.01 (0.99-4.08) | 0.049* | |
| SLC39A11 | Poorly differentiated | 121 | 0.85 (0.51-1.4) | 0.52 |
| Moderately differentiated | 67 | 2.04 (1.06-3.92) | 0.029* | |
| SLC39A12 | Poorly differentiated | 121 | 0.77 (0.45-1.33) | 0.36 |
| Moderately differentiated | 67 | 0.73 (0.37-1.44) | 0.36 | |
| SLC39A13 | Poorly differentiated | 121 | 1.4 (0.79-2.49) | 0.25 |
| Moderately differentiated | 67 | 1.96 (0.85-4.49) | 0.11 | |
| SLC39A14 | Poorly differentiated | 165 | 1.35 (0.89-2.04) | 0.16 |
| Moderately differentiated | 67 | 0.62 (0.3-1.28) | 0.19 | |
| Well differentiated | 32 | 0.39 (0.16-0.99) | 0.04* |
The * is mean significant p value.
Table 4.
The association between SLC family 39 genes expression and Lauren classification of GC patients
| Gene | Lauren classification | Cases | HR (95% CI) | p-value |
|---|---|---|---|---|
| SLC39A1 | Intestinal | 320 | 1.35 (0.97-1.87) | 0.071 |
| Diffuse | 241 | 1.41 (0.98-2.01) | 0.061 | |
| Mixed | 32 | 2.76 (0.86-8.87) | 0.077 | |
| SLC39A2 | Intestinal | 320 | 2.09 (1.51-2.88) | 4.5e-06* |
| Diffuse | 241 | 1.43 (1-2.05) | 0.048* | |
| Mixed | 32 | 4.83 (1.08-21.58) | 0.023* | |
| SLC39A3 | Intestinal | 269 | 1.68 (1.17-2.42) | 0.0047* |
| Diffuse | 240 | 1.5 (1.06-2.11) | 0.02* | |
| Mixed | 29 | 2.38 (0.52-10.86) | 0.25 | |
| SLC39A4 | Intestinal | 320 | 1.63 (1.18-2.26) | 0.0028* |
| Diffuse | 241 | 1.46 (0.96-2.21) | 0.075 | |
| Mixed | 32 | 2.74 (0.93-8.12) | 0.058 | |
| SLC39A5 | Intestinal | 269 | 1.31 (0.89-1.93) | 0.17 |
| Diffuse | 240 | 1.4 (0.95-2.08) | 0.09 | |
| Mixed | 29 | 5.57 (1.64-18.9) | 0.0021* | |
| SLC39A6 | Intestinal | 269 | 2.29 (1.59-3.31) | 5.4e-06* |
| Diffuse | 240 | 1.55 (1.08-2.22) | 0.015* | |
| Mixed | 29 | 0.34 (0.11-1.03) | 0.045* | |
| SLC39A7 | Intestinal | 320 | 0.48 (0.35-0.66) | 5.4e-06* |
| Diffuse | 241 | 0.6 (0.4-0.9) | 0.013* | |
| Mixed | 32 | 0.4 (0.11-1.41) | 0.14 | |
| SLC39A8 | Intestinal | 320 | 1.57 (1.15-2.16) | 0.0048* |
| Diffuse | 241 | 1.25 (0.89-1.75) | 0.2 | |
| Mixed | 32 | 0.56 (0.18-1.78) | 0.32 | |
| SLC39A9 | Intestinal | 320 | 1.88 (1.35-2.61) | 0.00013* |
| Diffuse | 241 | 0.64 (0.45-0.91) | 0.012* | |
| Mixed | 32 | 0.55 (0.15-1.94) | 0.34 | |
| SLC39A10 | Intestinal | 269 | 0.78 (0.54-1.12) | 0.18 |
| Diffuse | 240 | 1.57 (1.11-2.21) | 0.0097* | |
| Mixed | 29 | 0.28 (0.07-1.08) | 0.0498* | |
| SLC39A11 | Intestinal | 269 | 0.62 (0.43-0.89) | 0.0084* |
| Diffuse | 240 | 0.64 (0.44-0.93) | 0.018* | |
| Mixed | 29 | 1.87 (0.49-7.14) | 0.35 | |
| SLC39A12 | Intestinal | 269 | 0.79 (0.53-1.15) | 0.22 |
| Diffuse | 240 | 1.63 (1.16-2.29) | 0.0045* | |
| Mixed | 29 | 2.53 (0.83-7.7) | 0.092 | |
| SLC39A13 | Intestinal | 269 | 2.75 (1.91-3.97) | 1.5e-08* |
| Diffuse | 240 | 1.68 (1.1-2.55) | 0.014* | |
| Mixed | 29 | 2.11 (0.7-6.34) | 0.18 | |
| SLC39A14 | Intestinal | 320 | 0.39 (0.29-0.54) | 2.6e-09* |
| Diffuse | 241 | 0.5 (0.34-0.73) | 0.00027* | |
| Mixed | 32 | 2.55 (0.87-7.44) | 0.077 |
The * is mean significant p value.
