Abstract
Background
Long-chain free fatty acids (FFAs) are a type of backbone molecule that can react with alcohol to produce biodiesels. Various microorganisms have become potent producers of FFAs. Efforts have focused on increasing metabolic flux to the synthesis of either neutral fat or fatty acyl intermediates attached to acyl carrier protein (ACP), which are the source of FFAs. Membrane lipids are also a source of FFAs. As an alternative way of producing FFAs, exogenous phospholipase may be used after heterologous production and localization in the periplasmic space. In this work, we examined whether Rhodobacter sphaeroides, which forms an intracytoplasmic membrane, can be used for long-chain FFA production using phospholipase.
Results
The recombinant R. sphaeroides strain Rs-A2, which heterologously produces Arabidopsis thaliana phospholipase A2 (PLA2) in the periplasm, excretes FFAs during growth. FFA productivity under photoheterotrophic conditions is higher than that observed under aerobic or semiaerobic conditions. When the biosynthetic enzymes for FA (β-ketoacyl-ACP synthase, FabH) and phosphatidate (1-acyl-sn-glycerol-3-phosphate acyltransferase, PlsC) were overproduced in Rs-A2, the FFA productivity of the resulting strain Rs-HCA2 was elevated, and the FFAs produced mainly consisted of long-chain FAs of cis-vaccenate, stearate, and palmitate in an approximately equimolar ratio. The high-cell-density culture of Rs-HCA2 with DMSO in two-phase culture with dodecane resulted in an increase of overall carbon substrate consumption, which subsequently leads to a large increase in FFA productivity of up to 2.0 g L−1 day−1. Overexpression of the genes encoding phosphate acyltransferase (PlsX) and glycerol-3-phosphate acyltransferase (PlsY), which catalyze the biosynthetic steps immediately upstream from PlsC, in Rs-HCA2 generated Rs-HXYCA2, which grew faster than Rs-HCA2 and showed an FFA productivity of 2.8 g L−1 day−1 with an FFA titer of 8.5 g L−1.
Conclusion
We showed that long-chain FFAs can be produced from metabolically engineered R. sphaeroides heterologously producing PLA2 in the periplasm. The FFA productivity was greatly increased by high-cell-density culture in two-phase culture with dodecane. This approach provides highly competitive productivity of long-chain FFAs by R. sphaeroides compared with other bacteria. This method may be applied to FFA production by other photosynthetic bacteria with similar differentiated membrane systems.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1070-8) contains supplementary material, which is available to authorized users.
Keywords: Long-chain free fatty acids, Phospholipase A2, Periplasmic expression, Rhodobacter sphaeroides, Photoheterotrophic conditions, High-cell-density culture, Dodecane-overlaid two-phase culture
Background
Given the increasing concern over the past decade regarding the projected depletion of fossil-fuel reserves and the concomitant emissions of greenhouse gases, the exploration of renewable and sustainable alternative biofuels is imperative. Much attention has been given lately to the microbial production of biofuels [1–3], including bioalcohol and biodiesel.
Recombinant Escherichia coli engineered for enhanced metabolic flow to ethanol provides significantly increased ethanol yield and productivity [4, 5]. Ethanol is both an important fuel blender and a starting resource for other basic raw materials [6]. However, in terms of energy density, ethanol is inferior to other biofuels with longer carbon chains [7].
Biodiesel is a monoalkyl ester derived from reactions between FFAs (usually longer than C10) and alcohols such as methanol, ethanol, propanol, and butanol. Biodiesel can be produced using edible oils as a source of FFAs, but the availability of edible feedstock in many countries may be low owing to the high demand for food resources [7]. Therefore, nonedible plant oils are used as alternative feedstocks; however, their supply requires large areas of cultivated land.
Given the need for higher productivity in limited space, microorganisms have been used as potent producers of FFAs and biodiesel [8]. Recombinant E. coli is able to produce alkanes, fatty alcohols, FFAs, and fatty esters of varying alkyl-chain lengths [8, 9]. E. coli has been further manipulated to achieve FFA productivity in the range of approximately 3–4.5 g L−1day−1 [10–12]. To enhance FFA production by E. coli, FFA uptake and degradation were blocked by the interruption of long-chain fatty acid transporter, FadL [13, 14] and acyl-CoA synthetase, FadD [15]. Moreover, FFA-biosynthesis genes were overexpressed with the simultaneous expression of either exogenous (plant) [15] or endogenous [13] thioesterase; the overexpression of genes encoding acetyl-CoA carboxylase (AccABCD) [16], β-ketoacyl-ACP synthase III (FabH) [17], or malonyl-CoA:ACP transacylase (FabD) [18] also increased FFA production by E. coli. FFA efflux in E. coli is mainly mediated by the AcrAB-TolC multidrug pump [19], which comprises TolC in the outer membrane, AcrB in the inner membrane and AcrA in the periplasmic space [20].
FFA production was also demonstrated in recombinant Synechocystis sp. PCC6803 [21]. The FFA secretion pathways common to recombinant strains of both E. coli and Synechocystis sp. PCC6803 consist mainly of two steps: FFA hydrolysis by thioesterase from fatty acyl-ACP inside the cell, followed by its export out of the cell. As an alternative method for the production of long-chain FFAs, a differentiated membrane can be used as a substrate for exogenous phospholipase in the periplasmic space. The secretion of FFAs from the periplasm could be facilitated more effectively than secretion from the cytoplasm because the outer membrane is the only export barrier.
The purple nonsulfur photosynthetic bacterium Rhodobacter sphaeroides gratuitously forms an intracytoplasmic membrane (ICM) in addition to the cell membrane when the partial pressure of oxygen (pO2) is lowered [22]. This phenomenon increases the cellular lipid content by approximately two- to threefold, resulting in levels similar to those of photoheterotrophically grown cells [23].
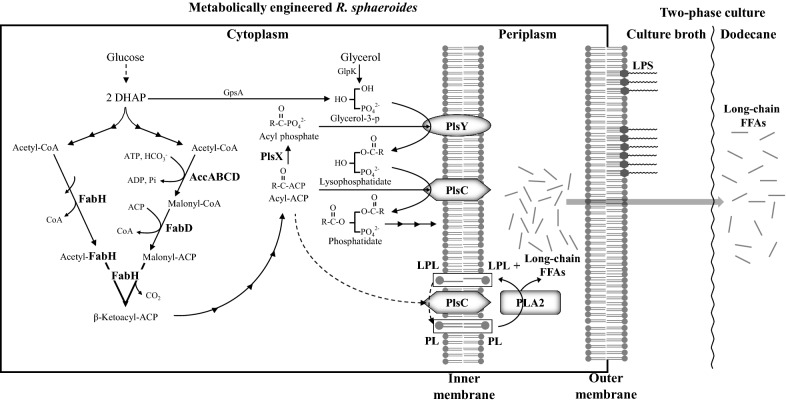
In this study, we examined whether R. sphaeroides heterologously producing phospholipase A2 (PLA2) of Arabidopsis thaliana, which is subsequently localized in the periplasmic space (Fig. 1), can produce long-chain FFAs during photoheterotrophic growth. R. sphaeroides was metabolically engineered further to enhance metabolic flux to phospholipid (PL) formation by increasing the production of enzymes for the synthesis of FA and phosphatidate. Moreover, we tried high-cell-density culture to further increase FFA productivity. Because FFAs in the culture broth may be reutilized by cells, a two-phase culture system with dodecane (Fig. 1) was employed to keep the FFAs in the layer of organic solvent, preventing their reuse. Because FFAs are derived from cell membranes, long-chain (C18 and C16) FFAs are expected to be main components. In fact, the FFAs cis-vaccenate, stearate, and palmitate were obtained. This approach resulted in highly competitive productivity of long-chain FFAs compared with that observed in other photosynthetic bacteria [21, 24].
Fig. 1.
Production of FFAs from metabolically engineered R. sphaeroides in a two-phase culture system with dodecane. R. sphaeroides was recombinantly engineered to overproduce FabH, PlsX, PlsY, and PlsC in the cytoplasm and A. thaliana PLA2 in the periplasm. The long-chain FFAs released by R. sphaeroides were directed and localized to the dodecane layer. FFA sequestration in the dodecane layer alleviated the inhibitory effect of FFAs on cell growth, further elevating the FFA productivity of the cells. Multiple biosynthetic steps are illustrated by a series of connecting arrows, whereas a putative LPL acyltransferase activity of PlsC, which may form PL from LPL using acyl-ACP, is shown by dotted arrows. The PLs and LPLs of the inner membrane, which are the substrate and product of PLA2, respectively, are highlighted in boxes; the PLs of the outer membrane are also thought to be used by PLA2. DHAP, dihydroxyacetone phosphate; ACP, acyl carrier protein; FabH, β-ketoacyl-ACP synthase; AccABCD, acetyl-CoA carboxylase; FabD, malonyl-CoA:ACP transacylase; GpsA, glycerol-3-phosphate dehydrogenase; GlpK, glycerol kinase; PlsX, phosphate acyltransferase; PlsY, glycerol-3-phosphate acyltransferase; PlsC, 1-acyl-sn-glycerol-3-phosphate acyltransferase; LPS, lipopolysaccharide; LPL, lysophospholipid; PL, phospholipid; PLA2, phospholipase A2 of A. thaliana
Materials and methods
Organisms and growth conditions
The wild-type (WT) strain R. sphaeroides KD131 [25] was cultured aerobically and photoheterotrophically at 28 °C in Sistrom’s succinate-based (Sis) minimal medium as described previously [25, 26]; Aerobic growth was performed by incubating 100 mL of culture in 1-L baffled flasks under vigorous shaking (250 rpm) on a gyratory shaker, while photoheterotrophic growth was obtained by incubating 100 mL of culture in completely filled vessels in the light (10 Watts/m2). Semiaerobic growth was achieved by incubating 100 mL of culture in 1-L baffled flasks under gentle agitation (50 rpm) on a gyratory shaker or by sparging (approximately 100 mL min−1 flow per 100 mL of culture) with a gas mixture of 2% O2, 1% CO2, and 97% N2. For high-cell-density culture, R. sphaeroides was grown in Sis minimal medium supplemented with succinate (up to 150 mM), glucose (100 mM), glycerol (150 mM), and yeast extracts (1% (w/v)) (SisH medium). When necessary, DMSO (10 mM) was added to the SisH medium (SisH-D medium). Dodecane (30%, v/v) was added to the bacterial broth (50–100 mL) for two-phase culture, which was mixed continuously with a magnetic stirring bar to ensure sufficient contact of dodecane with the cells. Cell aliquots were removed intermittently from the culture vessels for biochemical analyses; Cells grown photoheterotrophically were removed inside an anaerobic chamber (Model 10, COY, USA) to keep anaerobiosis, whereas cells grown aerobically and semiaerobically were removed under ambient conditions. Escherichia coli was grown at 37 °C in Luria–Bertani medium. Antibiotics for R. sphaeroides and E. coli cultures were used at concentrations previously indicated [27]. A. thaliana was grown photosynthetically under a light intensity of 110 ± 10 μmol m−2s−1 and a 16-h/8-h light–dark cycle at 22 ± 1 °C with 60 ± 10% humidity, as described previously [28].
