Abstract
Mesenchymal stem cells (MSCs) of the dental or craniofacial origin include Human periodontal ligament stem cells (hPDLSCs), which are able to readily differentiate into osteoblasts. Tanshinone IIA (TSA) is a diterpene quinone compound that is derived from Danshen (also known as Salvia miltiorrhiza) used frequently in the context of traditional Chinese medicine (TCM). This study sought to assess how TSA affects the osteogenic differentiation of hPDLSCs. We found that TSA promotes both this differentiation and hPDLSC maturation. This was dependent on TSA-mediated activation of the ERK1/2 signaling pathway, and ERK1/2 inhibition disrupted TSA-induced Runx2 expression. From these results, we conclude that TSA can induce hPDLSC osteogenesis through the ERK1/2-Runx2 axis, suggesting that TSA is a viable therapeutic option for regenerative medical approaches aimed at the treatment of periodontitis.
Keywords: Tanshinone IIA, human periodontal ligament stem cells, osteogenic differentiation, Runx2, ERK1/2
Introduction
Periodontitis is the infection of the gums and bone surrounding the teeth, leading to substantial local damage [1,2]. It is a major cause of gum inflammation and is the primary cause of adult tooth loss [3]. Periodontitis is also linked to the prevalence of other serious human diseases including pulmonary infections, chronic kidney disease and diabetes [4,5]. Whilst periodontal therapies are incapable of regenerating the normal functionality of damaged tissue and bone, stem cell-based tissue therapy offers promise in this area [6,7]. In this regard, human periodontal ligament stem cells (hPDLSCs) can be accessed more easily than can bone marrow stem cells, and can more readily differentiate into bone, cementum, and periodontal ligaments [8]. Clinical autologous hPDLSC based treatments used during periodontal therapy have been shown to be safe and effective [9,10]. Mechanisms to modulate the biological functions of hPDLSCs to improve their application to tissue engineering, now requires further investigation.
Tanshinone IIA (TSA) is a lipophilic compound that can be extracted from the roots of the herbal medicine Salvia miltiorrhiza Bunge [11]. TSA is widely employed in Eastern Asia medicine for cardiovascular disease, postmenopausal syndrome, and cerebrovascular disease treatment, primarily because it has antioxidant and anti-inflammatory capabilities [12,13]. Studies have demonstrated that the anti-inflammatory properties of TSA are mediated through its ability to suppress inflammatory cytokine expression (IL-6, IL-1β, and TNF-α), iNOS expression, and subsequent NO production [14,15]. Others have reported that in a rat model of ischemia-reperfusion injury can protect cardiomyocytes from oxidative stress-induced apoptosis [16,17]. Similar effects have been shown in other cell lines and in vivo animal models [14,18]. Numerous signaling pathways have been implicated in the mechanisms of TSA functionality, including p38, MAPK signaling, TGF-β, and NF-κB [19-22]. Whilst is it known that TSA exerts beneficial effects during the treatment of bone disease [23,24], its effects on the osteogenic differentiation of hPDLSCs remain to be studied fully. In this study, we have assessed how TSA affects hPDLSCs and investigated the underlying mechanisms that mediate its effects on osteoblast differentiation.
Materials and methods
hPDLSCs isolation and culture
Human periodontal ligament tissue was obtained from the premolars of 20 donors without oral or systematic diseases, during orthodontic procedures performed under informed consent. All donors were aged 12-25 years and consisted of 10 males and 10 females. Ethics Committee approval was provided by the The First Affliated Hospital of Harbin Medical University. hPDLSCs were harvested and cultured as previously described [25,26]. Briefly, tissue attached to the middle third of the tooth root was collected, sectioned (1 mm3 pieces) and digested for 1 hour at 37°C in Type I collagenase (3 mg/mL, Life Technologies, Carlsbad, CA, USA) and dispase (4 mg/mL, Life Technologies, Carlsbad, CA, USA). Tissues were cultured in complete α-minimum essential media (Gibco, Gaithersburg, MD, USA) plus 10% fetal bovine serum (FBS, Gibco, Gaithersburg, MD, USA), as well as penicillin and streptomycin (100 μg/mL) (HyClone, South Logan, UT, USA), and L-glutamine (5 mM) (Gibco, Gaithersburg, MD, USA). Media was replaced every 3-days. At 80% confluence, hPDLCs were split and sub-cultured.
Colony-forming efficiency assays
Colony-forming efficiency was assessed as previously described [25]. Briefly, hPDLSCs were seeded into 10 cm culture dishes (1×103 cells per dish) for 14 days and fixed in 4% paraformaldehyde (PFA) for 20 min. 0.1% (w/v) crystal violet (Sigma-Aldrich, St. Louis, MO, USA) was used to stain cells. Colony units, classed as clusters of ≥ 50 cells, could then be enumerated.
WST-1 proliferation assays
The toxicity of TSA (Sigma-Aldrich) on hPDLSCs was assessed via WST-1 cell cytotoxicity assays (Roche, Germany). Briefly, after plating cells in 96-well plate (5×103 cells/well) for 24 hrs and treated with varying drug concentrations. WST-1 was added each-day over a period of 7 days (10 µl per well) and a 4 hrs incubation was then performed in the dark in a humid environment. Microplates were mixed for 1 min via gentle vibration and read on a Bio-Rad 680 plate-reader at 450 nm with a 620 nm as a reference wavelength (Life Science Research, Japan).
Immunofluorescence analysis
4% PFA in PBS was used to fix hPDLSCs for 20 min and 5% BSA in PBS was used to block cells for 1 hr. Primary antibodies labeled cells overnight at 4°C and stained with AF488-conjugated secondary goat anti-mouse IgG (1:300; Life Technologies, Carlsbad, CA, USA) for 45 min. Cell nuclei were Hoechst stained (Life Technologies). A Zeiss Axio Observer Z1 (Cael Zeiss, Oberkochen, Germany) was used for all cell imaging.