Table 5.
The association between SLC family 39 genes expression and stages of GC patients
| Gene | Stage | Cases | HR (95% CI) | p-value |
|---|---|---|---|---|
| SLC39A1 | I | 67 | 0.71 (0.26-1.95) | 0.5 |
| II | 140 | 1.55 (0.82-2.94) | 0.17 | |
| III | 305 | 1.62 (1.17-2.25) | 0.0038* | |
| IV | 148 | 0.66 (0.44-0.99) | 0.041* | |
| SLC39A2 | I | 67 | 1.96 (0.63-6.11) | 0.23 |
| II | 140 | 2.32 (1.25-4.31) | 0.0058* | |
| III | 305 | 1.8 (1.32-2.45) | 0.00014* | |
| IV | 148 | 1.42 (0.91-2.23) | 0.12 | |
| SLC39A3 | I | 62 | 2.12 (0.69-6.53) | 0.18 |
| II | 135 | 1.73 (1.16-2.59) | 0.19 | |
| III | 197 | 1.73 (1.16-2.59) | 0.0065* | |
| IV | 140 | 1.68 (1.13-2.51) | 0.01* | |
| SLC39A4 | I | 67 | 2.69 (0.86-8.42) | 0.076 |
| II | 140 | 0.7 (0.38-1.26) | 0.23 | |
| III | 305 | 1.51 (1.13-2.01) | 0.0045* | |
| IV | 148 | 0.81 (0.54-1.22) | 0.31 | |
| SLC39A5 | I | 62 | 4.07 (0.9-18.38) | 0.048* |
| II | 135 | 2.38 (0.93-6.11) | 0.062 | |
| III | 197 | 1.39 (0.93-2.07) | 0.11 | |
| IV | 140 | 1.46 (0.98-2.18) | 0.063 | |
| SLC39A6 | I | 62 | 3.36 (0.9-12.49) | 0.057 |
| II | 135 | 1.74 (0.84-3.58) | 0.13 | |
| III | 197 | 1.91 (1.31-2.78) | 0.00068* | |
| IV | 140 | 2.17 (1.43-3.3) | 0.00021* | |
| SLC39A7 | I | 67 | 0.36 (0.12-1.06) | 0.055 |
| II | 140 | 0.66 (0.35-1.22) | 0.18 | |
| III | 305 | 0.78 (0.57-1.06) | 0.11 | |
| IV | 148 | 0.51 (0.33-0.78) | 0.0014* | |
| SLC39A8 | I | 67 | 4.12 (0.93-18.14) | 0.042* |
| II | 140 | 1.62 (0.87-3.01) | 0.13 | |
| III | 305 | 1.39 (1.05-1.85) | 0.022* | |
| IV | 148 | 0.69 (0.46-1.03) | 0.066 | |
| SLC39A9 | I | 67 | 2.08 (0.75-5.82) | 0.15 |
| II | 140 | 1.37 (0.72-2.58) | 0.34 | |
| III | 305 | 1.49 (1.1-2.03) | 0.01* | |
| IV | 148 | 0.83 (0.56-1.22) | 0.35 | |
| SLC39A10 | I | 62 | 0.5 (0.16-1.55) | 0.22 |
| II | 135 | 1.97 (1.05-3.72) | 0.032* | |
| III | 197 | 0.8 (0.55-1.16) | 0.23 | |
| IV | 140 | 0.66 (0.43-1.03) | 0.064 | |
| SLC39A11 | I | 62 | 0.43 (0.14-1.32) | 0.13 |
| II | 135 | 0.52 (0.27-1) | 0.048* | |
| III | 197 | 0.64 (0.43-0.98) | 0.037* | |
| IV | 140 | 0.66 (0.43-0.99) | 0.041* | |
| SLC39A12 | I | 62 | 5.23 (0.68-40.26) | 0.076 |
| II | 135 | 0.52 (0.27-1.01) | 0.05 | |
| III | 197 | 2 (1.37-2.92) | 0.00028* | |
| IV | 140 | 1.58 (1.04-2.4) | 0.032* | |
| SLC39A13 | I | 62 | 327998284.34 (0-Inf) | 0.03* |
| II | 135 | 1.78 (0.93-3.41) | 0.076 | |
| III | 197 | 2.6 (1.67-4.04) | 1.2e-05* | |
| IV | 140 | 1.88 (1.23-2.87) | 0.0031* | |
| SLC39A14 | I | 67 | 0.4 (0.15-1.11) | 0.071 |
| II | 140 | 0.55 (0.29-1.03) | 0.056 | |
| III | 305 | 0.48 (0.34-0.7) | 5.8e-05* | |
| IV | 148 | 0.45 (0.28-0.73) | 0.00091* |
The * is mean significant p value.