Preparation of DNA fragments encoding the enzymes for the metabolism of FAs and PLs
All DNA fragments containing genes encoding enzymes involved in the metabolism of FAs and PLs were PCR-amplified from R. sphaeroides genomic DNA, whereas the genes for phospholipases [phospholipase A1 (PLA1) and phospholipase A2 (PLA2)] were obtained from A. thaliana cDNA. The cDNA sequence of A. thaliana was obtained from http://www.arabidopsis.org/index.jsp, and cDNA was synthesized as follows. Total RNA was extracted from 2-week-old Arabidopsis plants using a Plant RNA Kit (Nanohelix, Daejeon, Korea). DNase I (New England Biolabs, Ipswich, MA, USA) was added to the total RNA to remove genomic DNA. Using the total RNA as templates, first-strand cDNA was synthesized by PrimeScript reverse transcriptase (Takara Bio, Shiga, Japan) according to the manufacturer’s protocol. Briefly, total RNA (5 μg) was mixed with oligo dT primer (5 μM) and dNTP (2 mM) in 10 μL of H2O and incubated at 65 °C for 5 min, followed by mixing with 20 U RNase inhibitor and 100 U reverse transcriptase in PrimeScript buffer (Takara). The reaction proceeded at 42 °C for 60 min, and the reverse transcriptase was heat-inactivated at 70 °C for 5 min to stop the reaction.
Construction of plasmids and recombinant strains
pINDA′-PLA1, pIND-PLA2 and pINDA′-PLA2
The 2.0-kb SmaI/StuI transcription and translation stop Ω [streptomycin/spectinomycin-resistant (Smr/Spr)] DNA was inserted into the SmaI site of the kanamycin-resistant (Kmr) gene of pIND4 [29] to obtain pIND4S, a recombinant plasmid that could then be maintained in a cell using Sm/Sp compatibly with the plasmid derived from pBBR1MCS-2 (Kmr) (Table 1). Among the fourteen phospholipases A1 (PLA1) of A. thaliana, At1g06800.1 was chosen for further cloning due to its broad specificity for PL head groups [30, 31]. To construct a pIND4S-derived plasmid carrying an insert DNA encoding PLA1 fused to the CycA signal peptide sequence (CycA′) of R. sphaeroides, the 63-bp NcoI–XbaI fragment encoding CycA′ was PCR-amplified from R. sphaeroides genomic DNA using primers CycA-F and CycA-R (Additional file 1: Table S1). Then, a 1.5-kb XbaI–HindIII fragment encoding PLA1, extending from its start codon to the termination codon, was PCR-amplified from A. thaliana cDNA using primers PLA1-F and PLA1-R (Additional file 1: Table S1). The two DNA fragments were digested with NcoI–XbaI and XbaI–HindIII, respectively, and cloned into the NcoI/HindIII sites of pIND4S to generate pINDA′-PLA1 (Table 1).
Table 1.
Strains and plasmids used in this study
| Strains/plasmids | Relevant characteristics | References/description |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αphe | phe::Tn10dCm of DH5α | [32] |
| HB101 | F− mcrB mrr hsdS20(r−B m−B) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(SmR) glnV44 λ− | [33] |
| S17-1 | C600::RP4 2-(Tc::Mu)(Km::Tn7) thi pro hsdR hsdM+ recA | [34] |
| R. sphaeroides | ||
| KD131 | Type strain | [25] |
| Rs-nA2 | R. sphaeroides containing pIND-PLA2 | This study |
| Rs-A2 | R. sphaeroides containing pINDA′-PLA2 | This study |
| Rs-nA2-phoA | R. sphaeroides containing pIND-PLA2-phoA | This study |
| Rs-A2-phoA | R. sphaeroides containing pINDA′-PLA2-phoA | This study |
| Rs-C | R. sphaeroides containing pLVplsC | This study |
| Rs-DC | R. sphaeroides containing pRKfabD and pLVplsC | This study |
| Rs-AccC | R. sphaeroides containing pRKaccABCD and pLVplsC | This study |
| Rs-HC | R. sphaeroides containing pRKfabH and pLVplsC | This study |
| Rs-AccCA2 | R. sphaeroides containing pRKaccABCD, pLVplsC and pINDA′-PLA2 | This study |
| Rs-CA2 | R. sphaeroides containing pLVplsC and pINDA′-PLA2 | This study |
| Rs-DCA2 | R. sphaeroides containing pRKfabD, pLVplsC and pINDA′-PLA2 | This study |
| Rs-HCA2 | R. sphaeroides containing pRKfabH, pLVplsC and pINDA′-PLA2 | This study |
| Rs-HXYCA2 | R. sphaeroides containing pRKfabH, pBBRplsX, pLVplsCY and pINDA′-PLA2 | This study |
| Rs-tol | A tolC mutant; Tpr | This study |
| Rs-A2tol | A tolC mutant containing pINDA′-PLA2; Tpr | This study |
| Plasmids | ||
| pBBR1MCS-2 | ori pBBR1 lacZa; Kmr | [35] |
| pBBRplsX | pBBR + 1.3-kb HindIII–XbaI fragment containing PlsX; Kmr | This study |
| pBS(−) | ori f1 lacZ′; Apr | Stratagene |
| pBSaccA | pBS + 1.1-kb XbaI–PstI fragment containing accA; Apr | This study |
| pBSaccD | pBS + 1.3-kb XbaI–PstI fragment containing accD; Apr | This study |
| pBSfabD | pBS + 1.2-kb XbaI–PstI fragment containing fabD; Apr | This study |
| pMD20 | ori f1 lacZ′; Apr | Takara |
| pMDaccBC | pMD + 2.9-kb HindIII–XbaI fragment containing accBC; Apr | This study |
| pIND4 | ori ColE1 IPTG-inducible promoter; Kmr | [29] |
| pIND4S | ori ColE1 IPTG-inducible promoter; Sm/Spr | This study |
| pIND-PLA2 | pIND4S + 387-bp NcoI–HindIII fragment containing PLA2; Sm/Spr | This study |
| pINDA′-PLA2 | pIND4S + 456-bp NcoI–HindIII fragment containing cycA′-PLA2; Sm/Spr | This study |
| pINDA′-PLA1 | pIND4S + 1.9-kb NcoI–HindIII fragment containing cycA′-PLA1; Sm/Spr | This study |
| pIND-PLA2-phoA | pIND4S + 1.9-kb NcoI–HindIII fragment containing PLA2-phoA; Sm/Spr | This study |
| pINDA′-PLA2-phoA | pIND4S + 2.0-kb NcoI–HindIII fragment containing cycA′-PLA2-phoA; Sm/Spr | This study |
| pLV106 | IncQ pUI511 containing the polylinker of EcoRI, SmaI, KpnI, and XbaI restriction sites inserted at PstI site | [36] |
| pLV-Tp | pLV106 + 1.6-kb HindIII fragment containing Tpr gene | This study |
| pLVplsC | pLV-Tp + 1.0-kb XbaI–KpnI fragment containing plsC; Tpr | This study |
| pLVplsCY | pLV-Tp + 1.0-kb XbaI–KpnI fragment containing plsC and 0.8-kb KpnI-EcoRI fragment containing plsY; Tpr | This study |
| pRK2013 | ori ColE1; Kmr | [37] |
| pRK415 | ori IncP-1 Mob RP4 lacZa; Tcr | [38] |
| pRKfabH | pRK415 + 1.8-kb KpnI–XbaI fragment containing fabH; Tcr | This study |
| pRKfabD | pRK415 + 1.2-kb XbaI–KpnI fragment containing fabD; Tcr | This study |
| pRKaccABCD | pRK415 + 5.3-kb XbaI–KpnI fragment containing accABCD; Tcr | This study |
| pSUP202 | ori p15A Mob RP4; Tcr Cmr Apr | [39] |
| pSUPTolC | pSUP202 + 2.6-kb DNA of tolC::Tp; tolC interrupted by 1.6-kb Tpr DNA; Tpr Tcr | This study |
Among the four instances of phospholipase A2 (PLA2) in A. thaliana, At2g06925 was chosen for further cloning due to its broad optimal pH range and relatively weak preference for PL head groups [40]. To construct a pIND4S-derived plasmid carrying an insert DNA encoding PLA2, a 387-bp NcoI–HindIII fragment encoding PLA2 without its signal peptide of twenty N-terminal residues was PCR-amplified from A. thaliana cDNA using primers PLA2-F and PLA2-R (Additional file 1: Table S1). The DNA fragment was digested with NcoI/HindIII and cloned into the NcoI/HindIII sites of pIND4S to generate pIND-PLA2 (Table 1), in which signal peptide-less PLA2 was oriented in-frame to the start codon at the multiple cloning sites of the plasmid.