Surface marker expression
For the assessment of surface markers, 5×105 cells were PBS washed and subsequently labeled with PE-conjugated antibodies against CD44, CD90, CD105, CD166, CD34, CD45, and HLA-DR (1:10; BD Biosciences, Franklin Lakes, NJ, USA). Fluorescent intensities were assessed using a BD Accuri C6 (BD Biosciences) immediately after staining. The CF Low Plus Software (BD Biosciences) was used for analyses.
Osteoblast and adipocyte differentiation
For osteoblast differentiation, hPDLSCs seeding at 1×105 per-well was conducted in 6 well plates. After adhering, treatment with osteogenic medium containing 10% FBS, 10 mM β-glycerophosphate, 10 nM dexamethasone and 50 μg/mL ascorbic acid was performed. Every 3 days this media was replaced.
For adipocyte differentiation, hPDLSCs seeding at 1×105 per-well of a 6-well plate was conducted, and cells were grown in adipogenic medium (Cyagen Biosciences, Santa Clara, CA, USA). After 2 weeks, 4% PFA was used to fix cells which were then stained for 15 min with Oil red O. Cells were imaged using the Zeiss Axio Observer Z1.
Alizarin red staining
Following culture in osteogenic medium, 4% PFA was used to fix hPDLSCs and cells were then stained with Alizarin red S (Sigma-Aldrich) for 15 min. Cells were washed, imaged and areas positive for Alizarin red were recognized as mineralized nodules. 10% cetylpyridinium chloride (Sigma-Aldrich) was used to dissolve nodules, followed by 562 nm absorbance being read with a spectrophotometer.
Alkaline phosphatase activity assay
hPDLSCs were lysed and alkaline phosphatase (ALP) activity assessed (Sigma-Aldrich). Absorbance was measured at 405 nm on a spectrophotometer.
qRT-PCR
Trizol (Life Technologies) was used for RNA extraction, and RNA concentrations were determined by spectrophotometry. For mRNA quantification, 1 μg of RNA was reverse-transcribed using the Reverse Transcriptase M-MLV Kit (Sigma-Aldrich). SYBR green qPCR was performed as followed protocal: 95°C for 10 min; 35 cycles of denaturation: 95°C, 30 s, annealing: 60°C, 30 s, extension at 72°C for 60 s. β-Actin served as the control gene. RNA (1μg) was reverse-transcribed and qPCR was performed on a Roche Light Cycler. The sequences of primers are listed as followed: ALP, Forward: 5’-AACCCAGACACAAGCATTCC-3’, Reverse: 5’-GCCTTTGAGGTTTTGGTCA-3’; OCN, Forward: 5’-ATGAGAGCCCTCACACTCCTC-3’, Reverse: 5’-GCCGTAGAAGCGCCGATAGGC-3’; OPN, Forward: 5’-CGAGGTGATAGTGTGGTTTATGGA-3’, Reverse: 5’-CGTCTGTAGCATCAGGGTACTG-3’; Runx2, Forward: 5’-CCAAATTTGCCTAACAGAATG-3’, Reverse: 5’-GAGGCTGTGGTTTCAAAGCA-3’; LPL, Forward: 5’-AGAGCCAAAAGAAGCAG-3’, Reverse: 5’-GGCAGCGTGAATGGGAT-5’; PPARγ, Forward: 5’-TTCAGCTCTGGGATGACCTT-3’, Reverse: 5’-CGAAGTTGGTGGGCCAGAAT-3’; β-actin, Forward: ACCTCCTACAATGAGCTGC Reverse: TGCCAATAGTGATGACCT.
Apoptosis assays
For the analysis of hPDLSC apoptosis, TSA was used to treat cells for 48 hrs in the absence of serum after first serum starving the cells for a 24 hrs period. Control cells were cultured in 10% FBS (positive controls). Apoptosis was analyzed by nuclear staining with Hoechst (Invitrogen). Annexin V-FITC/propidium iodide (PI) staining was performed using annexin V-FITC (Cell signaling technology) and propidium iodide (PI) (Life Technologies). A flow cytometer (CytoFLEX; Beckman Coulter, Brea, CA, USA) was used to analyze these staining conditions.
Western blot analysis
RIPA buffer was used to lyses cells, and the BCA assay (Pierce, Appleton, WI, USA) was used to determine protein concentraiton. 50 μg of total protein was run on a 10% SDS-Page gel and transferred to a nitrocellulose membrane that was then blocked using 5% non-fat milk in PBS containing 0.1% Tween-20 (PBS-T) for 1 hr prior to ALP (Sigma-Aldrich), OCN (Cell signaling technology), OPN (Santa Cruz Biotechnology), Runx2 (Boster Bio, Pleasanton, CA, USA), p-ERK (Cell signaling technology), ERK (Cell signaling technology), p-p38 (Cell signaling technology), p38 (Cell signaling technology), Cleaved-PARP (Cell signaling technology), Cleaved caspase 3 (Cell signaling technology), Ki67 (Cell signaling technology), PCNA (Cell signaling technology), p-JNK (Abcam), JNK (Abcam), LPL (Sigma-Aldrich), and β-actin (Sigma-Aldrich) antibodies labeling overnight at 4°C. Secondary antibodies were then used to label proteins for 30 min, and blots were imaged via odyssey two-color infrared laser imaging system (LI-COR Biosciences, USA). Blots were repeated a minimum of three times and the Image-ProPlus 5.0 software (Media Cybernetics Inc., Rockvill, MD, USA) was used for quantification.
