Abstract
Little is known about the role of long non-coding RNA SNHG7-1 in the development of recurrent spontaneous abortion (RSA). The aim of the present study was to investigate the levels of SNHG7-1 in villi of RSA, and explore its underlying mechanism. The qRT-PCR assay SNHG7-1 showed that the expression level was downregulated in RSA and HTR-8/SVneo cells. In addition, the growth rate of cells transfected with si-SNHG7-1 was significantly decreased compared to that with si-NC, which was reversed by miR-34a targeted with 3’-UTR. Moreover, miR-34a suppressed the expression of WNT1 by binding with the 3’-UTR, which interact with WNT1 to inhibit the proliferation of cells. Furthermore, miR-34a inhibitor rescued the dysregulation of WNT1, p-β-catenin, β-catenin, cyclinD and c-Myc by si-SNHG7A-1. In short, the current study suggests SNHG7-1 plays as an important role in RSA progression targeted by miR-34a via Wnt/β-catenin signaling pathway, providing a novel insight for the pathogenesis and underlying therapeutic target for RSA.
Keywords: Recurrent spontaneous abortion, SNHG7, miR-34a, Wnt/β-catenin signaling pathway
Introduction
Recurrent spontaneous abortion (RSA) is one of the common pregnancy complications, which refers to three or more consecutive pregnancy losses prior to the 20th week of gestation with about 1-3% incidence [1,2]. Many studies have reported that the causes of RSA involve chromosomal abnormality [3], anatomic deformation [4], endocrine disorder [5] and immunodeficiency [6]. However, there are still about 50% of recurrent abortions couldn’t be explained [7]. Thus, more research is needed to understand potential mechanisms and therapies of RSA.
Long noncoding RNAs (lncRNAs) are a type of evolutionarily highly conserved non-coding RNAs, with the length of more than 200 nucleotides [8]. LncRNAs play an important role in diverse cellular processes, such as regulation of gene expression, posttranslational processing and tumorigenesis [9,10].
SNHG7 (small nucleolar RNA host gene 7) is one of the identified lncRNAs, which is located on chromosome 9q34.3, with a length of 2176 bp [11]. SNHG7 have been reported to involve in multiple types of physiological processes, such as alternative splicing, nuclear organization, epigenetic modulating of gene expression, and a number of evidences indicate that SNHG7 also closely relate to various pathological processes in many cancers, such as lung cancer [12], hepatocellular carcinoma [13], gastric cancer [14] and renal cell carcinoma [15]. Nevertheless, the role of SNHG7 in RSA remains elusive.
MicroRNAs are short endogenous non-coding molecules (about 19-23 nucleotides) that regulate gene expression by binding to the 3’-untranslated regions (3’-UTRs) of target mRNAs, which play an important role in maintaining the homeostasis [16,17]. Multiple studies have reported microRNAs plays vital role in RSA. For example, miR-10a impairs expression of Bim to regulate RSA progression [18], the polymorphisms of miRNA-27a rs895819 A/G and Leptin rs7799039 G/A may contribute to an increased risk of RSA [19]. Whether miR-34a acts on RSA progression is still not very clear.
Taken together, our study revealed that SNHG7 was aberrantly down-regulated in RSA tissue and cells. The current study aims to investigate the levels of SNHG7 in the progression of RSAs, and explore its underlying mechanism.