To construct a pIND4S-derived plasmid carrying an insert DNA encoding PLA2 fused to CycA′ of R. sphaeroides, the 63-bp NcoI–XbaI fragment encoding CycA′ was PCR-amplified from R. sphaeroides genomic DNA as described above, and a 387-bp XbaI–HindIII fragment encoding PLA2 without its signal peptide was PCR-amplified from A. thaliana cDNA, using primers PLA2-F´ and PLA2-R (Additional file 1: Table S1). The two DNA fragments encoding CycA′ and signal peptide-less PLA2 were digested with NcoI–XbaI and XbaI–HindIII, respectively, and cloned into the NcoI/HindIII sites of pIND4S to generate pINDA′-PLA2 (Table 1).
pIND-PLA2-phoA and pINDA′-PLA2-phoA
A 1.5-kb PstI–HindIII fragment encoding alkaline phosphatase (PhoA) without its thirty-three N-terminal residues (signal peptide-less PhoA (PhoA′)) was PCR-amplified from E. coli genomic DNA using primers PhoA-F and PhoA-R (Additional file 1: Table S1) and then cloned into the PstI/HindIII sites of pBS(−) (Stratagene, San Diego, CA, USA) to generate pBSPhoA. A 384-bp NcoI–PstI fragment encoding PLA2, extending from the 21st codon of PLA2 to the second from the termination codon, was PCR-amplified from pIND-PLA2 using primers PLA2-t-F and PLA2-t-R (Additional file 1: Table S1). Likewise, a 453-bp NcoI–PstI fragment encoding CycA′-PLA2, extending from the start codon of CycA′ to the second from the termination codon of PLA2, was PCR-amplified from pINDA′-PLA2 using primers CycA-F and PLA2-t-R (Additional file 1: Table S1). These two DNA fragments were digested with NcoI–PstI, and each was cloned with a 1.5-kb PstI–HindIII fragment of PhoA′ of pBSPhoA into the NcoI/HindIII sites of pIND4S to yield pIND-PLA2-phoA and pINDA′-PLA2-phoA, respectively (Table 1).
pRKaccABCD
A 1.1-kb XbaI–PstI fragment extending from 171 bp upstream of the start codon of AccA to its termination codon was PCR-amplified using primers AccA-F and AccA-R (Additional file 1: Table S1). The resulting DNA fragment was digested with XbaI and PstI and ligated into the XbaI/PstI sites of pBS(−) (Stratagene) to generate pBSaccA (Table 1). Similarly, a 1.3-kb XbaI-PstI fragment extending from 363 bp upstream of AccD to its termination codon was PCR-amplified using primers AccD-F and AccD-R (Additional file 1: Table S1). The resulting DNA fragment was digested with XbaI and PstI and ligated into the XbaI/PstI sites of pBS(−) to yield pBSaccD (Table 1). The 2.9-kb HindIII/XbaI accBC DNA extending from 712 bp upstream of the AccB start codon to 233 bp downstream of AccC was PCR-amplified using primers AccBC-F and AccBC-R (Additional file 1: Table S1). The resulting fragment was ligated into T-vector pMD20 (Takara) to generate pMD-accBC (Table 1). The 1.1-kb XbaI–HindIII fragment containing accA, the 2.9-kb HindIII–XbaI fragment containing accBC, and the 1.3-kb XbaI–KpnI fragment containing accD were digested from pBSaccA, pMDaccBC, and pBSaccD, respectively, with restriction enzymes, followed by sequential ligation into the XbaI/KpnI sites of pRK415 to generate pRKaccABCD (Table 1).
pRKfabD
A 1.2-kb XbaI–PstI DNA fragment extending from 228 bp upstream of the FabD start codon to its termination codon was PCR-amplified using primers FabD-F and FabD-R (Additional file 1: Table S1). The resulting DNA fragment was digested with XbaI and PstI and ligated into the XbaI/PstI sites of pBS(−) to generate pBSfabD (Table 1). The 1.2-kb XbaI–KpnI DNA containing fabD was digested from pBSfabD by restriction enzymes and cloned into the XbaI/KpnI sites of pRK415 to generate pRKfabD (Table 1).
pRKfabH
A 1.8-kb KpnI–XbaI fragment extending from 586 bp upstream of the FabH start codon to 194 bp downstream of the termination codon was PCR-amplified using primers FabH-F and FabH-R (Additional file 1: Table S1). The resulting DNA fragment was digested with KpnI and XbaI and ligated into the KpnI/XbaI sites of pRK415 to generate pRKfabH (Table 1).
pLVplsC and pLVplsCY
A 970-bp XbaI–KpnI fragment extending from 177 bp upstream of the PlsC start codon to 78 bp downstream of the termination codon was PCR-amplified using primers PlsC-F and PlsC-R (Additional file 1: Table S1). The resulting DNA fragment was digested with XbaI and KpnI and ligated into the XbaI/KpnI sites of pLV106 to generate pLVplsC (Table 1). A 0.8-kb KpnI-EcoRI fragment extending from 220 bp upstream of the PlsY start codon to 21 bp downstream of the termination codon was PCR-amplified using primers PlsY-F and PlsY-R (Additional file 1: Table S1). The resulting DNA fragment was digested with KpnI and EcoRI and ligated into the KpnI/EcoRI sites of pLVplsC to generate pLVplsCY (Table 1), in which plsC and plsY are oriented in the same direction.
pBBRplsX
A 1.3-kb HindIII–XbaI DNA fragment extending from 167 bp upstream of the PlsX start codon to 42 bp downstream from the termination codon was PCR-amplified using primers PlsX-F and PlsX-R (Additional file 1: Table S1). The resulting DNA fragment was digested with HindIII and XbaI and ligated into the HindIII/XbaI sites of pBBR1MCS-2 to generate pBBRplsX (Table 1).
Construction of the R. sphaeroides tolC mutant Rs-tol
A 550-bp PstI–BamHI fragment upstream of the codon encoding the 2nd residue of TolC was PCR-amplified from R. sphaeroides genomic DNA using primers TolC-UF and TolC-UR (Additional file 1: Table S1). A 549-bp HindIII-EcoRI fragment downstream of the 465th residue of TolC was PCR-amplified using primers TolC-DF and TolC-DR (Additional file 1: Table S1). In parallel, a 1.6-kb BamHI/HindIII fragment containing the trimethoprim (Tp)-resistance (Tpr) gene was obtained from pLV-Tp (Table 1). The three fragments, including the 550-bp PstI–BamHI fragment, the 1.6-kb BamHI–HindIII Tpr gene, and the 549-bp HindIII–EcoRI fragment, were ligated sequentially into the PstI/EcoRI sites of pSUP202 [39] to generate pSUPtolC (Table 1), from which the tolC DNA encoding the polypeptide between the 2nd and 465th residue was deleted. pSUPtolC was mobilized into R. sphaeroides, and a Tpr and Tcs double-crossover recombinant was isolated as previously described [41]. The chromosome structure of the Rs-tol mutant (Table 1) was confirmed by Southern hybridization analysis.
Mobilization of recombinant plasmids into R. sphaeroides
Recombinant plasmids were mobilized into R. sphaeroides using E. coli S17-1 (Table 1), as described previously [41]. For plasmids carrying the Tpr gene, E. coli DH5αphe (Table 1) was used for conjugation [42].
Alkaline phosphatase (PhoA) assay
PhoA activity was detected using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) solution (Sigma-Aldrich, St. Louis, MO, USA), as described previously [43]. Aliquots of the BCIP/NBT solution (one tablet in 3 mL of H2O) were dropped onto colonies on Sis agar plates and incubated at 30 °C for 5 min.
Preparation of cytoplasmic and periplasmic fractions
The cytoplasmic and periplasmic fractions of R. sphaeroides were prepared as described previously [44]. Cells were grown under photoheterotrophic conditions and harvested at the exponential growth phase by centrifugation at 3000g and 4 °C for 20 min. The cell pellet was gently suspended in 1 mL of Tris-sucrose-EDTA (200 mM Tris–HCl, pH 8.0, 500 mM sucrose, 1 mM EDTA) buffer and incubated on ice for 30 min, followed by centrifugation at 16,000g and 4 °C for 30 min. The supernatant was recovered, representing the periplasmic fraction. The pellet containing spheroplasts was suspended in 20 mM sodium borate buffer (pH 9.0) and sonicated on ice, then centrifuged at 12,000g and 4 °C for 10 min. The resulting supernatant represented the cytoplasmic fraction.
Phospholipase assay
The phospholipase assay was carried out as described previously [45]. Membrane PLs were extracted from photoheterotrophically grown R. sphaeroides [46] and used as substrates for the assay. The reaction mixture contained an aliquot (35 µg protein) of either the cytoplasmic or periplasmic fraction (“Preparation of cytoplasmic and periplasmic fractions” section) and 400 µg of PLs in 1 mL of 20 mM sodium borate buffer (pH 9.0) containing 1 mM CaCl2. The reaction was performed at 30 °C for 15 min, and terminated by heating at 100 °C for 5 min. The reaction mixture was then centrifuged at 12,000g and 4 °C for 10 min, and the supernatant (50 µL) was examined for FFA using the FFA Quantification Colorimetric/Fluorometric Kit (BioVision, Milpitas, CA, USA). The reaction was performed with no cell fraction aliquot as a control, and a standard curve was prepared with varying concentrations of palmitate. One enzyme unit was defined as 1 µmol of FFAs released per min from PLs.
RNA isolation and quantitative reverse transcription PCR (RT-qPCR)
Cells were harvested during exponential growth, and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNase-free DNase (Qiagen) was used to remove DNA. The PrimeScript RT Reagent Kit (Takara Bio, Shiga, Japan) was used to reverse transcribe RNA using primers specific to each gene (Additional file 1: Table S1), and the cDNA obtained from each sample (1 μg) was used for further analysis. Relative mRNA quantification was performed using the LightCycler 480 real-time PCR system (Roche, Basel, Switzerland) with the SYBR Premix Ex Taq (Tli RNaseH Plus) Kit (Takara Bio). The amplification efficiencies of interest and reference genes were equal to 100%. Final mRNA levels were normalized to that of the housekeeping 16S rRNA by the 2−ΔΔCT method [47]. All the quantifications were independently repeated three times, and data are shown as the mean ± standard deviation (SD).
Thin-layer chromatography
Total lipids were extracted from cells as described previously [48]. The extracted lipids were dissolved in chloroform and loaded onto activated silica thin-layer chromatography (TLC) plates (Z185329, Sigma-Aldrich). The plate was developed with a solvent mixture of chloroform–methanol–acetic acid–water (85:15:10:3.5, v/v) [48], then air-dried and exposed to iodine vapor for approximately 20 min at room temperature until the bands appeared. To determine the lysophospholipids (LPLs) in the cell membrane, a mixture of chloroform–methanol–acetic acid–acetone–water (35:25:4:14:2, v/v) [49] was used as the development solvent instead.
Spectral complex determination
Cells were grown under photoheterotrophic conditions and harvested at the exponential growth phase. Cell-free lysates were prepared and analyzed for spectral complexes as described previously [22, 50].
Malate dehydrogenase assay
Cells were harvested and cell-free lysates were prepared as described in “Spectral complex determination” section. The culture supernatant was concentrated approximately tenfold by centrifugation with Amicon® Ultra4 centrifugal Filters-3K (Merck, Ireland). The enzyme reaction was performed at 25 °C by mixing 12 μg of protein, from either concentrated culture supernatant or cell-free extracts, with substrates (NADH and oxaloacetate at final concentrations of 0.5 mM and 0.2 mM, respectively) in 0.1 M potassium phosphate buffer (pH 7.4) at a final volume of 0.5 mL. Malate dehydrogenase activity was determined by measuring the decrease in absorbance of NADH at 340 nm, as described previously [51].