In conclusion, SLC39A1 high expression was closely related with worse OS in 5-FU based adjuvant treatment, poorly differentiated and in stage III GC patients, while better OS in stage IV GC patients. SLC39A2 high expression was significantly correlated with poor OS in surgery alone treatment, HER2 positive or negative, moderately differentiated, three Lauren classification subtypes including intestinal, diffuse, or mixed type, and in stage II/III GC patients. High expression of SLC39A3 indicated shorter OS in surgery alone treatment, HER2 positive, moderately differentiated, intestinal and diffuse subtypes, stage III/IV GC patients. Furthermore, overexpression of SLC39A4 was revealed worse OS in 5-FU based adjuvant treatment, HER2 negative, two differentiations including poorly differentiated and moderately differentiated, intestinal subtype and in stage III GC patients, however, better OS in well differentiated and other adjuvant treatment of GC patients. SLC39A5 high expression was significantly related with poor OS in surgery alone, mixed type and in stage I GC patients. Besides, low SLC39A6 expression was correlated with longer OS in surgery alone treatment, HER2 positive or negative, intestinal, diffuse, or mixed type and in stage III/IV GC patients. High expression level of SLC39A7 was found to be relevant to favorable OS in other adjuvant treatment, HER2 positive or negative, intestinal and diffuse types, stage IV GC patients, on the contrary, poor OS in poorly differentiated GC patients. GC patients with high expression of SLC39A8 was associated with better OS in poorly differentiated, while worse OS in surgery alone treatment, HER2 positive or negative, intestinal type and stage I/III. Upregulation of SLC39A9 was decreased the OS for GC patients in 5-FU based adjuvant treatment, HER2 negative, intestinal type and stage III, whereas increased the OS for GC patients in diffuse type. As for SLC39A10, the higher expression, the shorter OS in 5-FU based adjuvant and other treatments, poorly differentiated and moderately differentiated, diffuse type and stage II GC patients. SLC39A11 high expression was significantly indicated longer OS for GC patients in surgery alone and other treatments, HER2 positive or negative, intestinal and diffuse type, stage II/III/IV, while shorter OS in moderately differentiated. Downregulation of SLC39A12 was strongly associated with poor OS in HER2 negative, diffuse type, stage III/IV GC patients. Low expression level of SLC39A13 was found to be related with favorable OS for GC patients in different treatments including surgery alone, 5-FU based adjuvant, other adjuvant, HER2 positive or negative, intestinal and diffuse type, stage I/III/IV. What’s more, SLC39A14 high expression was closely correlated with favorable OS for GC patients in surgery alone and other adjuvant treatments, HER2 positive or negative, well differentiated, intestinal and diffuse types and stage III/IV.