Materials and methods
Patients and clinical samples collection
The villi tissues were obtained from patients with RSA and clinically normal pregnancies terminated for non-medical reasons, who visited Linyi Central Hospital from January 2017 to December 2017. The demographic characteristics and clinical profiles of RSA patients and control subjects are shown in Table 1. The RSA group consisted of 30 childless women aged 25-30 years with a history of three or more spontaneous abortions. Criteria for inclusion in RSA group: normal chromosomes of both husband and wife; normal reproductive endocrine; no diabetes, thyroid dysfunction and other systemic diseases; no organic deformity of uterus and genital tract; anti-endometrial test was negative; anti-cardiolipin antibody was negative; there was no inflammation in the genital tract; the sperm quality and activity of the husband were normal; toxoplasma, herpes simplex virus, cytomegalovirus and rubella virus antibodies IgM and IgG were all negative. The control group includes 30 pregnant women aged 25-30 years with request for termination of pregnancy because of unplanned pregnancy. Criteria for inclusion in control group: ultrasonic examination indicates the primitive cardiac tube pulsation; a history of live-birth deliveries; this pregnancy with no abdominal pain, no vaginal bleeding and other symptoms. The average gestational week of two groups was 8 weeks. Under sterile conditions, all chorionic villus samples were acquired by vacuum aspiration and washed with sterile saline to remove the excess blood, mucus and deciduas. All samples were snap-frozen in liquid nitrogen overnight and then stored at -80°C for further analysis. The study was approved by the ethics committee of Linyi Central Hospital and written informed consent was obtained from each participant with the following reference number: 20161201.
Table 1.
Clinical profiles of RSA patients and control subjects
| Subject | Control (n=30) | RSA (n=30) | P |
|---|---|---|---|
| Age (years) | 28.43 ± 5.97 | 27.93 ± 6.41 | 0.912 |
| BMI (kg/m2) | 21.89 ± 4.23 | 22.43 ± 4.01 | 0.653 |
| Number of miscarriages | NA | 4.89 ± 1.01 | |
| Live birth | 1.81 ± 0.57 | NA | |
| Gestation age (weeks) | 8.45 ± 0.23 | 8.57 ± 0.35 | 0.896 |
Note: RSA = recurrent spontaneous abortion; BMI = body mass index; NA = not applicable; values are mean ± standard deviation. P values were calculated using the two-sided t-test for continuous variables.
Cell culture and transfection
The HTR-8/SVneo cell lines (purchased from Jilin University) were cultured in RPMI1640 (Gibco, Invitrogen, USA) supplemented with 10% fetal bovine serum, heat inactivated, 1% penicillin/streptomycin, and 1% L-glutamine at 37°C in a humidified, 5% CO2 atmosphere. For transfection, cells were seeded into plates and transfected with si-SNHG7, miR-34a mimic and miR-34a inhibitor (Gene Pharm, Shanghai, China) mixed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, MA, USA) according to the manufacturer’s protocols.
Real time quantitative PCR analysis
The total RNA was extracted from cells using Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using a reverse transcription kit (Takara Biotechnology, Dalian, China). Relative quantity of cDNA was assessed by quantitative PCR. Relative levels of gene expression was expressed relative to GAPDH or U6 and calculated using the 2-ΔΔCt method.
MTT assay
Cells (3×103) were cultured in 96-well plates and incubated for 24 h and stained with 0.5 mg/ml MTT for 4 h. Supernatant was discarded and 200 μl of dimethylsulfoxide (DMSO) was added to dissolve precipitates. Samples were measured at 490 nm using an ELISA reader.
Colony formation assay
For the colony formation assay, cells were seeded into 6-well plates at a density of 5×103 cells per well. Then, cells were cultured at 37°C with 5% CO2 and cultured for 2 weeks. Finally, colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 30 min. Then, the number of colonies was counted on light microscopy.
Cell invasion assay
For invasion assay, the members of the upper chamber were precoated with Matrigel (Sigma), and then, HTR-8/SVneo cells at density of 1×105 per well were respectively seeded into the upper chamber filled with serum-free DMEM. The lower chamber was filled with culture medium with 10% FBS medium. After 24 hours incubation, cells adhered on the upper surface of membrane were removed, while the cells invaded into the lower chamber were stained with crystal violet (Sigma).
Luciferase reporter assay
The 3’-UTR of SNHG7 and WNT1 containing miR-34a binding sites was amplified and cloned into the pGL3-basic luciferase vector (Promega, USA), respectively. Cells were seeded into plates and transfected with involved oligonucleotides (the SNHG7 and WNT1 wild-type or mutant reporter vector, miR-34a mimics or negative control) mixed with Lipofectamine 2000 reagent according to the manufacturer’s protocols. Luciferase activity was detected after transfection 24 h using Dual-Luciferase Reporter System (Promega, Madison, WI, USA). Renilla luciferase was used as an internal reference.