Statistical analysis
Values are given as mean ± SD from a minimum of three replicates. SPSS 20.0 was employed for all statistical testing. Student’s t-test and ANOVA with post-hoc tests were conducted for inter-group comparisons. A p-value < 0.05 was the threshold for statistical significance.
Results
Characterization of hPDLSCs
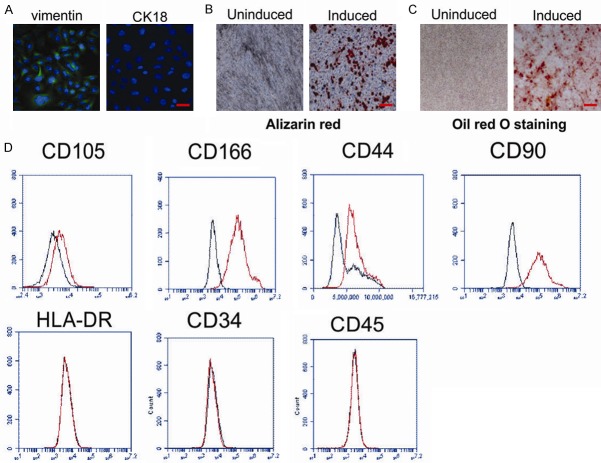
hPDLSCs were collected from the root surface of healthy premolars and mixed to decrease individual differences. Immunocytochemical staining was used to detect markers of mesenchymal and epithelial cells. Cells sub-cultured to a third passage displayed positive expression of vimentin, a marker of mesenchymal stem cells (MSCs), and negatively expressed CK18, an epithelium marker (Figure 1A). Next, hPDLSCs were grown in osteogenic and adipogenic induction media for 21 days and assessed. Cells displayed Alizarin red-positive calcium deposition (Figure 1B) and Oil red O-positive cytoplasmic lipid accumulation (Figure 1C). We further observed that treated hPDLSCs expressed markers of MSCs including CD105, CD166, CD44, and CD90, and negative for CD34, CD45, and HLA-DR (Figure 1D). Taken together, these results demonstrate that isolated hPDLSCs are stem cells of mesenchymal origin with powerful multipotency.
Figure 1.
Characteristics of human periodontal ligament stem cells (hPDLSCs). (A) Vimentin and CK18 expression was analyzed by immunocytochemical staining in the 3rd passage of hPDLSCs. Scale bar, 20 μm. (B, C) Multiple differentiation ability of hPDLSCs was demonstrated by Alizarin red (B) and Oil red O staining (C) under specific differentiation conditions for osteoblasts or adipocytes. Scale bar, 100 μm. (D) Cell surface markers including CD105, CD166, CD44, CD90, HLA-DR, CD34, and CD45 of hPDLSCs were detected by flow cytometry. Scale bar, 100 μm.
TSA enhances hPDLSC proliferation
We assessed the effects of TSA on hPDLSC viability using WST-1 assays. Our results demonstrate that PDLSCs treated with 2.5 or 5 μM TSA display higher proliferation rates compared to untreated controls (Figure 2A). TSA treatment at these concentrations also markedly enhanced the number of EdU-positive hPDLSCs (Figure 2B and 2C), and the expression of Ki67 and PCNA (Figure 2D). We selected 5 μM TSA for subsequent experiments as this concentration was the most potent stimulator of proliferation.
Figure 2.
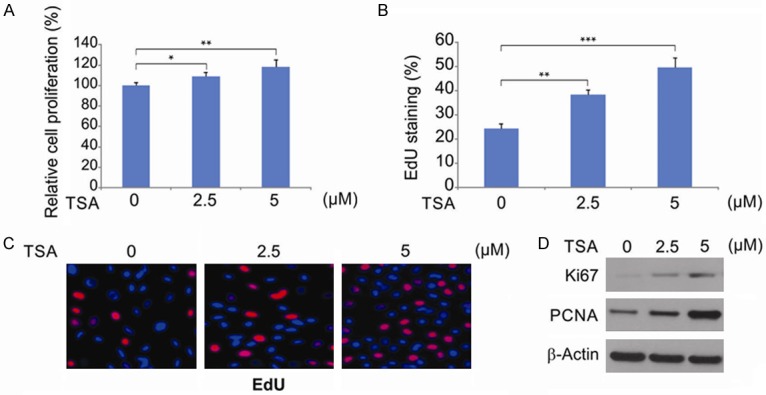
Effects of TSA on hPDLSCs proliferation. A. hPDLSCs were treated with difference concentration of TSA for 48 hrs, the number of viable hPDLSCs was analyzed by WST-1 assay. Data were presented as the mean ± SD. *, P < 0.05, **, P < 0.01 (ANOVA with Tukey’s post hoc test). B. Quantification of EdU-positive hPDLSCs was expressed by the percentage of DAPI-positive cells, and the data were presented as mean ± SD. **, P < 0.01, ***, P < 0.001 (ANOVA with Tukey’s post hoc test). C. EdU labelling analysis showed the fluorescence images of PDLSCs stimulated with TSA. Scale bar: 100 μm. D. hPDLSCs were treated with difference concentration of TSA for 48 hrs. Indicated protein level was analyzed by western blotting.
TSA influences the apoptotic induction of hPDLSCs
The effects of TSA on hPDLSC apoptosis were analyzed by nuclear staining and flow cytometry after staining of cells with FITC-annexin V and PI. In our research, the apoptosis rates of positive control (culture medium with 10% FBS) were significantly lower than negative control (culture medium without FBS and TSA). In addition, treated the hPDLSCs with 2.5 and 5 μM TSA attenuated FBS deprivation-induced apoptosis, while there was no significant difference between 5 μM TSA and negative control (Figure 3A and 3B). In addition, we detected cleaved-PARP1 and cleaved-caspase 3 in hPDLSCs, and found that FBS deprivation-induced apoptosis was attenuated by TSA treatment (Figure 3C).