Discussion
Notably, Zinc acts as a tumor suppressive agent. Moreover, the solute carrier family 39 (SLC39), or Zrt- and Irt-like protein (ZIP), controls the afflux of zinc into the cytoplasm from vesicles and from outside the cell. Previous studies have reported that SLC39A9, a member of subfamily I in vertebrates, could regulate zinc homeostasis in the secretory pathway [20]. Yu Y et al. proved that Zip11 acted as a cellular zinc transporter, which expression was partially regulated by zinc through hMTF-1 binding to MREs of the Zip11 promoter. Besides, murine Zip11 mRNA is highly expressed in the stomach [21]. The downregulation of ZIP protein negatively regulated EGFR pathway, thereby participating in cell proliferation, migration and progression in breast carcinoma [22]. Hence, SLC39 genes may play complicated roles in the pathogenesis and progression of cancer.
In the present study, we analyzed the expression profile of SLC family members in GC and explored their associations with survival outcomes in patients with GC by using available databases. Our results showed that SLC39A1, SLC39A2, SLC39A3, SLC39A4, SLC39A5, SLC39A6, SLC39A8, SLC39A9, SLC39A12, SLC39A13 were significantly higher in GC tissues than in corresponding normal tissues, and indicated worse OS in GC patients. However, high expression of SLC39A4, SLC39A5, SLC39A6, SLC39A10 in GC tissues indicated favorable OS in GC patients, and SLC39A10 has no significance with survival outcomes in GC patients. Collectively, these data suggest that SLC39A1, SLC39A2, SLC39A3, SLC39A4, SLC39A5, SLC39A6, SLC39A8, SLC39A9, SLC39A12 and SLC39A13 show better prospect for utilizing as prognostic biomarkers in patients with GC. All those members have a close relationship with clinicopathological features including status of human epidermal growth factor receptor 2 (HER2), differentiation, Lauren classification and clinical stages.
Recent studies have reported that SLC39A1, SLC39A2, SLC39A3 were downregulated in prostate cancer, as well as decreased the expression level of Zinc, that delayed the progression of the prostate cancer [23,24]. Besides, the expression of SLC39A1 was decreased the malignant potential of the cell renal cell carcinoma [25]. Moreover, in the early stage of pancreatic adenocarcinoma, SLC39A3 expression was obviously downregulated [26]. While SLC39A4 expression was upregulated in pancreatic adenocarcinoma tissues compared with in corresponding normal tissues, and ZIP4 could suppress apoptosis and promote invasiveness in hepatocellular carcinoma (HCC), as a poor prognosis factor in HCC patients after liver transplantation therapy [27,28]. In addition, higher SLC39A4 expression was associated with shorter OS and disease-free survival (DFS) in non-small cell lung cancer [29]. SLC39A5 and SLC39A6 both impose poor effects during the progression of esophageal squamous-cell carcinoma (ESCC), and higher SLC39A6 protein expression was found to be significantly related with worse survival in patients with advanced ESCC [30-32]. Knockdown the expression of SLC39A8 inhibited neuroblastoma metastasis and progression in vitro [33]. In addition, study has shown that SLC39A9 expression was upregulated in malignant prostate and breast tissues, suggesting that it may offer a novel target for prostate and breast cancer therapy [34]. SLC39A14 was reported to be an independent and important factor for recurrence-free survival analysis of prostate cancer patients [35]. However, researches about SLC39A12, SLC39A13 in cancer are limited.
It is obviously to see that dysfunction of SLC family 39 genes are closely correlated with the progression of many different types of cancer, and many of them may be strongly involved in the prognosis of patients with cancer, while at present litter is known about its potential mechanism, function and prognostic value in gastric cancer, more studies about of SLC family 39 genes are need to be further mined for GC therapy.
Conclusion
We evaluated the prognostic values of SLC family 39 genes in GC patients by using bioinformatic analysis. We identified that high mRNA expression level of SLC39A7, SLC39A11, SLC39A14 were associated with better OS, while SLC39A1, SLC39A2, SLC39A3, SLC39A4, SLC39A5, SLC39A6, SLC39A8, SLC39A9, SLC39A12, SLC39A13 were significantly related with worse OS in GC patients. Besides, the analysis between SLC family 39 genes expression and clinicopathological characteristics in GC indicated that SLC family 39 genes are promising prognostic biomarkers in GC patients, may offer novel targets for GC therapy. But the research of SLC family 39 genes function in GC is still insufficient. More trials need to be further launched in the future. We believe that SLC39 family genes will be effective prognostic factors for GC patients.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (81372462, 81572987) and Department of Science and Technology of Zhejiang Province (Grant No. 2014C03012).