Determination of cellular FAs (CFAs) and FFAs in culture broth
CFAs of R. sphaeroides were determined as described previously [46]. Standard fatty acid methyl esters (FAMEs) (37-component FAME; Sigma-Aldrich) were used to determine the retention times of sample FAMEs in gas-chromatographic analysis. cis-vaccenic acid (Sigma-Aldrich) was esterified for use as a control because it is not included as a standard FAME. An aliquot of pentadecanoic acid (Sigma-Aldrich) was used as an internal standard. To determine the FFAs in culture broth, 3 mL of culture supernatant was mixed with 10 µg of pentadecanoic acid, followed by solvent extraction with chloroform and methanol (2:1 [v/v]) for 2 h with shaking at room temperature. The chloroform layer was evaporated under a nitrogen stream, and the FFAs were determined as described previously [46]. When cells were cultured in two-phase system with dodecane, an aliquot of the solvent layer was removed and centrifuged to obtain 100 µL of dodecane, which was analyzed for FFAs in the same way as the culture supernatant sample.
FFA productivity was determined from the FFA level in culture supernatant (and in the dodecane in dodecane-overlaid culture) harvested at several time points from exponential phase to the beginning part of the stationary phase. The FFA productivity was derived from the FFA levels at a given sampling point divided by the time at that sampling point, which had elapsed after inoculation. The maximal FFA productivities at exponential growth phase, which remained unchanged to the beginning part of the stationary phase, were regarded as a representative FFA productivity. In contrast, FFA titer was determined from the maximal level of FFAs in culture supernatant (and in the dodecane in dodecane-overlaid culture) during the stationary phase.
Preparation of potassium cis-vaccenate
One gram of cis-vaccenic acid (Sigma-Aldrich, USA) was dissolved in 10 mL of alcoholic KOH (40 g L−1), followed by boiling for 1 h. The solution was cooled to room temperature, neutralized with 0.5 N HCl, and then completely dried at 65 °C and used as potassium cis-vaccenate.
Determination of glucose, glycerol and succinate
Aliquots of cell culture were centrifuged to obtain the supernatant, which was stored at 4 °C for further analysis. The glucose concentration was examined using a Glucose Assay Kit (Sigma-Aldrich). A Free Glycerol Colorimetric/Fluorometric Assay Kit (BioVision, Milpitas, CA, USA) was used to determine glycerol levels, and a Succinate (Succinic Acid) Colorimetric Assay Kit (BioVision) was used to examine the succinate levels in the culture supernatant.
Results and discussion
FFAs are released from the photoheterotrophically grown recombinant R. sphaeroides (Rs-A2) heterologously producing Arabidopsis thaliana PLA2, which is localized in the periplasm
The CFA content of R. sphaeroides KD131 grown under photoheterotrophic and semiaerobic conditions is approximately twofold higher than that of cells grown aerobically (Additional file 1: Fig. S1), which is attributed to the formation of ICM under O2-limited conditions [22]. Cells grow photoheterotrophically with a doubling time half that observed under semiaerobic conditions (Additional file 1: Fig. S1), and the resulting maximal turbidity is nearly doubled in the same comparison (Additional file 1: Fig. S1). In this work, we heterologously produced A. thaliana phospholipases, which are genetically engineered to be localized to the periplasmic space, in R. sphaeroides, and then addressed the question of whether the recombinant strain can produce FFAs (Fig. 1). Given that the cell membrane is a barrier to FFA translocation, the export of FFAs from the periplasm may be easier than secretion from the cytoplasm.
PLA1 and PLA2 of A. thaliana have been biochemically characterized in detail [31, 40] and used in this study. PLA1 of A. thaliana was fused in-frame to the signal peptide sequence (CycA′) of the periplasmic protein cytochrome c2 (CycA) of R. sphaeroides to generate the plasmid pINDA′-PLA1 (Table 1). When pINDA′-PLA1 was mobilized into WT cells, we failed to obtain any stable exconjugant, even from several independent trials. The reaction product of PLA1 is 2-acyl LPL, and its accumulation in the membrane might be toxic to R. sphaeroides. In contrast, pINDA′-PLA2 (Table 1), in which signal peptide-less PLA2 from A. thaliana was fused in-frame to CycA′, was successfully mobilized into R. sphaeroides to yield the exconjugant strain Rs-A2 (Table 1), which was used in the subsequent experiments. The cloning of signal peptide-less PLA2 without CycA′ generated pIND-PLA2 (Table 1), and its exconjugant Rs-nA2 (Table 1) was used as a control.
Rs-A2 was grown aerobically, semiaerobically and photoheterotrophically and harvested intermittently at several time points extending from exponential to stationary growth phases (Additional file 1: Fig. S2, A1, B1, and C1). FFA productivity usually reaches the maximal level at the end of exponential growth and remained unchanged to the beginning part of stationary phase under aerobic and photoheterotrophic conditions (Additional file 1: Fig. S2, A1 and C1), whereas the FFA productivity at low O2 reaches the maximal level at the exponential growth phase and gradually decreased during stationary phase (Additional file 1: Fig. S2, B1). The maximal FFA productivities at the exponential growth phase was regarded as the representative value of the recombinant strain. On the other hand, FFA titer usually reaches the maximal level during the stationary phase (Additional file 1: Fig. S2, A1, B1 and C1). Thus, Rs-A2 showed an FFA productivity of approximately 0.15 ~ 0.16 g L−1 day−1 under photoheterotrophic conditions (Fig. 2b, Additional file 1: Fig. S2, C1 and Fig. S3), which was approximately twofold higher than that of the aerobically grown cells (Additional file 1: Fig. S2, A1 and Fig. S3). Semiaerobically grown cells showed intermediate FFA productivity between those of the cells grown aerobically and photoheterotrophically (Additional file 1: Fig. S2, B1 and Fig. S3). Thus, cells were grown photoheterotrophically for higher FFA productivity in the subsequent experiments, unless specified otherwise.
Fig. 2.
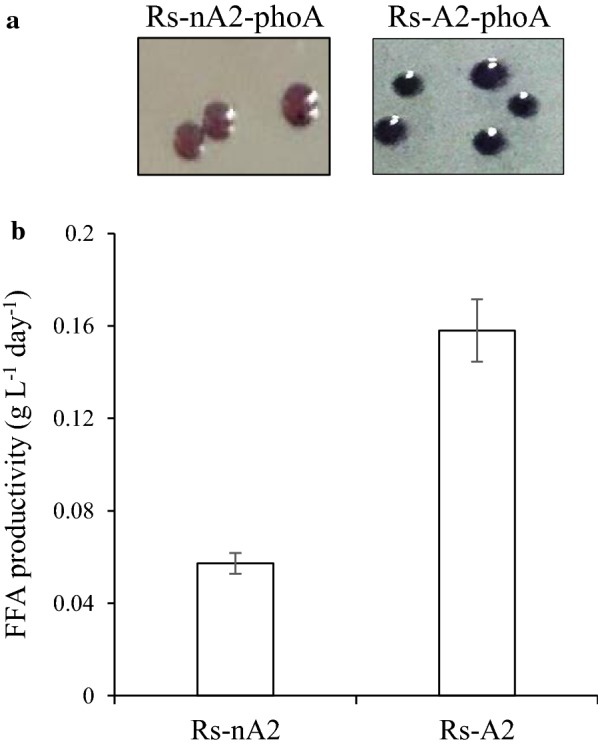
Heterologous production of PLA2 in the periplasm of Rs-A2 and its FFA productivity. a PhoA activity of Rs-A2-phoA was examined with BCIP/NBT solution, which was dropped onto colonies grown aerobically on Sis minimal medium agar plates and incubated at 30 °C for 5 min (right). Rs-nA2-phoA colonies were used as a control (left). b R. sphaeroides heterologously producing PLA2 in either the cytoplasm (Rs-nA2) or periplasm (Rs-A2) was grown photoheterotrophically and harvested at the exponential growth phase (OD660 of 2.3). The FFA level in the culture supernatant was measured, and the FFA productivity is shown with error bars from three independent experiments
FFA efflux in E. coli is mainly mediated by the AcrAB-TolC multidrug pump [19], which comprises TolC in the outer membrane and AcrB and AcrA in the inner membrane and periplasmic space [20], respectively. Likewise, Salmonella enterica serovar Typhimurium employs TolC, which has 94% similarity (90% identity) to E. coli TolC, to excrete long-chain FFAs into the culture medium [52]. We investigated whether FFAs from Rs-A2 utilized a similar export system. The R. sphaeroides TolC homolog (Rsp_1576), which shows approximately 41–43% similarity (23–24% identity) to the TolCs of E. coli and Salmonella, was interrupted to yield the Rs-tol strain (Table 1). Plasmid pINDA′-PLA2 was then mobilized into Rs-tol to yield the exconjugant Rs-A2tol (Table 1), and its FFA productivity was compared with that of Rs-A2. There was no difference in FFA productivity between Rs-A2 and its isogenic tolC-interruption mutant (Additional file 1: Fig. S4). Accordingly, our results suggest that FFAs from Rs-A2 are exported from the periplasmic space through a TolC-independent pathway.
To determine the cellular location of PLA2, PhoA′ was fused in-frame to the PLA2 of pINDA′-PLA2, followed by mobilization into WT cells to generate Rs-A2-phoA (Table 1). Likewise, pIND-PLA2-phoA was constructed and mobilized into WT cells to generate Rs-nA2-phoA (Table 1), in which PLA2 was produced without the signal peptide, CycA′. Activity staining of Rs-A2-phoA colonies with BCIP/NBT solution revealed blue staining, indicating PhoA activity in the periplasmic space, whereas colonies of Rs-nA2-PhoA showed no blue staining (Fig. 2a). Consistently, Rs-nA2 showed FFA productivity one-third that of Rs-A2 (Fig. 2b). Approximately 90% of the phospholipase activity was localized to the periplasmic fraction of Rs-A2, and the remaining 10% of phospholipase activity was found in the cytoplasmic fraction (Fig. 3). Thus, the results indicate that PLA2 of Rs-A2 is heterologously produced and localized in the periplasmic space. Heterologous production of PLA2 did not affect the photoheterotrophic growth of Rs-A2, as shown by the doubling time and maximal cell growth, which are similar to the corresponding values of WT cells (Fig. 4c).