Western blotting analysis
The cultured cells were washed with ice-cold PBS and lysed in RIPA buffer supplemented with protease inhibitor mixture. After electrophoresis, the protein samples were incubated with the primary antibody purchased from Abcam Campany (anti-WNT1, anti-β-catenin, anti-p-β-catenin, anti-cyclinD and anti-cMyc 1:1000 dilution). Samples were incubated with secondary antibodies conjugated by horseradish peroxidase (HRP). Bands were quantified using ImageJ software.
Apoptosis analysis by flow cytometry
At 24 h after transfection, the cells were examined for apoptotic status by double-staining with Annexin V and PI using an Annexin V-FITC/PI Apoptosis Detection Kit (KeyGen, Nanjing, China), followed by flow cytometric analysis.
Statistical analysis
Results are expressed as the mean ± standard deviation (SD). All calculations were performed using SPSS 17.0 software (IBM Software, Chicago, IL, USA) and GraphPad (vision 6.0, USA). A value of P<0.05 indicated a statistically significant difference.
Results
Reduced expression of SNHG7 in RSA
To investigate the potential role of SNHG7 in RSA pathogenesis, we first measured expression of SNHG7 in villi of RSA patients and normal pregnancy patients. As expected, SNHG7 level was significantly lower in RSA villi than it in normal pregnancy villi (Figure 1).
Figure 1.
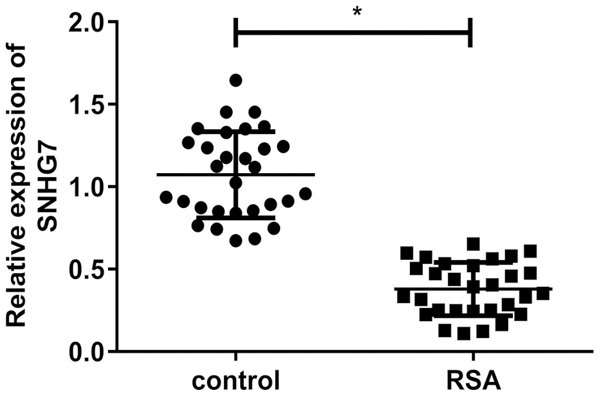
Reduced expression of SNHG7 in RSA. Expression of SNHG7 in RSA and normal pregnancy villi (*P<0.05).
Knockdown of SNHG7 suppressed proliferation and invasion, induced apoptosis in HTR-8/SVneo cells
To verify the role of SNHG7 in the development of RSA, we stably established SNHG7 silencing via siRNA transfection in HTR-8/SVneo cell line in vitro (Figure 2A). MTT assay showed that SNHG7 silencing inhibited the proliferation of HTR-8/SVneo cells significantly (Figure 2B). Quantitative analysis of apoptosis by flow cytometry revealed that silencing of SNHG7 markedly augmented apoptosis in cells compared with the non-transfected cells (Figure 2C). Colony formation assay revealed that SNHG7 silencing could induce HTR-8/SVneo cells formed significantly fewer colonies than control cells (Figure 2D). As demonstrated by transwell assay, SNHG7 silencing inhibited the invasion ability of cells (Figure 2E).
Figure 2.