Disclosure of conflict of interest
None.
References
- 1.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–13. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 4.You X, Ma M, Hou G, Hu Y, Shi X. Gene expression and prognosis of NOX family members in gastric cancer. Onco Targets Ther. 2018;11:3065–3074. doi: 10.2147/OTT.S161287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin L, Yee SW, Kim RB, Giacomini KM. SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov. 2015;14:543–60. doi: 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai X, Moraes TF, Reithmeier R. Structural biology of solute carrier (SLC) membrane transport proteins. Mol Membr Biol. 2017;34:1–32. doi: 10.1080/09687688.2018.1448123. [DOI] [PubMed] [Google Scholar]
- 7.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–8. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 8.Schlessinger A, Khuri N, Giacomini KM, Sali A. Molecular modeling and ligand docking for solute carrier (SLC) transporters. Curr Top Med Chem. 2013;13:843–56. doi: 10.2174/1568026611313070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T, Takeda TA, Takagishi T, Fukue K, Kambe T, Fukada T. Physiological roles of zinc transporters: molecular and genetic importance in zinc homeostasis. J Physiol Sci. 2017;67:283–301. doi: 10.1007/s12576-017-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhutia YD, Babu E, Ramachandran S, Yang S, Thangaraju M, Ganapathy V. SLC transporters as a novel class of tumour suppressors: identity, function and molecular mechanisms. Biochem J. 2016;473:1113–24. doi: 10.1042/BJ20150751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorsen K, Mansilla F, Schepeler T, Øster B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR, Laurberg S, Andersen CL, Ørntoft TF. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol Cell Proteomics. 2011;10:M110.002998. doi: 10.1074/mcp.M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu DM, Liu T, Deng SH, Han R, Xu Y. SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer. Sci Rep. 2017;7:7211. doi: 10.1038/s41598-017-07830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui XB, Shen YY, Jin TT, Li S, Li TT, Zhang SM, Peng H, Liu CX, Li SG, Yang L, Li N, Hu JM, Jiang JF, Li M, Liang WH, Li Y, Wei YT, Sun ZZ, Wu CY, Chen YZ, Li F. SLC39A6: a potential target for diagnosis and therapy of esophageal carcinoma. J Transl Med. 2015;13:321. doi: 10.1186/s12967-015-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu XM, Wang CG, Zhu YD, Chen WH, Shao SL, Jiang FN, Liao QD. Decreased expression of SLC 39A14 is associated with tumor aggressiveness and biochemical recurrence of human prostate cancer. Onco Targets Ther. 2016;9:4197–205. doi: 10.2147/OTT.S103640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, Andrews GK. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng N, Yan L, You W, Tan G, Gong J, Chen H, Yang Y, Hu L, Wang Z. Knockdown of SLC39A7 inhibits cell growth and induces apoptosis in human colorectal cancer cells. Acta Biochim Biophys Sin (Shanghai) 2017;49:926–934. doi: 10.1093/abbs/gmx094. [DOI] [PubMed] [Google Scholar]
- 17.Xu C, Wallace MB, Yang J, Jiang L, Zhai Q, Zhang Y, Hong C, Chen Y, Frank TS, Stauffer JA, Asbun HJ, Raimondo M, Woodward TA, Li Z, Guha S, Zheng L, Li M. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: a systemic comparison between EUS-FNA and surgical specimens. Curr Mol Med. 2014;14:309–15. doi: 10.2174/1566524013666131217112921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagara N, Tanaka N, Noguchi S, Hirano T. Zinc and its transporter ZIP10 are involved in invasive behavior of breast cancer cells. Cancer Sci. 2007;98:692–7. doi: 10.1111/j.1349-7006.2007.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuura W, Yamazaki T, Yamaguchi-Iwai Y, Masuda S, Nagao M, Andrews GK, Kambe T. SLC39A9 (ZIP9) regulates zinc homeostasis in the secretory pathway: characterization of the ZIP subfamily I protein in vertebrate cells. Biosci Biotechnol Biochem. 