Fig. 3.
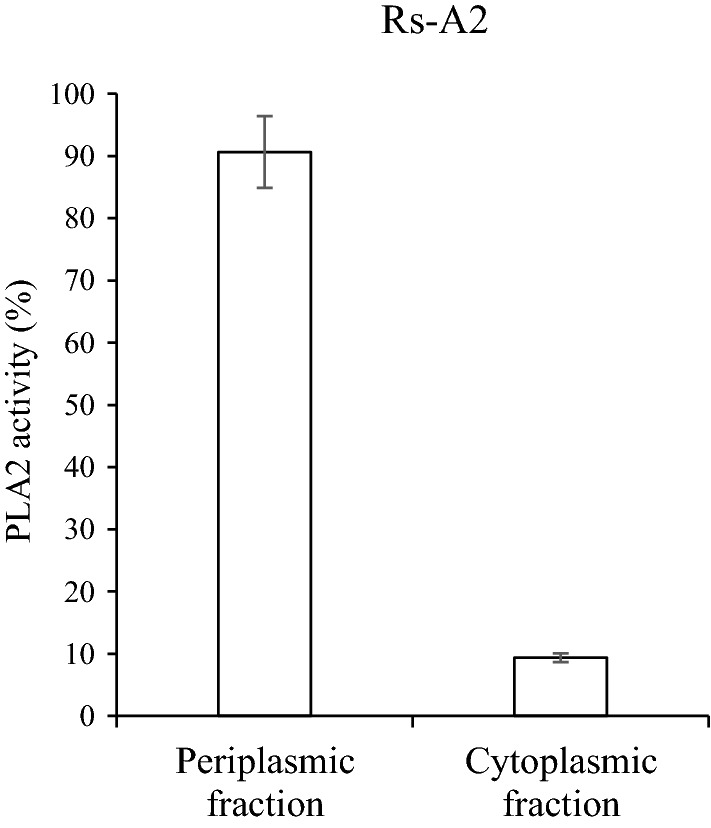
PLA2 activities of the periplasmic and cytoplasmic fractions of Rs-A2. Rs-A2 was grown photoheterotrophically in Sis minimal medium. Periplasmic and cytoplasmic fractions were prepared from cells harvested at the exponential growth phase (OD660 of 2.3). The PLA activity in each fraction was determined with membrane lipids from the photoheterotrophically grown WT cells and expressed as the percentage of the total activity, which amounted to approximately 111.5 U mg−1 protein. One enzyme unit was defined as 1 µmol of FFAs released per min from PLs under the assay conditions examined. PLA2 activities are shown with error bars from three independent experiments
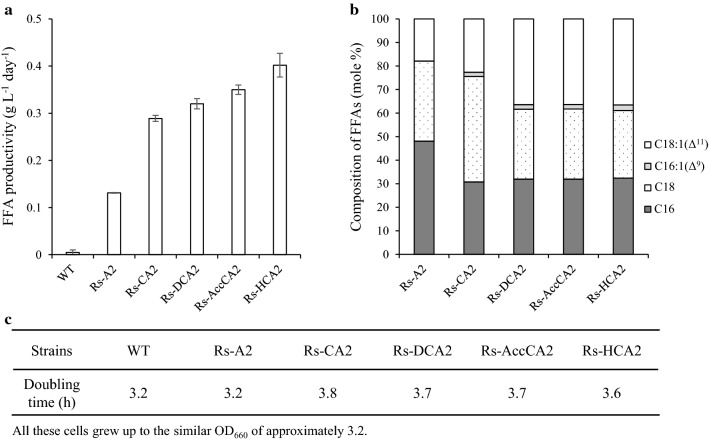
Fig. 4.
FFA productivity, FFA composition, and growth of Rs-A2, Rs-CA2, Rs-DCA2, Rs-AccCA2, and Rs-HCA2. a The recombinant R. sphaeroides strains were grown photoheterotrophically in Sis minimal medium and harvested at the exponential growth phase (OD660 of 2.5, 2.3, 2.3, 2.4, 2.3 and 2.2 for WT, Rs-A2, Rs-CA2, Rs-DCA2, Rs-AccCA2 and Rs-HCA2, respectively). FFAs in the culture supernatant were determined, and the FFA productivity is shown with error bars from three independent experiments. b The composition of FFAs is illustrated as the percentage of the total moles of FFAs. The experiments were independently repeated three times; the data shown are for one of three representative experiments. c Doubling times of recombinant R. sphaeroides strains are shown with WT cells as a control. The data presented here were reproduced within SDs of 10–15%. The experiments were independently repeated three times, and the data shown are for one of three representative experiments
Monounsaturated FA is the best component for biodiesel because it provides low-temperature fluidity and appropriate oxidative stability [53, 54]. R. sphaeroides harbors CFAs consisting of approximately 70% cis-vaccenic acid [55], and unsaturated FA is expected to be found most frequently at the sn2 carbon of PL. Analysis of the FFAs in the culture supernatant of Rs-A2, however, revealed approximately 20% cis-vaccenate at most (Fig. 4b). The remaining 80% of the FFAs mainly consist of stearate (~ 35%) and palmitate (~ 45%). The composition of FFAs did not vary during cell growth (Additional file 1: Fig. S2, A2, B2 and C2). Likewise, the heterologous expression levels of pla2 remained unchanged during the growth (Additional file 1: Fig. S2, A3, B3 and C3). The lower-than-expected cis-vaccenic acid content among the FFAs suggests its importance in the membrane for the viability and growth of R. sphaeroides.
Overproduction of FabH and PlsC in Rs-A2, generating Rs-HCA2, results in approximately a threefold increase in FFA productivity
We then examined whether the FFA productivity can be further increased by overproducing enzymes that catalyze the metabolic steps to form phosphatidate, a precursor of PL (Fig. 1). Because PLA2 hydrolyzes FAs from the sn2 carbon of PLs, the resulting LPL may need to be converted into PL again by LPL acyltransferase. However, no ORF of the R. sphaeroides genome has been specifically annotated as LPL acyltransferase. The 1-acyl-sn-glycerol-3-phosphate acyltransferase PlsC is known to exhibit LPL acyltransferase activity [56, 57]. Rsp_0735 has approximately 50% amino acid sequence identity with PlsC of Rhodobacter capsulatus (Additional file 1: Fig. S5), which was shown to complement the plsC mutant of E. coli [58]. When the gene encoding putative PlsC (Rsp_0735) was overexpressed in Rs-A2, the FFA productivity of the resulting recombinant strain Rs-CA2 (Table 1) was increased approximately twofold (Fig. 4a). We examined whether LPLs are present in the membranes of Rs-A2 and Rs-CA2 (Additional file 1: Fig. S6). Some LPLs, including lysophosphatidylethanolamine (lysoPE), were found to be associated with the ICM of R. capsulatus [59]. Interestingly, there is no difference in the PL and lysoPE profiles of the membranes of WT, Rs-A2 and Rs-CA2 cells (Additional file 1: Fig. S6), suggesting the rapid conversion of LPLs into PLs in these recombinant cells.
We further overexpressed the genes encoding enzymes that catalyze either the formation of malonyl-ACP (malonyl-CoA:ACP transacylase, FabD; acetyl-CoA carboxylase, AccABCD) or the initial condensation to form acetoacetyl-ACP (β-ketoacyl-ACP synthase, FabH) in Rs-CA2. FFA productivities of the resulting recombinant strains Rs-DCA2, Rs-AccCA2, and Rs-HCA2 were not less than Rs-CA2 (Fig. 4a). Unsaturated FAs constituted approximately 20% of FFAs of Rs-A2 and Rs-CA2 and twice as high a proportion in those of Rs-DCA2, Rs-AccCA2, and Rs-HCA2 (Fig. 4b). All the recombinant strains grew similarly to WT cells (Fig. 4c). Because Rs-HCA2 showed the highest FFA productivity, it was used in the subsequent experiments.
The CFA contents of R. sphaeroides overexpressing plsC alone or plsC with either fabD, accABCD, or fabH did not differ from those of WT cells (Additional file 1: Fig. S7A). The same was true for light-harvesting complexes (Additional file 1: Fig. S7B) and cell growth (Additional file 1: Fig. S7C). These results suggest that the FFAs of the recombinant strains (Fig. 4a) do not originate from the ready-made surplus membrane. The heterologous expression levels of pla2 in Rs-CA2, Rs-DCA2, Rs-AccCA2, and Rs-HCA2 did not differ from those in Rs-A2 (Additional file 1: Table S2). Thus, it would appear that the FFA productivity of the recombinant strains is not determined by the heterologous production of PLA2 but rather by the overproduced biosynthetic enzymes, which determine the conversion rate of the PLA2-generated LPLs into PLs.
High-cell-density culture of Rs-HCA2 grown in SisH-D medium in a two-phase system with dodecane increases FFA productivity up to 2.0 g L−1 day−1
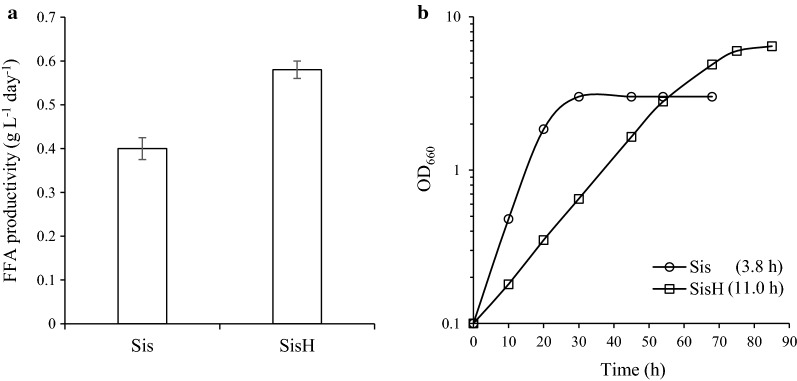
To enhance the FFA productivity of Rs-HCA2, cells were cultured at higher density in SisH medium (Sis minimal medium supplemented with 150 mM succinate, 100 mM glucose, 150 mM glycerol, and 1% (w/v) yeast extracts). The maximal turbidity of cells grown in SisH medium was almost twice that of cells grown in Sis minimal medium (Fig. 5b). Although the doubling time of Rs-HCA2 in SisH medium is threefold longer than in Sis minimal medium, the high-cell-density culture increases FFA productivity by approximately 50% (Fig. 5a).
Fig. 5.