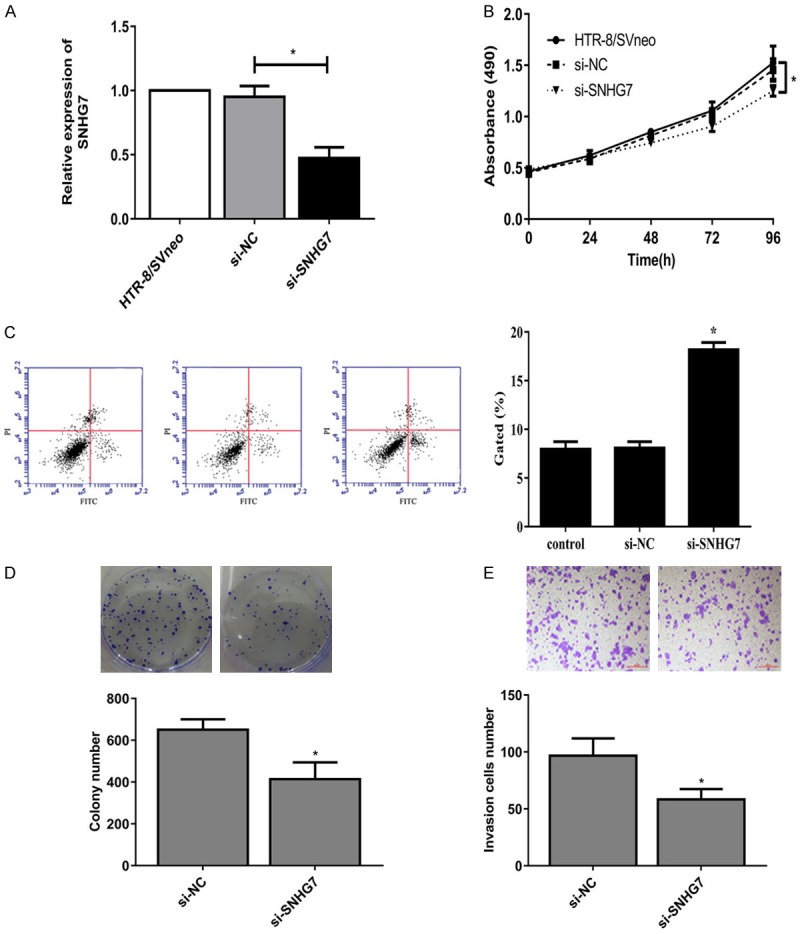
Knockdown of SNHG7 suppressed proliferation and invasion, induced apoptosis in HTR-8/SVneo cells. A: SNHG7 expression in HTR-8/SVneo cells transfected with siRNA or si-NC. B: MTT assay showed the proliferation ability. C: Flow cytometry showed apoptotic rate of HTR-8/SVneo cells. D: Colony formation assay showed the clone number in HTR-8/SVneo cells. E: Transwell assay showed the invasion of HTR-8/SVneo cells (*P<0.05).
SNHG7 acts as a molecular sponge for miR-34a
Increasing evidence had illustrated that lncRNAs function as sponges to bind specific miRNAs. Bioinformatics analysis was used to predict the candidate targets of microRNAs binding with SNHG7. Results found 3’UTR of SNHG7 was highly conserved to bind with miR-34a. The 3’-UTR binding sites can be seen in Figure 3A. MiR-34a level was significantly higher in RSA villi than it in normal pregnancy villi (Figure 3B). Luciferase reporter assay confirmed the binding with the decreasing fluorescence within miR-34a mimic and SNHG7 wild type (Figure 3C). Moreover, after HTR-8/SVneo cells were transfected with si-SNHG7, miR-34a expression levels were significantly up-regulated which was reversed by miR-34a inhibitor (Figure 3D, 3E). MTT assay revealed that miR-34a inhibitor could rescue the suppression by si-SNHG7 in the proliferation of HTR-8/SVneo cells 96 hours later (Figure 3F).
Figure 3.