2009;73:1142–8. doi: 10.1271/bbb.80910. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y, Wu A, Zhang Z, Yan G, Zhang F, Zhang L, Shen X, Hu R, Zhang Y, Zhang K, Wang F. Characterization of the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J Nutr Biochem. 2013;24:1697–708. doi: 10.1016/j.jnutbio.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Zhang H, Yu W, Chen Y, Gui B, Liang J, Wang Y, Sun L, Yang X, Zhang Y, Shi L, Li Y, Shang Y. ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 2009;28:2763–76. doi: 10.1038/emboj.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello LC, Franklin RB, Zou J, Feng P, Bok R, Swanson MG, Kurhanewicz J. Human prostate cancer ZIP1/zinc/citrate genetic/metabolic relationship in the TRAMP prostate cancer animal model. Cancer Biol Ther. 2011;12:1078–84. doi: 10.4161/cbt.12.12.18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desouki MM, Geradts J, Milon B, Franklin RB, Costello LC. hZip2 and hZip3 zinc transporters are down regulated in human prostate adenocarcinomatous glands. Mol Cancer. 2007;6:37. doi: 10.1186/1476-4598-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X, Kong C, Zhang Z, Liu X, Zhan B, Chen Z, Shi D. hZIP1 that is down-regulated in clear cell renal cell carcinoma is negatively associated with the malignant potential of the tumor. Urol Oncol. 2014;32:885–92. doi: 10.1016/j.urolonc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Costello LC, Zou J, Desouki MM, Franklin RB. Evidence for changes in RREB-1, ZIP3, and Zinc in the early development of pancreatic adenocarcinoma. J Gastrointest Cancer. 2012;43:570–8. doi: 10.1007/s12029-012-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang SM, Cousins RJ, Fisher WE, Brunicardi FC, Logsdon CD, Chen C, Yao Q. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci U S A. 2007;104:18636–41. doi: 10.1073/pnas.0709307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Guo HJ, Xie HY, Li J, Zhuang RZ, Ling Q, Zhou L, Wei XY, Liu ZK, Ding SM, Chen KJ, Xu ZY, Zheng SS. ZIP4, a novel determinant of tumor invasion in hepatocellular carcinoma, contributes to tumor recurrence after liver transplantation. Int J Biol Sci. 2014;10:245–56. doi: 10.7150/ijbs.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu DM, Liu T, Deng SH, Han R, Xu Y. SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer. Sci Rep. 2017;7:7211. doi: 10.1038/s41598-017-07830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin J, Li Z, Liu J, Wu Y, Gao X, He Y. Knockdown of zinc transporter ZIP5 (SLC39A5) expression significantly inhibits human esophageal cancer progression. Oncol Rep. 2015;34:1431–9. doi: 10.3892/or.2015.4097. [DOI] [PubMed] [Google Scholar]
- 31.Wu C, Li D, Jia W, Hu Z, Zhou Y, Yu D, Tong T, Wang M, Lin D, Qiao Y, Zhou Y, Chang J, Zhai K, Wang M, Wei L, Tan W, Shen H, Zeng Y, Lin D. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet. 2013;45:632–8. doi: 10.1038/ng.2638. [DOI] [PubMed] [Google Scholar]
- 32.Cui XB, Shen YY, Jin TT, Li S, Li TT, Zhang SM, Peng H, Liu CX, Li SG, Yang L, Li N, Hu JM, Jiang JF, Li M, Liang WH, Li Y, Wei YT, Sun ZZ, Wu CY, Chen YZ, Li F. SLC39A6: a potential target for diagnosis and therapy of esophageal carcinoma. J Transl Med. 2015;13:321. doi: 10.1186/s12967-015-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei Z, Yan P, Wang Y, Liu S, He F. Knockdown of zinc transporter ZIP8 expression inhibits neuroblastoma progression and metastasis in vitro. Mol Med Rep. 2018;18:477–485. doi: 10.3892/mmr.2018.8944. [DOI] [PubMed] [Google Scholar]
- 34.Thomas P, Pang Y, Dong J, Berg AH. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology. 2014;155:4250–65. doi: 10.1210/en.2014-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu XM, Wang CG, Zhu YD, Chen WH, Shao SL, Jiang FN, Liao QD. Decreased expression of SLC 39A14 is associated with tumor aggressiveness and biochemical recurrence of human prostate cancer. Onco Targets Ther. 2016;9:4197–205. doi: 10.2147/OTT.S103640. [DOI] [PMC free article] [PubMed] [Google Scholar]