FFA productivity of Rs-HCA2 grown at high cell density. a Rs-HCA2 was grown photoheterotrophically in Sis minimal and SisH (Sis minimal medium supplemented with 150 mM succinate, 100 mM glucose, 150 mM glycerol, and 1% (w/v) yeast extracts) media and harvested at the exponential growth phase (OD660 of 2.0 in Sis minimal medium; OD660 of 6.0 in SisH medium). FFAs in the culture supernatants were measured, and the FFA productivity is illustrated with error bars from three independent experiments. b Rs-HCA2 was cultured in Sis minimal and SisH media under photoheterotrophic conditions; data shown are for one of three representative experiments
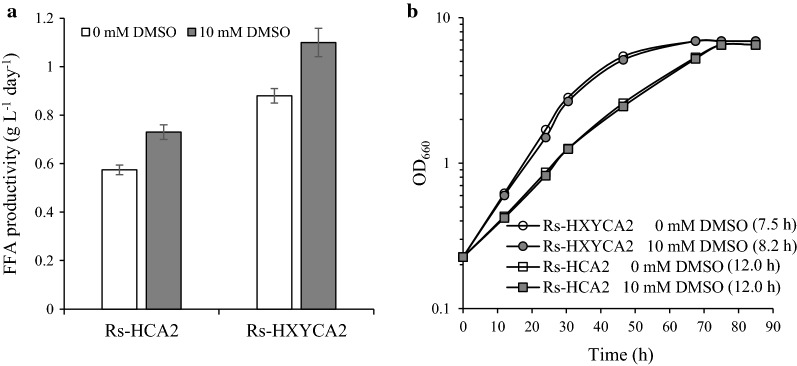
Dimethyl sulfoxide (DMSO) stimulates PL biosynthesis in yeast cells [60]. The CFA content of WT cells was reproducibly elevated by approximately 20% in several independent experiments as DMSO was increased to 10 mM (Additional file 1: Fig. S8A). At higher DMSO concentrations (≧ 25 mM), however, cell growth was affected (Additional file 1: Fig. S8C), and both CFAs (Additional file 1: Fig. S8A) and PLs (Additional file 1: Fig. S8D) decreased to lower levels than in control cells grown without DMSO. The compositions of both CFAs (Additional file 1: Fig. S8B) and PLs (Additional file 1: Fig. S8D) were not changed by DMSO. Thus, 10 mM DMSO (SisH-D medium) was used to stimulate the membrane synthesis of R. sphaeroides. When Rs-HCA2 was grown in SisH-D medium, the FFA productivity increased by approximately 25% (Fig. 6a) without affecting cell growth (Fig. 6b). Accordingly, Rs-HCA2 was grown photoheterotrophically in SisH-D medium to achieve higher FFA productivity in the subsequent experiments, unless otherwise specified.
Fig. 6.
FFA productivities of Rs-HCA2 and Rs-HXYCA2 grown at high cell densities in the presence of DMSO. a Rs-HCA2 and Rs-HXYCA2 were grown photoheterotrophically in SisH medium with or without DMSO (10 mM) and harvested at the exponential growth phase (OD660 of 5.2 and 5.1 for Rs-HCA2 and Rs-HXYCA2, respectively). FFAs in culture supernatants were measured, and the FFA productivity was illustrated with error bars from three independent experiments. b Rs-HCA2 and Rs-HXYCA2 were cultured in SisH medium with or without DMSO (10 mM) under photoheterotrophic conditions; data shown are for one of three representative experiments
FFAs produced by PLA2 in the periplasmic space may cross either the outer membrane for secretion or the inner membrane for reutilization by the cell. FFAs in the culture broth may also be utilized again after uptake by cells. Moreover, some unsaturated FAs, such as palmitoleic acid, oleic acid, linolenic acid, and arachidonic acid, affect bacterial growth by inhibiting enoyl-ACP reductase [61]. Likewise, the growth of R. sphaeroides was strongly affected by the presence of cis-vaccenate (Additional file 1: Fig. S9). Saturated FAs such as stearate and palmitate slightly slowed down the growth when present at 3 mg mL−1 (Additional file 1: Fig. S9A, S9B). On the other hand, the presence of cis-vaccenate at the same level (3 mg mL−1) resulted in the complete inhibition of cell growth (Additional file 1: Fig. S9C), and even a lower level (1 mg mL−1) of cis-vaccenate had a strong effect (Additional file 1: Fig. S9C). To keep these three FAs, which are the main constituents of FFAs (Fig. 4b), away from cells, a two-phase culture system with dodecane (Fig. 1) was employed. The growth of WT cells was not affected by the presence of 30% dodecane (v/v) (Additional file 1: Fig. S10). Consistently, no activity of the cytoplasmic enzyme malate dehydrogenase was detected in the culture supernatant of WT or Rs-HCA2 cells grown in the presence of 30% dodecane (Additional file 1: Fig. S11). Most enzyme activity was associated with cell lysate (Fig. S11), indicating that dodecane caused no lysis of cells. Furthermore, dodecane was shown not to be used as a sole carbon source for cell growth (Additional file 1: Fig. S10).
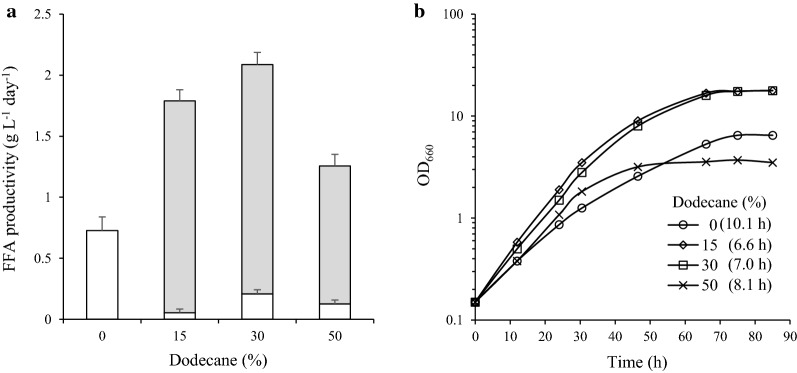
Rs-HCA2 grew faster in the two-phase culture system as dodecane levels increased up to 30% (Fig. 7b). The maximal cell density at 30% dodecane was threefold higher than that of cells grown without dodecane (Fig. 7b). Both the doubling time and maximal cell density were affected at higher (50%) dodecane levels (Fig. 7b). The FFA productivity of Rs-HCA2 was the highest, approximately 2.0 g L−1 day−1, at 30% dodecane (Fig. 7a). More than 90% of FFAs were localized to the dodecane layer (Fig. 7a), and there was no difference in composition between the FFAs in the culture broth and those in the dodecane layer (Additional file 1: Fig. S12). Thus, FFA productivity was elevated by keeping the FFAs in the dodecane layer. Accordingly, Rs-HCA2 was cultured in a two-phase system with 30% dodecane to increase FFA productivity in the subsequent experiments, unless otherwise specified.
Fig. 7.
FFA productivity of Rs-HCA2 grown at high cell density in a two-phase system with dodecane. a Rs-HCA2 was grown photoheterotrophically in SisH-D medium. Dodecane was added to the culture at concentrations of 15%, 30%, and 50% (v/v). The culture broth and dodecane layer were harvested at the exponential growth phase (OD660 of 5.3 for Rs-HCA2 in the absence of dodecane; OD660 of 16.0, 16.0 and 3.2 for Rs-HCA2 in the presence of 15%, 30% and 50% dodecane, respectively). FFAs in the culture supernatants (open bar) and in the dodecane layer (closed bar) were measured, and the FFA productivity is illustrated with error bars from three independent experiments. b Rs-HCA2 was cultured in SisH-D medium in the presence of varying levels of dodecane (0, 15, 30 and 50%) under photoheterotrophic conditions; data shown are for one of three representative experiments
The FFAs produced by Rs-HCA2 (Fig. 4b, Additional file 1: Fig. S12) consist mainly of cis-vaccenate, stearate, and palmitate in an approximately equimolar ratio. As in the FFAs produced by Rs-A2 (Fig. 4), the lower-than-expected proportion of cis-vaccenate among the FFAs indicates its importance in the membrane for the viability and growth of R. sphaeroides. The proportion of cis-vaccenate in the CFAs of Rs-HCA2 is approximately 60% (Additional file 1: Fig. S13A), which is less than that in WT cells by 10% (Additional file 1: Fig. S13A). The sum of stearate and palmitate comprises the remaining 40% of CFAs (Fig. S13A). The reason for the change in the CFA composition of Rs-HCA2 remains to be determined, but it would appear that the lower unsaturated FA in the Rs-HCA2 membranes might reflect the response to stress from the hydrolysis of the membrane by PLA2. The PL composition in Rs-HCA2, however, was not different from that of WT cells (Additional file 1: Fig. S13B).
Overproduction of both PlsX and PlsY in Rs-HCA2, generating Rs-HXYCA2, leads to accelerated cell growth
The expression levels of fabH and plsC in Rs-HCA2 were examined by RT-qPCR and shown as the mean ratio to the expression of the corresponding gene in WT cells (Table 2). As expected, both fabH and plsC were overexpressed in Rs-HCA2 compared with WT cells. Interestingly, plsX and plsY of Rs-HCA2, which encode phosphate acyltransferase and glycerol-3-phosphate acyltransferase, respectively, catalyzing the two consecutive metabolic steps immediately upstream of PlsC (Fig. 1), were expressed to a lesser extent than in WT cells (Table 2). The transcriptional network between plsX, plsY, and plsC in the presence of extra copies of pla2 remains to be defined, but the overexpression of plsC in Rs-HCA2 may result in a higher demand for acyl-ACP, which is also a substrate of PlsX (Fig. 1). As a result, the steady-state level of acyl-ACP may be lowered, which in turn may affect the expression of plsX and plsY (Fig. 1).
Table 2.