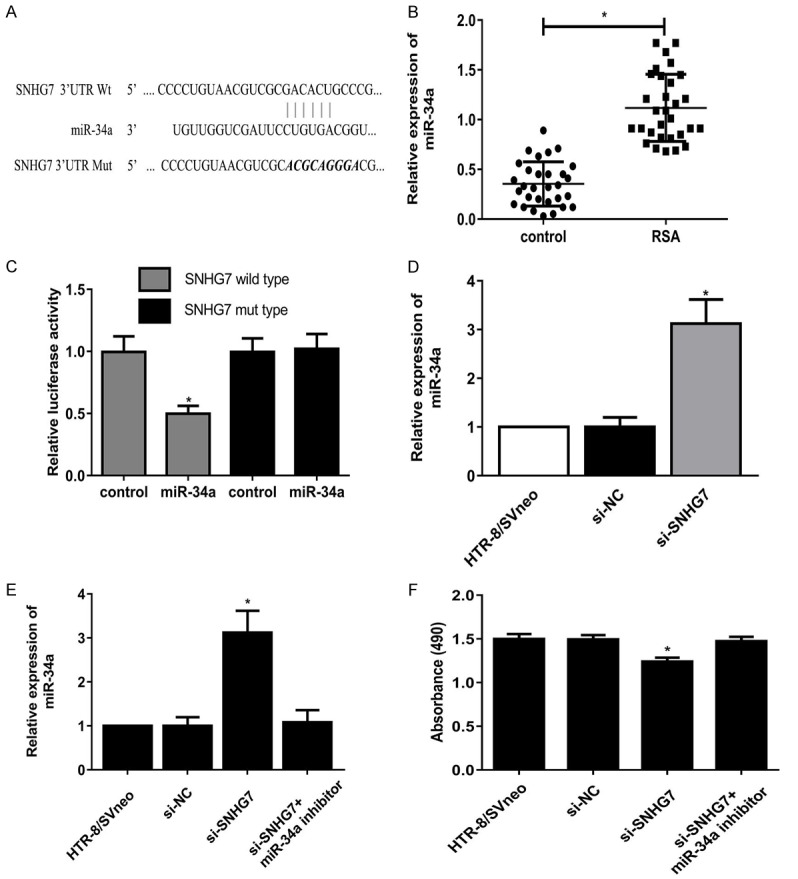
SNHG7 acts as a molecular sponge for miR-34a. A: The predicted binding sites within miR-34a and 3’-UTR of SNHG7. B: Expression of miR-34a in RSA and normal pregnancy villi. C: Luciferase reporter assay showed the decreasing fluorescence within miR-34a mimics and SNHG7 wild type. D, E: After HTR-8/SVneo cells were transfected with si-SNHG7 and miR-34a inhibitor, miR-34a expression levels in cells. F: After HTR-8/SVneo cells were transfected with si-SNHG7 and miR-34a inhibitor, MTT assay showed the proliferation ability of cells (*P<0.05).
miR-34a directly targets with 3’-UTR of WNT1
We examined computationally predicted targets of miR-34a using bioinformatics analysis. Results revealed 3’UTR of WNT1 was highly conserved to bind with miR-34a. The 3’-UTR binding sites can be seen in Figure 4A. WNT1 level was significantly lower in RSA villi than it in normal pregnancy villi (Figure 4B). Luciferase reporter assay showed transfection of miR-34a could significantly restrict the relative luciferase activity in cells, suggesting miR-34a has inhibitory effects on WNT1 expression via interaction with the 3’-UTR of WNT1 (Figure 4C). Moreover, as shown in Figure 4D and 4E, overexpression of miR-34a level obviously downregulated WNT1 expression in HTR-8/SVneo cells at the mRNA and protein levels. Overall, our study discovered that miR-34a suppressed the expression of WNT1 by binding with the 3’-UTR.
Figure 4.