Figure 3.
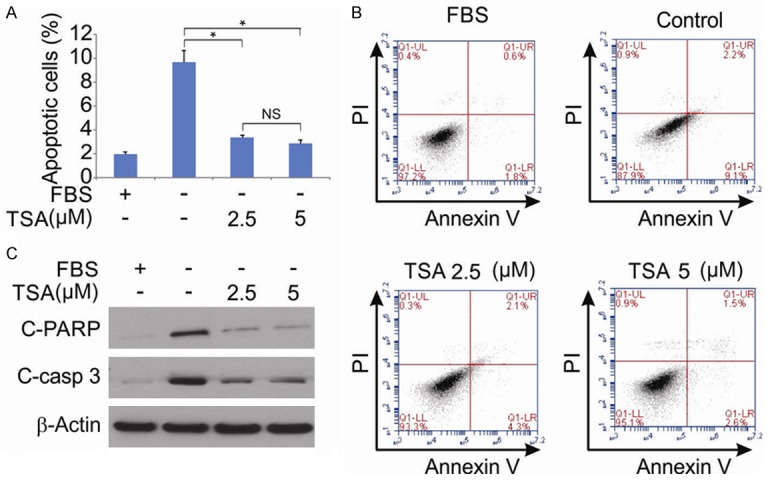
Effect of TSA on hPDLSCs apoptosis. A. hPDLSCs were treated with 10% FBS, indicated concentration of TSA for 48 hrs after serum starvation for 24 hrs, and cell apoptosis was determined by a nuclear fragmentation assay. Data were presented as mean ± SD. *, P < 0.05 (ANOVA with Tukey’s post hoc test). B. hPDLSCs were treated with 10% FBS, indicated concentration of TSA for 48 hrs after serum starvation for 24 hrs, and cell apoptosis was determined by flow cytometry using annexin V-FITC and propidium iodide (PI) staining. C. hPDLSCs were treated with 10% FBS, indicated concentration of TSA for 48 hrs after serum starvation for 24 hrs. Cleaved-PARP and cleaved-caspase 3 were determined by western blotting.
TSA induces osteogenic differentiation of PDLSCs
To assess influence of TSA on osteogenesis, hPDLSCs were treated with varying concentrations of TSA and ALP activity (an early osteogenic differentiation marker) assessed. In hPDLSCs treated with TSA at day 21, ALP activity, transcription and translation significantly increased, with the largest increase in activity evident for 5 μM TSA (Figure 4A-C), suggesting that TSA promotes osteogenic differentiation. To further confirm these findings, we investigated how TSA affects osteogenesis-related gene expression. At day 14 post-TSA stimulation, the expression of OCN and OPN were significantly upregulated, confirming its effects on osteogenesis (Figure 4D and 4E). Runx2 regulates the expression of osteogenic genes in osteoblasts, making it the central key factor regulating the differentiation of these cells [27,28]. Following 48 hrs of TSA treatment, Runx2 expression increased in a concentration dependent manner (Figure 4F and 4G). From this data, we concluded that TSA induces hPDLSC osteogenic differentiation via Runx2 upregulation.
Figure 4.
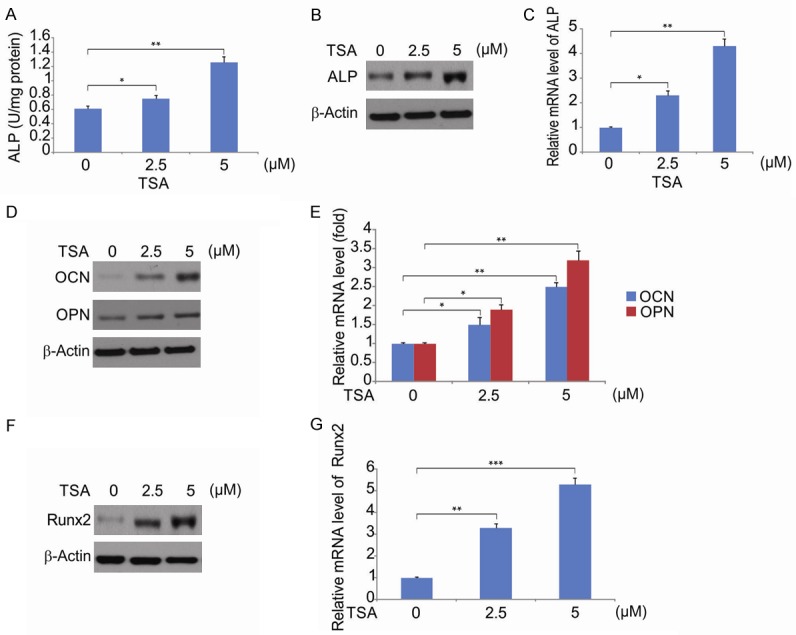
Effect of TSA on osteogenesis differentiation of hPDLSC. A. hPDLSCs were treated with difference concentration of TSA for 48 hrs, ALP activity assay was measured. Data were presented as the mean ± SD of three independent experiments. *, P < 0.05, **, P < 0.01 (ANOVA with Tukey’s post hoc test). B. The expression of ALP in control, TSA-treated hPDLSCs was examined using western blotting. C. The mRNA level of ALP in control, TSA-treated hPDLSCs was examined using qRT-PCR. Data were presented as the mean ± SD of three independent experiments. *, P < 0.05, **, P < 0.01 (ANOVA with Tukey’s post hoc test). D. The expression of OPN and OCN in control or TSA-treated hPDLSCs was examined using western blotting. E. The mRNA level of OPN and OCN in control or TSA-treated hPDLSCs was examined using qRT-PCR. Data were presented as the mean ± SD of three independent experiments. *, P < 0.05, **, P < 0.01 (ANOVA with Tukey’s post hoc test). F. hPDLSCs were treated with TSA. The relative gene expression of Runx2 was measured by western blotting. G. hPDLSCs were treated with TSA. The relative gene expression of Runx2 was measured by qRT-PCR. Data were presented as the mean ± SD of three independent experiments. **, P < 0.01, ***, P < 0.001 (ANOVA with Tukey’s post hoc test).