Expression of fabH, plsX, plsY, plsC, and pla2 in Rs-HCA2 and Rs-HXYCA2
| Genesb | Strainsa | ||
|---|---|---|---|
| WT | Rs-HCA2 | Rs-HXYCA2 | |
| fabH | 1.0 ± 0.7 | 64.7 ± 0.2d (0.007)e | 52.4 ± 0.1d (0.032)e |
| plsX | 1.0 ± 0.1 | 0.6 ± 0.1d (0.04)e | 15.1 ± 0.7d (0.001)e |
| plsY | 1.0 ± 0.1 | 0.2 ± 0.1d (0.012)e | 89.0 ± 0.7d (0.001)e |
| plsC | 1.0 ± 0.1 | 6.4 ± 0.4d (0.004)e | 6.2 ± 0.2d (0.004)e |
| pla2 | NAc | 1.0 ± 0.4 | 1.1 ± 0.1f (0.3)e |
aCells were grown photoheterotrophically in SisH medium and harvested at the exponential phase (OD660 of 9.5, 5.3 and 5.4 for WT, Rs-HCA2 and Rs-HXYCA2, respectively)
bExpression levels were examined by RT-qPCR
cNot applicable
dMean ratio (± SD) to the expression of the corresponding gene in WT cells, which was set to 1, from three independent experiments with 16S rRNA as a reference gene
eSignificance level regarded as significant when the value is less than 0.05
fMean ratio (± SD) to the expression of pla2 in Rs-HCA2, which was set to 1, from three independent experiments with 16S rRNA as a reference gene
To elevate the low expression of plsX and plsY, both genes were overexpressed in Rs-HCA2, generating the strain Rs-HXYCA2 (Table 2). RT-qPCR analysis revealed elevated expression of both plsX and plsY in Rs-HXYCA2 compared with the corresponding expression levels in Rs-HCA2 (Table 2). The expression of pla2 remained unchanged in the same comparison (Table 2). Interestingly, Rs-HXYCA2 grew faster than Rs-HCA2, irrespective of the presence of DMSO (Fig. 6b). The CFA composition of Rs-HXYCA2 was not different from that of Rs-HCA2 (Additional file 1: Fig. S13A). The same was true for the PLs of these two strains (Additional file 1: Fig. S13B). Thus, it would appear that the overexpression of plsX and plsY in Rs-HCA2 led to the efficient synthesis of phosphatidate as a precursor of PL and subsequently to accelerated cell growth. The FFA productivity of Rs-HXYCA2 was approximately 40% higher than that of Rs-HCA2 (Fig. 6a), and the addition of 10 mM DMSO to the culture broth stimulated the FFA productivity of Rs-HXYCA2 to an extent similar to that observed in Rs-HCA2 (Fig. 6a).
High-cell-density culture of Rs-HXYCA2 in SisH-D medium in a two-phase system with dodecane further elevates FFA productivity up to 2.8 g L−1 day−1 with an FFA titer of approximately 8.5 g L−1
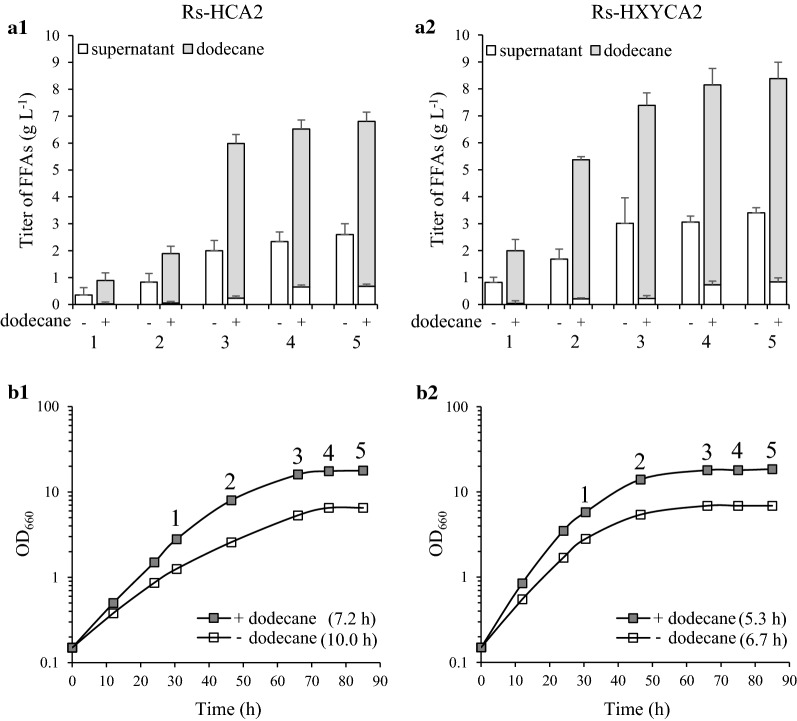
We then examined the FFA productivity of Rs-HXYCA2 grown in SisH-D medium in a two-phase culture system with dodecane (Fig. 8a2, b2). Rs-HCA2 was grown in the same conditions and examined as a control (Fig. 8a1, b1). The FFA productivity of Rs-HCA2 in a two-phase culture system with dodecane was 2.0 g L−1 day−1 with a maximal FFA titer of 6.8 g (Fig. 8a1 and Table 3). When Rs-HCA2 was grown without dodecane, the FFA productivity and the maximal FFA titer were approximately 40% of the corresponding values in the cells grown with dodecane (Fig. 8a1 and Table 3).
Fig. 8.
FFA productivities of Rs-HCA2 and Rs-HXYCA2 grown at high cell densities in a two-phase system with dodecane. a1 Rs-HCA2 and a2 Rs-HXYCA2 were grown photoheterotrophically in SisH-D medium with or without 30% dodecane (v/v). The culture broth and dodecane layer were harvested during growth, and the FFAs in the culture supernatant (open bar) and the dodecane layer (closed bar) were measured. The FFA productivity is illustrated with error bars from three independent experiments. The numbers 1–5 correspond to the time points of harvesting the culture broth and dodecane layer from b1 and b2. b1 Rs-HCA2 and b2 Rs-HXYCA2 were cultured in SisH-D medium with or without 30% dodecane (v/v) under photoheterotrophic conditions; data shown are for one of three representative experiments
Table 3.
Carbon-to-lipid conversion yield, FFA titer, and FFA productivity of Rs-HCA2 and Rs-HXYCA2
| Rs-HCA2 | Rs-HXYCA2 | |||
|---|---|---|---|---|
| Dodecane | ||||
| – | + | – | + | |
| Carbon-to-lipid conversion yield (g g−1)a | 0.14 ± 0.02d | 0.17 ± 0.01 | 0.18 ± 0.01 | 0.20 ± 0.01 |
| Titer (g L−1)b | 2.6 ± 0.2 | 6.8 ± 0.5 | 3.4 ± 0.5 | 8.5 ± 0.7 |
| Productivity (g L−1 day−1)c | 0.7 ± 0.2 | 2.0 ± 0.1 | 1.0 ± 0.2 | 2.8 ± 0.2 |
aRatio of total lipids to the carbon sources (glucose, glycerol, and succinate) consumed by the recombinant strains (Additional file 1: Fig. S16), calculated as described previously [62]
bMaximum FFA level per liter of culture medium
cProductivity at the exponential growth phase (OD660 of 6.5 and 17.5 for Rs-HCA2 in the absence and the presence of dodecane, respectively; OD660 of 6.9 and 14.0 for Rs-HXYCA2 in the absence and the presence of dodecane, respectively)
dMean ± SD
Rs-HXYCA2 grown in SisH-D medium in a two-phase culture system with dodecane showed higher FFA productivity of 2.8 g L−1 day−1 with a maximal FFA titer of approximately 8.5 g L−1 (Fig. 8a2 and Table 3). The FFA productivity and maximal FFA titer without dodecane were also approximately 40% of the corresponding values in the cells grown with dodecane (Fig. 8a2 and Table 3). Rs-HXYCA2 grew faster than Rs-HCA2 irrespective of the presence of dodecane (Fig. 8b1, b2). As observed with Rs-HCA2 (Fig. 7 and Fig. 8a1), more than 90% of FFAs were localized to the dodecane layer (Fig. 8a2). There was no difference in compositions between the FFAs in the culture broth and those in the dodecane (Additional file 1: Fig. S12). Furthermore, the FFA composition did not vary during growth of Rs-HCA2 and Rs-HXYCA2 (Additional file 1: Fig. S14). Interestingly, the expression levels of the genes carried on plasmids were elevated when Rs-HXYCA2 was grown with dodecane, compared with those in cells grown without dodecane (Additional file 1: Fig. S15, A2, B2, C2 and D2). The same was true for Rs-HCA2 (Additional file 1: Fig. S15, A1, B1, C1 and D1). Thus, keeping the FFAs in dodecane results in enhanced cell growth and higher FFA productivity, which is consistent with the elevated expression of genes encoding enzymes for the formation of CFAs and phosphatidate.
The residual carbons in the culture broth were determined (Additional file 1: Fig. S16), and the conversion yields from carbon to lipid by Rs-HCA2 and Rs-HXYCA2 were calculated as described previously [62]. Rs-HCA2 showed conversion yields in the range from 0.14 without dodecane to 0.17 with dodecane (Table 3). The conversion yields of Rs-HXYCA2 were in the range from 0.18 without dodecane to 0.20 with dodecane (Table 3). Thus, although the overexpression of plsX and plsY in Rs-HCA2 together with the sequestration of FFAs in the dodecane layer had a positive effect on the carbon-to-lipid conversion yield, the increase of FFA productivity in the two-phase system is due to an increase in overall carbon substrate consumption rather than due to an increased efficiency of carbon substrate conversion to FFA (Table 3, Fig. 8 and Additional file 1: Fig. S16).
In summary, the photosynthetic bacteria R. sphaeroides can be used as an ideal oleaginous host source for the production of long-chain FFAs based on its intrinsic high lipid contents. Because R. sphaeroides is capable of using various carbon and nitrogen sources as substrates to grow in a wide variety of conditions, additional genetic modification can also be applied to enhance the potential of this bacterium for the production of FFAs.
Additional file
Additional file 1. Additional figures and tables.