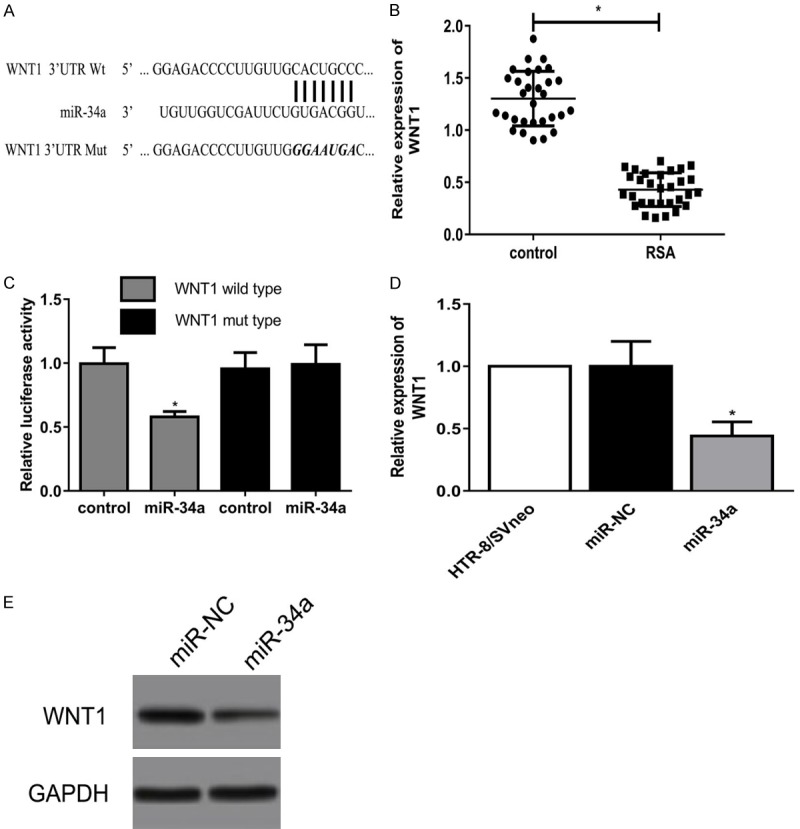
MiR-34a directly targets with 3’-UTR of WNT1. A: The predicted binding sites within miR-34a and 3’-UTR of WNT1. B: Expression of WNT1 in RSA and normal pregnancy villi. C: Luciferase reporter assay showed the decreasing fluorescence within miR-34a mimics and WNT1 wild type. D, E: WNT1 expression in HTR-8/SVneo cells transfected with miR-34a mimics at the mRNA and protein levels (*P<0.05).
SNHG7 silencing repressed the Wnt/β-catenin signaling pathway
As shown in Figure 5A, statistically significant differences were found in the expressions of WNT1, p-β-catenin and β-catenin among the different groups 48 h after transfection. Compared with the control, si-SNHG7 had significantly suppressed WNT1 and β-catenin expressions but promoted p-β-catenin expression, which was reversed by miR-34a inhibitor. PCR and western blot demonstrated significant differences in the expression of cyclinD and c-Myc, which were target genes of β-catenin. Si-SNHG7 had significantly inhibited cyclinD and c-Myc expressions, which were rescued by miR-34a inhibitor (Figure 5B).
Figure 5.
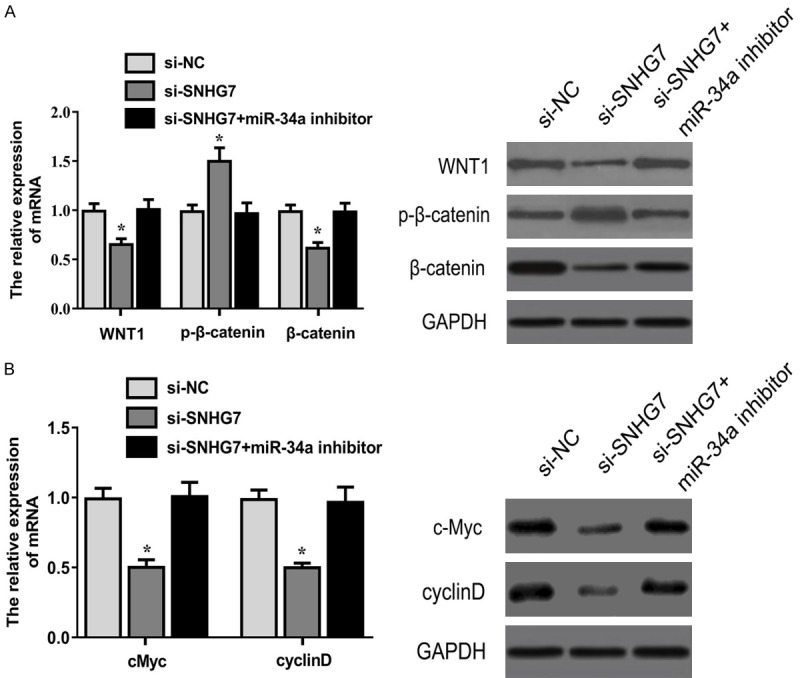
SNHG7 silencing repressed the Wnt/β-catenin signaling pathway. Expressions of WNT1, p-β-catenin, β-catenin, c-Myc and cyclinD in HTR-8/SVneo cells 48 h after transfection was detected by qRT-PCR and western blot (*P<0.05).