TSA decreases adipogenic differentiation
To investigate the effects of TSA during adipogenesis, hPDLSCs treated with TSA were induced into adipogenesis and Oil red O labeling and qRT-PCR performed. Staining results revealed that, 2-weeks post-induction, cells treated with TSA showed weaker adipogenesis (Figure 5A). Western blotting and qRT-PCR data that assesses the expression of adipogenic marker genes including LPL and PPARγ 1-week post-induction, confirmed these findings (Figure 5B and 5C).
Figure 5.
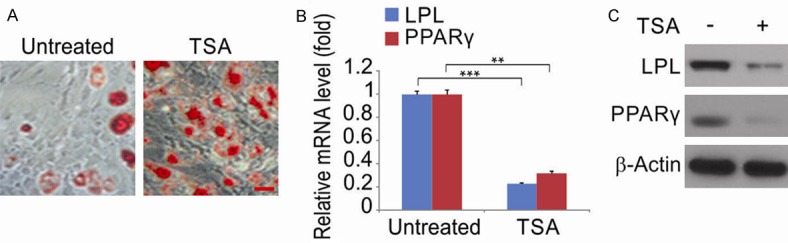
TSA regulates adipogenic differentiationof hPDLSCs. A. Oil red O staining of hPDLSCs treated with 5 μM TSA. Scale bar, 50 μm. B. hPDLSCs were treated with 5 μM TSA. The mRNA level of adipogenic marker genes LPL and PPARγ were evaluated by qRT-PCR. Data were presented as the mean ± SD of three independent experiments. **, P < 0.01, ***, P < 0.001 (Student’s t-test). C. hPDLSCs were treated with 5 μM TSA. The protein level of adipogenic marker genes LPL and PPARγ were evaluated by western blotting.
The role of ERK1/2 signaling during TSA-induced osteogenic differentiation
Runx2 expression is regulated by a myriad of signaling cascades during osteogenic differentiation [29,30]. Of these pathways, MAPK signaling is the most extensively studied. We thus next investigated if MAPK activation mediates TSA-induced osteogenic differentiation. Runx2 levels were assessed following treatment of hPDLSCs with 5 µM TSA for 48 hrs in the presence or absence of specific MAPK inhibitors. As shown in Figure 6A, the TSA mediated increase in Runx2 expression was inhibited by the ERK1/2 inhibitors UO126 and PD98059, whilst the inhibition of p38 and JNK (using SB203580 and SP600125, respectively) had no effect. When the expression of these MAPK signaling components was assessed at the protein level following 5 μM TSA treatment, we observed increased ERK1/2 phosphorylation, whilst the levels of p-p38 showed no changes and p-JNK could not be detected (Figure 6B). We next treated hPDLSCs with 5 μM TSA in the presence or absence of UO126 and PD98059 for 14 days. We observed that inhibition of ERK1/2 significantly disrupted the mineralization induced by TSA-induced (Figure 6C). Thus, we have identified a requirement for ERK1/2 activation during TSA-induced Runx2 expression and hPDLSC differentiation.
Figure 6.
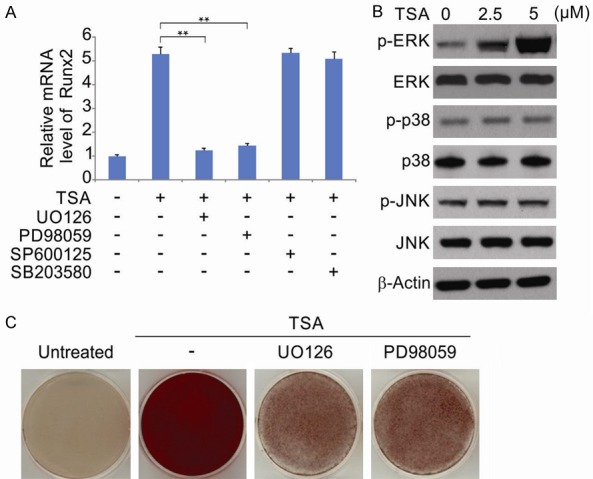
TSA induces ERK1/2 phosphorylation and the activity of ERK1/2 is required for TSA-induced osteogenesis of hPDLSCs. A. hPDLSCs were treated with 5 μM TSA with or without UO126, PD98059, SP600125 and SB203580 for 48 hrs. The relative gene expression of Runx2 was detected by qRT-PCR. The data were presented as the mean ± SD of three independent experiments. **, P < 0.01 (ANOVA with Tukey’s post hoc test). B. Cells were treated with TSA. The phosphorylated ERK1/2 (p-ERK1/2), phosphorylated p38 (p-p38), phosphorylated JNK (p-JNK), ERK1/2, p38, JNK and β-actin were detected by western blotting. C. Cells were treated with TSA with or without UO126, PD98059 for 14 days. The mineralized nodules formation was detected by Alizarin Red S staining.