Authors’ contributions
JKL conceived and supervised the study. XT and EKO performed the experiments. XT and BHL analyzed the data. XT and JKL prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this article and its additional file.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the Pioneer Research Center Program through NRF (No. 2013M3C1A3064325) of the Ministry of Science and ICT.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 2.Rude MA, Schirmer A. New microbial fuels: a biotech perspective. Curr Opin Microbiol. 2009;12:274–281. doi: 10.1016/j.mib.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Guo D, Cheng Y, Zhu F, Deng Z, Liu T. Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnol Bioeng. 2014;111:1841–1852. doi: 10.1002/bit.25239. [DOI] [PubMed] [Google Scholar]
- 4.Ingram LO, Conway T, Clark DP, et al. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munjal N, Mattam AJ, Pramanik D, Srivastava PS, Yazdani SS. Modulation of endogenous pathways enhances bioethanol yield and productivity in Escherichia coli. Microb Cell Fact. 2012;11:145. doi: 10.1186/1475-2859-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah YR, Sen DJ. Bioalcohol as green energy—a review. Int J Curr Sci Res. 2011;1:57–62. [Google Scholar]
- 7.Lee SY, Kim HM, Cheon S. Metabolic engineering for the production of hydrocarbon fuels. Curr Opin Biotechnol. 2015;33:15–22. doi: 10.1016/j.copbio.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152:2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 9.Choi YJ, Lee SY. Microbial production of short-chain alkanes. Nature. 2013;502:571–574. doi: 10.1038/nature12536. [DOI] [PubMed] [Google Scholar]
- 10.Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc Natl Acad Sci USA. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu P, Gu Q, Wang W, Wong L, Bower AGW, Collins CH, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Eng. 2010;12:378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Yu C, Feng D, Cheng T, Meng X, Liu W, et al. Production of extracellular fatty acid using engineered Escherichia coli. Microb Cell Fact. 2012;11:41. doi: 10.1186/1475-2859-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black PN, Dirusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev. 2003;67:454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol Bioeng. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Jeon E, Yun HS, Lee J. Improvement of fatty acid biosynthesis by engineered recombinant Escherichia coli. Biotechnol Bioprocess Eng. 2011;16:706–713. [Google Scholar]
- 18.Zhang X, Agrawal A, San KY. Improving fatty acid production in Escherichia coli through the overexpression of malonyl coA-Acyl carrier protein transacylase. Biotechnol Prog. 2012;28:60–65. doi: 10.1002/btpr.716. [DOI] [PubMed] [Google Scholar]
- 19.Lennen RM, Politz MG, Kruziki MA, Pfleger BF. Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. J Bacteriol. 2013;195:135–144. doi: 10.1128/JB.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu XY, Sheng J, Curtiss R. Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chory J, Donohue TJ, Varga AR, Staehelin LA, Kaplan S. Induction of the photosynthetic membranes of Rhodopseudomonas sphaeroides: biochemical and morphological studies. J Bacteriol. 1984;159:540–554. doi: 10.1128/jb.159.2.540-554.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmer KC, Dohnalkov AC, Noguera DR, Donohue TJ. Oxygen-dependent regulation of bacterial lipid production. J Bacteriol. 2015;197:1649–1658. doi: 10.1128/JB.02510-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmer KC, Donohue TJ. Mutations that alter the bacterial cell envelope increase lipid production. mBio. 2017;8:1–15. doi: 10.1128/mBio.00513-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee IH, Park J, Kho D, Kim MS, Lee J. Reductive effect of H2 uptake and poly-β-hydroxybutyrate formation on nitrogenase-mediated H2 accumulation of Rhodobacter sphaeroides according to light intensity. Appl Microbiol Biotechnol. 2003;60:147–153. doi: 10.1007/s00253-002-1097-2. [DOI] [PubMed] [Google Scholar]
- 26.Donohue TJ, McEwan AG, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1986;168:962–972. doi: 10.1128/jb.168.2.962-972.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim EJ, Kim JS, Kim MS, Lee JK. Effect of changes in the level of light harvesting complexes of Rhodobacter sphaeroides on the photoheterotrophic production of hydrogen. Int J Hydrogen Energy. 2006;31:531–538. [Google Scholar]
- 28.Preuss D. How to grow Arabidopsis. In: Weigel D, Glazebrook D, editors. Arabidopsis: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2002. pp. 1–18. [Google Scholar]
- 29.Ind AC, Porter SL, Brown MT, Byles ED, De Beyer JA, Godfrey SA, et al. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl Environ Microbiol. 2009;75:6613–6615. doi: 10.1128/AEM.01587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo YS, Kim EY, Kim JH, Kim WT. Enzymatic characterization of class I DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Lett. 2009;583:2301–2307. doi: 10.1016/j.febslet.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Greer MS, Weselake RJ. Plant phospholipase A: advances in molecular biology, biochemistry, and cellular function. Biomol Concepts. 2013;4:527–532. doi: 10.1515/bmc-2013-0011. [DOI] [PubMed] [Google Scholar]
- 32.Eraso JM, Kaplan S. PrrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyer HW, Rovlland-dvssoix D, Francisco S, Prancisco B. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 35.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 36.Lee JK, Kaplan S. Cis-acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1146–1157. doi: 10.1128/jb.174.4.1146-1157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figurski DH, Helinski DR. Replication of an origin containing derivative of plasmid RK2 dependent. Proc Natl Acad Sci. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keen NT, Tamaki S, Kobayashi D, Troilinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, O’Connell M, Labes M, Puhler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 40.Ryu SB, Hyoung YL, Doelling JH, Palta JP. Characterization of a cDNA encoding Arabidopsis secretory phospholipase A2-a, an enzyme that generates bioactive lysophospholipids and free fatty acids. Biochim Biophys Acta Mol Cell Biol Lipids. 2005;1736:144–151. doi: 10.1016/j.bbalip.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Davis J, Donohue TJ, Kaplan S. Construction, characterization, and complementation of a Puf-mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narro ML, Adams CW, Cohen SN. Isolation and characterization of Rhodobacter capsulatus mutants defective in oxygen regulation of the puf operon. J Bacteriol. 1990;172:4549–4554. doi: 10.1128/jb.172.8.4549-4554.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panicker IS, Kanci A, Chiu C-J, Veith PD, Glew MD, Browning GF, et al. A novel transposon construct expressing PhoA with potential for studying protein expression and translocation in Mycoplasma gallisepticum. BMC Microbiol. 2012;12:138. doi: 10.1186/1471-2180-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan S, Hiniker A, Collet J, Bardwell JCA. Isolation of bacteria envelope proteins. Methods Mol Biol. 2013;966:359–366. doi: 10.1007/978-1-62703-245-2_22. [DOI] [PubMed] [Google Scholar]
- 45.Soccio M, Trono D, Laus MN, Pastore D. An improved spectrophotometric phospholipase A2 assay using 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine as substrate and lipoxygenase as coupled enzyme. J Korean Soc Appl Biol Chem. 2013;56:369–376. [Google Scholar]
- 46.Kim EJ, Lee JK. Effect of changes in the composition of cellular fatty acids on membrane fluidity of Rhodobacter sphaeroides. J Microbiol Biotechnol. 2015;25:162–173. doi: 10.4014/jmb.1410.10067. [DOI] [PubMed] [Google Scholar]
- 47.Gürgan M, Erkal NA, Özgür E, Gündüz U, Eroglu I, Yücel M. Transcriptional profiling of hydrogen production metabolism of Rhodobacter capsulatus under temperature stress by microarray analysis. Int J Mol Sci. 2015;16:13781–13797. doi: 10.3390/ijms160613781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Leo V, Catucci L, Ventrella A, Milano F, Agostiano A, Corcelli A. Cardiolipin increases in chromatophores isolated from Rhodobacter sphaeroides after osmotic stress: structural and functional roles. J Lipid Res. 2009;50:256–264. doi: 10.1194/jlr.M800312-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Xu G, Waki H, Kon K, Ando S. Thin-layer chromatography of phospholipids and their lyso forms: application to determination of extracts from rat hippocampal CA1 region. Microchem J. 1996;53:29–33. [Google Scholar]
- 50.Kiley J, Meinhardt W, Crofts AR. Characterization of light-harvesting mutants of Rhodopseudomonas. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center to synthetic units emphasizes that reaction centers. Arch Biochem Biophys. 1985;236:130–139. doi: 10.1016/0003-9861(85)90612-5. [DOI] [PubMed] [Google Scholar]
- 51.Hayer-Hartl M. Assay of malate dehydrogenase. Methods Mol Biol. 2000;140:127–132. doi: 10.1385/1-59259-061-6:127. [DOI] [PubMed] [Google Scholar]
- 52.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio. 2016;7:1–9. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y, Liu W, Xu X, Zhang H, Wang J, Xian M. Production of free monounsaturated fatty acids by metabolically engineered Escherichia coli. Biotechnol Biofuels. 2014;7:59. doi: 10.1186/1754-6834-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akoh CC. Oxidative stability of fat substitutes and vegetable oils by the oxidative stability index method. J Am Oil Chem Soc. 1994;71:211–216. [Google Scholar]
- 55.Onishi JC, Niederman RA. Rhodopseudomonas sphaeroides membranes: alterations in phospholipid composition in aerobically and phototrophically grown cells. J Bacteriol. 1982;149:831–839. doi: 10.1128/jb.149.3.831-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez-Chacón G, Astudillo AM, Balgoma D, Balboa MA, Balsinde J. Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim Biophys Acta Mol Cell Biol Lipids. 2009;1791:1103–1113. doi: 10.1016/j.bbalip.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Ogawa T, Tanaka A, Kawamoto J, Kurihara T. Purification and characterization of 1-acyl-sn-glycerol-3-phosphate acyltransferase with a substrate preference for polyunsaturated fatty acyl donors from the eicosapentaenoic acid-producing bacterium Shewanella livingstonensis Ac10. J Biochem. 2018;164:33–39. doi: 10.1093/jb/mvy025. [DOI] [PubMed] [Google Scholar]
- 58.Aygun-Sunar S, Bilaloglu R, Goldfine H, Daldal F. Rhodobacter capsulatus OlsA is a bifunctional enyzme active in both ornithine lipid and phosphatidic acid biosynthesis. J Bacteriol. 2007;189:8564–8574. doi: 10.1128/JB.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pucheu NL, Kerber NL, Rivas EA, Cortez N, Garcia AF. Association of LHIα(B870) polypeptide with phospholipids during insertion in the photosynthetic membrane of an LHII-mutant of Rhodobacter capsulatus. Curr Microbiol. 1997;34:155–161. doi: 10.1007/s002849900161. [DOI] [PubMed] [Google Scholar]
- 60.Murata Y, Watanabe T, Sato M, Momose Y, Nakahara T, Oka S, et al. Dimethyl sulfoxide exposure facilitates phospholipid biosynthesis and cellular membrane proliferation in yeast cells. J Biol Chem. 2003;278:33185–33193. doi: 10.1074/jbc.M300450200. [DOI] [PubMed] [Google Scholar]
- 61.Zheng CJ, Yoo JS, Lee TG, Cho HY, Kim YH, Kim WG. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 2005;579:5157–5162. doi: 10.1016/j.febslet.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 62.Gong Z, Shen H, Zhou W, Wang Y, Yang X, Zhao ZK. Efficient conversion of acetate into lipids by the oleaginous yeast Cryptococcus curvatus. Biotechnol Biofuels. 2015;8:1–9. doi: 10.1186/s13068-015-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional figures and tables.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its additional file.