Discussion
Pregnancy is a complex and multicomponent process, in which embryonic implantation is the one of key factors [20]. In humans, embryonic implantation is the stage of pregnancy at which the already embryo enters the uterine cavity and adheres to the wall of the uterus. The controlled invasion of trophoblast cells plays an important role in embryonic implantation [21]. When the capacity of invasion and metastasis of trophoblast cells was decreased, fertilized egg development was retarded which could result in miscarriage [22].
Increasing researches has indicated that lncRNAs are involved in important biological processes, such as genomic imprinting [23], cell differentiation and proliferation, modulation of migration [24], and apoptosis [25]. In addition, abnormal expression of lncRNAs has been implicated in the pathogenesis many diseases [26]. SNHG7, which was first identified in lymphoblastoid cells [27], was found to be associated with the development of many cancers. As expected, SNHG7 level was significantly lower in RSA villi than it in normal pregnancy villi in our study, implying SNHG7 could promote the proliferation and invasion of trophoblast cells in vivo.
Resembling the behavior of trophoblast cells, many studies have been conducted based on the use of HTR-8/SVneo cell line in order to simulate the behavior of trophoblast cells in pregnancy [28]. We stably established SNHG7 silencing via siRNA transfection in thsi cell line in vitro. Results showed that SNHG7silencing inhibited the proliferation and invasion of HTR-8/SVneo cells significantly, suggesting the underlying role of SNHG7in RSA in vitro.
lncRNAs function as sponges for common miRNAs and abolished the endogenous suppressive effect of these miRNAs on their targeted transcripts [29]. Utilizing bioinformatics prediction program, we found 3’UTR of SNHG7 was highly conserved to bind with miR-34a. After cells were transfected with si-SNHG7, miR-34a expression levels were significantly up-regulated. And miR-34a inhibitor could rescue the suppression by si-SNHG7 in the proliferation of HTR-8/SVneo cells. These data illustrated SNHG7 could regulate the proliferation of trophoblast cells by targeting miR-34a.
Accumulating data indicated that abnormal expression of Wnt/β-catenin signaling way might be associated with pathogenesis of RSA [30,31]. As the central effector of the signaling pathway, translocation of β-catenin to the nucleus leads to expression of growth- and invasion associated genes, such as cyclin D and cMyc [32]. Bioinformatics analysis showed that miR-34a closely target with 3’-UTR of WNT1 in our study. Moreover, over-expression of miR-34a could lower the WNT1 mRNA and related protein production. Furthermore, si-SNHG7 suppressed WNT1 and β-catenin expressions but promoted p-β-catenin expression significantly, which was reversed by miR-34a inhibitor. PCR and western blot demonstrated significant differences in the expression of cyclinD and c-Myc, which were target genes of β-catenin. Si-SNHG7 had significantly inhibited cyclinD and c-Myc expressions, which were rescued by miR-34a inhibitor. These results demonstrated si-SNHG7 regulated the proliferation and invasion of trophoblast cells through inactivation of Wnt/β-catenin signaling pathway in vitro.
Conclusion
The current study suggests SNHG7 plays as an important role in RSA progression targeted by miR-34a via Wnt/β-catenin signaling pathway, providing a novel insight for the pathogenesis and underlying therapeutic target for RSA.
Acknowledgements
This work was supported by Linyi Central Hospital.
Disclosure of conflict of interest
None.