Discussion
In recent years, stem cell-based periodontal therapy has shown promising therapeutic potential, and stem cells are a key aspect of such therapy [7,31]. The type of stem cell to be used is a major contributor to the success of stem-cell based periodontal therapy [7]. In this regard, early work showed that human bone marrow mesenchymal stem cells (hBMMSCs) can regenerate periodontal tissue [32].
hPDLSCs are postnatal stem cells that can differentiate into adipocytes, neuronal-like cells, osteoblasts and odontoblasts [33,34]. Compared to hBMMSCs, hPDLSCs are more advantageous. Firstly, they are more accessible and can be collected during orthodontic procedures whilst hBMMSCs must be collected from bone marrow [35]. Secondly, hPDLSCs have a higher proliferation rate and express higher levels of specific transcription factors compared to hBMMSCs and other types of dental stem cells including those of the dental-pulp and periapical-follicles [36]. Thirdly, as the major aim of this study was to regenerate periodontal structures, hPDLSCs are preferable as they form cementum- and periodontal ligament-like structures in immunocompromised mice post-transplantation, whilst other types of MCSs are not reported to possess this ability [37]. Finally, hPDLSCs can restrict inflammatory and immune responses, which are remarkable since the majority of clinical patients who require periodontal regeneration are under inflammatory conditions [38,39]. Based on these unique characteristics, hPDLSCs were considered as the best candidates for dental clinical applications. Here, we demonstrate that TSA modulates the MAPK-Runx2 axis to regulate hPDLSC osteogenic differentiation.
TSA has been shown to induce heat shock protein 70 (HSP 70) and Bcl-2 expression, and disrupt Bax induction, leading to inhibition of apoptosis in neurons following ischemia/reperfusion injury [17]. Concerning the mechanism(s) for the effects of TSA during osteogenic differentiation of hPDLSCs, we found that TSA treatment increased the expression of Runx2 and OSX in hPDLSCs. Several studies highlight the role of these proteins during osteoblast function as both exhibit high levels of expression during the early stages of tooth development and bone formation [40,41]. Co-expression of Runx2 and OSX enhances the downstream transcription of osteogenic genes, particularly Runx2, which is suggested to be the key transcription factor for MSC osteogenic differentiation [42]. Accordingly, mice lacking Runx2 display a lack of osteoblasts and impaired bone formation [43]. Our results revealed that TSA treatment of hPDLSCs enhances Runx2 expression, suggesting it as the mechanism of its ability to promote osteoblast differentiation.
To better understand the osteogenic mechanism(s) of TSA, we investigated its influence on ERK1/2, p38, and JNK signaling. Previous study has shown that FGF-2 activation is involved in TSA-induced ERK1/2 activation [44]. In the current study, we found TSA actives ERK1/2 signaling pathway. The activation of ERK1/2 has been shown to be essential for Runx2 upregulation and transcriptional activity [45]. This is consistent with our findings, as we saw that ERK1/2 was activated upon TSA-induced osteogenic differentiation, while ERK inhibitors reduced the TSA-mediated upregulation of Runx2 expression. These findings suggest that ERK1/2 activation is the mechanism by which TSA induces Runx2 expression. The mechanism(s) by which TSA upregulates ERK1/2 activity were not identified and now warrant further investigation.
In conclusion, we demonstrate that TSA promotes hPDLSC differentiation via ERK activation. Our data increases our understanding of the molecular mechanism(s) that underpin hPDLSC differentiation and suggest that TSA has therapeutic potential during hPDLSC clinical applications.
Acknowledgements
This work was supported by Heilongjiang Provincial Health Department Project (2013-029).
Disclosure of conflict of interest
None.
References
- 1.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigbede AO, Babatope BO, Bamidele MK. Periodontitis and systemic diseases: a literature review. J Indian Soc Periodontol. 2012;16:487–491. doi: 10.4103/0972-124X.106878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maderal AD, Lee Salisbury P 3rd, Jorizzo JL. Desquamative gingivitis: clinical findings and diseases. J Am Acad Dermatol. 2018;78:839–848. doi: 10.1016/j.jaad.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 4.Scannapieco FA, Cantos A. Oral inflammation and infection, and chronic medical diseases: implications for the elderly. Periodontol 2000. 2016;72:153–175. doi: 10.1111/prd.12129. [DOI] [PubMed] [Google Scholar]
- 5.Ariyamuthu VK, Nolph KD, Ringdahl BE. Periodontal disease in chronic kidney disease and end-stage renal disease patients: a review. Cardiorenal Med. 2013;3:71–78. doi: 10.1159/000350046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangal B, Sugandhi A, Kumathalli KI, Sridhar R. Alternative medicine in periodontal therapy--a review. J Acupunct Meridian Stud. 2012;5:51–56. doi: 10.1016/j.jams.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Bassir SH, Wisitrasameewong W, Raanan J, Ghaffarigarakani S, Chung J, Freire M, Andrada LC, Intini G. Potential for stem cell-based periodontal therapy. J Cell Physiol. 2016;231:50–61. doi: 10.1002/jcp.25067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu W, Liang M. Periodontal ligament stem cells: current status, concerns, and future prospects. Stem Cells Int. 2015;2015:972313. doi: 10.1155/2015/972313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su F, Liu SS, Ma JL, Wang DS, E LL, Liu HC. Enhancement of periodontal tissue regeneration by transplantation of osteoprotegerin-engineered periodontal ligament stem cells. Stem Cell Res Ther. 2015;6:22. doi: 10.1186/s13287-015-0023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan J, Xu X, Lin J, Fan L, Zheng Y, Kuang W. Dental stem cell in tooth development and advances of adult dental stem cell in regenerative therapies. Curr Stem Cell Res Ther. 2015;10:375–383. doi: 10.2174/1574888x09666141110150634. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Liu P. Tanshinone II-A: new perspectives for old remedies. Expert Opin Ther Pat. 2013;23:149–153. doi: 10.1517/13543776.2013.743995. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Yu Y, Aori G, Wang Q, Kong D, Yang W, Guo Z, Zhang L. Tanshinone IIA attenuates diabetic peripheral neuropathic pain in experimental rats via inhibiting inflammation. Evid Based Complement Alternat Med. 2018;2018:2789847. doi: 10.1155/2018/2789847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XJ, Qian JX, Wei Y, Guo Q, Jin J, Sun X, Liu SL, Xu CF, Zhang GX. Tanshinone IIA sodium sulfonate attenuates LPS-induced intestinal injury in mice. Gastroenterol Res Pract. 2018;2018:9867150. doi: 10.1155/2018/9867150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H, Zhai C, Qian G, Gu A, Liu J, Ying F, Xu W, Jin D, Wang H, Hu H, Zhang Y, Tang G. Protective effects of tanshinone IIA on myocardial ischemia reperfusion injury by reducing oxidative stress, HMGB1 expression, and inflammatory reaction. Pharm Biol. 2015;53:1752–1758. doi: 10.3109/13880209.2015.1005753. [DOI] [PubMed] [Google Scholar]
- 15.Yin X, Yin Y, Cao FL, Chen YF, Peng Y, Hou WG, Sun SK, Luo ZJ. Tanshinone IIA attenuates the inflammatory response and apoptosis after traumatic injury of the spinal cord in adult rats. PLoS One. 2012;7:e38381. doi: 10.1371/journal.pone.0038381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y, Qian JX, Lu SQ, Chen JW, Zhao XD, Jiang Y, Wang LH, Zhang GX. Protective effects of tanshinone IIA sodium sulfonate on ischemia-reperfusion-induced myocardial injury in rats. Iran J Basic Med Sci. 2017;20:308–315. doi: 10.22038/ijbms.2017.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu YM, Ding GH, Huang J, Xiong Y. Tanshinone IIA pretreatment attenuates ischemia/reperfusion-induced renal injury. Exp Ther Med. 2016;12:2741–2746. doi: 10.3892/etm.2016.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei B, Li WW, Ji J, Hu QH, Ji H. The cardioprotective effect of sodium tanshinone IIA sulfonate and the optimizing of therapeutic time window in myocardial ischemia/reperfusion injury in rats. Atherosclerosis. 2014;235:318–327. doi: 10.1016/j.atherosclerosis.2014.05.924. [DOI] [PubMed] [Google Scholar]
- 19.Cheng L, Zhou S, Zhao Y, Sun Y, Xu Z, Yuan B, Chen X. Tanshinone IIA attenuates osteoclastogenesis in ovariectomized mice by inactivating NF-kB and Akt signaling pathways. Am J Transl Res. 2018;10:1457–1468. [PMC free article] [PubMed] [Google Scholar]
- 20.Guan R, Wang J, Li Z, Ding M, Li D, Xu G, Wang T, Chen Y, Yang Q, Long Z, Cai Z, Zhang C, Liang X, Dong L, Zhao L, Zhang H, Sun D, Lu W. Sodium tanshinone IIA sulfonate decreases cigarette smoke-induced inflammation and oxidative stress via blocking the activation of MAPK/HIF-1alpha signaling pathway. Front Pharmacol. 2018;9:263. doi: 10.3389/fphar.2018.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M, Luo Y, Hu P, Dou L, Huang S. Tanshinone IIA inhibits AGEs-induced proliferation and migration of cultured vascular smooth muscle cells by suppressing ERK1/2 MAPK signaling. Iran J Basic Med Sci. 2018;21:83–88. doi: 10.22038/IJBMS.2017.20100.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui H, Zhao J, Zhou L, Wen H, Deng W, Li C, Ji Q, Liu X, Feng Y, Chai N, Zhang Q, Cai J, Li Q. Tanshinone IIA inhibits beta-catenin/VEGF-mediated angiogenesis by targeting TGF-beta1 in normoxic and HIF-1alpha in hypoxic microenvironments in human colorectal cancer. Cancer Lett. 2017;403:86–97. doi: 10.1016/j.canlet.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Ren BX, Ji Y, Tang JC, Sun DP, Hui X, Yang DQ, Zhu XL. Effect of Tanshinone IIA intrathecal injections on pain and spinal inflammation in mice with bone tumors. Genet Mol Res. 2015;14:2133–2138. doi: 10.4238/2015.March.20.24. [DOI] [PubMed] [Google Scholar]
- 24.Tong Y, Xu W, Han H, Chen Y, Yang J, Qiao H, Hong D, Wu Y, Zhou C. Tanshinone IIA increases recruitment of bone marrow mesenchymal stem cells to infarct region via up-regulating stromal cell-derived factor-1/CXC chemokine receptor 4 axis in a myocardial ischemia model. Phytomedicine. 2011;18:443–450. doi: 10.1016/j.phymed.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Kang W, Du L, Ge S. Rho-kinase inhibitor Y-27632 facilitates the proliferation, migration and pluripotency of human periodontal ligament stem cells. J Cell Mol Med. 2017;21:3100–3112. doi: 10.1111/jcmm.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao S, Zhao W, Ou Q, Liang L, Lin X, Wang Y. MicroRNA-214 suppresses osteogenic differentiation of human periodontal ligament stem cells by targeting ATF4. Stem Cells Int. 2017;2017:3028647. doi: 10.1155/2017/3028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Li Z, Hou Y, Fang W. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am J Transl Res. 2015;7:2527–2535. [PMC free article] [PubMed] [Google Scholar]