References
- 1.Rai R, Regan L. Recurrent miscarriage. Lancet. 2006;368:601–611. doi: 10.1016/S0140-6736(06)69204-0. [DOI] [PubMed] [Google Scholar]
- 2.Delabaere A, Huchon C, Deffieux X, Beucher G, Gallot V, Nedellec S, Vialard F, Carcopino X, Quibel T, Subtil D, Barasinski C, Gallot D, Vendittelli F, Laurichesse-Delmas H, Lemery D. [Epidemiology of loss pregnancy] . J Gynecol Obstet Biol Reprod (Paris) 2014;43:764–775. doi: 10.1016/j.jgyn.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Caseiro AL, Regalo A, Pereira E, Esteves T, Fernandes F, Carvalho J. Implication of sperm chromosomal abnormalities in recurrent abortion and multiple implantation failure. Reprod Biomed Online. 2015;31:481–485. doi: 10.1016/j.rbmo.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Li TC, Makris M, Tomsu M, Tuckerman E, Laird S. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. 2002;8:463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 5.Kaur R, Gupta K. Endocrine dysfunction and recurrent spontaneous abortion: an overview. Int J Appl Basic Med Res. 2016;6:79–83. doi: 10.4103/2229-516X.179024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patriarca A, Piccioni V, Gigante V, Benedetto C. The use of intravenous immunoglobulin in sine causa or alloimmune recurrent spontaneous abortion (RSA) Panminerva Med. 2000;42:193–195. [PubMed] [Google Scholar]
- 7.Kutteh WH, Hinote CD. Antiphospholipid antibody syndrome. Obstet Gynecol Clin North Am. 2014;41:113–132. doi: 10.1016/j.ogc.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 12.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 13.Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med. 2017;6:2932–2941. doi: 10.1002/cam4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, Xia TS. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–4622. [PubMed] [Google Scholar]
- 15.He HT, Xu M, Kuang Y, Han XY, Wang MQ, Yang Q. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. Onco Targets Ther. 2016;9:6399–6406. doi: 10.2147/OTT.S116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 17.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Wang XQ, Zhang L, Lv XD, Su X, Tian S, Liu CM, Ma X, Xia HF. A SNP in pri-miR-10a is associated with recurrent spontaneous abortion in a Han-Chinese population. Oncotarget. 2016;7:8208–8222. doi: 10.18632/oncotarget.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CY, Wang SG, Wang JL, Zhou LY, Liu HJ, Wang YF. Effect of miRNA-27a and leptin polymorphisms on risk of recurrent spontaneous abortion. Med Sci Monit. 2016;22:3514–3522. doi: 10.12659/MSM.897147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knofler M, Pollheimer J. Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet. 2013;4:190. doi: 10.3389/fgene.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hustin J, Jauniaux E, Schaaps JP. Histological study of the materno-embryonic interface in spontaneous abortion. Placenta. 1990;11:477–486. doi: 10.1016/s0143-4004(05)80193-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Li H, Zhang L, Jia L, Wang P. Differential expression of beta-catenin and Dickkopf-1 in the third trimester placentas from normal and preeclamptic pregnancies: a comparative study. Reprod Biol Endocrinol. 2013;11:17. doi: 10.1186/1477-7827-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 25.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. Adv Exp Med Biol. 2014;825:129–158. doi: 10.1007/978-1-4939-1221-6_4. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. 2013;14:9099–9110. doi: 10.3390/ijms14059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson T, Risto O, Knutsson A, Wahlstrom O. Heterotopic ossification following internal fixation or arthroplasty for displaced femoral neck fractures: a prospective randomized study. Int Orthop. 2001;25:223–225. doi: 10.1007/s002640100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raisz LG. Osteoporosis: current approaches and future prospects in diagnosis, pathogenesis, and management. J Bone Miner Metab. 1999;17:79–89. doi: 10.1007/s007740050069. [DOI] [PubMed] [Google Scholar]
- 30.Lein T, Bula P, Jeffries J, Engler K, Bonnaire F. Fractures of the femoral neck. Acta Chir Orthop Traumatol Cech. 2011;78:10–19. [PubMed] [Google Scholar]
- 31.Luo D, Zou W, He Y, Xian H, Wang L, Shen J, Lian K, Lin D. Modified dynamic hip screw loaded with autologous bone graft for treating Pauwels type-3 vertical femoral neck fractures. Injury. 2017;48:1579–1583. doi: 10.1016/j.injury.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 32.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]