- 28.Teplyuk NM, Zhang Y, Lou Y, Hawse JR, Hassan MQ, Teplyuk VI, Pratap J, Galindo M, Stein JL, Stein GS, Lian JB, van Wijnen AJ. The osteogenic transcription factor runx2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol Endocrinol. 2009;23:849–861. doi: 10.1210/me.2008-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruderer M, Richards RG, Alini M, Stoddart MJ. Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater. 2014;28:269–286. doi: 10.22203/ecm.v028a19. [DOI] [PubMed] [Google Scholar]
- 30.Zou L, Kidwai FK, Kopher RA, Motl J, Kellum CA, Westendorf JJ, Kaufman DS. Use of RUNX2 expression to identify osteogenic progenitor cells derived from human embryonic stem cells. Stem Cell Reports. 2015;4:190–198. doi: 10.1016/j.stemcr.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SH, Seo BM, Choung PH, Lee YM. Adult stem cell therapy for periodontal disease. Int J Stem Cells. 2010;3:16–21. doi: 10.15283/ijsc.2010.3.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moshaverinia A, Xu X, Chen C, Ansari S, Zadeh HH, Snead ML, Shi S. Application of stem cells derived from the periodontal ligament or gingival tissue sources for tendon tissue regeneration. Biomaterials. 2014;35:2642–2650. doi: 10.1016/j.biomaterials.2013.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Feng C, Gu X, He Q, Wei F. Effect of cryopreservation on proliferation and differentiation of periodontal ligament stem cell sheets. Stem Cell Res Ther. 2017;8:77. doi: 10.1186/s13287-017-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato H, Taguchi Y, Tominaga K, Kimura D, Yamawaki I, Noguchi M, Yamauchi N, Tamura I, Tanaka A, Umeda M. High glucose concentrations suppress the proliferation of human periodontal ligament stem cells and their differentiation into osteoblasts. J Periodontol. 2016;87:e44–51. doi: 10.1902/jop.2015.150474. [DOI] [PubMed] [Google Scholar]
- 35.Hu L, Liu Y, Wang S. Stem cell-based tooth and periodontal regeneration. Oral Dis. 2018;24:696–705. doi: 10.1111/odi.12703. [DOI] [PubMed] [Google Scholar]
- 36.Elahi KC, Klein G, Avci-Adali M, Sievert KD, MacNeil S, Aicher WK. Human mesenchymal stromal cells from different sources diverge in their expression of cell surface proteins and display distinct differentiation patterns. Stem Cells Int. 2016;2016:5646384. doi: 10.1155/2016/5646384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira NM, Zucconi E, Bueno CR Jr, Secco M, Suzuki MF, Bartolini P, Vainzof M, Zatz M. Human multipotent mesenchymal stromal cells from distinct sources show different in vivo potential to differentiate into muscle cells when injected in dystrophic mice. Stem Cell Rev. 2010;6:560–566. doi: 10.1007/s12015-010-9187-5. [DOI] [PubMed] [Google Scholar]
- 38.Zayed MN, Schumacher J, Misk N, Dhar MS. Effects of pro-inflammatory cytokines on chondrogenesis of equine mesenchymal stromal cells derived from bone marrow or synovial fluid. Vet J. 2016;217:26–32. doi: 10.1016/j.tvjl.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Rashed L, Gharib DM, Hussein RE, Tork O, Abusree A. Combined effect of bone marrow derived mesenchymal stem cells and nitric oxide inducer on injured gastric mucosa in a rat model. Tissue Cell. 2016;48:644–652. doi: 10.1016/j.tice.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang Y, Sun X, Zhang X, Wang X, Zhang C, Zheng S. RUNX2 mutation reduces osteogenic differentiation of dental follicle cells in cleidocranial dysplasia. Mutagenesis. 2018;33:203–214. doi: 10.1093/mutage/gey010. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Zhang C, Li X, Lv Z, Chen Y, Zhao J. Isoquercitrin promotes the osteogenic differentiation of osteoblasts and BMSCs via the RUNX2 or BMP pathway. Connect Tissue Res. 2018:1–11. doi: 10.1080/03008207.2018.1483358. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Campo FM, Santurtun A, Garcia-Ibarbia C, Pascual MA, Valero C, Garces C, Sanudo C, Zarrabeitia MT, Riancho JA. Osterix and RUNX2 are transcriptional regulators of sclerostin in human bone. Calcif Tissue Int. 2016;99:302–309. doi: 10.1007/s00223-016-0144-4. [DOI] [PubMed] [Google Scholar]
- 43.McGee-Lawrence ME, Carpio LR, Bradley EW, Dudakovic A, Lian JB, van Wijnen AJ, Kakar S, Hsu W, Westendorf JJ. Runx2 is required for early stages of endochondral bone formation but delays final stages of bone repair in Axin2-deficient mice. Bone. 2014;66:277–286. doi: 10.1016/j.bone.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen JL, Chen YS, Lin JY, Tien YC, Peng WH, Kuo CH, Tzang BS, Wang HL, Tsai FJ, Chou MC, Huang CY, Lin CC. Neuron regeneration and proliferation effects of danshen and tanshinone IIA. Evid Based Complement Alternat Med. 2011;2011:378907. doi: 10.1155/2011/378907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanno T, Takahashi T, Tsujisawa T, Ariyoshi W, Nishihara T. Mechanical stress-mediated Runx2 activation is dependent on Ras/ERK1/2 MAPK signaling in osteoblasts. J Cell Biochem. 2007;101:1266–1277. doi: 10.1002/jcb.21249. [DOI] [PubMed] [Google Scholar]